Abstract
The mouse mot-2 protein was earlier shown to bind to the tumor suppressor protein, p53. The mot-2 binding site of p53 was mapped to C-terminal amino acid residues 312–352, which includes the cytoplasmic sequestration domain. In the present study, we have found that both mot-1 and mot-2 bind to p53 in vitro. By using His-tagged deletion mutant proteins, the p53-binding domain of mot-2 was mapped to its N-terminal amino acid residues 253–282, which are identical in mot-1 and mot-2 proteins. Some peptides containing the p53-binding region of mot-2 were able to compete with the full-length protein for p53 binding. The data provided rationale for in vitro binding of mot-1 and mot-2 proteins to p53 and supported the conclusion that inability of mot-1 protein to bind p53 in vivo depends on secondary structure or its binding to other cellular factors. Most interestingly, the p53-binding region of mot-2 was common to its MKT-077, a cationic dye that exhibits antitumor activity, binding region. Therefore it is most likely that MKT-077-induced nuclear translocation and restoration of wild-type p53 function in transformed cells takes place by a competitional mechanism.
Keywords: mot, p53, binding domain, MKT-077
Introduction
Mot-2 is an hsp70 family member that was shown to bind to p53 [1]. Its overexpression results in malignant transformation of NIH 3T3 cells [2], growth advantage and attenuation of differentiation HL-60 promyelocytic leukemia cells [3], and extension of life span of normal human fibroblasts [4]. Mouse mot-1 protein differs from mot-2 by two amino acids in the C-terminus; it has pancytoplasmic cellular distribution, causes senescence in NIH 3T3 cells and is allelic to mot-2 [5–8]. Mot-2 binds to and represses p53 activity [1]. Mot-1 neither colocalizes nor coprecipitates nor affects p53 function [Refs. [1,9]; unpublished observations]. In contrast to mouse cells, human cells have only one kind of mortalin protein that has activity similar to mouse mot-2 [2] and is therefore called hmot-2. It has been suggested that there are at least two mechanisms operating for differential distributions of the mot protein. One is by distinct cDNAs, mot-1 and mot-2 found in mouse, and the other by yet undefined protein modifications or cellular factors found in mouse and human cells.
Mot-2 was also identified as PBP74, mtHSP70, and GRP75 and has been assigned roles in antigen processing, in vivo nephrotoxicity, and radioresistance [10]. In different studies, it has been localized to mitochondria, ER, plasma membrane, and cytoplasmic vesicles [11–13]. Expression level of mot-2/mthsp70/GRP75 level correlates with muscle activity [14], mitochondrial activity, and biogenesis [15], and is induced by low levels of ionizing radiations [16], glucose deprivation [17], calcium ionophore [18], and ozone [19]. Whereas the expression of mot-2/mthsp70/GRP75 is upregulated in human transformed and tumor cell [2,20,21], it decreased during induction of differentiation in HL-60 promyelocytic leukemia cells. Mot-2-overexpressing derivatives were markedly impaired for induction of differentiation [3].
The yeast homologue of mot-2/mthsp70/GRP75, SSC1p, is essential for cell viability [22] and has indispensable functions in mitochondrial import [23,24]. It binds to Tim-44, an inner mitochondrial membrane anchor, and is an essential component of mitochondrial import machinery [24,25]. Mutations in Tim-44 that result in inefficient recruitment of mthsp70/SSC1 are lethal in Saccharomyces cerevisiae [25,26]. Based on the studies in yeast, at least three kinds of activities can be hypothesized for mot-2/mthsp70/GRP75. These include (i) unfolding of proteins outside mitochondria, (ii) unidirectional translocation across mitochondrial membranes that is initiated by membrane potential MΨ, and (iii) completion of import by acting as an ATP-driven motor. It is also required for degradation of misfolded peptides by m-AAA and PIM1 proteases in mitochondria [27]. There is some evidence to suggest that it cooperates with mthsp60 and CPN 10 chaperonins for folding of imported proteins to functionally competent forms in mitochondria and for yet undefined roles of mthsp60 at extra-mitochondrial sites [28]. It is likely but not shown so far that mitochondrial importer and chaperonin functions of mot-2/mthsp70 as predicted above contribute to the phenotypes described above. We have shown that inactivation of wild-type p53 by mot-2 mediates life span extension and malignant transformation phenotypes of normal human (MRC-5) and immortal murine (NIH 3T3) cells, respectively [1,2,4]. Most recently, we have shown that MKT-077, a water-soluble delocalized lipophilic cationic dye that exhibits significant antitumor activity binds to mot-2. It results in abrogation of cytoplasmic sequestration and reactivation of wild-type p53 function in transformed cells [29].
The present study was undertaken to map the domain of mot-2 that is responsible for its interaction with p53. Both mot-1 and mot-2 coimmunoprecipitated with p53 in in vitro pull-down assays. Mortalin residues 253–282 that are identical in mot-1 and mot-2 were found critical for its binding to p53. Based on these data, the inability of mot-1 to bind p53 in vivo can be attributed to either some other mot-1 binding cellular factors or the secondary structure of the proteins. The p53-binding region of mot defined in this study overlapped with the MKT-077-binding region of mot [29]. MKT-077 was shown to abrogate mot-2-p53 interactions and cytoplasmic sequestration of p53 resulting in reactivation of wild-type p53 function and growth arrest of transformed cells [29]. Binding of p53 and MKT-077 to the same region of mot elucidates a competitional mechanism for abrogation of cytoplasmic retention of p53 by MKT-077 [29].
Materials and Methods
Cell Culture
Monkey kidney cells (COS 7) were cultured in Dulbec-co's modified Eagle's minimal essential medium supplemented with 10% fetal bovine serum.
In Vitro Pull-Down Assays
Murine p53 (pSP65/p53, a kind gift from Dr. Antony Braithwaite) [30] was transcribed (Transprobe T kit, Pharmacia) and translated in vitro using rabbit reticulocyte lysate (Stratagene, La Jolla, CA) supplemented with l-[35S]methionine for 1 h. The translation products were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by autoradiography using a phosphorimager. In vitro translated p53 protein was immunoprecipitated with anti-p53 antibodies (PAb421 and PAb 1620, Calbiochem; PAb122, Boehringer Mannheim) and was used for in vitro binding assays as below.
For preparation of recombinant mortalins, the ORF of mortalin cDNA and its various deletions were amplified by polymerase chain reaction (PCR) with sense and antisense primers with BamHI and SalI sites, respectively. These were then cloned into pQE30 vector (Qiagen) to yield His-tagged proteins that were purified as described earlier [31]. The purity of preparations was examined by SDS-PAGE. Aliquots of the purified protein were stored at -20°C in small volumes to avoid repeated freeze-thaw cycles. Purified recombinant His-tagged mortalin proteins (0.5–1 µg) were mixed with in vitro translated 35S-labeled wild-type p53 in the presence of either 1–2 mg bovine serum albumin or COS 7 cell lysate (400 µg protein) in 400 µl Nonidet P-40 lysis buffer [4]. The mixture was incubated with an antimortalin (mthsp70, Affinity Bioreagents) or anti-p53 (CM-1, Novocastra) antibody at 4°C for 1–2 h. Immunocomplexes were separated by incubation with protein-A/G sepharose (Gibco) (20 µl) for 30 min followed by centrifugation. After heating at 95°C in SDS sample buffer, proteins were resolved on SDS-PAGE, transferred to nitrocellulose membrane by semidry transfer and Western blotted with the indicated antibodies and detected by ECL-chemiluminescence (Amersham Pharmacia Biotech, Piscataway, NJ). For competition assays, the indicated amounts of the competitor peptide were incubated with COS 7 lysates for 30 min at 4°C following which His-tagged mortalins were added. Anti-p53 antibody was added 30 min after the addition of mortalin proteins. COS 7 cells transfected with expression plasmids (Invitrogen, San Diego, CA) encoding various V5-tagged mortalin deletion mutants were used for p53 immunoprecipitation. The detection of coprecipitating mortalin was detected by anti-V5 tag antibody (Invitrogen).
Results and Discussion
In Vitro Binding of mot-1 to p53
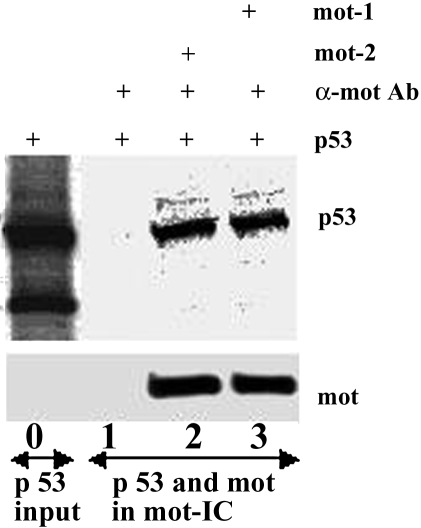
In vitro translated 35S-labeled murine p53 was mixed with recombinant His-tagged mot-1 and mot-2 proteins and was immunoprecipitated from the mixture with anti-mortalin antibody. Immunoprecipitated mortalins and p53 were detected by Western blotting with anti-His tag antibody and autoradiography, respectively. As shown in Figure 1, p53 was immunoprecipitated along with mot-2 as well as mot-1 proteins. Controls such as incubation of p53 alone with anti-mortalin antibody or mortalin-p53 mixture with control isotype matched IgG did not result in precipitation of p53. In a reciprocal assay, glutathione S-transferase (GST)-wild-type p53 fusion protein (Santa Cruz Biotechnology, Santa Cruz, CA) or GST alone was incubated with either recombinant His-tagged mot-1 or mot-2 protein. Proteins were pulled down using glutathione-sepharose and coprecipitating mortalins were detected with anti-mortalin or anti-His antibodies. p53 was detected with anti-GST antibodies (Santa Cruz Biotechnology). Both mot-1 and mot-2 were seen to coimmunoprecipitate with p53 protein (data not shown). This suggested that the region harboring the two amino acid differences between mot-1 and mot-2 may not be involved in its binding to p53 in vitro. These data showed that in contrast to the in vivo situation in which only mot-2 was seen to affect p53 function both mot-1 and mot-2 bind to p53 protein in vitro.
Figure 1.
In vitro binding of mot-1 and mot-2 to p53. In vitro translated 35S-labeled murine p53 (53-kDa band) and a small-sized (43-kDa) product (lane 0) were mixed with His-tagged mot-2 (lane 2) or mot-1 (lane 3) recombinant proteins and were immunoprecipitated with anti-mortalin antibodies. Immunoprecipitated mot-1 and mot-2 were detected by Western blotting with anti-His tag antibody and coimmunoprecipitated p53 was detected by autoradiography. p53, but not its carboxy-terminal truncated form coprecipitated with both mot-1 and mot-2 (lanes 2–3).
p53 Interacting Domain of Mortalin
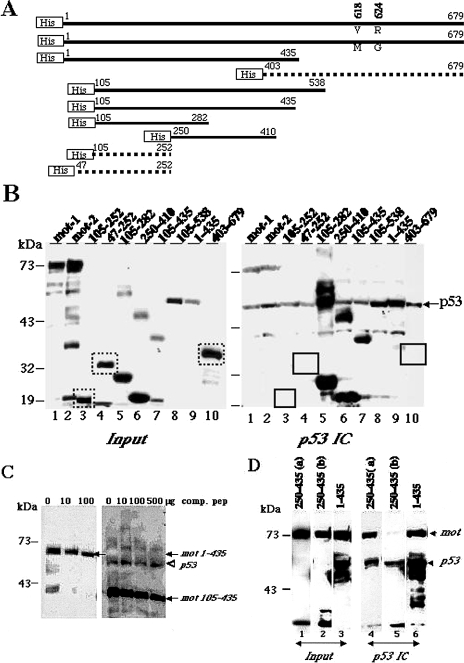
The above data led us to define p53-binding domain of mot-2. Various His-tagged deletion mutants of mot protein (Figure 2A) were expressed in bacteria and were purified as described in Materials and Methods. These were detected as expected size bands by Western blotting with anti-His tag antibody; some of the constructs also gave low- and high-molecular-weight bands (most likely the degraded and aggregated products, respectively). COS 7 cell lysate (400 µg) that provided a complex mixture of proteins with wild-type p53 for pull-down assays, was mixed with 0.5–1 µg of bacterially derived recombinant 6 X His-tagged full-length or variously deleted mortalins. p53 immunocomplexes from the mixture were analyzed for coprecipitated recombinant mortalins by Western blotting with anti-His tag antibody (Clontech, Palo Alto, CA). As seen in Figure 2B, mot 403–679 a. a. residues of mortalin did not show any coimmunoprecipitation with p53. Mortalin fragments 105–538 a. a., 105–435 a. a., 105–282 a. a. and 250–410 a. a. showed binding to p53 to various extents. Interestingly, two of the fragments spanning 47–252 a. a. and 105–252 a. a. did not show any binding. The extra bands of high molecular weight in lanes 5 and 6 (Figure 2B) most likely are the aggregated proteins that reacted to anti-His antibody and were coprecipitated with p53. Similarly, the unexpected low molecular weight bands in lanes 1, 2, 7, and 10 (Figure 2B) are degraded by-products and did not show appreciable coprecipitation with p53. These data implied that the mortalin amino acid residues 253–282 are critically required for its binding to p53 (Figure 2, A and B). These regions are identical in mot-1 and mot-2, in agreement with the data that both proteins bind to p53 in vitro (Figure 1).
Figure 2.
Mapping the p53 binding domain of the mot. (A) Schematic representation of His-tagged mortalin deletion mutants. The two amino acids (residues 618 and 624) that differ between mot-1 and mot-2 are indicated. The deletion mutants that did not bind to p53 are shown as dotted lines. (B) His-tagged mortalin deletion mutants were added to COS 7 cell lysates and were detected by Western blotting with anti-His tag antibody (input (10%), left panel). p53 immunocomplexes (p53 IC) were analyzed for coprecipitated mortalin by Western blotting with anti-His tag antibody (right panel). The bands corresponding to the deletion mutants that did not show binding are boxed in the input as well as p53 immunocomplexes panels Immunoprecipitated p53 was detected by probing the same membrane with anti-p53 (monoclonal) antibody, indicated by an arrowhead. Extra bands of high molecular weight in lanes 5 and 6 most likely are the aggregated proteins that also coprecipitated with p53. Low-molecular-weight bands in lanes 1, 2, 7, and 10 are the degraded products and did not coprecipitate with p53. (C) Addition of increasing amounts of a peptide (comp. pep) corresponding to a. a. residues 253–282 of mortalin reduced the amount of mot 1–435 a. a. and 105–435 a. a. p53 was immunoprecipitated by a polyclonal anti-p53 antibody (CM-1, Novocastra). Coimmunoprecipitated recombinant mortalin was detected by Western blotting with anti-His tag antibody (indicated by arrows). Immunoprecipitated p53 along with mot 105–435 is shown by an arrow. (D) A high level of expression of mot 250–435 a. a. prevents binding of p53 to endogenous mortalin. COS 7 cells were transfected with expression constructs encoding V5-tagged mortalin deletion mutant proteins spanning residues 250–435 (a — 5 µg DNA and b — 15 µg DNA per 10-cm dish) and 1–435 (15 µgDNA), and the lysates were immunoprecipitated with an anti-p53 antibody (CM-1, Novocastra). p53 immunocomplexes (lanes 4–6) and input protein (lanes 1–3, 10% of the amount of lysate used for immunoprecipitation) were Western blotted with anti-mortalin and anti-V5 tag antibodies to detect endogenous mot (indicated by arrow) and V5-tagged mot fragments, respectively. The p53 immunocomplexes (IC) were also Western blotted with a monoclonal anti-p53 antibody (Pab 421) (indicated by arrowhead) to detect immunoprecipitated p53. Immunoprecipitated p53 and mot 1–435 protein are overlapped in lane 6. Note that only negligible amount of endogenous mot was coprecipitated with p53 from cells that were transfected with a high amount of DNA (lanes 2 and 5).
A peptide corresponding to mortalin amino acid residues 253–282 was next used to compete out the binding of recombinant His-tagged mortalin to p53. Increasing amounts of the competitor peptide were incubated with COS 7 lysates before the addition of His-tagged mortalin proteins, mot 1–435, 105–435 and 105–538 (not shown) that contained p53-binding region and did not show either the degradation or aggregation (based on data in Figure 2B). It resulted in decreased amounts of coimmunoprecipitated His-tagged mots (Figure 2C). The detection of immunoprecipitated p53 along with mot 1–435 a. a. and mot 105–538 a. a. (not shown) was obscured because of the very similar sizes of p53 and these mortalin deletion mutants. Western detection of p53 on the same blot with 105–435 a. a. ruled out the possibility that the decreasing amount of immunoprecipitated mot protein was due to a decreased amount of immunoprecipitated p53 (Figure 2C). Although the peptide corresponding to 253–282 a. a. residues was able to compete out binding of larger mortalin fragments, the large amount of peptide required suggested that its competition effect is weak. Such inefficiency of the peptide may be due to its binding-incompetent secondary or tertiary structures. Therefore, we next expressed V5-tagged mortalin deletion mutants expressing amino acid residues 250–435, 252–679 (not shown) and 1–435 in COS 7 cells. Their presence in p53 immunocomplexes was analyzed by Western blotting with anti-V5 tag antibody (Figure 2D). Each of the mortalin deletion mutants was detected in the p53 immunocomplexes (as a single band in the case of a. a. 250–435, and as multiple bands (degraded products) in the case of a. a. 1–435 indicating their binding to p53. Notably, only a negligible amounts of endogenous mortalin was coprecipitated with p53 from lysates expressing mortalin residues 250–435 (Figure 2D, lane 5) demonstrating that this region can compete efficiently with full-length mot for its binding to p53. Furthermore, competition effect was seen only in lysates that expressed higher amount of the 250–435 protein (compare lanes 1 and 2 for expression of mot 250–435 and lanes 4 and 5 for mot in p53 immunocomplexes, respectively). The other two constructs (252–679 and 1–435 a. a.) that also contained p53-binding residues as defined by in vitro pull-down assays did not compete out the binding of full-length mortalin for the reasons that remain undefined in this study. Nevertheless, these data defined mortalin residues 253–282 as essential for its binding to p53 and residues 250–435 as an efficient competitor of the binding of full-length mortalin to p53. Because the residues 253–282 are identical in mot-1 and mot-2 proteins it implies that there are yet unknown in vivo factor(s) that prevent mouse mot-1 from binding to p53.
Most interestingly, the p53-binding region of mot (residues 253–282) defined in this study overlapped with the region that is defined as MKT-077-binding region (residues 252–310) [29]. We have shown that MKT-077 binds to these residues of mot. Treatment of transformed cells with increasing concentrations of MKT-077 resulted in abrogation of mot-p53 complexes and concurrent nuclear translocation and transcriptional activation of p53 [29]. The present study has shown that p53 and MKT-077 bind to the same region of mot, and therefore MKT-077 could compete out mot-p53 interactions resulting in abrogation of the cytoplasmic retention of p53.
Acknowledgements
We thank Antony Braithwaite for the kind gift of pSP65/p53 plasmid. The help of M. Robert in the initial pull-down assays is acknowledged.
References
- 1.Wadhwa R, Takano S, Robert M, Yoshida A, Nomura H, Reddel RR, Mitsui Y, Kaul SC. Inactivation of tumor suppressor p53 by mot-2, a hsp70 family member. J Biol Chem. 1998;273:29586–29591. doi: 10.1074/jbc.273.45.29586. [DOI] [PubMed] [Google Scholar]
- 2.Kaul SC, Duncan EL, Englezou A, Reddel RR, Mitsui Y, Wadhwa R. Malignant transformation of NIH 3T3 cells by overexpression of mot-2 protein. Oncogene. 1998;17:907–911. doi: 10.1038/sj.onc.1202017. [DOI] [PubMed] [Google Scholar]
- 3.Xu J, Xiao HH, Sartorelli AC. Attenuation of the induced differentiation of HL-60 leukemia cells by mitochondrial chaperone HSP70. Oncol Res. 1999;11:429–435. [PubMed] [Google Scholar]
- 4.Kaul SC, Reddel RR, Sugihara T, Mitsui Y, Wadhwa R. Inactivation of p53 and life span extension of human diploid fibroblasts by mot-2. FEBS Lett. 2000;474:159–164. doi: 10.1016/s0014-5793(00)01594-5. [DOI] [PubMed] [Google Scholar]
- 5.Wadhwa R, Kaul SC, Ikawa Y, Sugimoto Y. Identification of a novel member of mouse hsp 70 family: its association with cellular mortal phenotype. J Biol Chem. 1993;268:6615–6621. [PubMed] [Google Scholar]
- 6.Wadhwa R, Kaul SC, Sugimoto Y, Mitsui Y. Induction of cellular senescence by transfection of cytosolic mortalin cDNA in NIH 3T3 cells. J Biol Chem. 1993;268:22239–22242. [PubMed] [Google Scholar]
- 7.Wadhwa R, Pereira-Smith OM, Reddel R, Sugimoto Y, Mitsui Y, Kaul SC. Correlation between complementation group for immortality and the cellular distribution of mortalin. Exp Cell Res. 1995;216:101–106. doi: 10.1006/excr.1995.1013. [DOI] [PubMed] [Google Scholar]
- 8.Kaul SC, Duncan E, Sugihara T, Reddel RR, Mitsui Y, Wadhwa R. Structurally and functionally distinct mouse hsp70 family members mot-1 and mot-2 proteins are encoded by two alleles. DNA Res. 2000;7:229–231. doi: 10.1093/dnares/7.3.229. [DOI] [PubMed] [Google Scholar]
- 9.Kaul SC, Takano S, Reddel RR, Mitsui Y, Wadhwa R. Transcriptional inactivation of p53 by deletions and single amino acid changes in mouse mot-1 protein. Biochem Biophys Res Commun. 2000 doi: 10.1006/bbrc.2000.3986. (in press) [DOI] [PubMed] [Google Scholar]
- 10.Kaul SC, Mitsui Y, Wadhwa R. Molecular insights to cellular mortality and immortalization. Ind J Exp Biol. 1998;36:345–352. [PubMed] [Google Scholar]
- 11.Dahlseid JN, Lill R, Green JM, Xu X, Qiu Y, Pierce SK. PBP74, a new member of the mammalian 70-kDa heat shock protein family, is a mitochondrial protein. Mol Biol Cell. 1994;5:1265–1275. doi: 10.1091/mbc.5.11.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ran Q, Wadhwa R, Kawai R, Kaul SC, Sifers RN, Bick RJ, Smith JR, Pereira-Smith OM. Extra-mitochondrial localization of mortalin/mthsp70/PBP74/GRP75. Biochem Biophys Res Communn. 2000;275:174–179. doi: 10.1006/bbrc.2000.3237. [DOI] [PubMed] [Google Scholar]
- 13.Soltys BJ, Gupta RS. Mitochondrial proteins at unexpected cellular locations: export of proteins from mitochondria from an evolutionary perspective. Int Rev Cytol. 2000;194:133–196. doi: 10.1016/s0074-7696(08)62396-7. [DOI] [PubMed] [Google Scholar]
- 14.Mattson JP, Ross CR, Kilgore JL, Musch T. Induction of mitochondrial stress proteins following treadmill running. Med Sci Sports Exercise. 2000;32:365–369. doi: 10.1097/00005768-200002000-00016. [DOI] [PubMed] [Google Scholar]
- 15.Ornatsky OI, Connor MK, Hood DA. Expression of stress proteins and mitochondrial chaperonins in chronically stimulated skeletal muscle. Biochem J. 1995;311:119–123. doi: 10.1042/bj3110119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sadekova S, Lehnert S, Chow TY. Induction of PBP74/mortalin/Grp75, a member of the hsp70 family, by low doses of ionizing radiation: a possible role in induced radioresistance. Int J Radiat Biol. 1997;72:653–660. doi: 10.1080/095530097142807. [DOI] [PubMed] [Google Scholar]
- 17.Merrick BA, Walker VR, He C, Patterson RM, Selkirk JK. Induction of novel Grp75 isoforms by 2-deoxyglucose in human and murine fibroblasts. Cancer Lett. 1997;119:185–190. doi: 10.1016/s0304-3835(97)00270-x. [DOI] [PubMed] [Google Scholar]
- 18.Massa SM, Longo FM, Zuo J, Wang S, Chen J, Sharp FR. Cloning of rat grp75, an hsp70-family member, and its expression in normal and ischemic brain. J Neurosci Res. 1995;40:807–819. doi: 10.1002/jnr.490400612. [DOI] [PubMed] [Google Scholar]
- 19.Wu R, Zhao YH, Plopper CG, Chang MM, Chmiel K, Cross JJ, Weir A, Last JA, Tarkington B. Differential expression of stress proteins in nonhuman primate lung and conducting airway after ozone exposure. Am J Physiol. 1999;277:L511–L522. doi: 10.1152/ajplung.1999.277.3.L511. [DOI] [PubMed] [Google Scholar]
- 20.Takano S, Wadhwa R, Yoshii Y, Nose T, Kaul SC, Mitsui Y. Elevated levels of mortalin expression in human brain tumors. Exp Cell Res. 1997;237:38–45. doi: 10.1006/excr.1997.3754. [DOI] [PubMed] [Google Scholar]
- 21.Bini L, Magi B, Marzocchi B, Arcuri F, Tripodi S, Cintorio M, Sanchez JC, Frutiger S, Hughes G, Pallini V, Hochstrasser DF, Tosi P. Protein expression profiles in human breast ductal carcinoma and histologically normal tissue. Electrophoresis. 1997;18:2832–2841. doi: 10.1002/elps.1150181519. [DOI] [PubMed] [Google Scholar]
- 22.Craig EA, Kramer J, Shilling J, Werner-Washburne M, Holmes S, Kosic-Smithers J, Nicolet CM. SSC1, an essential member of the yeast HSP70 multigene family, encodes a mitochondrial protein. Mol Cell Biol. 1989;9:3000–3008. doi: 10.1128/mcb.9.7.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voos W, Martin H, Krimmer T, Pfanner N. Mechanisms of protein translocation into mitochondria. Biochim Biophys Acta. 1999;16:235–254. doi: 10.1016/s0304-4157(99)00007-6. [DOI] [PubMed] [Google Scholar]
- 24.Krimmer T, Rassow J, Kunau WH, Voos W, Pfanner N. Mitochondrial protein import motor: the ATPase domain of matrix hsp70 is crucial for binding to Tim44, while the peptide binding domain and the carboxy-terminal segment play a stimulatory role. Mol Cell Biol. 2000;20:5879–5887. doi: 10.1128/mcb.20.16.5879-5887.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulke N, Sepuri NB, Gordon DM, Saxena S, Dancis A, Pain D. A multisubunit complex of outer and inner mitochondrial membrane protein translocases stabilized in vivo by translocation intermediates. J Biol Chem. 1999;274:22847–22854. doi: 10.1074/jbc.274.32.22847. [DOI] [PubMed] [Google Scholar]
- 26.Merlin A, Voos W, Maarse AC, Meijer M, Pfanner N, Rassow J. The J-related segment of tim44 is essential for cell viability: a mutant Tim44 remains in the mitochondrial import site, but inefficiently recruits mtHsp70 and impairs protein translocation. J Cell Biol. 1999;145:961–972. doi: 10.1083/jcb.145.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savel'ev AS, Novikova LA, Kovaleva IE, Luzikov VN, Neupert W, Langer T. ATP-dependent proteolysis in mitochondria, m-AAA protease and PIM1 protease exert overlapping substrate specificities and cooperate with the mtHsp70 system. J Biol Chem. 1998;273:20596–20602. doi: 10.1074/jbc.273.32.20596. [DOI] [PubMed] [Google Scholar]
- 28.Cechetto JD, Soltys BJ, Gupta RS. Localization of mitochondrial 60-kD heat shock chaperonin protein (Hsp60) in pituitary growth hormone secretory granules and pancreatic zymogen granules. J Histochem Cytochem. 2000;48:45–56. doi: 10.1177/002215540004800105. [DOI] [PubMed] [Google Scholar]
- 29.Wadhwa R, Sugihara T, Yoshida A, Nomura H, Reddel RR, Simpson R, Maruta H, Kaul SC. Selective toxicity of MKT-077 to cancer cells is mediated by its binding to hsp70 family protein, mot-2, and reactivation of p53 function. Cancer Res. 2000;60 [PubMed] [Google Scholar]
- 30.Braithwaite AW, Jenkins JR. Ability of p53 and the adenovirus E1b. 58-kilodalton protein to form a complex is determined by p53. J Virol. 1989;63:1792–1799. doi: 10.1128/jvi.63.4.1792-1799.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wadhwa R, Sugihara T, Yoshida A, Duncan EL, Hardeman EC, Nomura H, Reddel RR, Kaul SC. Cloning and characterization of a novel gene, Striamin, that interacts with the tumor suppressor protein p53. J Biol Chem. 1999;274:14948–14955. doi: 10.1074/jbc.274.21.14948. [DOI] [PubMed] [Google Scholar]