Abstract
Lactobacilli are nonpathogenic gram-positive inhabitants of microflora. At least some Lactobacillus strains have been postulated to have health beneficial effects, such as the stimulation of the immune system. Here we examined the stimulatory effects of lactobacilli on mouse immune cells. All six heat-killed Lactobacillus strains examined induced the secretion of tumor necrosis factor alpha (TNF-α) from mouse splenic mononuclear cells, albeit to various degrees. When fractionated subcellular fractions of Lactobacillus casei were tested for NF-κB activation and TNF-α production in RAW264.7, a mouse macrophage cell line, the activity was found to be as follows: protoplast > cell wall ≫ polysaccharide-peptidoglycan complex. Both crude extracts and purified lipoteichoic acids (LTAs) from two Lactobacillus strains, L. casei and L. fermentum, significantly induced TNF-α secretion from RAW264.7 cells and splenocytes of C57BL/6, C3H/HeN, and C3H/HeJ mice but not from splenocytes of C57BL/6 TLR2−/− mice. Lactobacillus LTA induced activation of c-Jun N-terminal kinase activation in RAW264.7 cells. Furthermore, in HEK293T cells transected with a combination of CD14 and Toll-like receptor 2 (TLR2), NF-κB was activated in response to Lactobacillus LTA. Taken together, these data suggest that LTAs from lactobacilli elicit proinflammatory activities through TLR2.
Mucosal defense against bacteria is essential for the homeostasis of the host. In the case of colonized intestine, protection against enteropathogens partly depends on the indigenous microflora (15). The importance of the microflora is evident in the susceptibility of germfree animals to intestinal infections (3, 7). Lactobacilli are nonpathogenic gram-positive inhabitants of the normal human intestine. Lactobacilli are known for their health-promoting effects such as nonspecific enhancement of the immune system, protection against intestinal infection, decreasing cholesterol levels in serum, and anticarcinogenic activity (5). In the intestinal immune system, macrophages play important roles in the maintenance of health by their ability to eliminate invading microorganisms, defective cells, and poisonous substances. Macrophages also secrete a large number of cytokines and chemokines, thus orchestrating the immune responses of the host. Both live and dead lactobacilli stimulate macrophage functions, and this stimulating activity has been reported to vary among Lactobacillus strains. For example, when an antigen (Chikungunya virus) was given to mice with oral lactobacilli, the number of cytokine-producing cells in the duodenum increased in a Lactobacillus strain-dependent manner (18). As for the downstream signals of macrophage activation by lactobacilli, it has recently been reported that live Lactobacillus rhamnosus GG induces NF-κB, STAT1, and STAT3 DNA-binding activity in human macrophages (23). However, the activation of STATs does not seem to be directly mediated by lactobacilli, since it is blocked by cycloheximide, a protein synthesis inhibitor, indicating that this activation is mediated secondarily by secreted cytokines. Although these findings have clearly suggested that at least some Lactobacillus strains can directly activate host immune cells, the exact mechanisms how lactobacilli can activate these macrophage functions have remained obscure.
Toll, first identified as a protein controlling dorsoventral pattern formation in the early development of Drosophila melanogaster (2), has been shown to participate in antimicrobial immune responses (16). Toll homologues are conserved throughout various species. They encode transmembrane proteins containing intracellular domains that are homologous to that of the interleukin-1 (IL-1) receptor family proteins and repeating leucine-rich motifs in their extracellular regions (2). Recently, several mammalian Toll homologues have been identified and termed Toll-like receptors (TLRs) (31). In the past few years two of the mammalian TLRs, TLR2 and TLR4, have been shown to mediate the lipopolysaccharide (LPS) responsiveness in in vitro transfection systems (6). However, the role of TLR2 as an LPS receptor in vivo has been questioned due to recent findings: two mouse strains (C3H/HeJ and C57BL10/ScCr) that exhibit impaired ability to respond to many types of LPS have different mutations in the TLR4 gene (28, 30). Furthermore, gene-disrupted mice of TLR4, but not TLR2, demonstrate phenotypes similar to LPS-hyporesponsive strains (38). These findings suggest that TLR4 is the dominant receptor for at least most types of LPS, whereas TLR2 is dispensable. Quite recently, however, TLR2 has been reported to be the essential receptor for LPS of some bacterial species: Leptospira interrogans and Porphyromonas gingivalis (13, 40). On the other hand it has recently been reported, using transient-transfection experimental system, that TLR2 mediates signals from other bacterial components, including lipoteichoic acid (LTA), peptidoglycan, and lipoproteins and/or lipopeptides (12). The role of TLR2 as the LTA receptor, however, remains ambiguous since TLR4 gene-disrupted mice have been demonstrated to be unresponsive to LTA (38).
In the present study, we examined the stimulatory effects of six Lactobacillus strains on mouse immune cells. All of the six heat-killed Lactobacillus strains induced the secretion of tumor necrosis factor alpha (TNF-α) from mouse splenic mononuclear cells, albeit to various degrees. When fractionated subcellular components of L. casei were tested, the protoplast fraction most efficiently induced NF-κB activation and TNF-α production in RAW264.7, a mouse macrophage cell line. Purified LTA, a component of protoplast, from L. casei and L. fermentum significantly induced TNF-α secretion from RAW264.7 cells and splenocytes of C57BL/6, C3H/HeN, and C3H/HeJ mice, but not splenocytes of C57BL/6 TLR2−/− mice. Lactobacillus LTA also induced the activation of c-Jun N-terminal kinase (JNK) in RAW264.7 cells. Furthermore, NF-κB was activated in response to Lactobacillus LTAs in HEK293T cells transected with a combination of CD14 and TLR2 but not TLR4. Taken together, these data suggest that LTA from at least some Lactobacillus strains elicit proinflammatory activies through TLR2.
MATERIALS AND METHODS
Bacterial strains.
L. casei YIT 9029, L. fermentum YIT 0159 (FERM P-13859), L. rhamnosus YIT 0232 (ATCC 53103), L. acidophilus YIT 0070 (ATCC 4356), L. plantarum YIT 0102 (ATCC 14917), and L. reuteri YIT 0197 (JCM 1112) were all provided by Yakult Central Institute for Microbiological Research (Kunitachi, Japan).
Reagents and antibodies.
LPS from Escherichia coli serotype B6:O26, was obtained from Sigma Chemical Co. (St. Louis, Mo.). RPMI 1640 medium and Dulbecco modified Eagle medium were from Gibco-BRL (Rockville, Md.). Synthetic E.coli-type lipid A, compound 506, was a generous gift from T. Ogawa and was previously described (20, 26). Fetal calf serum (FCS) was purchased from Sigma Chemical Co. The anti-JNK1 polyclonal antibody was obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, Calif.). This antibody is specific to JNK1 and does not cross-react with either JNK2 or JNK3.
Plasmids.
A mammalian expression vector pcDNA3.1(+) was purchased from Invitrogen Corporation (Carlsbad, Calif.). The mouse CD14 expression plasmid [pcDNA3.1(+)-mCD14], a C-terminal Flag-tagged mouse TLR2 expression plasmid (pEFBOS-Flag/mTLR2), and an NF-κB responsive reporter (pGL3-NFκB-Luc) used here have been described previously (21). A C-terminal Flag-tagged mouse TLR4 (mTLR4) expression plasmid, p3XFlag-CMV14-TLR4, was generated by inserting the whole mTLR4 coding region cDNA into the p3XFlag-CMV14 vector (Sigma Chemical Co.). A C-terminal Flag-tagged mTLR6 expression vector, pEFBOS-Flag/mTLR6, was prepared by inserting the whole mTLR6 coding region cDNA into the AscI site of the pEFBOS-Flag vector.
Mice.
The mutant mouse strain deficient in TLR2 was kindly provided by S. Akira (Osaka University, Osaka, Japan). Female BALB/c, C3H/HeN, and C3H/HeJ mice were purchased from Japan SLC (Shizuoka, Japan) and were kept under germfree conditions. All experiments were conducted with sex-matched, 6- to 1-week-old mice.
Cells.
All cell lines were grown in tissue culture flasks at 37°C in 5% CO2 and 95% air and then passaged every 2 or 3 days to maintain logarithmic growth. A mouse macrophage cell line, RAW264.7, and a human embryonic kidney cell line, HEK293T, were obtained from the Riken cell bank (Tsukuba, Japan) and maintained in Dulbecco modified Eagle medium with 10% FCS (Sigma Chemical Co.). Splenic mononuclear cells from mice were purified by a density gradient separation with Percoll.
Preparation of L. casei intact cell walls.
Intact cell walls were prepared by the method of Sekine et al. with minor modifications (33). Briefly, heat-killed cells (100°C, 20 min) were suspended in 0.3% sodium dodecyl sulfate (SDS) solution and boiled for 15 min. After centrifugation (20,000 × g, 30 min), precipitate was collected and washed three times with distilled water. The precipitate was washed with acetone and resuspended in 50 mM Tris-HCl buffer (pH 7.5). The suspension was treated with pronase (Roche Molecular Biochemicals) at 37°C for 20 h. Lipids were removed from the digested residue by successive refluxing with methanol and methanol-chloroform-water (1:1:1). These materials were then treated with Benzon nuclease (Merck) at 37°C for 20 h and digested with pronase for 72 h in 50 mM Tris-HCl buffer (pH 7.2). The insoluble material was washed three times with distilled water and then lyophilized.
Preparation of the L. casei protoplast and PS-PG fractions.
The procedure used to prepare the L. casei protoplast and polysaccharide-peptidoglycan (PS-PG) fractions was a modification of a previously described method (25). Briefly, 5 g of heat-killed cells (20 min at 100°C) was suspended and treated with N-acetylmuramidase SG and Benzon nuclease at 39°C for 20 h. After centrifugation at 20,000 rpm for 25 min, the precipitate was washed three times with doubly distilled H2O and then collected as the protoplast fraction. The supernatant was collected and treated with 5 mg of pronase at 38°C for 8 h, followed by digestion with trypsin at 37°C for 16 h. The digested supernatant was filtered, followed by dialysis. The dialyzed material was lyophilized as the PS-PG fraction.
Purification of LTA.
LTA was prepared from L. fermentum and L. casei by the method of Fischer et al. (9). Briefly, the protoplast fraction was suspended and broken by sonication. The sonicated material was extracted with hot phenol and treated with nuclease. LTA was purified by column chromatography of Macroprep High Q and octyl-Sepharose CL-4B. Neither protein amino acids nor polysaccharides were detectable in this preparation.
Transient transfection.
Cells were plated onto 35-mm plates at 7.5 × 105 cells/plate on the day before transfection. Combinations of expression plasmid DNAs (2 μg total/plate) were transfected by using Lipofectamine (Gibco-BRL) according to the manufacturer's instructions. Cells were harvested 48 h later with phosphate-buffered saline and used for further analyses.
Luciferase reporter assay.
At 48 h after transfection, some cells were stimulated with the indicated reagents for 8 h. Cells were lysed, and the lysates were used for luciferase activity measurements by using the dual luciferase reporter assay system (Toyo Ink Co., Tokyo, Japan) according to the manufacturer's instructions. The results were shown as relative luciferase activities (luciferase activity/seapansy activity). All of the luciferase assays shown in the current study were repeated at least three times, and a typical result was shown for each experiment.
Immune complex kinase assay.
Cells were lysed in PLC lysis buffer (50 mM HEPES, pH 7.0; 150 mM NaCl; 10% glycerol; 1% Triton X-100; 1.5 mM MgCl2; 1 mM EGTA; 100 mM NaF; 10 mM NaPPi; 1 mM Na3VO4; 1 mM phenylmethylsulfonyl fluoride; 10 mg of aprotinin/ml; 10 mg of leupeptin/ml) at 108 cells/ml. Cell lysates (107 cells/sample) were incubated with 0.4 μg of polyclonal anti-JNK1 antibody for 2 h at 4°C, followed by protein A-Sepharose beads (Amersham Pharmacia Biotech, Inc.) for additional 1 h. The beads were washed three times in PLC lysis buffer and then once in kinase buffer (20 mM Tris-HCl, pH 7.4; 20 mM MgCl2; 2 mM EGTA; 0.5 mM sodium vanadate; 10 mM β-glycerophosphate; 1 mM dithiothreitol). The kinase reaction was initiated by the addition of 40 μl of kinase buffer with 20 μM ATP, 5 μCi of [γ-32P]ATP, and 0.5 μg of glutathione S-transferase (GST)-c-Jun(5-89) (20); this step was allowed to proceed for 15 min at 30°C. The reaction was terminated by the addition of 2× SDS sample buffer. Samples were boiled and resolved by SDS-polyacrylamide gel electrophoresis, and the fixed gel was then exposed to an X-ray film.
ELISA.
RAW264.7 cells (5 × 105 cells/ml) were incubated for 8 h with or without 1 ng of LPS/ml. Cell culture supernatants were collected, and the levels of mouse TNF-α were quantitated by using a commercial enzyme-linked immunosorbent assay (ELISA) kit (TECHNE Corp., Minneapolis, Minn.) according to the manufacturer's instructions.
RESULTS
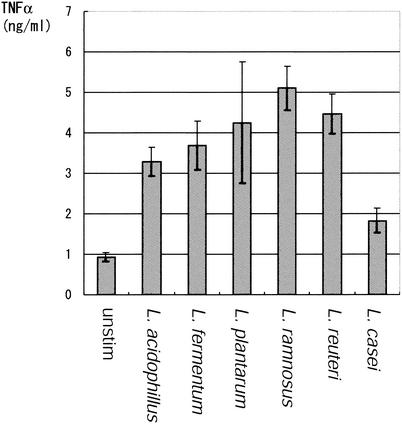
Cytokine secretion is induced by various Lactobacillus strains. Six different Lactobacillus strains were examined for their abilities to induce cytokine secretion from mononuclear cells isolated from a BALB/c mouse spleen. Splenic mononuclear cells were incubated with 1 μg of heat-killed bacteria/ml, and TNF-α concentrations in the culture supernatants were measured after 12 h of incubation. The experiment was repeated three times with similar results, and a typical result is shown in Fig. 1. All six strains significantly increased TNF-α secretion from the spleen cells. The average TNF-α-inducing activities were as follows: L. ramnosus > L. reuteri > L. plantarum > L. fermentum > L. acidophilus > L. casei.
FIG. 1.
TNF-α secretion from mouse mononuclear cells induced by various Lactobacillus strains. Mononuclear cells were isolated from spleens of 6-week-old female BALB/c mice. The mononuclear cells (2 × 106/ml) were incubated in RPMI 1640 plus 10% FCS containing 1 μg of heat-killed Lactobacillus bacteria/ml of six different strains. The TNF-α concentration in the culture supernatants was measured by ELISA after 12 h of incubation. The assays were done in triplicate, and the averages and the the standard deviation (SD) values are shown. The same experiment was repeated three times, yielding similar results, and a typical result is shown.
The protoplast fraction of Lactobacillus has the highest activity for TNF-α induction and NF-κB activation in a macrophage cell line.
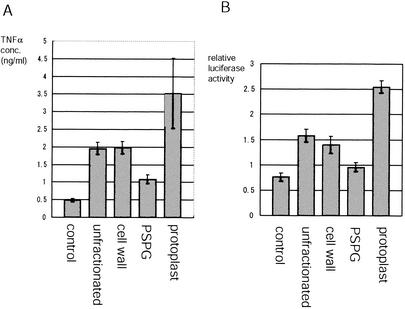
In order to examine which components of Lactobacillus have the TNF-α-inducing activities, RAW264.7, a mouse macrophage cell line, was stimulated with subcellular fractions isolated from L. casei. L. casei was separated into three fractions: cell wall, protoplast, and PS-PG, as described in Materials and Methods. Each bacterial fraction was added to the culture medium at 1 μg/ml, and the TNF-α concentration in the culture supernatants was measured after 12 h of incubation. Although both cell wall and protoplast fractions significantly increased TNF-α levels, the protoplast fraction showed the highest TNF-α-inducing activity (Fig. 2A). In contrast, PS-PG only modestly induced the cytokine secretion.
FIG. 2.
Proinflammatory activities of Lactobacillus subcellular fractions. Various subcellular fractions of L. casei were prepared as described in Materials and Methods. (A) RAW264.7 cells (5 × 105/ml) were stimulated with subcellular fractions isolated from L. casei. Each bacterial fraction was added to the culture medium at 1 μg/ml, and the TNF-α concentration in the culture supernatants was measured by ELISA after 12 h of incubation. (B) RAW264.7 cells were plated at 7.5 × 105 cells/plate and on the next day were transiently transfected with 1 μg each of a reporter plasmid containing tandem NF-κB consensus binding sites (pGL3-NF-kB-luc) and pRL-SV40 (as an internal control). At 48 h after the transfection, Lactobacillus subcellular fractions were added at 1 μg/ml, and the cell lysates were collected after 8 h of incubation to measure the relative luciferase activities. The experiment was repeated three times with similar results. A typical result is shown.
Two transcriptional factors, NF-κB and AP-1, are essential for the TNF-α expression in macrophages. In order to assess the NF-κB activity in RAW264.7 cells stimulated with each Lactobacillus fraction, RAW264.7 cells were transiently transfected with a reporter plasmid containing tandem NF-κB consensus binding site. At 48 h after the transfection Lactobacillus fractions were added, and the cell lysates were collected after 8 h of incubation. Consistent with the TNF-α secretion, the protoplast fraction most significantly increased the NF-κB activity in RAW264.7 cells (Fig. 2B).
TLR2, but not TLR4, is essential for the induction of TNF-α by Lactobacillus LTA.
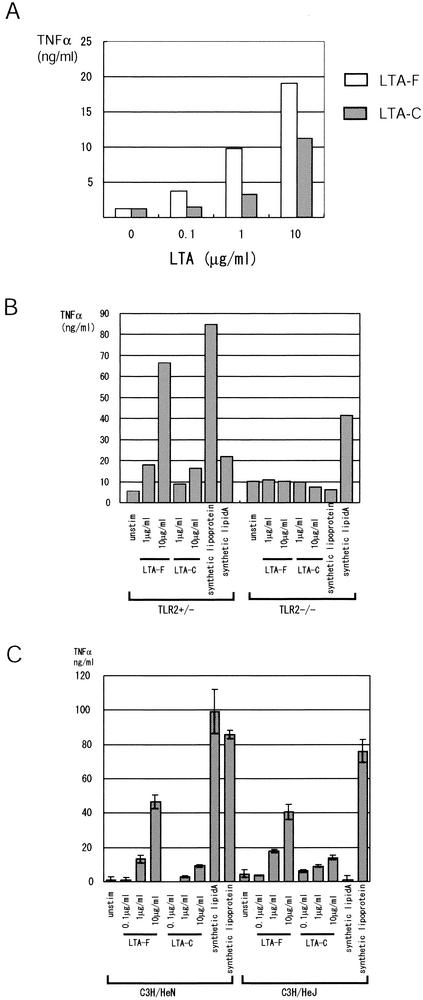
With our method of fractionation, 99% of LTA is contained in the protoplast fraction (data not shown). Since LTAs from other gram-positive bacteria induce the activation of macrophages, including the secretion of TNF-α (22), we purified the LTAs from two Lactobacillus strains, L. fermentum and L. casei, and examined their effects on RAW264.7 cells. As shown in Fig. 3, LTA from both strains induced a large amount of TNF-α from RAW264.7 cells. The activity was significantly higher for the LTA from L. fermentum, which was consistent with the result with the heat-killed bacteria (Fig. 1).
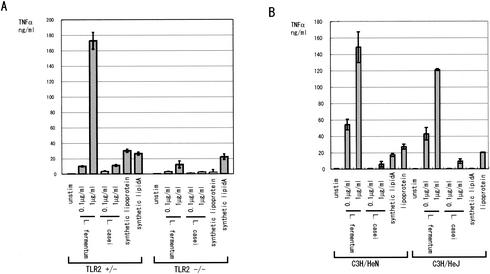
FIG. 3.
TLR2 is essential for TNF-α secretion induced by Lactobacillus LTA. (A) RAW264.7 cells (5 × 105 cells/ml) were stimulated with increasing amounts of LTA purified from L. fermentum (LTA-F) or L. casei (LTA-C). The culture supernatants were collected after 12 h, and the TNF-α concentration was measured by ELISA. The same experiment was repeated three times producing similar results, and a typical result is shown. (B) Splenic mononuclear cells were isolated from TLR2 gene-disrupted mice along with the control (TLR2+/−). The cells (2 × 106 cells/ml) were stimulated with the indicated amounts of purified LTAs from L. fermentum and L. casei, synthetic lipoprotein, or synthetic lipid A. After 12 h of incubation, the TNF-α concentration in the supernatants was measured by ELISA. The experiment was repeated three times, yielding similar results, and a typical result is shown. (C) Splenic mononuclear cells from C3H/HeN and its variant, C3H/HeJ. The cells were treated as in panel B. The TNF-α concentration in the supernatants was measured in triplicate, and the averages and the SD values are shown. The experiment was repeated three times, yielding similar results, and a typical result is shown.
The role of TLR in the recognition of bacterial LTA is controversial. The LTAs from the gram-positive bacteria Bacillus subtilis, Streptococcus pyogenes, and Streptococcus sanguis induce NF-κB activation in HEK293 cells overexpressing TLR2 but not in cells overexpressing TLR4 (32). On the other hand, TLR4 gene-disrupted mice, but not TLR2-gene disrupted mice, have been reported to lack responses to LTA from Staphylococcus aureus or Streptococcus sanguis (38). To assess the role of TLR2 in mediating signals from Lactobacillus LTA, we isolated the splenic mononuclear cells from TLR2 gene-disrupted mice along with the control (TLR2+/−) mice and stimulated the cells with LTAs from L. fermentum and L. casei. In TLR2+/− mice, LTA from both Lactobacillus strains induced TNF-α secretion in dose-dependent manners (Fig. 3B). The activity was ca. 10-fold higher for L. fermentum LTA. As controls, both lipoprotein, which signals through TLR2, and lipid A, which signals through TLR4, induced significant amount of TNF-α in the TLR2+/− mice. In contrast, neither LTA induced significant amounts of TNF-α from splenic cells of TLR2−/− mice. In TLR2−/− mice, lipoprotein also failed to induce TNF-α secretion, whereas lipid A induced a significant amount of TNF-α.
Furthermore, we explored the effects of Lactobacillus LTAs on splenic mononuclear cells from C3H/HeN and its variant, C3H/HeJ, which has a nonfunctional mutation in the TLR4 gene and is hyporesponsive to LPS. As shown in Fig. 3C, splenocytes from C3H/HeJ mice responded to Lactobacillus LTAs as well as splenocytes from C3H/HeN mice. As controls, synthetic lipoprotein, which signals through TLR2, induced significant TNF-α secretion from C3H/HeJ splenocytes, whereas lipid A failed to do so. Taken together, these results suggest that LTAs from the two Lactobacillus strains need TLR2, but not TLR4, for their TNF-α-inducing activities.
TLR2 mediates NF-κB activation by Lactobacillus LTA.
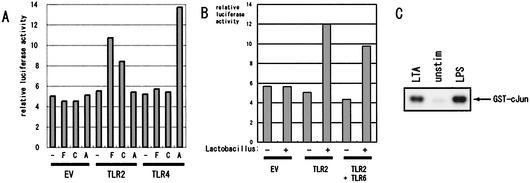
Mouse TLR2 mediates the activation of a transcription factor, NF-κB, and a mitogen-activated protein kinase, JNK, in response to its ligands. To directly show the involvement of TLR2 in Lactobacillus LTA signals, expression plasmids for mouse TLR2 or TLR4 and CD14 were transfected into HEK293T cells, which do not express detectable amounts of endogenous TLR2, TLR4, TLR6, or CD14, along with a luciferase reporter plasmid for NF-κB. At 48 h after transfection, protein expression of CD14, TLR2, and TLR4 were confirmed by Western blotting (data not shown). HEK293T cells transfected with CD14 alone did not increase NF-κB activity in response to Lactobacillus LTAs (Fig. 4A). When TLR2 was cotransfected, however, NF-κB activity significantly increased with the LTA treatments. In contrast, the expression of TLR4 did not endow responsiveness to Lactobacillus LTAs. These results confirmed the results with the gene-disrupted mice, strongly indicating that TLR2, but not TLR4, is essential for the responsiveness to Lactobacillus LTAs.
FIG. 4.
Signal transduction induced by Lactobacillus spp. (A) HEK293T cells were transiently transfected with 0.5 μg of each of pcDNA3.1(+)-mCD14, pGL3-NF-kB-luc, and pRL-SV40 (as an internal control), plus 0.5 μg of one of the following: pcDNA3.1(+) (EV), pEFBOS-mTLR2/Flag (TLR2), or p3XFlag-mTLR4 (TLR4). At 48 h after the transfection, the cells were untreated, treated with LTA (1 μg/ml) of L. fermentum (F) or L. casei (C) or with synthetic lipid A (1 μg/ml) (A) for 8 h, and then lysed to measure the luciferase activities. The experiment was repeated three times, yielding similar results, and a typical result is shown. (B) HEK293T cells were transiently transfected with 0.5 μg each of pcDNA3.1(+)-mCD14, pGL3-NF-kB-luc, and pRL-SV40, plus 1 μg of one of the following: pcDNA3.1(+) (EV), pEFBOS-mTLR2/Flag (TLR2) plus pcDNA3/1(+) (0.5 μg each), or the combination of pEFBOAS-mTLR2/Flag and pEFBOS-mTLR6/Flag (0.5 μg/each) (TLR2 + TLR6). At 48 h after the transfection, the cells were either not treated or treated with heat-killed L. fermentum (1 μg/ml) for 8 h and then lysed for the measurement of the luciferase activities. The experiment was repeated three times, yielding similar results, and a typical result is shown. (C) Lactobacillus LTA induces JNK activation in RAW264.7 cells. RAW264.7 cells were either not treated or treated with 1 μg of either LPS or purified LTA from L. fermentum/ml for 30 min. Cells were lysed, and the JNK1 kinase activity was measured by using GST-c-Jun as the substrate.
In a recent study, Ozinsky et al. reported that TLR6 plays an essential role by forming heterodimers with TLR2 in recognizing gram-positive bacteria (27). To explore the possible role of TLR6 in responding to lactobacilli, TLR6 was cotransfected with TLR2 into HEK293T cells, and then the cells were treated with the heat-killed L. fermentum. As shown in Fig. 4B, the expression of TLR6 did not further increase the NF-κB activation by L. fermentum, indicating that TLR6 is not necessary for the recognition of lactobacilli.
Furthermore, we measured the JNK activation in RAW264.7 cells in response to L. fermentum LTA. RAW264.7 cell lysate was prepared at 30 min after the LTA stimulation, and the JNK1 immunoprecipitate was measured for its kinase activity by using GST-c-Jun as the substrate. As shown in Fig. 4C, L. fermentum LTA significantly increased JNK1 activity in RAW264.7 cells. LPS from E. coli was also used as a control.
TLR2, but not TLR4, is essential for the responses to Lactobacillus strains.
To explore the possibilities that bacterial components other than LTA may induce immune responses through TLRs other than TLR2, we examined the heat-killed Lactobacillus strains for their effects on TLR2 gene-disrupted cells. As shown in Fig. 5A, heat-killed extracts of L. fermentum and L. casei induced significant amounts of TNF-α secretion from splenic mononuclear cells of the control TLR2+/− mice in dose-dependent manners. In contrast, neither extracts induced meaningful amounts of TNF-α from splenic cells of TLR2−/− mice.
FIG. 5.
TLR2 is essential for TNF-α secretion induced by Lactobacillus spp. (A) Splenic mononuclear cells were isolated from TLR2 gene-disrupted mice, along with the control (TLR2+/−). The cells (2 × 106 cells/ml) were stimulated with the indicated amounts of heat-killed L. fermentum and L. casei, synthetic lipoprotein, or synthetic lipid A. After 12 h of incubation, the TNF-α concentration in the supernatants was measured by ELISA. Measurements were done in triplicate, and the averages and the SD values are shown. The experiment was repeated three times, yielding similar results, and a typical result is shown. (B) Splenic mononuclear cells from C3H/HeN and its variant, C3H/HeJ. The cells were treated as in panel A. The TNF-α concentration in the supernatants was measured in triplicate, and the averages and the SD values are shown. The experiment was repeated three times producing similar results, and a typical result is shown.
Significantly, splenocytes from C3H/HeJ mice responded to heat-killed Lactobacillus as well as splenocytes from C3H/HeN mice (Fig. 5B). These results suggest that TLR2, but not TLR4, is essential for the TNF-α production in response to lactobacilli.
DISCUSSION
Although the importance of microflora in the mucosal immunity has been well documented, molecular mechanisms of the proinflammatory activities of nonpathogenic bacteria are not clearly known. In the present study, we examined the proinflammatory activities of Lactobacillus components on macrophages. When fractionated Lactobacillus extracts were used, the protoplast fraction most potently induced NF-κB activation and TNF-α production in a macrophage cell line, RAW264.7. Both heat-killed bacteria and highly purified LTA, a protoplast component in Lactobacillus, from two different strains, L. fermentum and L. casei, mediated NF-κB activation and TNF-α secretion in a TLR2-dependent manner. Furthermore, treatment with Lactobacillus LTA significantly increased the JNK1 activity in RAW264.7 cells. Since both NF-κB and JNK are important mediators of various functions of macrophages, including the production of TNF-α, IL-1β, IL-6, and inducible NO synthase, our results are consistent with the idea that LTA-TLR2 interaction plays an important role in the proinflammatory activities of Lactobacillus spp.
When heat-killed bacteria were used, all six Lactobacillus strains induced significant amounts of TNF-α secretion from mouse mononuclear cells (Fig. 1). However, there was a clear difference among strains. The most active L. ramnosus induced approximately four times more TNF-α than the least active L. casei. This result was consistent with the previous reports that proinflammatory activity was considerably different among Lactobacillus strains (18, 19, 39). Maassen et al. (18) compared several strains of Lactobacillus for their abilities to induce cytokines in gut villi when the strains were orally administered into mice immunized with Chikungunya virus. These researchers found that L. reuteri most potently induced both IL-1β and TNF-α (ca. fourfold greater compared to L. casei) (18). In contrast to the present study, Tejada-Simon et al. (39) found that L. casei induced approximately three times more TNF-α from RAW264.7 cells than did L. reuteri. The reason for the differences among these reports is unclear. These differences could be due to minor strain differences. It is also possible that various doses of bacteria might lead to different results. For example, Tejada-Simon et al. (39) used as much as 50 μg of heat-killed bacteria/ml, whereas we used heat-killed bacteria at 1 μg/ml.
Our experiment with NF-κB reporter plasmid indicated that the activity to induce NF-κB binding is strongest in the protoplast fraction (Fig. 2B). This finding is consistent with the TNF-α production data (Fig. 2A), since NF-κB binding is important for the induction of TNF-α promoter activity (17). A good candidate for this activity in the protoplast fraction is LTA. LTA is a class of surface glycolipid similar to LPS. Many of the clinical features observed during pathogenic gram-positive bacteria have been related to LTA (4). LTAs from pathogenic gram-positive bacteria have been shown to activate macrophages, including the secretion of cytokines such as TNF-α, IL-6, and IL-8 (4, 22, 35). Although the immunostimulatory effects of LTA from nonpathological gram-positive bacteria have not been studied in depth, LTAs from L. casei and L. fermentum are effective for protection against Pseudomonas aeruginosa (34).
In the current study, we demonstrated that purified LTAs from two Lactobacillus strains effectively induced TNF-α secretion from RAW264.7 cells in a dose-dependent fashion (Fig. 3A). Since we used an established macrophage cell line in the assay, this finding showed the direct interaction of LTAs with macrophages. Recently, it has been shown that a newly identified family of proteins, TLRs, play an essential role in innate immunity as pattern recognition receptors for various bacterial components. Of 11 TLR members reported thus far two, TLR2 and TLR4, have been the most extensively studied. Studies with TLR4-mutated mouse strains and TLR4 gene-disrupted mice have identified TLR4 to be an essential receptor for LPS, whereas bacterial lipoproteins signal through TLR2 (28, 30, 38). We previously reported that RAW264.7 cells express both TLR2 and TLR4 (20).
The identification of the responsible TLR in the recognition of bacterial LTA is controversial. LTA from the gram-positive bacteria Bacillus subtilis, Streptococcus pyogenes, and Streptococcus sanguis induces NF-κB activation in HEK293 cells overexpressing TLR2 but not in cells overexpressing TLR4 (32). On the other hand, TLR4 gene-disrupted mice, but not TLR2 gene-disrupted mice, are reported to lack responses to LTA from Staphylococcus aureus or Streptococcus sanguis (38). It is possible that LTA is recognized by different TLRs depending on minor structural differences among bacterial species. It is noteworthy that LPSs from different bacterial species seem to be recognized by either TLR4 or TLR2 (13, 40). To assess the role of TLRs in mediating signals from Lactobacillus LTA, we isolated the splenic mononuclear cells from TLR2 gene-disrupted mice, along with the control (TLR2+/−) mice, and stimulated the cells with LTAs from L. fermentum and L. casei. Significantly, neither LTA induced meaningful amounts of TNF-α from splenic cells of TLR2−/− mice, whereas they effectively induced the cytokine from TLR2+/− spleen cells (Fig. 3B). On the other hand, Lactobacillus LTAs were almost equally effective on splenic mononuclear cells from C3H/HeN and its variant, C3H/HeJ, which has nonfunctional TLR4 for TNF-α induction (Fig. 3C). These results clearly indicate that LTAs from at least two Lactobacillus strains signal through TLR2 but not through TLR4.
It is noteworthy that the cell wall fraction of L. casei had less proinflammatory activity than the protoplast fraction (Fig. 2). Compared to the proloplast fraction, the cell wall fraction induced much less TNF-α secretion and NF-κB activity. This result was somewhat surprising since the bacterial cell wall fraction, which contains peptidoglycan as the main component, has strong proinflammatory activities for some gram-positive bacteria, such as Bacillus subtilis, Streptococcus pyogenes, and Streptococcus sanguis, through TLR2 (32). There have been controversial results on the proinflammatory fractions of Lactobacillus. For example, de Ambrosini et al. (8) examined peptidoglycan purified from four Lactobacillus strains and found that only one could activate phagocytosis by mouse peritoneal macrophages. It may be beneficial to the host that cell wall components of intestinal microflora do not induce strong proinflammatory responses.
It has recently been reported that TLR6 is essential for the recognition of various gram-positive bacteria strains by forming heterodimers with TLR2 (27). In our cotransfection experiment, however, TLR6 did not further increase the TLR2-mediated NF-κB activation by L. fermentum. In the study by Ozinsky et al. it was demonstrated that TLR6 is essential for the recognition of peptidoglycan, but these authors did not examine the role of TLR6 in the recognition of LTA. We presume that, since the strongest TNF-α-inducing activity of the Lactobacillus strains resides in LTA but not peptidoglycan, TLR6 may not be essential for the responses to these bacterial strains.
It was previously reported that the cytokine-inducing activity of the LTA fractions from Enterococcus hirae did not reside in the major structure of LTA but in the contaminating glycolipids (11, 36). In a recent study of LTA from Staphylococcus aureus, however, it was demonstrated that some alanine substituents are lost during standard purification, resulting in the attenuated cytokine-inducing activity of LTA, indicating that LTA itself is a potent cytokine inducer (24). We could not detect any contaminating glycolipids or proteins in the purified LTAs from Lactobacillus strains by standard detection methods, although we could not totally exclude the possibility that small amounts of contaminating substances contributed to the proinflammatory activities. Interestingly, in a preliminary study, alanine ester but no glucose unit was linked to LTA of L. casei (unpublished result). Thus, the structure seems somewhat different from Entelococcus LTA, to which glucose units are also linked (11).
It has not been long since the importance of probiotics was widely accepted. There is increasing information about the role of probiotic bacteria (most typically, Lactobacillus spp.) to modulate proinflammatory responses of the host. For the clinical usage of Lactbacillus (or its components), it is absolutely necessary to better understand the molecular mechanisms of the proinflammatory responses. Our current findings may indicate that purified LTA may be a better candidate for the clinical usage than the whole bacteria, since it does not contain other bacterial components that may cause various side effects. Our findings also indicate that Lactobacillus LTA signals through TLR2 differently from more pathogenic gram-positive bacteria, which may utilize TLR4. Interestingly, recent reports showed that the different downstream signals between TLR2 and TLR4 mediate different downstream signals, indicating that these TLRs probably induce distinct sets of immune responses (1). For example, as for LPS, it has been reported that TLR4-dependent E. coli LPS induces a Th1-type response, whereas Porphyromonas gingivalis LPS, which signals through TLR2, induces a Th2-type response (29). This is at least partly due to a recently identified molecule called Mal or TIRAP, which is, in addition to MyD88, also involved in TLR4 signals (10, 14), whereas TLR2 seems to be dependent on MyD88 for its signals (37). Thus, Lactobacillus spp. may be unique among gram-positive bacteria since Lactobacillus LTA may induce distinct proinflammatory activities in the host.
Acknowledgments
We thank S. Akira (Osaka University, Osaka, Japan) for providing us with TLR2 gene-disrupted mice. We also thank K. Itano and A. Nishikawa for their technical assistance.
This work was supported in part by grants from Ono Pharmaceutical Company, the Yokoyama Research Foundation for Clinical Pharmacology, and the Naito Foundation (to T.M.); by a grant from the Ministry of Education, Science, and Culture of the Japanese Government (JSPS-RFTF97L00703); and by a grant from the Yakult Bioscience Foundation (to T.M. and Y.Y.).
REFERENCES
- 1.Akira, S., K. Hoshino, and T. Kaisho. 2000. The role of Toll-like receptors and MyD88 in innate immune responses. J. Endotoxin Res. 6:383-387. [PubMed] [Google Scholar]
- 2.Belvin, M. P., and K. V. Anderson. 1996. A conserved signaling pathway: the Drosophila Toll-dorsal pathway. Annu. Rev. Cell Dev. Biol. 12:393-416. [DOI] [PubMed] [Google Scholar]
- 3.Berg, R. D., and D. C. Savage. 1975. Immune responses of specific pathogen-free and gnotobiotic mice to antigens of indigenous and nonindigenous microorganisms. Infect. Immun. 11:320-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhakdi, S., T. Klonisch, P. Nuber, and W. Fischer. 1991. Stimulation of monokine production by lipoteichoic acids. Infect. Immun. 59:4614-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloksma, N., E. de Heer, H. van Dijk, and J. M. Willers. 1979. Adjuvanticity of lactobacilli. I. Differential effects of viable and killed bacteria. Clin. Exp. Immunol. 37:367-375. [PMC free article] [PubMed] [Google Scholar]
- 6.Chow, J. C., D. W. Young, D. T. Golenbock, W. J. Christ, and F. Gusovsky. 1999. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J. Biol. Chem. 274:10689-10692. [DOI] [PubMed] [Google Scholar]
- 7.Collins, F. M., and P. B. Carter. 1978. Growth of salmonellae in orally infected germfree mice. Infect. Immun. 21:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Ambrosini, V. M., S. Gonzalez, G. Perdigon, A. P. de Ruiz Holgado, and G. Oliver. 1996. Chemical composition of the cell wall of lactic acid bacteria and related species. Chem. Pharm. Bull. 44:2263-2267. [DOI] [PubMed] [Google Scholar]
- 9.Fischer, W., H. U. Koch, and R. Haas. 1983. Improved preparation of lipoteichoic acids. Eur. J. Biochem. 133:523-530. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald, K. A., E. M. Palsson-McDermott, A. G. Bowie, C. A. Jefferies, A. S. Mansell, G. Brady, E. Brint, A. Dunne, P. Gray, M. T. Harte, D. McMurray, D. E. Smith, J. E. Sims, T. A. Bird, and L. A. O'Neill. 2001. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature 413:78-83. [DOI] [PubMed] [Google Scholar]
- 11.Hashimoto, M., J. Yasuoka, Y. Suda, H. Takada, T. Yoshida, S. Kotani, and S. Kusumoto. 1997. Structural feature of the major but not cytokine-inducing molecular species of lipoteichoic acid. J. Biochem. 121:779-786. [DOI] [PubMed] [Google Scholar]
- 12.Hirschfeld, M., C. J. Kirschning, R. Schwandner, H. Wesche, J. H. Weis, R. M. Wooten, and J. J. Weis. 1999. Cutting edge: inflammatory signaling by Borrelia burgdorferi lipoproteins is mediated by Toll-like receptor 2. J. Immunol. 163:2382-2386. [PubMed] [Google Scholar]
- 13.Hirschfeld, M., J. J. Weis, V. Toshchakov, C. A. Salkowski, M. J. Cody, D. C. Ward, N. Qureshi, S. M. Michalek, and S. N. Vogel. 2001. Signaling by Toll-like receptor 2 and 4 agonists results in differential gene expression in murine macrophages. Infect. Immun. 69:1477-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horng, T., G. M. Barton, and R. Medzhitov. 2001. TIRAP: an adapter molecule in the Toll signaling pathway. Nat. Immunol. 2:835-841. [DOI] [PubMed] [Google Scholar]
- 15.Kraehenbuhl, J. P., and M. R. Neutra. 1992. Molecular and cellular basis of immune protection of mucosal surfaces. Physiol. Rev. 72:853-879. [DOI] [PubMed] [Google Scholar]
- 16.Lemaitre, B., E. Nicolas, L. Michaut, J. M. Reichhart, and J. A. Hoffmann. 1996. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86:973-983. [DOI] [PubMed] [Google Scholar]
- 17.Liu, H., P. Sidiropoulos, G. Song, L. J. Pagliari, M. J. Birrer, B. Stein, J. Anrather, and R. M. Pope. 2000. TNF-α gene expression in macrophages: regulation by NF-κB is independent of c-Jun or C/EBPβ. J. Immunol. 164:4277-4285. [DOI] [PubMed] [Google Scholar]
- 18.Maassen, C. B., C. van Holten-Neelen, F. Balk, M. J. den Bak-Glashouwer, R. J. Leer, J. D. Laman, W. J. Boersma, and E. Claassen. 2000. Strain-dependent induction of cytokine profiles in the gut by orally administered Lactobacillus strains. Vaccine 18:2613-2623. [DOI] [PubMed] [Google Scholar]
- 19.Marin, M. L., M. V. Tejada-Simon, J. H. Lee, J. Murtha, Z. Ustunol, and J. J. Pestka. 1998. Stimulation of cytokine production in clonal macrophage and T-cell models by Streptococcus thermophilus: comparison with Bifidobacterium sp. and Lactobacillus bulgaricus. J. Food Protein 61:859-864. [DOI] [PubMed] [Google Scholar]
- 20.Matsuguchi, T., T. Musikacharoen, T. Ogawa, and Y. Yoshikai. 2000. Gene expressions of Toll-like receptor 2, but not Toll-like receptor 4, is induced by LPS and inflammatory cytokines in mouse macrophages. J. Immunol. 165:5767-5772. [DOI] [PubMed] [Google Scholar]
- 21.Matsuguchi, T., K. Takagi, T. Musikacharoen, and Y. Yoshikai. 2000. Gene expressions of lipopolysaccharide receptors, Toll-like receptors 2 and 4, are differently regulated in mouse T lymphocytes. Blood 95:1378-1385. [PubMed] [Google Scholar]
- 22.Mattsson, E., L. Verhage, J. Rollof, A. Fleer, J. Verhoef, and H. van Dijk. 1993. Peptidoglycan and teichoic acid from Staphylococcus epidermidis stimulate human monocytes to release tumour necrosis factor-alpha, interleukin-1β and interleukin-6. FEMS Immunol. Med. Microbiol. 7:281-287. [DOI] [PubMed] [Google Scholar]
- 23.Miettinen, M., A. Lehtonen, I. Julkunen, and S. Matikainen. 2000. Lactobacilli and streptococci activate NF-κB and STAT signaling pathways in human macrophages. J. Immunol. 164:3733-3740. [DOI] [PubMed] [Google Scholar]
- 24.Morath, S., A. Geyer, and T. Hartung. 2001. Structure-function relationship of cytokine induction by lipoteichoic acid from Staphylococcus aureus. J. Exp. Med. 193:393-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagaoka, M., M. Muto, K. Nomoto, T. Matuzaki, T. Watanabe, and T. Yokokura. 1990. Structure of polysaccharide-peptidoglycan complex from the cell wall of Lactobacillus casei YIT9018. J. Biochem. 108:568-571. [DOI] [PubMed] [Google Scholar]
- 26.Ogawa, T., Y. Suda, W. Kashihara, T. Hayashi, T. Shimoyama, S. Kusumoto, and T. Tamura. 1997. Immunobiological activities of chemically defined lipid A from Helicobacter pylori LPS in comparison with Porphyromonas gingivalis lipid A and Escherichia coli-type synthetic lipid A (compound 506). Vaccine 15:1598-1605. [DOI] [PubMed] [Google Scholar]
- 27.Ozinsky, A., D. M. Underhill, J. D. Fontenot, A. M. Hajjar, K. D. Smith, C. B. Wilson, L. Schroeder, and A. Aderem. 2000. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between Toll-like receptors. Proc. Natl. Acad. Sci. USA 97:13766-13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poltorak, A., X. He, I. Smirnova, M. Y. Liu, C. V. Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, M. Freudenberg, P. Ricciardi-Castagnoli, B. Layton, and B. Beutler. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in tlr4 gene. Science 282:2085-2088. [DOI] [PubMed] [Google Scholar]
- 29.Pulendran, B., K. Palucka, and J. Banchereau. 2001. Sensing pathogens and tuning immune responses. Science 293:253-256. [DOI] [PubMed] [Google Scholar]
- 30.Qureshi, S. T., L. Lariviere, G. Leveque, S. Clermont, K. J. Moore, P. Gros, and D. Malo. 1999. Endotoxin-tolerant mice have mutations in Toll-like receptor 4 (Tlr4). J. Exp. Med. 189:615-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rock, F. L., G. Hardiman, J. C. Timans, R. A. Kastelein, and J. F. Bazan. 1998. A family of human receptors structurally related to Drosophila Toll. Proc. Natl. Acad. Sci. USA 95:588-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwandner, R., R. Dziarski, H. Wesche, M. Rothe, and C. J. Kirschning. 1999. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by Toll-like receptor 2. J. Biol. Chem. 274:17406-17409. [DOI] [PubMed] [Google Scholar]
- 33.Sekine, K., T. Toida, M. Saito, M. Kuboyama, T. Kawashima, and Y. Hashimoto. 1985. A new morphologically characterized cell wall preparation (whole peptidoglycan) from Bifidobacterium infantis with a higher efficacy on the regression of an established tumor in mice. Cancer Res. 45:1300-1307. [PubMed] [Google Scholar]
- 34.Setoyama, T., K. Nomoto, T. Yokokura, and M. Mutai. 1985. Protective effect of lipoteichoic acid from Lactobacillus casei and Lactobacillus fermentum against Pseudomonas aeruginosa in mice. J. Gen. Microbiol. 131:2501-2503. [DOI] [PubMed] [Google Scholar]
- 35.Standiford, T. J., D. A. Arenberg, J. M. Danforth, S. L. Kunkel, G. M. VanOtteren, and R. M. Strieter. 1994. Lipoteichoic acid induces secretion of interleukin-8 from human blood monocytes: a cellular and molecular analysis. Infect. Immun. 62:119-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suda, Y., H. Tochio, K. Kawano, H. Takada, T. Yoshida, S. Kotani, and S. Kusumoto. 1995. Cytokine-inducing glycolipids in the lipoteichoic acid fraction from Enterococcus hirae ATCC 9790. FEMS Immunol. Med. Microbiol. 12:97-112. [DOI] [PubMed] [Google Scholar]
- 37.Takeuchi, O., K. Hoshino, and S. Akira. 2000. Cutting edge: TLR2-deficient and MyD88-deficient mice are highly susceptible to Staphylococcus aureus infection. J. Immunol. 165:5392-5396. [DOI] [PubMed] [Google Scholar]
- 38.Takeuchi, O., K. Hoshino, T. Kawai, H. Sanjo, H. Takada, T. Ogawa, K. Takeda, and S. Akira. 1999. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 11:443-451. [DOI] [PubMed] [Google Scholar]
- 39.Tejada-Simon, M. V., and J. J. Pestka. 1999. Proinflammatory cytokine and nitric oxide induction in murine macrophages by cell wall and cytoplasmic extracts of lactic acid bacteria. J. Food Protein 62:1435-1444. [DOI] [PubMed] [Google Scholar]
- 40.Werts, C., R. I. Tapping, J. C. Mathison, T. H. Chuang, V. Kravchenko, I. Saint Girons, D. A. Haake, P. J. Godowski, F. Hayashi, A. Ozinsky, D. M. Underhill, C. J. Kirschning, H. Wagner, A. Aderem, P. S. Tobias, and R. J. Ulevitch. 2001. Leptospiral lipopolysaccharide activates cells through a TLR2-dependent mechanism. Nat. Immunol. 2:346-352. [DOI] [PubMed] [Google Scholar]