Abstract
Mechanosensitive (MS) channels that provide protection against hypoosmotic shock are found in the membranes of organisms from the three domains of life: bacteria, archaea, and eucarya. Two families of ubiquitous MS channels are recognized, and these have been designated the MscL and MscS families. A high-resolution X-ray crystallographic structure is available for a member of the MscL family, and extensive molecular genetic, biophysical, and biochemical studies conducted in many laboratories have allowed postulation of a gating mechanism allowing the interconversion of a tightly closed state and an open state that controls transmembrane ion and metabolite fluxes. In contrast to the MscL channel proteins, which are of uniform topology, the much larger MscS family includes protein members with topologies that are predicted to vary from 3 to 11 α-helical transmembrane segments (TMSs) per polypeptide chain. Sequence analyses reveal that the three C-terminal TMSs of MscS channel proteins are conserved among family members and that the third of these three TMSs exhibits a 20-residue motif that is shared by the channel-forming TMS (TMS 1) of the MscL proteins. We propose that this C-terminal TMS in MscS family homologues serves as the channel-forming helix in a homooligomeric structure. The presence of a conserved residue pattern for the putative channel-forming TMSs in the MscL and MscS family proteins suggests a common structural organization, gating mechanism, and evolutionary origin.
OVERVIEW OF KEY DEVELOPMENTS
Mechanosensitive (MS) channels were first demonstrated in bacterial cells by using patch clamp analysis of giant bacterial protoplasts and by fusion of membranes with liposomes. Both approaches indicated the presence of high-conductance channels in the membranes of gram-positive and gram-negative bacteria (15, 29, 53, 60). Initially the data were greeted with scepticism, based on the similarity of the conductances of MS channels to those of porins and the recognized need of the cytoplasmic membrane to exhibit tight control over H+ permeability in order to effect energy transduction. Activation of MS channels by membrane-intercalating amphipathic compounds suggested that these channels are sensitive to mechanical perturbations in the lipid bilayer (22, 28). Support for the presence of channels was provided by the discovery and reconstitution of two distinct channel activities from Escherichia coli, each with unique properties (52). Further support came from the discovery that the efflux of solutes from E. coli cells in response to a lowering of the external osmolarity could be prevented by gadolinium ions, which are classical inhibitors of MS channels in higher organisms (6).
A landmark event was the purification and cloning of the first MS channel protein, MscL, from E. coli. This heroic piece of biochemistry required that each fraction derived from the solubilized and fractionated membrane be reconstituted into liposomes and the MS channel activity be measured (51). Availability of the amino-terminal sequence of the protein led to identification of the gene. Following this breakthrough, a new age of MS channel protein structure-function analysis dawned (7, 9-11, 42), culminating in the crystal structure of a mycobacterial MscL channel (13) (Fig. 1). Extensive genetic and biophysical analyses of MscL protein movement in real time, coupled with model building, electron paramagnetic resonance spectroscopy, and site-directed spin labeling studies, provided an explanation of how the protein can exist in at least two states—one tightly closed and the other creating a large pore in the membrane (23, 42, 48, 49) (Fig. 2). MS channels are now thought to be important to many bacteria (8) and archaea (20, 21, 24).
FIG. 1.
Three-dimensional structure of the homopentameric mechanosensitive channel MscL from M. tuberculosis as revealed by X-ray crystallography (13). The side view was rendered using Molscript (17) and Raster-3D (34). The monomers within the channel are individually colored. The NH2- and COOH-terminal ends of the cyan monomer are indicated, and the dimensions of the channel are shown. (Reproduced from reference 4 [PDB ID 1MSL, reference 13; http://www.pdb.org/].)
FIG. 2.
Ribbon representation of models of M. tuberculosis MscL in a closed (left), an intermediate (middle), and an open (right) conformation shown as viewed from outside the cell (top row) and from the side (bottom). Only one subunit is colored in the side view, so that the conformation of a single subunit can be visualized. Structural models of the MscL proteins have been developed in 13 different conformations with different size transmembrane pores. Three of these conformations are illustrated here. The color code is as follows: N, red; MI, orange; Loop, yellow; M2, light blue; Linker, aqua blue; S, dark blue. (Modified with permission from reference 49.)
The genetic advances with MscL posed a further problem—why does an mscL null mutant lack an apparent physiological phenotype? Patch clamp analysis had revealed the presence of at least two MS channels in E. coli membranes, and subsequent studies led to the possibility that five or more genetically distinct channels exist (5). Such apparent biochemical redundancy implied that observation of a phenotype might require the construction of a mutant lacking more than one channel protein. Preliminary support for the protective role of MscL was discovered by expressing the channel in Vibrio and observing protection from hypoosmotic shock (38). The discovery of the structural gene for MscS, the second major MS channel in E. coli, allowed this functional hypothesis to be tested (25). Through the genetic analysis of a missense mutation, called RQ2, which displayed a K+-specific phenotype at high osmolarity (33), a family of proteins was identified, two of which were demonstrated to have MS channel activity equivalent to the MscS signal previously detected by patch clamp analysis (25). The two MS channels, YggB (MscS) and KefA (MscK), could be deleted without a significant physiological phenotype. However, an mscS mscL double mutant exhibited decreased rates of K+ release on mild osmotic downshock (25). Exposure of the double mutant to a large drop in osmolarity (greater than 0.3 M NaCl difference between the growth medium and the shock medium [Fig. 3A ]) resulted in severe loss of viability and lysis of more than 90% of the cells (Fig. 3B). Expression of either mscL or mscS alone in the double deletant increased survival in response to the shock procedure (25). This observation confirmed the functional redundancy of the two channels.
FIG. 3.
Protection of E. coli cells against hypoosmotic shock by the action of MS channels. Cells were grown to the mid-exponential phase in minimal medium containing 0.5 M NaCl and then diluted 20 fold into prewarmed medium containing <0.2 M NaCl to ensure a hypoosmotic shock of ≥0.3 M NaCl. After a 30-min incubation, samples were withdrawn and culture viability and light scattering (optical density at 600 nm [OD600]) were recorded. The optical density of the culture supernatant was assayed at 260 nm to determine the extent of cell lysis (see reference 25). (A) Osmotic shock protocol; (B) survival and OD260 of the culture supernatant for the parent, Frag1, and the mscL mscS double mutant.
Analysis of the degree of hypoosmotic shock needed to activate the channels by using a novel combined acid and osmotic down shock assay revealed that MS channels are activated at a pressure differential just below that causing cell lysis in a channel-less mutant (12, 25). This was the first demonstration that MS channels function in adaptation to hypoosmotic shock. Although a further MS channel, MscM, has been characterized electrophysiologically and is predicted to have a substantial conductance (8), it appears to provide limited protection against hypoosmotic shock. The lack of genetic information about this channel has precluded analysis of its role in cell physiology.
The patch clamp assay for MS channel activity can reveal subtle changes in gating pressure, for example, by comparing the opening pressures of MscS and MscL activities (11, 36, 42). In addition, patch clamp techniques can reveal the existence of partially open states and alterations in channel kinetics. Finally, combined with microscopy, patch clamp analysis allows definition of the absolute relationships between pressure and channel gating (48, 49). Less sophisticated physiological methods of analysis of “in-cell” activity include growth inhibition associated with the expression of gain-of-function mutations (9, 42, 58), survival following hypoosmotic shock (25), and penetration to the cytoplasm of molecules that are usually excluded (30, 31) (Fig. 3). These methods have been used by a number of research groups to analyze mutants and for determination of the effects of chemical modification on channel activities (57). It is from these assays that our picture of channel regulation and structure-function relationships is emerging.
A number of archaeal channels have been characterized electrophysiologically, and they exhibit characteristics similar to those of the E. coli MscL and MscS proteins (22). One archaeon, Haloferax volcanii, exhibits MS channels similar in conductance and mass to YggB of E. coli, but the sequences of these channel proteins are not available (24). Two sequenced MscS homologues from Methanococcus jannaschii have recently been functionally characterized (20, 21), and they exhibit properties expected for MscS channels. A Synechocystis MscS homologue (slr1575) possesses a C-terminal domain homologous to the cyclic AMP-dependent protein kinase A regulatory subunit (40), suggesting that this MscS homologue may be a cyclic nucleotide-regulated channel.
In this review, we identify all currently available members of the MscL and MscS families and determine their organismal distributions. While sequenced members of the MscL family are currently restricted to one archaeon, a single fungus, and bacteria, the MscS family is much more widely distributed in the three domains of life. The sequences of the MscL and MscS homologues have been multiply aligned, and phylogenetic trees have been derived. We demonstrate considerable diversity in the MscS family compared with the MscL family. Thus, MscS family members vary in length (from less than 200 to over 1,100 residues) with topologies that vary from 3 to 11 putative transmembrane segments (TMSs). In spite of extensive sequence divergence of MscS family members, the 3 C-terminal TMSs are common to all family members and a 20-residue conserved motif in the third conserved TMS is shared by TMS 1 of MscL family proteins. This observation suggests that the conserved TMS 3 in MscS homologues is the channel-forming helix, as is established for TMS 1 in MscL homologues. Further, the similarities between the sequence conservation patterns of the MscL and MscS families may be fundamental to their organization and gating mechanisms. They may even suggest a common evolutionary origin for the channel-forming segments of these proteins. We summarize the currently available functional data for members of these two families of MS channel-forming proteins.
MscL CHANNEL FAMILY
Limited phylogenetic data have been published for MscL channels (46). Currently sequenced members of the MscL family (TC 1.A.22) are presented in Table 1 (45). These proteins are derived from bacteria, a single archaeon, Methanosarcina acetovorans, and a single fungus, Neurospora crassa. As expected, the archaeal and fungal proteins are the most divergent members of the MscL family, both in size and in sequence. The archaeal homologue is 20% smaller than the smallest bacterial MscL family member, and the fungal homologue is 120% larger than the largest bacterial homologue, in agreement with observations concerning the relative sizes of other homologous transport proteins in the three domains of life (14). The two predicted TMSs and a loop region of the fungal protein show strongest sequence similarity to the Clostridium perfringens MscL homologue of the bacterial proteins (33% identity, 56% similarity, E value of 2e−32). The loop between TMSs 1 and 2 in the fungal protein is 52 amino acids (aa) in length, in contrast to 36 aa for the largest bacterial MscL homologue, and its large glycine-rich carboxy-terminal domain exhibits sequence similarity to glycine-rich regions in animal and animal parasite proteins such as human trophinin (spQ12816), Caenorhabditis elegans RNA helicase GLH-2 (gbAAB03337), the sea urchin α-2 collagen fibrillar chain precursor (gbAAA30040), and the Plasmodium vivax circumsporozoite protein (pirA41156). The archaeal protein is of similar topology, most closely resembling the Lactococcus lactis homologue (40% identity, 60% similarity, E value of 2e−34). The archaeal homologue lacks the C-terminal hydrophilic extension present in the L. lactis protein. It is interesting that both the archaeal and the eukaryotic proteins most closely resemble low-G+C gram-positive bacterial homologues.
TABLE 1.
Sequenced proteins of the MscL family
| Abbreviation | Database description | Organism | Sizeb | gic number |
|---|---|---|---|---|
| Gram-negative bacteria | ||||
| Bme | Large-conductance MS channel | Brucella melitensis | 144 | 17987888 |
| Bja | MscL protein | Bradyrhizobium japonicum | 157 | 6136300 |
| Ccr | Large-conductance MS channel CC3585 | Caulobacter crescentus | 139 | 13425329 |
| Chi | Large-conductance MS channel | Clostridium histolyticum | 133 | 7674133 |
| Eco | Large-conductance MS channel | Escherichia coli | 136 | 547924 |
| Fnu | Large-conductance MS channel | Fusobacterium nucleatum subsp. | 142 | 19714305 |
| Hin | Large-conductance MS channel HI0626 | Haemophilus influenzae | 128 | 1171031 |
| Lin | Similar to large-conductance MS channel protein | Listeria innocua | 128 | 16801236 |
| Lmo | Similar to large-conductance MS channel protein | Listeria monocytogenes | 128 | 16804103 |
| Mlo1 | Large-conductance MS channel mlr5692 | Mesorhizobium loti | 157 | 13474738 |
| Mlo2 | Large-conductance MS channel mll4699 | Mesorhizobium loti | 144 | 13473940 |
| Mlo3 | Large-conductance MS channel mlr4713 | Mesorhizobium loti | 140 | 13473952 |
| Mlo4a | Large-conductance MS channel mlr5747 | Mesorhizobium loti | 140 | 13474781 |
| Pmu | Large-conductance MS channel | Pasteurella multocida | 133 | 13431698 |
| Pea | Large-conductance MS channel | Pectobacterium carotovorum | 137 | 6016603 |
| Pae | Large-conductance MS channel PA4614 | Pseudomonas aeruginosa | 137 | 13431712 |
| Pfl | Large-conductance MS channel | Pseudomonas fluorescens | 136 | 6016605 |
| Sty1 | Large-conductance MS channel | Salmonella enterica serovar Typhimurium | 136 | 8650506 |
| Sty2 | Large-conductance MS channel | Salmonella enterica serovar Typhi | 137 | 16762872 |
| Ssp | Large-conductance MS channel slr0875 | Synechocystis sp. | 145 | 6016607 |
| Vch | Large-conductance MS channel VCA0612 | Vibrio cholerae | 136 | 11280192 |
| Xfa | Large-conductance MS channel XF0039 | Xylella fastidiosa | 134 | 11280193 |
| Ype | MS ion channel | Yersinia pestis | 137 | 16120576 |
| Gram-positive bacteria | ||||
| Atu | AGR_C_934p | Agrobacterium tumefaciens | 142 | 15887877 |
| Bsu | Large-conductance MS channel | Bacillus subtilis | 130 | 6016602 |
| Cpe | Large-conductance MS channel | Clostridium perfringens | 152 | 1709120 |
| Cgl | COG1970: Large conductance MS channel | Corynebacterium glutamicum | 135 | 19552104 |
| Dra | Large-conductance MS channel DR2422 | Deinococcus radiodurans | 128 | 7473090 |
| Lla | Large-conductance MS channel | Lactococcus lactis | 122 | 12725155 |
| Mle | Large-conductance MS channel | Mycobacterium leprae | 154 | 7674135 |
| Mtu | Large-conductance MS channel | Mycobacterium tuberculosis | 151 | 6016604 |
| Rso | Probable large-conductance MS channel | Ralstonia solanacearum | 141 | 175476649 |
| Sme | Probable large-conductance MS channel | Sinorhizobium meliloti | 142 | 15964307 |
| Sau | Large-conductance MS channel | Staphylococcus aureus | 120 | 6016606 |
| Spn | Large-conductance MS channel SP1010 | Streptococcus pneumoniae | 125 | 14972483 |
| Spy | Large-conductance MS channel mscL | Streptococcus pyogenes | 120 | 13621957 |
| Sco | Putative MS channel SCE22.07 | Streptomyces coelicolor | 156 | 7799510 |
| Archaea | ||||
| Mac | Large-conductance MS channel protein | Methanosarcina acetivorans | 101 | 19916224 |
| Fungi | ||||
| Ncr | Related to glycine-rich cell wall structural protein | Neurospora crassa | 373 | 16945414 |
The sequence of this protein is different from that reported in the database due to a frameshift mutation which was corrected in the studies reported here. We could not ascertain if this mutation represented a sequencing error or a bona fide frame shift in the gene sequence.
Number of amino acids.
gi, GenBank index.
Many gram-negative and gram-positive bacteria possess MscL family members (Table 1), but only one bacterium, Mesorhizobium loti, has more than one MscL homologue. Mycoplasma and Ureaplasma species, Rickettsia prowazekii, Helicobacter pylori, Campylobacter jejuni, Aquifex aeolicus, Thermotoga maritima, and Neisseria meningitidis, all with fully sequenced genomes, are not represented, showing that MscL family members are not ubiquitous. Among the gram-negative bacterial homologues, most are from proteobacteria, with the exceptions of Fusobacterium nucleatum, Synechocystissp. strain PCC6803, and the unusual double-membrane-possessing but lipopolysaccharide-lacking organism Deinococcus radiodurans, sometimes classified as a gram-positive bacterium.
Phylogenetic Analyses
The phylogenetic tree of the MscL family proteins is shown in Fig. 4A while the corresponding 16S rRNA tree is shown in Fig. 4B. This tree is based on the MscL family multiple alignment shown on our ALIGN website (http://www.biology.ucsd.edu/∼msaier/align.html). Clustering of the proteins (Fig. 4A) is usually according to organismal type within experimental error (compare Fig. 4A and B). Thus, with the exception of the divergent Xylella fastidiosa protein, all of the γ-proteobacterial proteins cluster loosely together and in accordance with clustering patterns for the 16S rRNAs. Unexpectedly, the Vibrio cholerae protein clusters with the Pseudomonas proteins. Further, in contrast to expectation, the Caulobacter crescentus homologue does not cluster with the other α-proteobacterial proteins.
FIG. 4.
Phylogenetic trees for MscL homologues (A) and 16S rRNAs from the organisms represented (B). Protein abbreviations in panel A are as indicated in Table 1, and organismal abbreviations in panel B are essentially the same. The CLUSTAL X program (54) was used to generate the multiple alignment on which the trees were based. Organismal types are indicated where the Greek letters refer to the four subgroups of γ-proteobacteria (γ1 to γ4) as well as the α and β subdivisions of proteobacteria.
The high-G+C gram-positive bacterial proteins form a single coherent cluster, although the low-G+C gram-positive bacterial proteins do not (Fig. 4A). The D. radiodurans homologue clusters loosely with the former group of organisms, while the Synechocystis protein branches from a point at the base of the principal gram-positive bacterial cluster.
The 16S rRNA tree portrayed in Fig. 4B reveals the similarities and differences between the MscL protein and the 16S rRNA phylogenetic trees. Considering the small sizes of the MscL homologues, the configuration of the tree is consistent with the suggestion that most of these proteins are orthologues, serving a single function. It is interesting that in the only organism with multiple MscL paralogues, Mesorhizobium loti, the four paralogues cluster tightly together on the phylogenetic tree, suggesting that they arose by recent gene duplication events.
Sequence-Function Correlates
MscL of E. coli is the most extensively characterized bacterial MS channel, and limited functional studies have been performed on some of its homologues (10, 11, 13, 42, 48-51). The MscL protein of E. coli spans the membrane twice (M1 and M2) as α-helices (10, 11), a characteristic of all MscL family members. In addition, there is an amino-terminal α-helix (N), an inter-TMS loop (I) that connects M1 and M2, and a short but important carboxy-terminal helix (S) linked to M2 by a flexible linker (L) (Fig. 5).
FIG. 5.

Schematic depictions of the structure of MscL MS channels. (A) Linear depiction; (B) topological depiction. M1 and M2, N- and C-terminal transmembrane spanners; N, α-helical region N-terminal to M1; I, inter-TMS loop region between M1 and M2; S, carboxy-terminal α-helical domain following M2; L, flexible linker connecting M2 to S.
The three-dimensional structure of the Mycobacterium tuberculosis MscL has been solved to 3.5 Å resolution (Fig. 1), and the crystal structure has been shown to reflect the probable structure in the intact cell membrane (13, 37, 44, 48, 49). MscL forms a homopentameric channel (13) that is proposed to undergo extensive rearrangement when the closed channel opens (43, 48, 49) (Fig. 2). The carboxy-terminal S domains form a bundle when the channel is closed, and the amino-terminal N domains, which were not evident in the original crystal structure, may project just below the membrane surface. The tight seal in the MscL channel, which is essential to the closed state and is frequently disrupted in gain-of-function mutants, is formed by a ring of hydrophobic residues proximal to the membrane face of TMS1 (13). The first stage of channel opening involves small movements in M1 (43) and may require the participation of the N domains to seal the channel by relocating to the bottom of an otherwise open basket (48, 49), thereby forming a second gate. It is the springing of this second gate, swinging back to interact with the lumen of the channel, that leads to the open state. Tension is proposed to expand the 10 TMS/5 subunit transmembrane barrel structure near the cytoplasmic surface. Cross-linking between N segments prevents opening; N and M2 interact in the open channel, and cross-linking N to M2 impedes channel closure (48, 49). The length of the linker between N and M1 is critical for proper channel gating (48, 49). It is notable that variations in the size of the N domain (8 to 12 aa) should affect the length of the α-helix by one turn. This may have implications for the gating mechanisms. The massive rearrangements which accompany transition to the open state involve both M1 and M2 (43).
When the MscL channel of C. perfringens, with a short N domain, is expressed in E. coli, it exhibits conductance and pressure sensitivity similar to those of the E. coli MscL homologue but has shorter dwell times (36). Thus, the maintenance of a tight seal in the closed state and the formation of a high-conductance open channel require major reorganization of the protein in response to membrane tension. A requirement for such rearrangements probably imposed constraints on sequence divergence, which may explain the limited size and sequence variations of these proteins.
In contrast to the amino terminus and the linker between N and M1 (48, 49), the short linkers (L) between the M2 and S regions show extensive sequence variation. L is AP rich in some proteins but predominantly charged and hydrophilic in others. Although deletion of the S domain of the E. coli MscL homologue was originally reported not to impair channel activity (10), more recent analyses have shown that this region is required for proper MscL channel formation (2). Mutant E. coli MscL proteins truncated at residue 110, a residue that lies at the amino-terminal end of the S helix, form functional channels that can protect the double mscL yggB mutant of E. coli during hypoosmotic shock. However, the mutated channel gates at a lower membrane tension and leads to more extensive ATP loss at lower osmotic downshock than observed for the wild-type channel (2). The S region may therefore perform a function in maintaining the closed state of the channel (48, 49). Since the archaeal MscL protein (Mac) lacks the S domain altogether, it may prove to undergo the transition from the closed to the open state at very low membrane tension.
M1 comprises the pentameric, amphipathic, pore-forming element, while M2 faces the hydrophobic environment of the lipid bilayer (13). M1 of the M. tuberculosis protein is quite divergent in sequence from the E. coli homologue, but genetic analyses (37) have shown that equivalent mutations cause the same general changes in properties (i.e., lowered gating pressure and altered open states). Nevertheless, differences in the properties of the channels, e.g., their pressure sensitivities when expressed in E. coli, cannot always be explained by the sequence variation at strongly conserved positions. Thus, the M. tuberculosis channel expressed in E. coli requires a much higher pressure to gate than does the E. coli channel. Alanine at position 20 is found in the two channels from M. tuberculosis and Synechocystis which exhibit a high gating pressure, but conversion of Ala-20 in the M. tuberculosis protein to Gly, the residue in the equivalent position in the E. coli protein, has only subtle effects on the frequency with which channel activity is observed in patches, consistent with a mild lowering of the response to pressure (37). Gain-of-function mutations can be introduced at conserved positions (e.g., M. tuberculosis MscL G24S), but sequence divergence that has arisen over an extended evolutionary period is likely to be complex (32). Thus, gating of these channels should be considered to be a property of the whole protein, as has been indicated by the suggestion of two gates—the hydrophobic seal and the N helix (48, 49). It is likely that during evolution, changes that occurred at conserved positions have been compensated for by others that have occurred at nonconserved positions (18, 26, 39).
Size Variation among Bacterial MscL Homologues
The nature of the transition from the closed to the open state for MscL homologues, which may require insertion of S sequences into the membrane-cytoplasmic interface (43, 48, 49), may have imposed constraints on the size and sequence divergence of these proteins. The sizes of the identified bacterial homologues exhibit a strong clustering within different taxonomic groups (Table 1). The high-G+C gram-positive bacteria have large homologues (135 to 156 aa) while the low-G+C gram-positive bacteria have small homologues (120 to 133 aa), with the sole exception of the C. perfringens homologue (152 aa). The extra residues in the C. perfringens protein are in the loop between M1 and M2, part of which may insert into the membrane at the periplasmic face (48, 49), as well as in a poorly conserved linker between M2 and S (Fig. 5). By contrast, the mycobacterial proteins have an extension to S (Fig. 5), and the Streptomyces coelicolor protein has extra residues in the N, inter-TMS loop (I), linker (L), and S regions. The greater length of the S domain in the M. tuberculosis MscL protein may pose problems for the models of the open MscL structure since this segment is envisaged to form part of the channel wall (48, 49). Potentially this is an adaptation to the different lipid composition of the membranes of this organism and may account for the difficulties encountered in expressing channel activity in E. coli (37).
The gram-negative bacterial homologues fall into the size range of the gram-positive bacterial homologues (128 to 157 aa), with the Bradyrhizobium japonicum homologue and one M. loti paralogue being the largest. A longer loop between M1 and M2 is found in all M. loti paralogues as well as in the B. japonicum orthologue, but the M. loti paralogue, Mlo1, also has an expanded N region. The somewhat smaller Synechocystis protein has extensions in both N and S but no increase in the loop region. Thus, the core regions comprising M1, M2, and S are well conserved in both sequence and size, and the larger homologues have insertions in various nonconserved regions as well as possible extensions to S.
Patterns of Conservation within Bacterial MscL Homologues
Alignment of bacterial members of the MscL family has revealed the relative degree of conservation along the length of these proteins (Fig. 6). Most importantly, M1 and the linker between N and M1 are much better conserved than are N, M2, I, L, and S. Six residues that lie in or amino-terminal to M1 are found in all bacterial members of the family: F10, R/K13, G14, N15, A20, and F29 in the E. coli MscL. A strong periodicity in the quality fit determined using the Clustal X-derived alignment is noteworthy (Fig. 6). Approximately every third or fourth residue is poorly conserved, while the intervening residues show strong conservation, consistent with the α-helical arrangement of M1. In contrast, M2 exhibits limited conservation, where F85 in the E. coli MscL is the only fully conserved residue, and there is a less pronounced periodicity. F85 is thought to be important for the interaction between N and M2 in the open state of the channel. Modified MscL proteins, carrying I3C (N domain) and I96C (M2) substitutions, form cross-links in the presence of iodine that prevent closure of the channel. This and related evidence (48, 49) indicate that N and M2 are in close proximity in the open channel, consistent with the proposed interaction of F85 with F7 or F10 in the N domain. It is interesting that all the above-mentioned conserved residues except R/K13 are found in the fungal homologue but only F10, A20, and F85 are retained in the archaeal protein.
FIG. 6.
Relative residue conservation at each position in the multiple alignment of the bacterial MscL homologues. TMS positions were calculated using the WHAT (59) and MEMSAT (19; modified by us) programs. The two most prevalent residues at each alignment position within TMS 1 (M1) are presented. Red residues are shared by dominant residues found in conserved TMS 3 of the MscS proteins shown in Fig. 9. The relative conservation index was calculated using the CLUSTAL X program (Gonnet Pam 250 [3]).
MscS CHANNEL FAMILY
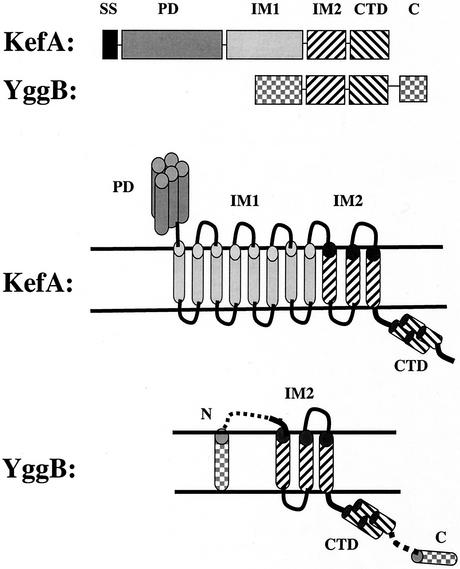
The MscS family (TC 1.A.23) is larger and much more variable in size and sequence than the MscL family (45) (Table 2). In E. coli there are two primary topological classes (Fig. 7). KefA (AefA) (1,120 aa), YjeP (1,107 aa), and YbiO (741 aa) are all large proteins that exhibit 11 putative TMSs. KefA has a cleavable amino-terminal signal sequence (SS), a large N-terminal periplasmic domain (PD) that is predicted to be a helical bundle (possibly with a coiled-coil structure) (residues 1 to 470 in KefA), a hydrophobic region including a total of 11 predicted TMSs (residues 480 to 940) with a linker (L) between TMSs 8 and 9, and a carboxy-terminal cytoplasmic domain (CTD) (residues 941 to 1120) (33). On the basis of the presence of the linker between TMSs 8 and 9, two membrane domains (IM1 and IM2) can be proposed (Fig. 7), with IM1 containing eight TMSs and IM2 containing three TMSs. While KefA and YjeP are similar in size, the principal size difference between KefA and YbiO arises from an in-frame deletion in the N-terminal periplasmic domain in the latter. This periplasmic domain is a common characteristic of the KefA subfamily of MscS homologues. Assuming that KefA is multimeric (33), this raises the possibility that this domain may form a supramolecular structure. KefA subfamily proteins are restricted to gram-negative bacteria. At least one organism, Magnetococcus, which lacks a full-length KefA homologue, has a separate secreted protein similar in sequence to the amino terminus of KefA. It has been suggested that the amino-terminal domain of KefA may form a link to the gram-negative bacterial outer membrane, as does TolC (33).
TABLE 2.
Sequenced proteins of the MscS family
| Abbreviation | Database description | Organism | Sized | gie number |
|---|---|---|---|---|
| Archaea | ||||
| Ape1 | Hypothetical protein APE1441 | Aeropyrum pernix | 285 | 7444698 |
| Ape2 | Hypothetical protein APE1867 | Aeropyrum pernix | 295 | 7516782 |
| Ape3 | Hypothetical protein APE2455 | Aeropyrum pernix | 283 | 7517184 |
| Afu | Hypothetical protein AF1546 | Archaeoglobus fulgidus | 283 | 11499141 |
| Hsp1 | Vng1164c | Halobacterium sp. strain NRC-1 | 300 | 10580699 |
| Hsp2 | Vng2113c | Halobacterium sp. strain NRC-1 | 274 | 10581527 |
| Hsp3c | Vng1388h | Halobacterium sp. strain NRC-1 | 259 | 10580895 |
| Mja1 | Hypothetical protein MJ0170 | Methanococcus jannaschii | 350 | 2501530 |
| Mja2 | Hypothetical protein MJ1143 | Methanococcus jannaschii | 361 | 2501531 |
| Mja3 | Hypothetical protein MJ0700 | Methanococcus jannaschii | 324 | 2833543 |
| Mth | Conserved hypothetical protein MTH1830 | Methanobacterium thermoautotrophicum | 248 | 7482176 |
| Pab | Hypothetical protein PAB1281 | Pyrococcus abyssi | 346 | 14521824 |
| Pho | Hypothetical protein PH0336 | Pyrococcus horikoshii | 335 | 7518590 |
| Sso1 | Conserved hypothetical protein SSO2769 | Sulfolobus solfataricus | 308 | 13816106 |
| Sso2 | Conserved hypothetical protein SSO2829 | Sulfolobus solfataricus | 291 | 13816184 |
| Sso3c | Hypothetical protein SSO0550 | Sulfolobus solfataricus | 314 | 6015922 |
| Sso4a | Hypothetical protein SSO2786 | Sulfolobus solfataricus | 205 | 13816129 |
| Tac1 | Conserved hypothetical membrane protein Ta0796 | Thermoplasma acidophilum | 286 | 10640074 |
| Tac2 | Conserved hypothetical membrane protein Ta0909 | Thermoplasma acidophilum | 297 | 10640186 |
| Tvo1 | Small-conductance MS channel TVN0705 | Thermoplasma volcanium | 288 | 13541536 |
| Tvo2 | Small-conductance MS channel TVN0821 | Thermoplasma volcanium | 297 | 13541652 |
| Gram-negative bacteria | ||||
| Aae1 | Hypothetical protein AQ_812 | Aquifex aeolicus | 368 | 6136577 |
| Aae2 | Conserved hypothetical protein AQ_1013 | Aquifex aeolicus | 436 | 7514431 |
| Bbu | Conserved hypothetical protein BB0453 | Borrelia burgderi | 280 | 7444701 |
| Bsp | Hypothetical protein BU452 | Buchnera sp. strain APS | 305 | 11387303 |
| Bma | YggB protein | Burkholderia mallei | 290 | 13446682 |
| Bps | YggB protein | Burkholderia pseudomallei | 271 | 13932325 |
| Cje1 | Probable integral membrane protein Cj0238 | Campylobacter jejuni | 627 | 11346911 |
| Cje2 | Probable membrane protein Cj1007c | Campylobacter jejuni | 523 | 11282200 |
| Ccr1 | Conserved hypothetical protein CC3612 | Caulobacter crescentus | 328 | 13425360 |
| Ccr2 | Conserved hypothetical protein CC3000 | Caulobacter crescentus | 341 | 13424636 |
| Ctea | AefA protein (fragment) | Chlorobium tepidum | 230 | 10198122 |
| Eic | Hypothetical 30.6-kDa protein | Edwardsiella ictaluri | 286 | 6686193 |
| KefA (Eco) | Potassium efflux system KefA | Escherichia coli | 1120 | 2501527 |
| YggB (Eco2) | Hypothetical 30.9-kDa protein | Escherichia coli | 286 | 140687 |
| YjeP (Eco3) | Hypothetical 123.8-kDa protein | Escherichia coli | 1107 | 2851660 |
| Eco4 | Hypothetical 38.8-kDa protein | Escherichia coli | 343 | 6176597 |
| YbiO (Eco5) | Hypothetical 81.9-kDa protein | Escherichia coli | 741 | 3915948 |
| Eco6 | Hypothetical 46.6-kDa protein | Escherichia coli | 415 | 2506616 |
| Hin | Protein HI0195.1 precursor | Haemophilus influenzae | 1111 | 2501528 |
| Hpy1 | Hypothetical integral membrane protein HP0983 | Helicobacter pylori | 274 | 7444700 |
| Hpy2 | Hypothetical integral membrane protein HP0284 | Helicobacter pylori | 523 | 7463934 |
| Hpy3 | Hypothetical protein HP0415 | Helicobacter pylori | 623 | 6136524 |
| Mlo1 | Hypothetical protein mll1272 | Mesorhizobium loti | 850 | 13471329 |
| Mlo2 | Hypothetical protein mlr4006 | Mesorhizobium loti | 283 | 13473416 |
| Mlo3 | Hypothetical proteins mll3287 | Mesorhizobium loti | 739 | 13472860 |
| Mlo4c | Probable integral membrane protein mlr0973 | Mesorhizobium loti | 410 | 13471089 |
| Nme1 | Conserved hypothetical protein NMB0042 | Neisseria meningitidis MC58 | 282 | 11278435 |
| Nme2 | Hypothetical protein NMB0213 | Neisseria meningitidis MC58 | 328 | 11282692 |
| Neua | ORF2 (fragment) | Nitrosomonas europaea | 109 | 3777488 |
| Pmu | Unknown | Pasteurella multocida | 1113 | 12720603 |
| Pch1 | BspA protein | Pectobacterium chrysanthemi | 1106 | 11691630 |
| Pch2a | Hypothetical protein; BspB (fragment) | Pectobacterium chrysanthemi | 999 | 15027208 |
| Pae1 | Conserved hypothetical protein PA5022 | Pseudomonas aeruginosa | 1118 | 11348278 |
| Pae2 | Conserved hypothetical protein PA4394 | Pseudomonas aeruginosa | 278 | 11348170 |
| Pae3 | Conserved hypothetical protein PA4925 | Pseudomonas aeruginosa | 283 | 11348259 |
| Pae4 | Hypothetical protein PA1408 | Pseudomonas aeruginosa | 807 | 11349198 |
| Pae5 | Hypothetical protein PA5121 | Pseudomonas aeruginosa | 735 | 11350368 |
| Pae6 | Conserved hypothetical protein PA3468 | Pseudomonas aeruginosa | 442 | 11348032 |
| Pae7c | Conserved hypothetical protein PA1775 | Pseudomonas aeruginosa | 274 | 11347832 |
| Pae8c | Hypothetical protein PA5251 | Pseudomonas aeruginosa | 192 | 11350399 |
| Pfl | CmpX | Pseudomonas fluorescens | 274 | 5668604 |
| Rpr | Hypothetical protein RP047 | Rickettsia prowazekii | 388 | 7467637 |
| Styc | Hypothetical protein | Salmonella enterica serova Typhimurium | 377 | 2337947 |
| Sme1 | Conserved hypothetical protein SMa1582 | Sinorhizobium meliloti | 432 | 14523992/PICK> |
| Sme2a | Conserved hypothetical membrane protein SMa0630 | Sinorhizobium meliloti | 523 | 14523416 |
| Sme3 | Conserved hypothetical protein SMa0937 | Sinorhizobium meliloti | 719 | 14523607 |
| Sme4b | Hypothetical protein SMa1678 (partial homologue) | Sinorhizobium meliloti | 93 | 14524059 |
| Ssp1 | Hypothetical 33.2-kDa protein slr0639 | Synechocystis sp. strain PCC6803 | 296 | 2501529 |
| Ssp2 | Hypothetical protein slr1575 | Synechocystis sp. strain PCC6803 | 479 | 7469751 |
| Ssp3 | Hypothetical protein slr0510 | Synechocystis sp. strain PCC6803 | 505 | 7469671 |
| Ssp4 | Hypothetical protein sll0985 | Synechocystis sp. strain PCC6803 | 704 | 7469967 |
| Ssp5 | Hypothetical protein slr0765 | Synechocystis sp. strain PCC6803 | 617 | 7470274 |
| Ssp6 | Hypothetical protein sll1040 | Synechocystis sp. strain PCC6803 | 765 | 7469981 |
| Ssp7 | Hypothetical protein slr0109 | Synechocystis sp. strain PCC6803 | 318 | 7469704 |
| Ssp8c | Hypothetical protein sll0590 | Synechocystis sp. strain PCC6803 | 564 | 7469688 |
| Tma | Conserved hypothetical protein TM1563 | Thermotoga maritima | 268 | 7444699 |
| Tpa | Conserved hypothetical protein TP0822 | Treponema pallidum | 301 | 7514621 |
| Vch1 | Conserved hypothetical protein VC0480 | Vibrio cholerae | 287 | 11278432 |
| Vch2 | Conserved hypothetical protein VC1751 | Vibrio cholerae | 292 | 11278433 |
| Vch3 | Conserved hypothetical protein VC0265 | Vibrio cholerae | 412 | 11354507 |
| Vch4 | Hypothetical protein VCA0817 | Vibrio cholerae | 194 | 11345735 |
| Vch5 | Hypothetical protein VCA0181 | Vibrio cholerae | 291 | 11355375 |
| Xfa1 | Small-conductance MS ion channel XF1258 | Xylella fastidiosa | 305 | 11362678 |
| Xfa2 | Conserved hypothetical protein XF0437 | Xylella fastidiosa | 431 | 11360698 |
| Zmo1 | Hypothetical protein | Zymomonas mobilis | 618 | 5354203 |
| Zmo2c | Unknown | Zymomonas mobilis | 376 | 4378179 |
| Gram-positive bacteria | ||||
| Bha1 | BH2683, unknown conserved protein | Bacillus halodurans | 379 | 10175304 |
| Bha2 | BH2666, unknown conserved protein | Bacillus halodurans | 276 | 10175287 |
| Bsu1 | Hypothetical 42.5-kDa protein | Bacillus subtilis | 371 | 6136721 |
| Bsu2 | Conserved hypothetical protein YkuT | Bacillus subtilis | 267 | 7474634 |
| Bsu3 | Conserved hypothetical protein YfkC | Bacillus subtilis | 280 | 14523156 |
| Dra1 | Conserved hypothetical protein DR0211 | Deinococcus radiodurans | 368 | 7471627 |
| Dra2 | Conserved hypothetical protein DR1995 | Deinococcus radiodurans | 426 | 7471720 |
| Lla | Conserved hypothetical protein YncB | Lactococcus lactis | 248 | 12724266 |
| Mtu1b,c | Cyclic nucleotide-binding protein Rv2434c | Mycobacterium tuberculosis | 481 | 7478360 |
| Mtu2 | Hypothetical protein Rv3104c | Mycobacterium tuberculosis | 308 | 7477495 |
| Sau | Conserved hypothetical protein SAV0361 | Staphylococcus aureus | 293 | 13700275 |
| Spn | MS ion channel SP1752 | Streptococcus pneumoniae | 201 | 14973248 |
| Spy | Conserved hypothetical protein SPy1897 | Streptococcus pyogenes | 281 | 13622927 |
| Sco1 | Hypothetical protein SCF43A.26c | Streptomyces coelicolor | 333 | 7479987 |
| Sco2 | Putative membrane protein SC8E4A.26 | Streptomyces coelicolor | 408 | 6900955 |
| Sco3 | Probable membrane protein SC4A10.03 | Streptomyces coelicolor | 382 | 7480980 |
| Sco4c | Putative integral membrane protein SC8F11.27c | Streptomyces coelicolor | 353 | 7649646 |
| Eukaryotes | ||||
| Ath1c | Unknown protein | Arabidopsis thaliana | 676 | 12321837 |
| Ath2 | Similarity to putative protein T30F21.6 | Arabidopsis thaliana | 881 | 8671879 |
| Ath3 | Hypothetical protein | Arabidopsis thaliana | 856 | 4836872 |
| Ath4 | Hypothetical protein | Arabidopsis thaliana | 849 | 6598357 |
| Ath5 | Hypothetical protein F14F18.230 | Arabidopsis thaliana | 734 | 11357421 |
| Ath6 | GblAAD30575.1 ∼gene_id:T21E2.7∼ | Arabidopsis thaliana | 881 | 11994592 |
| Ath7a | Hypothetical protein | Arabidopsis thaliana | 698 | 6598358 |
| Ath8 | Hypothetical protein F12B17.160 | Arabidopsis thaliana | 447 | 7474572 |
| Ath9 | Hypothetical protein A_IG005I10.9 (partial homologue) | Arabidopsis thaliana | 353 | 7485270 |
| Ath10c | Hypothetical protein F12B17.160 | Arabidopsis thaliana | 477 | 11357358 |
| Spo1 | Conserved hypothetical protein SPAC2C4.17c | Schizosaccharomyces pombe | 840 | 7490286 |
| Spo2 | Hypothetical protein SPCC1183.11 | Schizosaccharomyces pombe | 1011 | 7491774 |
These proteins are thought to be fragments and therefore were not included in the study reported in Table 3.
These proteins were more divergent in sequence than the other homologues and were therefore omitted from our study.
These proteins were omitted from Fig. 11 (see our ALIGN website) because the conserved motif could not be established throughout their sequences.
Number of amino acids.
gi, GenBank index.
FIG. 7.
Schematic depictions of the proposed structures of two structural types present within the MscS family represented by the E. coli KefA and YggB proteins. The illustrations at the top are linear depictions, while those at the bottom are topological models. SS, signal sequence; PD, periplasmic domain; IM1 and IM2, membrane-spanning domains 1 and 2 (of eight and three putative TMSs, respectively); CTD, carboxy-terminal domain; N and C, amino-terminal and carboxy-terminal regions, respectively, of YggB and structurally related proteins. The IM2 and CTD domains (hatched) are similar in organization, structure, and sequence between the KefA and YggB subfamilies. The smaller members of the MscS family may have extensions at the amino and carboxy termini (checkered regions in the bottom figure).
Proteins resembling E. coli YggB are generally much smaller than the E. coli KefA protein, but they are nevertheless heterogeneous in size. E. coli proteins YggB (286 aa), YbdG (415 aa), and YjcR (343 aa) are relatively short with a core sequence that corresponds to the IM2 domain plus the carboxy-terminal domain (CTD) of KefA (Fig. 7). The YggB-like proteins exhibit considerable diversity in size due to variations in the length of the IM1 domain (largely absent in YggB; only two or three spans in M. jannaschii MJ1143 [Mja2; 361 aa; spQ58543]) and in the length of the CTD. Examination of Table 2 reveals that close YggB homologues occur in archaea, in bacteria, and, within the eukaryotic domain, in both plants and yeasts but not in animals. Several organisms have multiple paralogues. For example, Arabidopsis thaliana has 10, P. aeruginosa has 9, Synechocystis sp. strain PCC6803 has 8, E. coli has 6, and V. cholerae has 5. Some archaea have three or four paralogues. However, several organisms with fully sequenced genomes do not encode recognizable MscS homologues. These organisms include the gram-negative chlamydias, the gram-positive clostridia, mycoplasmas and ureaplasmas, most of the archaeal methanogens, and animals. Thus, although more widespread than MscL homologues, the MscS family is by no means ubiquitous.
Phylogenetic Analyses
The phylogenetic tree for the MscS family is shown in Fig. 8A, and that for the 16S rRNAs from the same organisms is shown in Fig. 8B. Most of the eukaryotic proteins fall into a single cluster (cluster XVII), where six plant proteins segregate from the two yeast proteins. Two remaining plant proteins are found on an additional branch (cluster XI), while the third such protein (Ath8) is not closely related to any other family member. The archaeal proteins are found on seven very divergent branches, but the majority of these proteins cluster on just two of these branches (clusters IV and X). Just as no eukaryotic protein clusters with a prokaryotic protein, no archaeal protein clusters closely with a bacterial protein.
FIG. 8.
(A) Phylogenetic tree for MscS homologues; (B) 16S rRNA tree for organisms in panel A. Protein and organismal abbreviations are as indicated in Table 2. The various phylogenetic clusters I to XX are labeled by number. Their size variations and organismal types are reported in Table 3. The footnoted proteins listed in Table 2 may have incomplete sequences, and these proteins were not included in the phylogenetic tree.
The gram-positive bacterial proteins are found on just six deep-rooted branches, two of which include all of the low-G+C gram-positive bacterial proteins and four of which include the high-G+C gram-positive bacterial proteins. No gram-positive bacterial protein clusters closely with a gram-negative bacterial protein, although one of the two paralogues from D. radiodurans clusters loosely with the largest of the high-G+C gram-positive bacterial clusters. Finally, among the gram-negative bacterial proteins, the two spirochete proteins and those from evolutionarily divergent bacterial species (Tma, Aae, Dra, and Ssp) do not cluster with any of the proteobacterial proteins or with each other. This tree therefore argues against a relatively recent horizontal transfer of genes encoding MscS homologues between the three domains of life as well as between the evolutionarily divergent bacterial kingdoms.
Examination of the close clustering patterns shown in Fig. 8A for the MscS family reveals several cases of recent gene duplications within a single organism as well as highly probable orthologous relationships between proteins of different species, particularly within the proteobacteria. Within each phylogenetic cluster, there is little size variation even though there is tremendous size variation between clusters (Table 3). Thus, for example, cluster I gram-negative bacterial proteins have a size range of 705 ± 57 residues; cluster II high-G+C gram-positive bacterial proteins have a size range of 369 ± 57 residues; cluster III low-G+C gram-positive bacterial proteins have a size range of 274 ± 18 residues; cluster VII gram-negative bacterial proteins have a size range of 1,113 ± 6 residues; cluster VIII gram-negative bacterial proteins have a size range of 417 ± 20 residues; and cluster XVII eukaryotic proteins have a size range of 867 ± 89 residues. These considerations reveal that phylogenetic cluster correlates remarkably well with both size and organismal type. One can further suggest that phylogeny also predicts close functional relationships.
TABLE 3.
Size variation for twenty phylogenetic clusters of MscS family homologuesa
| Cluster no. | Organismal type | No. of proteins | Average sized ± SD | Average no. of TMS ± SD by:
|
|
|---|---|---|---|---|---|
| WHATb | TOPPRED2c | ||||
| I | Gram-negative bacteria | 9 | 710 ± 57 | 8.9 ± 3.0 | 9.0 ± 2.8 |
| II | High-G+C gram-positive bacteria | 4 | 370 ± 57 | 4.0 ± 1.2 | 4.0 ± 1.2 |
| III | Low-G+C gram-positive bacteria | 6 | 270 ± 18 | 3.0 ± 0 | 3.0 ± 0.5 |
| IV | Archaea | 7 | 290 ± 8.0 | 4.6 ± 0.8 | 4.3 ± 1.0 |
| V | Gram-negative bacteria | 5 | 290 ± 14 | 3.6 ± 0.6 | 3.6 ± 0.6 |
| VI | Gram-negative bacteria | 11 | 290 ± 16 | 3.4 ± 1.3 | 3.5 ± 0.5 |
| VII | Gram-negative bacteria | 7 | 1,100 ± 6.0 | 11 ± 0.5 | 12 ± 0.7 |
| VIII | Gram-negative bacteria | 6 | 440 ± 62 | 5.2 ± 2.8 | 4.7 ± 2.4 |
| IX | Archaea | 2 | 290 ± 18 | 3.0 ± 0 | 3.0 ± 0 |
| X | Archaea and spiroplasma | 7 | 300 ± 33 | 4.0 ± 1.2 | 3.6 ± 1.0 |
| XI | Plants | 2 | 580 ± 140 | 4.0 ± 1.4 | 3.0 ± 1.4 |
| XII | Archaea | 2 | 360 ± 8.0 | 5.0 ± 0 | 4.0 ± 0 |
| XIII | Gram-negative bacteria and low-G+C gram-positive bacteria | 4 | 370 ± 16 | 5.0 ± 0 | 5.3 ± 0.5 |
| XIV | Gram-negative bacteria | 2 | 630 ± 3.0 | 4.0 ± 0 | 5.5 ± 0.7 |
| XV | Archaea | 3 | 290 ± 33 | 4.3 ± 0.6 | 4.0 ± 1.0 |
| XVI | Gram-negative bacteria | 2 | 520 ± 0 | 5.0 ± 1.4 | 3.0 ± 0 |
| XVII | Eukaryotes | 8 | 870 ± 89 | 6.1 ± 1.1 | 6.4 ± 0.7 |
| XVIII | Gram-negative bacteria | 5 | 420 ± 20 | 3.9 ± 1.1 | 4.6 ± 0.9 |
| XIX | Gram-negative bacteria | 6 | 270 ± 44 | 3.8 ± 1.0 | 3.2 ± 1.5 |
| XX | Mixed | 3 | 360 ± 34 | 3.3 ± 2.5 | 4.0 ± 1.0 |
Proteins described in Table 2 footnote a are thought to be incompletely sequenced fragments and therefore were not included in this study.
Average number of TMSs within that cluster predicted using the WHAT program (59).
Average number of TMSs within that cluster predicted using the TOPPRED2 program (47).
Number of amino acids.
The multiple alignment of more than 100 homologues on which the tree shown in Fig. 8A was based utilized the most highly conserved portions of the longer MscS proteins and included the complete sequences of several of the shorter homologues (i.e., Ape1, Ape2, Ccr2, Eie, Eco2, and Pae2). This alignment includes 450-residue positions and can be viewed on our ALIGN website (http://www.biology.ucsd.edu/∼msaier/align.html). Based on the MscS family phylogenetic tree (Fig. 8A), most members of the MscS family were assigned to clusters and were analyzed for putative TMSs by using the WHAT and TOPPRED2 programs, as reported in Table 3 (47, 59). The mean of the numbers of TMSs within each cluster, along with the standard deviation is provided. In spite of the tremendous overall topological variation, little variation is observed within most of the clusters. Only clusters I, VIII, and XX show significant topological variation.
The topology of the YggB protein has been investigated using phoA fusion technology (10, 27, 33, 35). PhoA fusions were isolated in 13 positions, and the highest activity was associated with a fusion at residue A94. All other fusions gave low alkaline phosphatase activity and were unstable, consistent with a cytoplasmic location. The data agree with the locations of positive charges in YggB in accordance with the positive-inside rule (1, 56). A three-TMS topology is consistent with the results and would give the simplest conventional structure with the CTD (residues 175 to 286) in the cytoplasm (35). These data are in agreement with the proposed structure of the related protein, KefA (33).
Analysis of the YggB subfamily showed that in this core region, there are only three sites at which sequence insertions have occurred: at positions equivalent to ∼60, ∼150, and ∼200 in YggB of E. coli. Of these, the most significant is the one at ∼60, since this corresponds to the end of the first putative TMS. All of the insertions augment the already significant numbers of lysine and arginine residues in this putative loop (adding up to 10 basic residues in the largest insertion), which would anchor this region firmly at the cytoplasmic face of the membrane (1, 56).
Sequence Conservation and Comparisons with MscL Family Proteins
Kloda and Martinac speculated that the M. jannaschii MJ0170 protein has evolved through the fusion of ancestral MscL and YggB type sequences (20). These authors proposed a relationship between M1 in MscL and TMS 1 in MJ0170 that corresponds to TMS 1 in YggB. We found that the conservation of residue character in MscL M1 (see above) is not apparent in TMS 1 of MJ0170 or in the corresponding TMS in other MscS homologues. Further, although family trees can be drawn that appear to link any unrelated sequences such as the MscS and MscL sequences (22), there is little or no statistical evidence for the link proposed by Kloda and Martinac (20, 22). As shown below, if there is an evolutionary link between the MscS and MscL families, it is apparent only when the MscL TMS 1 is equated to the conserved MscS TMS 3.
The most highly conserved region in the entire MscS family encompasses the last common α-helical TMS shown in Fig. 7 (55). Although there are few strongly conserved residues common to the entire MscS family (see below), striking patterns of sequence conservation can be observed within individual clusters. Thus, it appears that within the subgroups, there are residues conserved for specific functions.
The pattern of conservation for the YggB subfamily of the MscS family is shown in Fig. 9. For proteins closely related to YggB (subcluster VI in Fig. 8A), strong periodicity of conservation is observed throughout the third transmembrane span, i.e., at intervals of about 3 or 4 residues (see Fig. 9). This is in marked contrast to the other two TMSs, which are relatively poorly conserved. This observation clearly suggests that the last TMS is of particular functional significance.
FIG. 9.
Relative residue conservation within the YggB subfamily (cluster VI) of the MscS family. The three TMSs displayed correspond to the three well-conserved TMSs common to all MscS family proteins. The format of presentation and the programs used are the same as for Fig. 6. The red residues within TMS 3 are the residues is shared with those in TMS 1 of MscL family proteins (Fig. 6).
Each of the 20 subfamilies within the MscS family was analyzed for sequence conservation in the region of conserved TMS 3. In Fig. 10, the results are summarized; only well-conserved residues for the subfamilies are shown, and fully conserved residues within each subfamily are noted by asterisks. Residues shown in red are well conserved between the subfamilies. The conserved consensus sequence for the entire family is presented at the bottom of the figure with the percent conservation indicated in parentheses after the residue. This consensus sequence is G X11 G D X [I V] X30 G X V X31 P N X9 N, where X is any residue; alternative residues at one alignment position are indicated in brackets. These observations lead to the conclusion that some structural and/or functional features are common to all members of the MscS family.
FIG. 10.
Multiple alignment of the most highly conserved portions of the 20 principal subfamilies of the MscS family. The numbers of the clusters are the same as those in Fig. 8A and Table 3. Red residues are those well conserved in most subfamilies; conserved residues in each subfamily are presented below that group of aligned sequences: ∗, fully conserved; :, only close conservative substitutions; •, more distant conservative substitutions. The pattern of conserved residues for the entire MscS family is provided at the bottom of the figure, with the percent identities indicated in parentheses following the most highly conserved residues. X, indicates any residue; alternative residues at any one position are indicated in brackets (e.g., [I V]). Note that cluster XI was excluded because the conserved motif could not be established in the alignment for this cluster.
As noted above, TMS 1 in MscL is known to be the channel-forming helix, and this TMS is far better conserved in sequence than is TMS 2 (Fig. 6). Moreover, of the three TMSs common to all MscS channels, TMS 3 is much better conserved than the other two (Fig. 9). In Fig. 11, we compare conserved residues in MscL TMS 1 (Fig. 11A) with those in MscS TMSs 3 (Fig. 11B). When these TMSs in the two families of channel proteins were aligned, significantly more identities and similarities were found by the GAP (16) and CLUSTAL X programs than when the MscS TMS 3 were compared with MscL TMS 2 (see the values to the right of the alignment shown in Fig. 11B). Moreover, the dominant residues in 8 of the 22 positions were identical between the two sequences, while in 19 of these 22 positions, predominantly conservative substitutions occurred (Fig. 11C). These results clearly argue that TMS 3 in the MscS proteins serves the same function as TMS 1 in the MscL proteins. We propose that both serve as channel-lining helices with a common generalized structure and possibly a common evolutionary origin. It is interesting that MscS TMS 3 is proposed to have an “out-to-in” orientation rather than the “in-to-out” orientation of MscL TMS 1. Partial conservation of the adjacent N- and C-terminal regions (Fig. 9) suggests that they might be involved in gating.
FIG. 11.
(A and B) Comparison of TMS 1 in MscL homologues (A) with conserved TMS 3 in MscS homologues (B). Protein abbreviations are as presented in Tables 1 and 2, respectively. The MscL consensus sequence can be seen under the multiple alignment in panel A (red letters). The numbers of identities (#Identities), the numbers of similarities (#Similarities), and the scores (an identity = 4 points; a similarity = 1 point) are presented to the right of the aligned sequences in panel B. The average values and the average control values (observed when the same MscL consensus sequence is compared with TMSs 1 or 2 of the MscS proteins) are presented. (C) Comparison of the one or two most highly conserved residues presented in panels A and B for the MscL TMS 1 and the MscS TMS 3, respectively. Remarkable positional conservation between the two sequences is apparent, as noted in the text.
CONCLUSIONS AND PERSPECTIVES
In this study, we have analyzed two families of MS channel proteins, designated MscL and MscS. On the basis of our analyses, we conclude that the two families of proteins are distinct and, at least in the recent past, have followed separate evolutionary pathways. If conservation of sequence and organization can be taken as a guide to function, then one can speculate that TMS 3 of the YggB subfamily proteins in the MscS family (and probably the corresponding TMSs of all MscS proteins) may be the functional equivalent of TMS 1 in MscL family proteins. In marked contrast to the situation with the MscL family, few gain-of-function mutants affecting MscS homologues have to be isolated to provide confirmation of this proposal. Recently, Blount and colleagues have identified a gain-of-function mutation in YggB (V40D) with similar characteristics to MscL mutations that have a modified hydrophobic seal (41). The mutation in YggB would be positioned close to the cytoplasmic face of TMS 1, which is in a position similar to gain-of-function mutations in MscL that cause a reduction in the gating pressure of this channel. The discovery of this YggB allele has led to the proposal that both channels require a hydrophobic seal in the closed state (41). A tight seal must be maintained in the YggB channel since the perpetual open state would be expected to cause profound growth inhibition. Analysis of TMS 1 of YggB reveals that this TMS is predominantly hydrophobic, unlike TMS 1 of MscL, which, as a classical pore-lining helix, is amphipathic. The V40D mutation probably causes a profound alteration in the conformation of YggB, suggesting an important role for TMS 1 in maintaining the closed state. However, a number of gain-of-function alleles have now been identified in YggB, and these lie in the periplasmic loop between TMS 2 and TMS 3 (T93R) as well as in TMS 3 (L109S and A102P) (35). The finding that V40D is a gain-of-function allele is an important piece of evidence that will ultimately bear on the structural changes associated with gating, but it is insufficient to lead to the conclusion that it is the residue at the hydrophobic seal in the manner observed for V23 of E. coli MscL.
The transport mechanisms and/or the modes of regulation for members of the MscS family may prove to vary in accordance with topology. Possible differences in function between KefA and YjeP and between YggB and YjcR have been suggested previously (25, 55). It was observed that E. coli mutants lacking YggB and KefA failed to exhibit significant MscS-type channel activity despite the presence of four homologues of KefA and YggB (25) (Table 2). In addition, overexpression of YjcR did not restore survival to an mscL yggB double mutant, suggesting either that this protein is not an Msc channel or that the gating pressure is too high to allow complementation of the defect in the double mutant (N. R. Stokes and I. R. Booth, unpublished data). In Erwinia chrysanthemi, a yjeP gene homologue (bspA) was identified following selection of mutants sensitive to high osmolarity in the presence of the compatible solute betaine (55). Loss of BspA caused a growth defect that was not seen when yjeP was deleted from E. coli. The reasons for this difference are not known, but given the absence of evidence for channel function associated with YjeP, it seems possible that the dominant function of this protein is not that of an MS channel. Alternatively, the yjeP gene may be expressed at too low a level to give rise to functional channel activity.
Several interesting observations have resulted from our studies. First, the MscS family is much larger and more widespread than the MscL family, frequently with numerous paralogues in any one organism. By contrast, the MscL family is largely restricted to bacteria, and only one bacterium was found to exhibit more than one MscL homologue. However, while all members of the MscL family tested except the M. tuberculosis homologue were positive for complementation of the double-channel mutant MJF455 (37), only the E. coli YggB channel protein within the MscS family has been shown to complement this mutant. Several close homologues showed no significant complementation (Stokes and Booth, unpublished). This may suggest that these proteins serve a diversity of functions. However, negative results of this kind are difficult to interpret.
Second, we found that in both families, protein phylogenetic clustering generally correlates with organismal type, suggesting orthologous relationships for all or most members of the family (in the case of the MscL family) or for members of specific subfamilies (in the case of the MscS family). It seems clear that in the latter family, early gene duplication events gave rise to sequence divergent paralogues while recent duplications gave rise to easily identifiable sequence-similar paralogues. In neither family was there evidence of lateral gene transfer between distantly related organisms.
Third, each phylogenetic cluster within the MscS family shows a characteristic size, topology and organismal origin even though two different clusters, including proteins of very different size, may be derived from the same group of organisms. This observation further leads to the suggestion that each cluster represents a group of topologically and functionally homogeneous proteins. A tendency of MscL proteins to cluster according to both organismal type and size was also noted, although this tendency was less pronounced than for the much larger and more diverse MscS family.
Finally, we found that both MscL and MscS channels are represented in the various organisms in ways that correlate roughly with genome size. Thus, the genome size and number of MscS paralogues correlate together as follows: A. thaliana > P. aeruginosa > E. coli, V. cholerae or Synechocystis sp. strain PCC6803 > most archaea and small-genome bacterial pathogens > Mycoplasma and Ureaplasma species. Moreover, MscL homologues are found in most large- and moderately sized genome bacteria and archaea but in only a few small-genome bacteria or archaea. One tends to find reduced numbers of MscS family members in organisms that lack MscL family members, suggesting that genome reduction, correlating with a diminished need for adaptive capacity, correlates with loss of MS channel function. When homeostasis is provided by a host organism, as with many human and animal pathogens, the need for quick adaptation in response to osmotic change may be lost. Such observations may provide a clue to the “Achilles heel” of certain pathogenic bacteria.
Acknowledgments
Work in the Saier laboratory was supported by NIH grants GM55434 and GM64368 from the National Institute of General Medical Sciences. Work in the Booth laboratory was supported by a Wellcome Trust Programme grant (040174); I. R. Booth is a Wellcome Trust Research Leave Fellow.
We thank Mary Beth Hiller for her assistance in the preparation of the manuscript and Salar Partovi for assistance with some of the computational analyses. We thank Paul Blount, Sergei Sukharev, Ching Kung, and Tarmo Roosild for helpful discussions, preprints of unpublished work, and communication of unpublished results.
REFERENCES
- 1.Andersson, H., and G. von Heijne. 1994. Membrane protein topology: effects of ΔμH+ on the translocation of charged residues explain the "positive inside' rule. EMBO J. 13:2267-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anishkin, A., N. Sharifi, and S. Sukharev. The C-terminal domain confers structural and functional stability to the bacterial mechanosensitive channel MscL. J. Gen. Physiol., in press. [DOI] [PMC free article] [PubMed]
- 3.Benner, S. A., M. A. Cohen, and G. H. Gonnet. 1994. Amino acid substitution during functionally constrained divergent evolution of protein sequences. Protein Eng. 7:1323-1332. [DOI] [PubMed] [Google Scholar]
- 4.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berrier, C., M. Benard, B. Ajouz, A. Coulombe, and A. Ghazi. 1996. Multiple mechanosensitive channels from Escherichia coli, activated at different thresholds of applied pressure. J. Membr. Biol. 151:175-187. [DOI] [PubMed] [Google Scholar]
- 6.Berrier, C., A. Coulombe, I. Szabo, M. Zoratti, and A. Ghazi. 1992. Gadolinium ion inhibits loss of metabolites induced by osmotic shock and large stretch-activated channels in bacteria. Eur. J. Biochem. 206:559-565. [DOI] [PubMed] [Google Scholar]
- 7.Blount, P., M. J. Schroeder, and C. Kung. 1997. Mutations in a bacterial mechanosensitive channel change the cellular response to osmotic stress. J. Biol. Chem. 272:32150-32157. [DOI] [PubMed] [Google Scholar]
- 8.Blount, P., S. Sukharev, P. C. Moe, B. Martinac, and C. Kung. 1999. Mechanosensitive channels in bacteria. Methods Enzymol. 294:458-482. [DOI] [PubMed] [Google Scholar]
- 9.Blount, P., S. I. Sukharev, P. C. Moe, S. K. Nagle, and C. Kung. 1996. Towards an understanding of the structural and functional properties of MscL, a mechanosensitive channel in bacteria. Biol. Cell. 87:1-8. [PubMed] [Google Scholar]
- 10.Blount, P., S. I. Sukharev, P. C. Moe, M. J. Schroeder, H. R. Guy, and C. Kung. 1996. Membrane topology and multimeric structure of a mechanosensitive channel protein of Escherichia coli. EMBO J. 15:4798-4805. [PMC free article] [PubMed] [Google Scholar]
- 11.Blount, P., S. I. Sukharev, M. J. Schroeder, S. K. Nagle, and C. Kung. 1996. Single residue substitutions that change the gating properties of a mechanosensitive channel in Escherichia coli. Proc. Natl. Acad. Sci. USA 93:11652-11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Booth, I. R., and P. Louis. 1999. Managing hypoosmotic stress: aquaporins and mechanosensitive channels in Escherichia coli. Curr. Opin. Microbiol. 2:166-169. [DOI] [PubMed] [Google Scholar]
- 13.Chang, G., R. H. Spencer, A. T. Lee, M. T. Barclay, and D. C. Rees. 1998. Structure of the MscL homolog from Mycobacterium tuberculosis: a gated mechanosensitive ion channel. Science 282:2220-2226. [DOI] [PubMed] [Google Scholar]
- 14.Chung, Y. J., C. Krueger, D. Metzgar, and M. H. Saier, Jr. 2001. Size comparisons among integral membrane transport protein homologues in bacteria, archaea, and eucarya. J. Bacteriol. 183:1012-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delcour, A. H., B. Martinac, J. Adler, and C. Kung. 1989. Modified reconstitution method used in patch-clamp studies of Escherichia coli ion channels. Biophys. J. 56:631-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esnouf, R. M. 1997. An extensively modified version of Molscript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model. 15:112-113, 132-134. [DOI] [PubMed] [Google Scholar]
- 18.Heinz, D. W., W. A. Baase, F. W. Dahlquist, and B. W. Matthews. 1993. How amino-acid insertions are allowed in an α-helix of T4 lysozyme. Nature 361:561.. [DOI] [PubMed] [Google Scholar]
- 19.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1994. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry 33:3038-3049. [DOI] [PubMed] [Google Scholar]
- 20.Kloda, A., and B. Martinac. 2001. Molecular identification of a mechanosensitive channel in archaea. Biophys. J. 80:229-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kloda, A., and B. Martinac. 2001. Structural and functional differences between two homologous mechanosensitive channels of Methanococcus jannaschii. EMBO J. 20:1888-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kloda, A., and B. Martinac. 2002. Common evolutionary origins of mechanosensitive ion channels in archaea, bacteria and cell-walled eukarya. Archaea 1:35-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong, Y., Y. Shen, T. E. Warth, and J. Ma. 2002. Conformational pathways in the gating of Escherichia coli mechanosensitive channel. Proc. Natl. Acad. Sci. USA 99:5999-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Dain, A. C., N. Saint, A. Kloda, A. Ghazi, and B. Martinac. 1998. Mechanosensitive ion channels of the archaeon Haloferax volcanii. J. Biol. Chem. 273:12116-12119. [DOI] [PubMed] [Google Scholar]
- 25.Levina, N., S. Tötemeyer, N. R., Stokes, P. Louis, M. A. Jones, and I. R. Booth. 1999. Protection of E. coli cells against extreme turgor by activation of MscS and MscL mechanosensitive channels: identification of genes required for MscS activity. EMBO J. 18:1730-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lockless, S., and R. Ranganathan. 1999. Evolutionary conserved pathways of energetic connectivity in protein families. Science 286:295-299. [DOI] [PubMed] [Google Scholar]
- 27.Manoil, C. 1992. Analysis of membrane protein topology using alkaline phosphatase and β-galactosidase gene fusions. Methods Cell Biol. 34:61-75. [DOI] [PubMed] [Google Scholar]
- 28.Martinac, B., J. Adler, and C. Kung. 1990. Mechanosensitive channels of E. coli activated by amphipaths. Nature 348:261-263. [DOI] [PubMed] [Google Scholar]
- 29.Martinac, B., M. Buechner, A. H. Delcour, J. Adler, and C. Kung. 1987. Pressure-sensitive ion channel in Escherichia coli. Proc. Natl. Acad. Sci. USA 84:2297-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maurer, J. A., and D. A. Dougherty. 2001. A high-throughout screen for MscL channel activity and mutational phenotyping. Biochim. Biophys. Acta Biomembr. 1514:165-169. [DOI] [PubMed] [Google Scholar]
- 31.Maurer, J. A., and D. A. Dougherty. 2002. Analysis of random mutations to the bacterial mechanosensitive channel of large conductance (MscL) using a high-throughout fluorescence screen. Biophys. J. 82:1302. [Google Scholar]
- 32.Maurer, J. A., D. E. Elmore, H. A. Lester, and D. A. Dougherty. 2000. Comparing and contrasting Escherichia coli and Mycobacterium tuberculosis mechanosensitive channels (MscL): new gain of function mutations in the loop region. J. Biol. Chem. 275:22238-22244. [DOI] [PubMed] [Google Scholar]
- 33.McLaggan, D., M. A. Jones, G. Gouesbet, N. Levina, S. Lindey, W. Epstein, and I. R. Booth. 2002. Analysis of the kefA2 mutation suggests that KefA is a cation-specific channel involved in osmotic adaptation in Escherichia coli. Mol. Microbiol. 43:521-536. [DOI] [PubMed] [Google Scholar]
- 34.Merritt, E. A., and D. J. Bacon. 1997. Raster-3D: photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 35.Miller, S., W. Bartlett, S. Chandrasekaran, S. Simpson, M. Edwards, and I. R. Booth. 2002. Domain organization of the MscS mechanosensitive channel of Escherichia coli. EMBO J. 22:36-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moe, P. C., P. Blount, and C. Kung. 1998. Functional and structural conservation in the mechanosensitive channel MscL implicates elements crucial for mechanosensation. Mol. Microbiol. 28:583-592. [DOI] [PubMed] [Google Scholar]
- 37.Moe, P. C., G. Levin, and P. Blount. 2000. Correlating protein structure with function of a bacterial mechanosensitive channel. J. Biol. Chem. 275:31121-31127. [DOI] [PubMed] [Google Scholar]
- 38.Nakamaru, Y., Y. Takahashi, T. Unemoto, and T. Nakamura. 1999. Mechanosensitive channel functions to alleviate the cell lysis of marine bacterium, Vibrio alginolyticus, by osmotic downshock. FEBS Lett. 444:170-172. [DOI] [PubMed] [Google Scholar]
- 39.Ness, L. S., and I. R. Booth. 1999. Different foci for the regulation of the activity of the KefB and KefC glutathione-gated K+ efflux systems. J. Biol. Chem. 274:9524-9530. [DOI] [PubMed] [Google Scholar]
- 40.Ochoa de Alda, J., and J. Houmard. 2000. Genomic survey of cAMP and cGMP signalling components in the cyanobacterium Synechocystis PCC 6803. Microbiology 146:3183-3194. [DOI] [PubMed] [Google Scholar]
- 41.Okada, K., P. C. Moe, and P. Blount. 2002. Functional design of bacterial mechanosensitive channels: comparisons and contrasts illuminated by random mutagenesis. J. Biol. Chem. 277:27682-27688. [DOI] [PubMed] [Google Scholar]
- 42.Ou, X., P. Blount, R. J. Hoffman, and C. Kung. 1998. One face of a transmembrane helix is crucial in mechanosensitive channel gating. Proc. Natl. Acad. Sci. USA 95:11471-11475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perozo, E., D. M. Cortes, P. Sompornpisut, A. Kloda, and B. Martinac. 2002. Open channel structure of MscL and the gating mechanism of mechanosensitive channels. Nature 418:942-948. [DOI] [PubMed] [Google Scholar]
- 44.Perozo, E., A. Kloda, D. M. Cortes, and B. Martinac. 2001. Site-directed spin-labeling analysis of reconstituted Mscl in the closed state. J. Gen. Physiol. 118:193-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saier, M. H., Jr. 2000. Families of proteins forming transmembrane channels. J. Membr. Biol. 175:165-180. [DOI] [PubMed] [Google Scholar]
- 46.Saier, M. H., Jr., B. H. Eng, S. Fard, J. Garg, D. A. Haggerty, W. J. Hutchinson, D. L. Jack, E. C. Lai, H. J. Liu, D. P. Nusinew, A. M. Omar, S. S. Pao, I. T. Paulsen, J. A. Quan, M. Sliwinski, T.-T. Tseng, S. Wachi, and G. B. Young. 1999. Phylogenetic characterization of novel transport protein families revealed by genome analyses. Biochim. Biophys. Acta 1422:1-56. [DOI] [PubMed] [Google Scholar]
- 47.Sipos, L., and G. von Heijne. 1993. Predicting the topology of eukaryotic membrane proteins. Eur. J. Biochem. 213:1333-1340. [DOI] [PubMed] [Google Scholar]
- 48.Sukharev, S., M. Betanzos, C. S. Chiang, and H. R. Guy. 2001. The gating mechanism of the large mechanosensitive channel MscL. Nature 409:720-724. [DOI] [PubMed] [Google Scholar]
- 49.Sukharev, S., S. R. Durell, and H. R. Guy. 2001. Structural models of the MscL gating mechanism. Biophys. J. 81:917-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sukharev, S., M. J. Schroeder, and D. R. McCaslin. 1999. Stoichiometry of the large conductance bacterial mechanosensitive channel of E. coli. A biochemical study. J. Membr. Biol. 171:183-193. [DOI] [PubMed] [Google Scholar]
- 51.Sukharev, S. I., P. Blount, B. Martinac, F. R. Blattner, and C. Kung. 1994. A large-conductance mechanosensitive channel in E. coli encoded by mscL alone. Nature 368:265-268. [DOI] [PubMed] [Google Scholar]
- 52.Sukharev, S. I., B. Martinac, V. Y. Arshavsky, and C. Kung. 1993. Two types of mechanosensitive channels in the Escherichia coli cell envelope: solubilization and functional reconstitution. Biophys. J. 65:177-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szabo, I., V. Petronilli, and M. Zoratti. 1993. A patch-clamp investigation of the Streptococcus faecalis cell membrane. J. Membr. Biol. 131:203-218. [DOI] [PubMed] [Google Scholar]
- 54.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Touzé, T., G. Gouesbet, C. Boiangiu, M. Jebbar, S. Bonnassie, and C. Blanco. 2001. Glycine betaine loses its osmoprotective activity in a bspA strain of Erwinia chrysanthemi. Mol. Microbiol. 42:87-99. [DOI] [PubMed] [Google Scholar]
- 56.von Heijne, G., and Y. Gavel. 1988. Topogenic signals in integral membrane proteins. Eur. J. Biochem. 174:671-678. [DOI] [PubMed] [Google Scholar]
- 57.Yoshimura, K., A. Batiza, and C. Kung. 2001. Chemically charging the pore constriction opens the mechanosensitive channel MscL. Biophys. J. 80:2198-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoshimura, K., A. Batiza, M. Schroeder, P. Blount, and C. Kung. 1999. Hydrophilicity of a single residue within MscL correlates with increased channel mechanosensitivity. Biophys. J. 77:1960-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhai, Y., and M. H. Saier, Jr. 2001. A web-based program (WHAT) for the simultaneous prediction of hydropathy, amphipathicity, secondary structure and transmembrane topology for a single protein sequence. J. Mol. Microbiol. Biotechnol. 3:501-502. [PubMed] [Google Scholar]
- 60.Zoratti, M., and V. Petronilli. 1990. Ion-conducting channels in a Gram-positive bacterium. FEBS Lett. 240:105-109. [DOI] [PubMed] [Google Scholar]