Abstract
Deficiencies of the early components of the classical complement pathway impair the actions of innate and humoral immunity and may lead to increased susceptibility to infections. We have studied the genetic basis of total C4B deficiency in a Finnish patient with recurrent meningitis, chronic fistulas and abscesses. The maternal chromosome carried a four-gene deletion including the C4B gene, and a conversion from C4B to C4A gene was found on the paternal chromosome resulting in complete deficiency of C4B. In the converted C4A gene, mutation screening did not reveal any amino acid changes or prominent mutations, yet a large number of nucleotide variations were found. Further, the patient was heterozygous for structural deficiency of mannan binding lectin (MBL) associating with medium levels of serum MBL. Our data provides new information on the genetic instability of the C4 gene region, and on the association of homozygous C4B deficiency and variant MBL genotype with increased susceptibility to recurrent and chronic infections. Importantly, plasma therapy induced a prompt clinical cure with long-term effects.
The four consecutive genes RP, C4, CYP21, and TNX form two modules in 69% of human major histocompatibility complex (MHC) haplotypes (8). Only the C4 gene is functional in both modules. RP, CYP21, and TNX genes can be either functional or nonfunctional. The size of the four-gene segment is either 32.7 or 26.4 kb, depending on the length polymorphism of the C4 gene caused by an endogenous retrovirus HERV-K(C4) in intron 9 (12, 40, 42, 43). The genes and their pseudogene counterparts in the C4 region share a great homology and they are susceptible to deletions and conversions through nonhomologous recombination, thus adding to the diversity.
The two isotypic forms of C4, C4A and C4B, have over 99% gene homology. The isotype specificity is determined by four amino acids; C4A carries P1101C1102L1105D1106, whereas the sequence L1101S1102I1105H1106 is specific for C4B (5, 23). The two isotypes also differ in their covalent binding activities. The binding reaction of C4B is very fast, with a half-life less than 1 s, compared to the 10 s for C4A (14). C4B has higher hemolytic activity than C4A, and it binds preferentially to free hydroxyl groups on bacterial surface polysaccharides. In contrast to C4A, C4B is found in all species of mammals studied and can be regarded as more important in defending against pathogens (13, 32). C4B deficiency, manifested as low serum concentration or total absence of the protein, may result from gene deletion or nonfunctionality of the gene due to conversion or mutation. In the North American Caucasian population, partial C4B deficiency is quite common, with a gene frequency of 18.3%, whereas total C4B deficiency was found only in two individuals among the 150 subjects studied (8). In Finland, the estimated frequency for total C4B deficiency is 8% (M.-L. Lokki, unpublished data).
Complete C4B deficiency predisposes for bacterial meningitis and impairs the host response for enveloped viruses (7, 33). Finnish patients with C4B deficiency manifest urticaria eruptions and vasculitis-related joint symptoms, indicating an inadequacy in immune complex clearance (S. Meri, personal communication). In viral infection, the complement activation and resulting complement cleavage products enhance the neutralizing function of immunoglobulin M (IgM) and IgG antibodies (30). Thus, complement provides an essential link between innate and adaptive immunity. Complement binding increases immunogenicity of an antigen and B cells become stimulated through complement receptors without T-cell-dependent antibody responses. Studies of C4-deficient mice show impaired B-cell priming, reduced number of germinal centers, and features of immune complex disease (E. Paul, O. O. Pozdnyakova, and M. C. Carroll, Abstr. 8th Eur. Meet. Complement Hum. Dis., Strasbourg, France, 2001) (16). Human mannan binding lectin (MBL) is involved in defense against bacterial, fungal and viral pathogens activating complement through the classical pathway. Low levels of MBL in serum due to variant alleles is associated with susceptibility to acute respiratory tract infections during childhood (22).
In this study, we report homozygous C4B deficiency and variant MBL genotype in a patient with increased susceptibility to severe infections. The patient suffered from recurrent meningitis during infancy and chronic draining fistulas and pararectal abscesses later on. Analysis of the C4 gene region revealed a deletion of CYP21A-TNXA-RP2-C4B genes on the maternal chromosome. On the paternal chromosome, cloning and sequencing showed the presence of C4A genes only due to the conversion of C4B to C4A gene resulting in total C4B deficiency. The patient also had a structurally heterozygous MBL genotype, which is known to have a diminishing effect on the activation of classical complement pathway through MBL binding. The data concur with our hypothesis of genetic instability as the reason for total C4B deficiency in this patient, and an increased susceptibility to infections due to defects of C4B and MBL.
CASE REPORT
A female baby was born in 1977 by normal vaginal delivery after a full-term and uncomplicated pregnancy. The infant's birth weight was 2,960 g. The histories of family members included viral type meningitis in the father and the mother's sister. The infant was well until 3 months of age, when she was admitted to hospital due to a fever. A bacterial type culture negative meningitis was detected (718 × 106 leukocytes/liter with 85% granulocytes in the spinal fluid). A good response to chloramphenicol treatment was recorded. At the age of 5 months a second purulent meningitis (3,390 × 106 leukocytes/liter with 89% granulocytes in the spinal fluid) was diagnosed, and at the age of 6 months a third purulent bacterial, culture-negative meningitis (7,000 × 106/l leukocytes with 89% granulocytes in the spinal fluid) was diagnosed. The clinical response to antibiotic treatments was delayed. A small sacral fistula was found and operated. At the age of 11 months chronic sacral fistulas were developed. The child suffered from persistent fever and had the erythrocyte sedimentation rates constantly between 60 to 100 mm/h. Pararectal abscesses with chronic draining were developed and she had poor weight gain. During the next 3 years the child was constantly ill, she was treated with many different antibiotics, and several operations to correct the draining fistulas were carried out. At the age 46 months, an intravenous fresh frozen plasma treatment (30 ml/kg) every 4 weeks was initiated with prompt clinical response. All fistulas were closed within 2 weeks after the first infusion. Fresh frozen plasma treatment was given for 1 year and the child did well. Therefore, the treatment was stopped. Except for occasional urinary tract infections, no other infections were recorded thereafter. At the age of 17 years she developed a sacral abscess which was operated and successfully treated with antibiotics and fresh frozen plasma (26 ml/kg) infusions.
Immunologic evaluations were carried out numerous times. Normal numbers of peripheral blood leukocytes as well as T lymphocytes (CD3+), B lymphocytes (CD19+) and NK cells (CD16/56+) were detected. Serum immunoglobulin levels, IgG subclasses, serum pneumococcal IgG antibodies, and antibody responses to tetanus toxoid were normal, showing intact B-cell function. T cells responded normally to phytohemagglutinin, concanavalin A, and pokeweed mitogen. The lymph node biopsy showed normal histology. Phagocytosis, killing test, and nitroblue tetrazolium test measuring granulocyte function were within normal limits when determined using routine clinical laboratory methods.
At the age of 17 years in 1994, total hemolytic complement of the patient's serum was undetectable three times. The dose of 500 ml of fresh frozen plasma made the total hemolytic complement detectable, 18 U/ml, while after the infusion of 1,000 ml (22 ml/kg) it was 28 U/ml (reference interval 50 to 150 U/ml). The hemolytic complement values of the mother, father, brother, and sister were 30, 72, 29, and 53 U/ml, respectively. The patient's C4 concentrations were persistently low, 0.11 to 0.16 g/liter, being approximately 60% of normal values. C4 concentrations for the mother, father, brother and sister were 0.18, 0.22, 0.16, and 0.29 g/liter, respectively. C4 allotyping showed total C4B deficiency in the patient, as well as in the mother and brother (Fig. 1). All other complement components (C1q, C1r, C1s, C2, C3, C5, C6, C7, C8, and C9) were normal when measured by nephelometry and electroimmunoassay at three different times (February 1980, May 1997, and December 1997).
FIG. 1.
C4 phenotypes based on allotyping of the patient (Pa), sister (Si), control (Co), father (Fa), brother (Br), and mother (Mo). The C4 concentrations for the patient, sister, father, brother, and mother were 0.16, 0.29, 0.22, 0.16, and 0.18 g/liter, respectively, corresponding to the absence of C4B protein in the patient, brother, and mother.
MATERIALS AND METHODS
Study subjects.
A patient having suffered from recurrent and severe infections and her parents and two siblings were studied. Informed consent was obtained from the patient and her family members by the attending physician at Turku University Central Hospital, Turku, Finland.
HLA and complement typing.
The HLA-A, -B, and -C typing was performed using the standard microlymphocytotoxicity test (1). The HLA-B locus was sequenced to determine the subtype of B35 alleles as previously described (6). The HLA-DRB1 alleles were genotyped using a LIPA HLA-DRB1 kit (Innogenetics, Zwijndrecht, Belgium). Complement factor B and C4 allotypes were determined by immunofixation electrophoresis as previously described (27, 34). Factor B alleles F and S, as well as the subtypes FA and FB, were identified using a PCR-based method utilizing restriction polymorphism in the codon seven of exon two (21).
DNA preparation.
Genomic DNA was extracted from acid-citrate dextrose anticoagulated peripheral blood samples by standard salting out method (28).
Analysis of MBL gene polymorphism.
Variants of MBL due to mutations at codon 52, 54, and 57 in exon 1 of the MBL gene was determined by PCR amplification and restriction enzyme digestion of amplified DNA according to the method of Madsen et al. (25). Promoter variants at position −550 (H/L) and −221 (X/Y) were determined by allele specific PCR amplification (26).
RFLP analysis.
Restriction fragment length polymorphism (RFLP) analysis was performed using probes for RP, C4, CYP21, and TNX genes and the Southern hybridization protocol described previously (19).
Isotype specific PCR.
C4A/C4B specific PCR was performed according to the method of Barba et al. using primers L3, A-UP, and B-UP (4). PCR products were analyzed on 2% agarose gel and visualized by ethidium bromide staining.
PCR amplification of the C4d-region.
A 2.9-kb fragment including the C4d-region was amplified with C4-specific primers C4E 22.5 and C4E 31.3 (24) using the Expand High-Fidelity PCR system (Boehringer Mannheim, Indianapolis, Ind.). The PCR conditions in a final volume of 100 μl were 0.2 mM concentrations of deoxynucleoside triphosphates (dNTPs), a 0.3 μM concentration of each primer, 500 ng of genomic DNA, and 2.6 U of High-Fidelity enzyme mix. The following PCR program was used: 1 cycle at 94°C for 2 min; 10 cycles at 94°C for 30 s, 66°C for 1 min, 72°C for 2 min; 20 cycles at 94°C for 30 s, 57°C for 1 min 72°C for 3 min; final extension at 72°C for 7 min, followed by a 4°C dwell. An aliquot of each PCR product was analyzed on agarose gel using ethidium bromide staining.
Cloning and sequencing the C4d-region.
The amplification products were purified with QIAquick spin columns (Qiagen, Chatsworth, Calif.) and cloned into a TA-cloning vector using the Original TA Cloning Kit (Invitrogen, Carlsbad, Calif.). The plasmid DNA was extracted from 5 ml overnight cultures using QIAprep Spin Plasmid kit (Qiagen). The C4d insert was amplified by PCR and sequenced using the BigDye Terminator cycle sequencing ready reaction kit. Electrophoresis was performed with ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Foster City, Calif.). The primers used in sequencing were C4E 25.3, C4E 26.5, C4E 27.3, C4E 28.5, and C4E 29.3 (24).
Restriction enzyme digestions with NlaIV and Eco0109I.
The NlaIV restriction enzyme digestion was performed to further determine C4 isotypes. PCR products encompassing the C4d region were purified with ethanol precipitation and restriction digested with 4 U of NlaIV. The restriction fragments were analyzed on 2% agarose gel and detected with ethidium bromide. A 467-bp fragment represents C4B, while two fragments, 276 and 191 bp, represent C4A. The NlaIV digestion was also used to identify C4A and C4B clones. In addition, the patient's C4d clones were digested with Eco0109I to evaluate the isotype specific differences.
Analysis of C2 gene.
Screening for the four known mutations of C2 gene in exons 2, 5, 6, and 11 was performed as described earlier (19).
Screening for known C4 mutations.
The recently found site of a novel cystein deletion at codon 522 in exon 13 was studied using a mutation specific primer C4del13 (5′ CATCACCTGGCACCCTCCTTTA 3′) and reverse primer C4E14.3R (5′ CTTGCCCATGTTGAGGGGCT 3′) (K. L. Rupert, J. M. Moulds, Y. Yang, R. Warren, J. Reveille, F. C. Arnett, and C. Y. Yu, abstract from the XVIIIth Int. Complement Workshop, Immunopharmacology, 49:29, 2000). The PCR conditions were 100 ng of genomic DNA, 0.2 μM (each) primer, 100 mM Tris-HCl, 500 mM KCl, 15 mM MgCl2, 0.01% gelatin, 0.1 mM dNTPs, and 2.5 U of AmpliTaq Gold DNA polymerase (Roche Molecular Systems, Branchburg, N.J.). The following PCR program was used: 1 cycle at 94°C for 10 min; 35 cycles at 94°C for 30 s, 64°C for 45 s, 72°C for 1 min; final extension at 72°C for 10 min, followed by a 4°C dwell. An aliquot of the PCR product was analyzed using 1% agarose gel and ethidium bromide staining.
The amplification of exon 20 was performed using primers XC-9 and C420up described by Nordin Fredrikson et al. to detect a 1-bp deletion which abolishes an AciI restriction site (29). The amplification product was purified with ethanol precipitation, digested with AciI and separated on 3% MetaPhor agarose (FMC BioProducts, Rockland, Maine) to reveal the presence or absence of a cleavage site in the fragment.
To detect a 2-bp insertion in exon 29 originally described by Barba et al., a mutation specific primer C4ins29 (5′ GCTCTGAGAACCAGTGACTGAGAG 3′) was designed to be used with isotype specific primers A-down and B-down (3, 4). PCR was performed under standard reaction conditions with AmpliTaq Gold DNA polymerase using the following program: 10 min at 94°C; 30 s at 94°C, 45 s at 62°C, 1 min at 72°C for 38 cycles; ten minute hold at 72°C. Screening for the mutation in exon 29 was also confirmed by the PCR method described by Sullivan et al. (36).
Screening for new C4 mutations.
PCR amplification of all 41 exons of C4 genes was performed using a 0.2 μM concentration of each primer, 100 ng of genomic DNA, 0.1 mM dNTPs and AmpliTaq, AmpliTaq Gold, or DyNAzyme EXT (Finnzymes, Espoo, Finland) DNA polymerase with the buffer recommended by the manufacturer (Triton-free buffer for samples to denaturing high-performance liquid chromatography [DHPLC] analysis). Samples for single-stranded conformation polymorphism analysis (SSCP) analysis were internally labeled by fluorescent [R110]dCTP (Applied Biosystems) with a final concentration of 0.5 μM. An aliquot of each amplification product was separated electrophoretically and visualized with ethidium bromide. Specific data for C4 primer sequences and thermal cycler conditions are available on request.
All 41 exons were subjected to SSCP analysis and 27 exons of them were additionally analyzed with DHPLC system. For SSCP the amplification products were purified by QIAquick PCR purification kit (Qiagen). Samples were prepared by combining 1 μl of PCR product, 0.5 μl of 0.3 M NaOH, 10 μl of deionized formamide and 0.5 μl of GeneScan [ROX] size standard (Applied Biosystems). The mixture was denatured for 5 min at 95°C and run under nondenaturing conditions using 5% GeneScan polymer with 10% glycerol on ABI PRISM 310 genetic analyzer. Tris-borate-EDTA (1×) with 10% glycerol was used as the electrode buffer. Injection time varied between 1 and 8 s at 15.0 kV, and samples were subjected to electrophoresis for 30 to 45 min at 13.0 kV at a temperature of 30°C. The change of a single base may cause a conformational change in the DNA molecule and affect the electrophoretic mobility.
DHPLC analysis was carried out using the Wave HS nucleic acid fragment analysis system (Transgenomic, Omaha, Nebr.). Prior to temperature modulated heteroduplex analysis (TMHA), PCR products were denatured at 95°C for 5 min and then gradually reannealed by slow cooling (−0.5°C/cycle) to 23°C. An aliquot of reannealed PCR product was automatically injected into the DNASeph column and eluted with constant flow rate of 0.9 ml/min and an acetonitrile gradient (2% increase in buffer B/min; buffer A consisting of 0.1 M triethylamine acetate and buffer B 0.1 M triethylamine acetate with 25% acetonitrile). Melting temperatures were estimated with WAVEmaker software (version 4.0.29) and the DHPLC Melt program (http://insertion.stanford.edu/melt.html). TMHA was performed with individual samples from patient and control DNA, and with a mixed sample in combined analysis. Equal amounts of their DNA was mixed and rehybridized in order to reveal possible differences in between their DNA sequences. PCR fragments with a change in the electrophoretic mobility (SSCP) or in the melting profile (DHPLC) were subjected to subsequent sequencing using the BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems) to determine the nature of the mutation. Electrophoresis was performed with ABI PRISM 310 or 3100 genetic analyzer.
RESULTS
MBL gene analysis.
Two MBL haplotypes HYA and LYB, located on chromosome 10q21, were found in the family. The normal wild-type structural allele is denoted A, whereas allele B has a nucleotide change at codon 54 resulting in an aspartic acid instead of a glycine residue (38). The sister was homozygous for the HYA haplotype and the brother for LYB, while the patient and both the parents were heterozygous. Thus, only the brother was MBL deficient. The patient had a variant MBL allele, which is known to associate with MBL serum concentration in the lower normal range, at average about 400 μg/liter according to Madsen et al. (26).
Southern blot analysis.
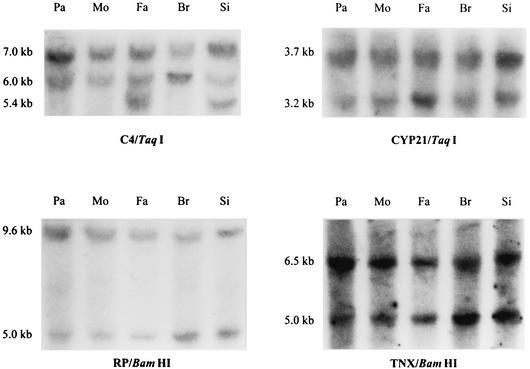
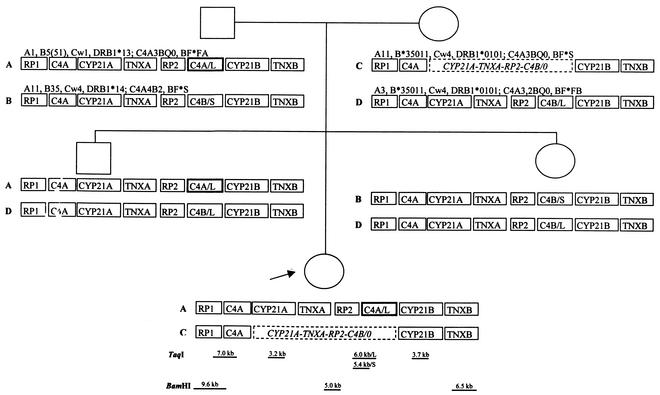
To determine the genetic basis of the total C4B deficiency, the organization of MHC class III genes RP1 and RP2, C4A and C4B, CYP21A and CYP21B, as well as TNXA and TNXB, was studied. DNA samples of all family members were subjected to restriction enzyme mapping (Fig. 2). The 3.7- and 3.2-kb TaqI fragments for CYP21B and CYP21A genes, respectively, could be seen in all family members. In the patient and the mother, the 3.7-kb fragment was twice as intense as the 3.2-kb fragment. Thus, they carried a heterozygous deletion of CYP21A gene. The intensities of the 6.5- and 5.0-kb BamHI fragments representing TNXB and TNXA genes, respectively, showed a deletion on TNXA gene in the patient and the mother. The intensity of the 5.0-kb BamHI fragment identifying RP2 gene was decreased in the patient and the mother, indicating a deletion of RP2 gene on the maternal chromosome inherited by the patient. The 7.0-kb TaqI fragment specifying RP1-C4 long gene locus was present in all family members, as well as the fragment of 6.0 kb corresponding to RP2-C4 long locus. The 5.4-kb fragment for RP2-C4 short locus was seen in father and sister only. According to the intensities of the TaqI fragments, the patient had a missing long C4B gene inherited from the mother. In conclusion, the restriction data indicate that the patient has a deletion of four genes, CYP21A, TNXA, RP2, and C4B, inherited on the maternal chromosome, and two long C4 genes on the paternal chromosome (Fig. 3).
FIG. 2.
Southern blot analysis with TaqI and BamHI restriction digests showing a deletion of CYP21A, TNXA, RP2, and C4B genes on maternal haplotype inherited by the patient. The 7.0-, 6.0-, and 5.4-kb TaqI fragments represent RP1-C4 long, RP2-C4 long, and RP2-C4 short, respectively. The fragment intensities suggested that the patient had a missing C4B gene inherited on the maternal chromosome. The 3.7-kb TaqI fragment for CYP21B gene and the 3.2-kb TaqI fragment for CYP21A gene could be seen in all family members. However, the weaker intensity of the 3.2-kb fragment in the patient and the mother showed that they carried a heterozygous deletion of CYP21A gene. The 9.6- and 5.0-kb BamHI fragments corresponding to RP1 and RP2 genes showed decreased intensity of the 5.0-kb fragment in the patient and the mother. The intensities of the 6.5- and 5.0-kb BamHI fragments representing TNXB and TNXA genes, respectively, showed a deletion on TNXA gene in the patient and the mother. The restriction data indicate a deletion of four genes encompassing CYP21A, TNXA, RP2, and C4B on the maternal chromosome inherited by the patient. The samples are presented in the following order: patient (Pa), mother (Mo), father (Fa), brother (Br), and sister (Si).
FIG. 3.
Pedigree showing the MHC haplotypes with HLA and complotypes. Deleted genes on maternal chromosome are outlined with a dashed line, and the converted C4 gene of paternal origin is marked with bold line. The approximate locations and sizes of the restriction fragments are indicated at the bottom.
Isotype-specific PCR and restriction analysis.
All family members had the C4A isotype verified by PCR and by NlaIV restriction. The C4B isotype was absent in the patient, and detected in father and sister only. The patient's C4d clones carried the Rg1 determinant identified by Eco0109I digestion. Hence, only C4 genes with C4A isotype were present, corresponding to the total C4B deficiency on protein level.
Sequence analysis of C4d region.
The known C4 allotype specific determinants are carried in exons 25, 28, and 29. In order to define the allotypes of two C4A genes on the chromosome inherited from the father, the C4d region of the patient and the father was cloned and sequenced. As a result, all the clones studied were of the same allotype C4A3a. Nucleotide alterations in intron 28 and exon 29 of these C4A3a clones specified one C4A3a gene and one long C4B gene converted to C4A3a-like.
Screening for mutations.
The converted C4B gene of the patient was screened for new mutations in the coding region by SSCP and sequencing. In all, 10 alterations were observed in the patient compared to the published C4A3 sequences M59816 and M59815 (42). However, none of these caused amino acid changes or prominent mutations (Table 1). Nucleotide 5494 in exon 28 was found to be either a T or a C, and a substitution, 5911G→A, in exon 29 was revealed. All the other alterations found were of intronic origin. The segments without variation in SSCP were subjected to further screening with DHPLC, TMHA, and sequencing, which revealed the presence of four additional alterations. One single nucleotide polymorphism (SNP) was found in exon 12, all other point mutations were in intronic regions (Table 1). The sequence variation detected in the patient has been recorded in Human Genome Variation Database HGVbase (9) (http://hgvbase.cgb.ki.se/) under identification numbers IND/SNP001026494-IND/SNP001026518. Previously described C2 and C4 mutations were not detected in this family.
TABLE 1.
Novel nucleotide alterations found in patient's C4 sequence
| Exon or intron | Nucleotide(s)a | Amino acid | Codon sequence | Variationb | Origin |
|---|---|---|---|---|---|
| Intron 2 | 598 | Deletion of T | |||
| 627 | Deletion of A | ||||
| 638 | Deletion of C | ||||
| 649 | G→T | ||||
| Intron 4 | 1314-1315 | Insertion of C | |||
| 1327 | Deletion of T | ||||
| Intron 8 | 2293 | C→G | |||
| Intron 11 | 626 | C/T | C, maternal; T, paternal | ||
| Exon 12 | 908 | A476 | GCC/GCT | C/T | |
| Intron 20 | 3432 | A/G | |||
| Intron 24 | 4622-4623 | Insertion of A | Paternal A | ||
| 4648 | Deletion of T | Paternal A | |||
| Exon 28 | 5494 | F1159 | TTT/TTC | T/C | |
| Intron 28 | 5654-5655 | Insertion of C | |||
| 5680-5681 | Insertion of C | Paternal A | |||
| 5690-5691 | Insertion of C | Paternal A | |||
| 5840 | T/C | T, maternal; C, paternal | |||
| Exon 29 | 5911 | S1223 | TCG/TCA | G/A | G, maternal; A, paternal |
| Intron 30 | 6428 | C→A | Paternal A | ||
| Intron 38 | 10657-10658 | Insertion of C | |||
| 10681 | C→T | ||||
| 10700 | Deletion of T | ||||
| 10718 | Deletion of T | ||||
| 10721 | Deletion of T | ||||
| Intron 39 | 10943 | Deletion of C |
DISCUSSION
The patient with severe infections had total deficiency of C4B. The MHC region on the maternal chromosome carried a common deletion with C4B gene and on the paternal chromosome the C4B gene was converted to C4A. The presence of two C4A genes on a chromosome, instead of one C4A and one C4B, has been observed in 13.3% of the Caucasian population (8). The most prevalent homoexpressed C4 is C4A3,3, and the resulting partial deficiency of C4B is among the most common variations in human genome. When the amount of C4A proteins in allotyping has been correlated to the number of C4A genes in RFLP analyses, the results indicate that the converted genes are indeed functional. In our study, the patient showed one C4A3 gene and one C4B gene converted to the C4A3-like gene on the paternal chromosome with HLA-B51. The maternal haplotype with CYP21A-TNXA-RP2-C4B deletion carried HLA-B*35011, as did the other maternal haplotype. HLA-B51 and HLA-B35 form a serologically cross-reacting antigen group, and three subtypes from this group have recently shown to predispose to rapid progression of AIDS. Subtypes HLA-B*3502 and HLA-B*3503 have the accelerating influence in whites, and subtype HLA-B*5301 has the accelerating influence in African Americans (17). HLA-B*3501 had no influence on AIDS progression, while susceptibility to other pathogenic microbes is not yet known.
In the studied family, the expressed C4A allotypes do not support strong protein expression of the converted C4B gene. Mutation screening revealed no mutations in the converted gene that would conclusively explain its nonfunctionality. However, several alterations in intronic regions were found. Insertion 1314-1315insC in intron 4 resides in a very close proximity to the branch point upstream of the 3′ splicing site. The replacement 626C→T in intron 11 is located right after the 5′ consensus sequence involved in the splicing reaction, and the 5840T/C heterozygosity in intron 28 is seen near the 3′ branch point. It is not fully known how sequence variation affects the splicing of introns. A retroposon-like insertion in intron 13 generating abnormal mRNA splicing has been shown to lower C4 production in mice (44). In neurofibromatosis type 1, an intronic Alu sequence insertion causes defective gene expression due to interference of branch point recognition (41). In a study describing nonexpressed C4A and C4B genes due to a frameshift mutation in exon 29, the size of mRNA was normal yet the amount was reduced (24). Even though greater flexibility in the consensus sequences have been observed in higher eukaryotes, intronic point mutations may impede splicing or lead to the generation of cryptic splicing signals and exceptional splicing of precursor RNA. In this patient, the alterations found in the intronic regions may reduce if not prevent the expression of the converted C4A gene.
Congenital absence of C4 is most often associated with systemic lupus erythematosus-like syndrome (37). Also, an increased risk for bacterial infections has been reported. Of 46 children with bacterial meningitis, 11% were homozygous deficient for C4B versus 3% in 223 controls (33). In 29 white patients with bacteremic Streptococcus pneumoniae, Haemophilus influenzae, or Neisseria meningitidis infections, 14% had homozygous C4B deficiency compared to 2% in race-matched controls (7). However, there are studies that do not indicate C4B deficiency as a significant predisposing factor in bacterial meningitis (11, 15). Our patient had suffered from recurrent meningitis and chronic draining sacral fistulas. Intensive antibiotic and surgical treatments were ineffective. After empirical treatment with fresh frozen plasma, correcting the complement function temporarily, a prompt clinical recovery was noted. Interestingly, the plasma dose of 20 to 30 ml/kg of body weight, also used in replacement therapy of primary hypogammaglobulinemia (10), made the total hemolytic complement just detectable. In a patient with C2 deficiency and systemic lupus erythematosus, plasma therapy has been shown to induce a full clinical remission for 6 to 8 weeks and a brief restoration of hemolytic complement activity (35). In our patient, plasma therapy was given for 12 months and the patient has stayed free of severe infections thereafter. The treatment may have induced substitutive mechanisms to accommodate immune responses as it has had long-term effects without a continuous supply of C4B or MBL. A similar observation has previously been demonstrated after intravenous gammaglobulin treatment (2). In C1q deficient mice, a recent study shows that a small proportion of bone marrow cells is sufficient to restore normal C1q levels in serum (31).
In northern Europe, N. meningitidis is the major cause of meningitis (20). In the present study, the patient had a structurally heterozygous MBL genotype in addition to the homozygous C4B deficiency. Besides the normal MBL allele, the patient had a variant allele carrying the codon 54 substitution. In in vitro studies, a recombinant protein of the variant allele has been proved unstable (39). Moreover, from the heterozygous MBL trimers seven in eight have been postulated abnormal (38), yet in other studies they possess normal biochemical and functional characteristics (18). Anyhow, the variant MBL genotype of the patient produces a structurally variant protein and is known to associate with medium levels of MBL serum concentrations (26). MBL has been shown to bind to N. meningitidis and further the activation of C4 (20). The partial deficiency of MBL may result in poor activation of complement on encapsulated organisms, particularly in combination with C4B deficiency. In conclusion, the complete C4B deficiency together with the heterozygous state of MBL is indicated as the cause for increased susceptibility to infections in this patient. The restricted repertoire of HLA-B molecules may play a critical role in epitope recognition and antigen presentation in immune defense against pathogenic microbes.
Acknowledgments
This study was supported by grants from the Foundation for Pediatric Research and the Medical Research Fund of Finnish Red Cross Blood Transfusion Service. Financial support for Turku Centre for Biotechnology was provided by the Academy of Finland, the National Technology Agency (TEKES), and Turku University Hospital Research Fund (EVO).
We thank Marjaana Mustonen for her technical assistance with complement typing and Tuija Kyrölä for DNA sequencing and upkeep of the Wave apparatus. We thank Yung Yu for providing C4 primers to our laboratory.
REFERENCES
- 1.Amos, D., H. Badir, W. Boyle, M. McQueen, and A. Tiilikainen. 1969. A simple microcytotoxicity test. Transplantation 7:220-223. [DOI] [PubMed] [Google Scholar]
- 2.Ballow, M. 1997. Mechanisms of action of intravenous immune serum globulin in autoimmune and inflammatory diseases. J. Allergy Clin. Immunol 100:151-157. [DOI] [PubMed] [Google Scholar]
- 3.Barba, G., C. Rittner, and P. M. Schneider. 1993. Genetic basis of human complement C4A deficiency. Detection of a point mutation leading to nonexpression. J. Clin. Invest 91:1681-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barba, G. M., L. Braun-Heimer, C. Rittner, and P. M. Schneider. 1994. A new PCR-based typing of the Rodgers and Chido antigenic determinants of the fourth component of human complement. Eur. J. Immunogenet. 21:325-339. [DOI] [PubMed] [Google Scholar]
- 5.Belt, K. T., M. C. Carroll, and R. R. Porter. 1984. The structural basis of the multiple forms of human complement component. Cell 36:907-914. [DOI] [PubMed] [Google Scholar]
- 6.Bettinotti, M. P., Y. Mitsuishi, K. Bibee, M. Lau, and P. I. Terasaki. 1997. Comprehensive method for the typing of HLA-A, B, and C alleles by direct sequencing of PCR products obtained from genomic DNA. J. Immunother. 20:425-430. [DOI] [PubMed] [Google Scholar]
- 7.Bishof, N. A., T. R. Welch, and L. S. Beischel. 1990. C4B deficiency: a risk factor for bacteremia with encapsulated organisms. J. Infect. Dis. 162:248-250. [DOI] [PubMed] [Google Scholar]
- 8.Blanchong, C. A., B. Zhou, K. L. Rupert, E. K. Chung, K. N. Jones, J. F. Sotos, W. B. Zipf, R. M. Rennebohm, and Y. C. Yung. 2000. Deficiencies of human complement component C4A and C4B and heterozygosity in length variants of RP-C4-CYP21-TNX (RCCX) modules in Caucasians: the load of RCCX genetic diversity on major histocompatibility complex-associated disease. J. Exp. Med. 191:2183-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brookes, A. J., H. Lehväslaiho, M. Siegfried, J. G. Boehm, Y. P. Yuan, C. M. Sarkar, P. Bork, and F. Ortigao. 2000. HGBASE: a database of SNPs and other variations in and around human genes. Nucleic Acids Res. 28:356-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buckley, R. H. 1972. Plasma therapy in immunodeficiency diseases. Am. J. Dis. Child 124:376-381. [DOI] [PubMed] [Google Scholar]
- 11.Cates, K. L., P. Densen, J. C. Lockman, and R. P. Levine. 1992. C4B deficiency is not associated with meningitis or bacteremia with encapsulated bacteria. J. Infect. Dis. 165:942-944. [DOI] [PubMed] [Google Scholar]
- 12.Dangel, A. W., A. R. Mendoza, B. J. Baker, C. M. Daniel, M. C. Carroll, L. C. Wu, and C. Y. Yu. 1994. The dichotomous size variation of human complement C4 genes is mediated by a novel family of endogenous retroviruses, which also establishes species-specific genomic patterns among Old World primates. Immunogenetics 40:425-436. [DOI] [PubMed] [Google Scholar]
- 13.Dodds, A. W., and S. K. Law. 1990. The complement component C4 of mammals. Biochem. J. 265:495-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dodds, A. W., X. D. Ren, A. C. Willis, and S. K. Law. 1996. The reaction mechanism of the internal thioester in the human complement component C4. Nature 379:177-179. [DOI] [PubMed] [Google Scholar]
- 15.Ernst, T., P. J. Spath, C. Aebi, U. B. Schaad, and M. G. Bianchetti. 1997. Screening for complement deficiency in bacterial meningitis. Acta Paediatr. 86:1009-1010. [DOI] [PubMed] [Google Scholar]
- 16.Fischer, M. B., M. Ma, S. Goerg, X. Zhou, J. Xia, O. Finco, S. Han, G. Kelsoe, R. G. Howard, T. L. Rothstein, E. Kremmer, F. S. Rosen, and M. C. Carroll. 1996. Regulation of the B cell response to T-dependent antigens by classical pathway complement. J. Immunol. 157:549-556. [PubMed] [Google Scholar]
- 17.Gao, X., G. W. Nelson, P. Karacki, M. P. Martin, J. Phair, R. Kaslow, J. J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, S. J. O'Brien, and M. Carrington. 2001. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N. Engl. J. Med. 344:1668-1675. [DOI] [PubMed] [Google Scholar]
- 18.Garred, P., S. Thiel, H. O. Madsen, L. P. Ryder, J. C. Jensenius, and A. Svejgaard. 1992. Gene frequency and partial protein characterization of an allelic variant of mannan binding protein associated with low serum concentrations. Clin. Exp. Immunol. 90:517-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaatinen, T., O. Ruuskanen, L. Truedsson, and M. L. Lokki. 1999. Homozygous deletion of the CYP21A-TNXA-RP2-C4B gene region conferring C4B deficiency associated with recurrent respiratory infections. Hum. Immunol. 60:707-714. [DOI] [PubMed] [Google Scholar]
- 20.Jack, D. L., A. W. Dodds, N. Anwar, C. A. Ison, A. Law, M. Frosch, M. W. Turner, and N. J. Klein. 1998. Activation of complement by mannose-binding lectin on isogenic mutants of Neisseria meningitidis serogroup B. J. Immunol. 160:1346-1353. [PubMed] [Google Scholar]
- 21.Jahn, I., J. E. Mejía, M. Thomas, C. Darke, H. Schröder, G. Geserick, and G. Hauptmann. 1994. Genomic analysis of the F subtypes of human complement factor B. Eur. J. Immunogenet. 21:415-423. [DOI] [PubMed] [Google Scholar]
- 22.Koch, A., M. Melbye, P. Sørensen, P. Homøe, H. O. Madsen, K. Mølbak, C. H. Hansen, L. H. Andersen, G. W. Hahn, and P. Garred. 2001. Acute respiratory tract infections and mannose-binding lectin insufficiency during early childhood. JAMA 285:1316-1321. [DOI] [PubMed] [Google Scholar]
- 23.Law, S. K. A., A. W. Dodds, and R. R. Porter. 1984. A comparison of the properties of two classes, C4A and C4B, of the human complement component C4. EMBO J. 3:1819-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lokki, M. L., A. Circolo, P. Ahokas, K. L. Rupert, C. Y. Yu, and H. R. Colten. 1999. Deficiency of human complement protein C4 due to identical frameshift mutations in the C4A and C4B genes. J. Immunol. 162:3687-3693. [PubMed] [Google Scholar]
- 25.Madsen, H. O., P. Garred, J. A. Kurtzhals, L. U. Lamm, L. P. Ryder, S. Thiel, and A. Svejgaard. 1994. A new frequent allele is the missing link in the structural polymorphism of the human mannan-binding protein. Immunogenetics 40:37-44. [DOI] [PubMed] [Google Scholar]
- 26.Madsen, H. O., P. Garred, S. Thiel, J. A. Kurtzhals, L. U. Lamm, L. P. Ryder, and A. Svejgaard. 1995. Interplay between promoter and structural gene variants control basal serum level of mannan-binding protein. J. Immunol. 155:3013-3020. [PubMed] [Google Scholar]
- 27.Marcus, D., and C. A. Alper. 1986. Methods for allotyping complement proteins, p. 185-196. In N. R. Rose, H. Friedman, and J. L. Fahey (ed.), Manual of clinical laboratory immunology. American Society for Microbiology, Washington, D.C.
- 28.Miller, S. A., D. D. Dykes, and H. F. Polesky. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16:1215.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nordin Fredrikson, G., B. Gullstrand, P. M. Schneider, K. Witzel-Schlömp, A. G. Sjöholm, C. A. Alper, Z. Awdeh, and L. Truedsson. 1998. Characterization of non-expressed C4 genes in a case of complete C4 deficiency: identification of a novel point mutation leading to a premature stop codon. Hum. Immunol. 59:713-719. [DOI] [PubMed] [Google Scholar]
- 30.Ochsenbein, A. F., D. D. Pinschewer, B. Odermatt, M. C. Carroll, H. Hengartner, and R. M. Zinkernagel. 1999. Protective T cell-independent antiviral antibody responses are dependent on complement. J. Exp. Med. 190:1165-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petry, F., M. Botto, R. Holtappels, M. J. Walport, and M. Loos. 2001. Reconstitution of the complement function in C1q-deficient (C1qa-/-) mice with wild-type bone marrow cells. J. Immunol. 167:4033-4037. [DOI] [PubMed] [Google Scholar]
- 32.Ren, X. D., A. W. Dodds, and S. K. Law. 1993. The thioester and isotypic sites of complement component C4 in sheep and cattle. Immunogenetics 37:120-128. [DOI] [PubMed] [Google Scholar]
- 33.Rowe, P. C., R. H. McLean, R. A. Wood, R. J. Leggiadro, and J. A. Winkelstein. 1989. Association of homozygous C4B deficiency with bacterial meningitis. J. Infect. Dis. 160:448-451. [DOI] [PubMed] [Google Scholar]
- 34.Sim, E., and S. J. Cross. 1986. Phenotyping of human complement component C4, a class-III HLA antigen. Biochem. J. 239:763-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steinsson, K., K. Erlendsson, and H. Valdimarsson. 1989. Successful plasma infusion treatment of a patient with C2 deficiency and systemic lupus erythematosus: clinical experience over forty-five months. Arthritis Rheum. 32:906-913. [PubMed] [Google Scholar]
- 36.Sullivan, K. E., N. A. Kim, D. Goldman, and M. A. Petri. 1999. C4A deficiency due to a 2 bp insertion is increased in patients with systemic lupus erythematosus. J. Rheumatol. 26:2144-2147. [PubMed] [Google Scholar]
- 37.Sullivan, K. E., and J. A. Winkelstein. 1999. Genetically determined deficiencies of the complement system, p. 397-416. In H. D. Ochs, C. I. E. Smith, and J. M. Puck (ed.), Primary immunodeficiency diseases. A molecular and genetic approach. Oxford University Press, New York, N.Y.
- 38.Sumiya, M., M. Super, P. Tabona, R. J. Levinsky, T. Arai, M. W. Turner, and J. A. Summerfield. 1991. Molecular basis of opsonic defect in immunodeficient children. Lancet 337:1569-1570. [DOI] [PubMed] [Google Scholar]
- 39.Super, M., S. D. Gillies, S. Foley, K. Sastry, J. E. Schweinle, V. J. Silverman, and R. A. Ezekowitz. 1992. Distinct and overlapping functions of allelic forms of human mannose binding protein. Nat. Genet. 2:50-55. [DOI] [PubMed] [Google Scholar]
- 40.Tassabehji, M., T. Strachan, M. Anderson, R. D. Campbell, S. Collier, and M. Lako. 1994. Identification of a novel family of human endogenous retroviruses and characterization of one family member, HERV-K(C4), located in the complement C4 gene cluster. Nucleic Acids Res. 22:5211-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallace, M. R., L. B. Andersen, A. M. Saulino, P. E. Gregory, T. W. Glover, and F. S. Collins. 1991. A de novo Alu insertion results in neurofibromatosis type 1. Nature 353:864-866. [DOI] [PubMed] [Google Scholar]
- 42.Yu, C. Y. 1991. The complete exon-intron structure of a human complement component C4A gene. DNA sequences, polymorphism, and linkage to the 21-hydroxylase gene. J. Immunol. 146:1057-1066. [PubMed] [Google Scholar]
- 43.Yu, C. Y., Z. Yang, C. A. Blanchong, and W. Miller. 2000. The human and mouse MHC class III region: a parade of 21 genes at the centromeric segment. Immunol. Today 21:320-328. [DOI] [PubMed] [Google Scholar]
- 44.Zheng, J. H., S. Natsuume-Sakai, M. Takahashi, and M. Nonaka. 1992. Insertion of the B2 sequence into intron 13 is the only defect of the H-2k C4 gene which causes low C4 production. Nucleic Acids Res. 20:4975-4979. [DOI] [PMC free article] [PubMed] [Google Scholar]