Abstract
The 26S proteasome is an ATP-dependent eukaryotic protease responsible for degrading many important cell regulators, especially those conjugated with multiple ubiquitins. Bound on both ends of the 20S core protease is a multisubunit regulatory particle that plays a crucial role in substrate selection by an as yet unknown mechanism(s). Here, we show that the RPN12 subunit of the Arabidopsis regulatory particle is involved in cytokinin responses. A T-DNA insertion mutant that affects RPN12a has a decreased rate of leaf formation, reduced root elongation, delayed skotomorphogenesis, and altered growth responses to exogenous cytokinins, suggesting that the mutant has decreased sensitivity to the hormone. The cytokinin-inducible genes CYCD3 and NIA1 are upregulated constitutively in rpn12a-1, indicating that feedback-inhibitory mechanisms also may be altered. rpn12a-1 seedlings also showed changes in auxin-induced growth responses, further illustrating the close interaction between auxin and cytokinin regulation. In yeast, RPN12 is necessary for the G1/S and G2/M transitions of the cell cycle, phases that have been shown to be under cytokinin control in plants. We propose that RPN12a is part of the Arabidopsis 26S proteasome that controls the stability of one or more of the factors involved in cytokinin regulation.
INTRODUCTION
Regulated protein turnover provides a mechanism to adjust rapidly to changing ligand concentrations and/or environmental conditions and is essential for many signal response pathways. In eukaryotes, the ubiquitin/26S proteasome pathway is particularly important, being responsible for removing most short-lived intracellular proteins (Hershko and Ciechanover, 1998; Callis and Vierstra, 2000). In this proteolytic pathway, proteins committed for degradation first are modified by the covalent attachment of multiple ubiquitins. This conjugation is directed by an ATP-dependent reaction cascade involving the sequential action of E1s, E2s, and E3s, which ultimately attach one or more ubiquitins to appropriate targets. In most cases, the ubiquitinated proteins then are recognized and degraded by the 26S proteasome, a multisubunit ATP-dependent protease with broad substrate specificity.
The 26S proteasome is a 2-MD complex assembled from two particles: the 20S core particle (CP) and the 19S regulatory particle (RP) (Voges et al., 1999). The proteolytic activities reside within the central chamber of the 28-subunit CP, whereas the functions that direct substrate recognition, unfolding, and subsequent entry into the 20S particle reside within the >18-subunit RP. The RP can be divided further in two subcomplexes, the lid and base (Glickman et al., 1998). The base contains six ATPase subunits, RPT1 to RPT6, which presumably use ATP hydrolysis to unfold target proteins, and three non-ATPase subunits, RPN1, RPN2, and RPN10. The lid contains nine additional RPN subunits (RPN3, RPN5 to RPN9, and RPN11 to RPN13). Many of the lid RPN subunits share sequence motifs with components of the COP9/signalosome and EIF3 complexes, implying a common ancestry (Glickman et al., 1998; Fu et al., 2001). To date, the roles of only two RPN subunits are known. RPN13 (also known as UCH37) can disassemble multiubiquitin chains, suggesting that it releases the ubiquitin moieties before target breakdown (Voges et al., 1999). RPN10 appears to help tether the lid to the base and may participate in recognizing multiubiquitinated proteins before digestion (Fu et al., 1998a; Glickman et al., 1998; Fu et al., 2001).
A growing body of evidence indicates that the ubiquitin/26S proteasome pathway controls the levels of many important fungal and animal regulatory processes. Targets include key checkpoint proteins within the cell cycle and components of numerous hormone signaling systems (Hershko and Ciechanover, 1998). Recent studies also have implicated the pathway in environmental and developmental responses in plants (Callis and Vierstra, 2000). Deletion of RPN10 in Physcomitrella patens blocks the transition from the vegetative to the reproductive phase of the protonema (Girod et al., 1999). A number of Arabidopsis mutations in ubiquitin ligation have been described that affect floral development, male gametogenesis, photomorphogenesis, and circadian rhythms (Callis and Vierstra, 2000; Dieterle et al., 2001). Two mutants of particular interest for hormone regulation are transport inhibitor response-1 (tir1) and coronatine insensitive-1 (coi1). They identify E3 genes that are required specifically for auxin and jasmonate sensitivity, respectively (Xie et al., 1998; Gray and Estelle, 2000). Furthermore, the detection of ubiquitin conjugates and the use of 26S proteasome inhibitors and dominant-negative ubiquitin mutants have implicated the pathway in leaf development, defense responses, abscisic acid signaling, and control of phytochrome A and cyclin A and B levels (Bachmair et al., 1990; Genschik et al., 1998; Clough et al., 1999; Becker et al., 2000; Lopez-Molina et al., 2001).
Another important signaling pathway in plants controls responses to the cytokinin family of hormones. Cytokinins regulate numerous developmental processes, including seed germination, photomorphogenesis, root/shoot differentiation, apical dominance, sink-to-source relations, senescence, and cell division (Mok and Mok, 1994). Although the phenotypic effects of exogenous cytokinins have been studied thoroughly, the underpinning sensory transduction chain(s) is not well understood. In Arabidopsis, cytokinins are detected by CRE1 and possibly other receptors related to bacterial two-component histidine kinases (Inoue et al., 2001). Several response regulators have been identified as cytokinin-inducible genes (Brandstatter and Kieber, 1998; Taniguchi et al., 1998) that could serve as the phospho-acceptors for the activated histidine kinases. Further downstream, cytokinin stimulates the G1/S transition of the cell cycle, which is possibly mediated in part by the transcriptional induction of the CYCD3 gene encoding a D-type cyclin (Riou-Khamlichi et al., 1999). Because cytokinins also are required for the G2/M transition, it is expected that they activate or repress additional cell cycle regulators (Mironov et al., 1999). The hormone auxin also is known to affect plant cell division. In fact, auxins and cytokinins often act synergistically, and several of the auxin response mutants also have altered responses to cytokinins (Coenen and Lomax, 1997).
In an attempt to define how the ubiquitin/26S proteasome pathway participates in plant growth and development, we have begun to systematically isolate and characterize Arabidopsis mutants affecting various components. In particular, we have focused on the 26S proteasome, given its central role in substrate breakdown. Initial studies identified all of the CP subunits and most of the RP subunits and showed that the Arabidopsis complex is remarkably similar in structure and function to those described in fungi and animals (Fu et al., 1999b). Here, we describe the RPN12a subunit within the lid of the Arabidopsis RP. RPN12 is potentially encoded by two genes, one that encompasses the entire polypeptide (RPN12a) and another that encodes an N-terminal truncated version (RPN12b). Studies on yeast rpn12 mutants indicate that this essential subunit plays a special role in the cell cycle by helping to selectively degrade the Clb-specific Cdc28 kinase inhibitor Sic1, whose loss is required for the G1/S phase transition (Bailly and Reed, 1999). From analysis of the T-DNA mutant rpn12a-1, we show that Arabidopsis RPN12 is important for numerous cytokinin-regulated growth responses. The phenotypes suggest that RPN12a in particular, and the 26S proteasome in general, is required to degrade one or more repressors of cytokinin action in plants.
RESULTS
Arabidopsis 26S Proteasome Subunit RPN12
Using the subunit sequences of the Saccharomyces cerevisiae 26S proteasome as queries, we have identified the genes encoding all of the corresponding subunits of the Arabidopsis complex in the nearly complete genomic sequence (Fu et al., 1998b, 1999a, 1999b; H. Fu, J. Smalle, and R.D. Vierstra, unpublished data). For clarity, we have adopted the nomenclature recently proposed for subunits of the yeast 26S proteasome complex (Finley et al., 1998). Similar to what was found for subunits of the CP, most of the Arabidopsis RP subunits are encoded by two genes (the exceptions being RPT3, RPN6, RPN7, and RPN10).
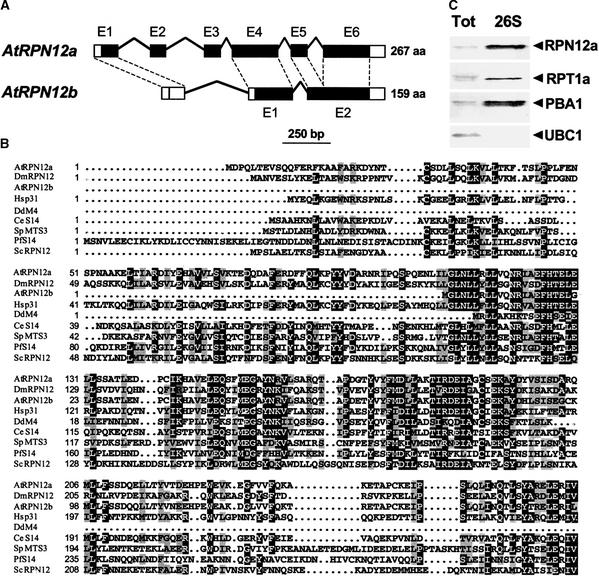
An unusual pair of Arabidopsis genes is related to yeast RPN12. In addition to one locus encoding a full-length version (RPN12a), a second gene appears to synthesize a truncated form (RPN12b) missing a substantial portion from the N-terminal end (Figure 1A). The predicted RPN12a gene contains five introns and encodes a 267–amino acid protein. Comparison of the amino acid sequence of RPN12a with orthologs from various fungi and animals revealed similarity over their entire lengths (Figure 1B). For example, the overall amino acid similarity of Arabidopsis RPN12a to its yeast and human orthologs is 35 and 47%, respectively. Close relatives of RPN12a also were found in the expressed sequence tag (EST) databases of other plant species, including tomato, cotton, sugar beet, maize, wheat, barley, rice, potato, soybean, Lotus japonicum, and Medicago truncatula (data not shown). The presence of numerous cDNAs in various Arabidopsis EST collections indicated that RPN12a is transcribed actively in many tissue types. This was confirmed by RNA gel blot analysis, which detected the corresponding RPN12a mRNA of 1.1 kb in all Arabidopsis organs tested (Figure 2B and data not shown).
Figure 1.
Organization and Derived Amino Acid Sequence Comparisons of the Arabidopsis RPN12a and RPN12b Genes and Association of the RPN12a Protein with the 26S Proteasome.
(A) Genomic organization of Arabidopsis RPN12a and predicted RPN12b. Boxes denote exons (designated E1 to E6), with open and black boxes indicating noncoding and coding regions, respectively. Lines show positions and lengths of introns. Dashed lines show exon relationships for RPN12a and RPN12b. aa, amino acids.
(B) Derived amino acid sequence comparison of RPN12a and RPN12b with possible orthologs from animals and fungi. Identical and similar residues are shown in reverse type and gray boxes, respectively. Sequences include those for Drosophila melanogaster DmRPN12, Homo sapiens Hsp31, Dictyostelium discoideum DdM4, Caenorhabditis elegans CeS14, Schizosaccharomyces pombe SpMTS3, Plasmodium falciparum PfS14, and S. cerevisiae ScRPN12.
(C) Copurification of RPN12a with other subunits of the Arabidopsis 26S proteasome. The Arabidopsis 26S proteasome was enriched by sequential polyethylene glycol precipitations followed by size exclusion fast protein liquid chromatography (FPLC). Both the total crude extract (Tot) and the FPLC fraction with highest CP peptidase activity (26S) were subjected to SDS-PAGE and immunoblot analysis with antibodies prepared against the Arabidopsis 26S proteasome subunits RPT1a, RPN12a, PBA1, and a control protein, the Arabidopsis E2 UBC1.
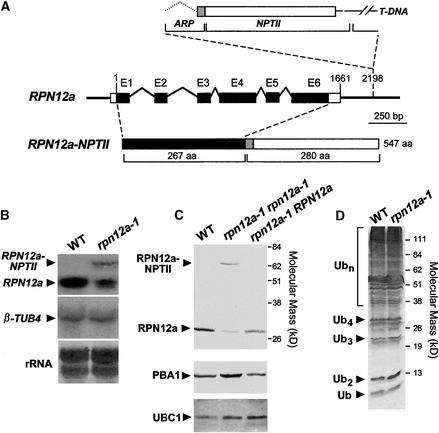
Figure 2.
Molecular and Biochemical Descriptions of the Arabidopsis rpn12a-1 Mutation.
(A) Scheme of the RPN12a gene and the predicted protein product of rpn12a-1, a RPN12a-NTPII translational fusion protein. Boxes and lines denote exons and predicted introns, respectively. Upon insertion of the T-DNA, the 3′ untranslated sequence of the RPN12a gene (bp 1661 to 2198) and the 5′ end of the ARP-NPTII gene from the T-DNA insertion form a chimeric intron that when spliced creates the translational fusion gene (RPN12a-NPTII) containing the full coding regions of RPN12a and the ARP-NPTII genes (Babiychuk et al., 1997). aa, amino acid.
(B) Detection of RPN12a transcripts in wild-type (WT) and rpn12a-1 seedlings by RNA gel blot analysis using a RPN12a gene probe. Arrowheads indicate the mRNA for wild-type RPN12a and the chimeric RPN12a-NPTII transcript. Equal amounts of total RNA were analyzed; loading was confirmed by staining for rRNA with methylene blue and by reprobing the blots with DNA encoding β-tubulin (β-TUB4).
(C) Immunoblot detection of the RPN12a protein. Total protein was extracted from wild-type seedlings and seedlings either hemizygous or homozygous for the rpn12a-1 mutation and subjected to SDS-PAGE and immunoblot analysis with anti-RPN12a antibodies. Arrowheads indicate RPN12a and the RPN12a-NPTII fusion protein. Control immunoblot analysis was performed with antibodies against the CP β-subunit PBA1 and UBC1.
(D) Levels of ubiquitin (Ub) and ubiquitin–protein conjugates in wild-type and rpn12a-1 seedlings. Equal amounts of total protein were subjected to SDS-PAGE and immunoblot analysis with anti-ubiquitin antibodies. The positions of the free ubiquitin, free multiubiquitin chains containing two to four monomers, and multiubiquitinated proteins are indicated.
To verify that the RPN12a protein is part of the Arabidopsis 26S proteasome, the complex was partially purified from young green Arabidopsis seedlings by sequential polyethylene glycol precipitations followed by size exclusion fast protein liquid chromatography (FPLC) and assayed with anti-RPN12a antibodies. In accordance with previous mass determinations of the complex (Peng et al., 2001), the Arabidopsis 26S proteasome behaved as a 2-MD particle during size exclusion chromatography. The RPN12a protein of the expected size (31 kD) could be detected in fractions also enriched for CP peptidase activity (using the substrate Suc-LLVY-MCA) and other subunits of the 26S proteasome, including the RP subunit RPT1a and the CP subunit PBA1 (Figure 1C and data not shown).
Compared with RPN12a, the RPN12b gene is missing the second and third exons along with two introns (Figure 1A). Conceptual translation of this gene suggested that the resulting RPN12b protein would start at a methionine 102 residues downstream from the start of RPN12a, thus generating a truncated 18-kD protein containing just 159 amino acids (Figure 1B). When this shorter sequence was aligned with RPN12a, the two proteins displayed 87% amino acid identity and 98% amino acid similarity. There is no direct evidence that RPN12b is expressed, because we have been unable to find RPN12b cDNAs in the Arabidopsis EST collections or to detect the RPN12b transcript and protein in green Arabidopsis seedlings (data not shown).
T-DNA Insertion Mutation Affecting RPN12a Expression
To help identify the function(s) of RPN12, we characterized a T-DNA insertion mutant affecting RPN12a that was created by exon-trap mutagenesis (Babiychuk et al., 1997). The exon-trap vector pSL4 used here contained a fusion of the first intron of the apurinic endonuclease gene (ARP) lacking its 5′ donor splice site followed by a coding region containing exon 2 of ARP appended in frame to that for neomycin phosphotransferase II (NPTII) lacking its translation initiation codon (Figure 2A). The ARP-NPTII fusion gene was placed immediately after the right border of the T-DNA. Previous studies showed that upon transformation of Arabidopsis with the pSL4 T-DNA, kanamycin resistance created by the expression of NPTII was possible only when the T-DNA integrated into a host gene that provided the promoter activity, the translation start site, and a 5′ donor intron splice site. This splice site enabled removal of the ARP intron to allow translation of the ARP-NPTII protein using the upstream, host-derived ATG codon. From 5′ rapid amplification of cDNA ends polymerase chain reaction (PCR) analysis of ∼400 independent kanamycin-resistant transgenic lines of Arabidopsis ecotype C24, ∼90% had the T-DNA integrated within known or predicted gene-coding loci (Babiychuk et al., 1997; data not shown).
Sequence analysis of the 5′ rapid amplification of cDNA ends product from the kanamycin-resistant line SL42-38 (now designated rpn12a-1) revealed the presence of a chimeric mRNA containing the ARP-NPTII coding region fused to the full coding region for RPN12a. RNA gel blots showed a single NPTII mRNA, indicating that rpn12a-1 contains only one transcriptionally active insertion. PCR analysis of genomic DNA using RPN12a- and NPTII-specific primers showed that the T-DNA was inserted 531-bp downstream of the RPN12a stop codon. We assume that this insertion created a chimeric intron in which the end of the RPN12a coding region provided the 5′ donor splice site (AG/GT) for the ARP intron (Figure 2A). RNA gel blot analysis of rpn12a-1 plants using RPN12a or NPTII probes detected the predicted fusion mRNA of 1.8 kb (Figure 2B and data not shown). The RPN12a probe also hybridized to a smaller, more abundant transcript indistinguishable in size from that of the wild-type RPN12a mRNA (Figure 2B). We assume that the two mRNAs were created by differential transcript processing and/or termination using either the polyadenylation signal within the 3′ untranslated region of the RPN12a gene or the signal downstream of the NPTII coding region.
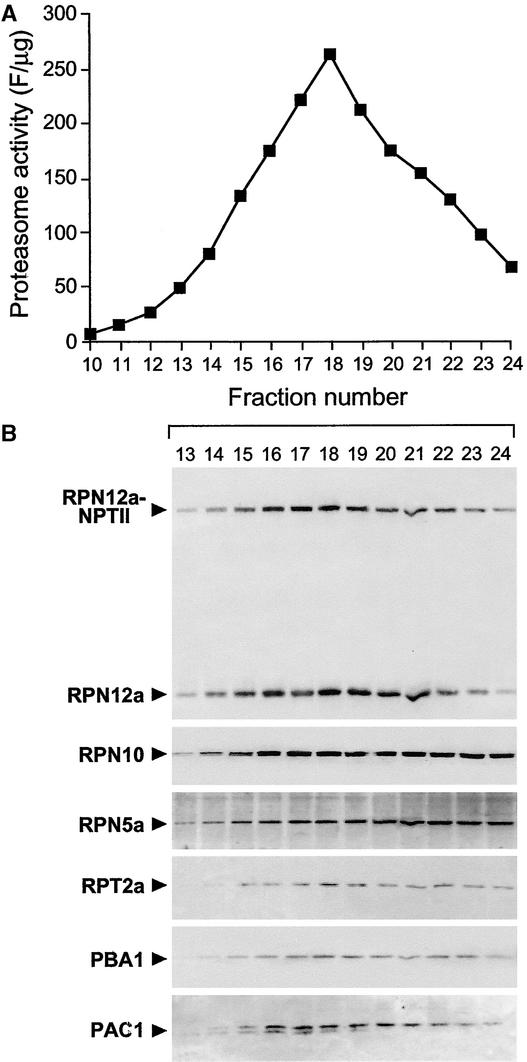
Immunoblot analysis with anti-RPN12a antibodies detected both the RPN12a-NPTII fusion and wild-type RPN12a proteins in homozygous rpn12a-1 plants (Figure 2C), consistent with the presence of two mRNAs. Despite an apparently lower level of the RPN12a-NPTII mRNA, the levels of the corresponding proteins were nearly equal. This similar abundance was observed in all tissues and developmental stages examined (data not shown), suggesting that the mutation affected most, if not all, cell types. Interestingly, the RPN12a-NPTII fusion protein assembled into the 26S proteasome. As can be seen in Figure 3, the fusion protein coeluted during size exclusion FPLC in a 2-MD complex with CP peptidase activity and with wild-type RPN12a and other subunits of the 26S complex, including RPN10, RPN5a, and RPT1a of the RP and PBA1 and PAC1 of the CP. (The slight tailing of RPN10 and RPN5a suggested that they partially dissociate from the lid during FPLC, an effect observed previously for yeast RPN10 [van Nocker et al., 1996b].) The fact that RPN12a-NPTII coeluted with both the lid (RPN12a and RPN5a) and base (RPT1 and RPN10) subunits and with the CP suggested that the rpn12a-1 mutation did not appreciably alter the structure and subunit composition of the allied 26S complex.
Figure 3.
Association of the RPN12a-1–NPTII Fusion Protein with the 26S Proteasome.
The 26S proteasome was partially purified from Arabidopsis seedlings homozygous for the rpn12a-1 mutation by sequential polyethylene glycol precipitations and subjected to size exclusion FPLC.
(A) Column fractions were assayed for CP peptidase activity. Only the portion of the FPLC elution profile around the peak of peptidase activity is shown.
(B) Column fractions subjected to SDS-PAGE and immunoblot analysis with antibodies against RPN12a, RPN10, RPN5a, RPT2a subunits of the RP, and PBA1 and PAC1 subunits of the CP. The positions of the wild-type RPN12a and the RPN12a-NPTII fusion protein are indicated by arrowheads.
Whereas both forms of RPN12a could be detected in homozygous rpn12a-1 plants, only the wild-type version could be seen in hemizygous plants (Figure 2C). Overexpression of wild-type RPN12a in plants homozygous for rpn12a-1 also resulted in the disappearance of the RPN12a-NPTII fusion protein (see below). These observations suggested that wild-type RPN12a can competitively block the association of the RPN12a-NPTII fusion protein with the 26S complex; the free fusion protein then is subjected to rapid degradation. However, even hemizygous rpn12a-1 plants are kanamycin resistant, indicating that these plants retained sufficient amounts of NPTII activity even though the RPN12a- NPTII fusion protein was undetectable.
The incorporation of both forms of RPN12a into the 26S proteasome of homozygous rpn12a-1 plants implied that two populations of 26S complexes existed. Thus, instead of causing a strong 26S proteasome defect, it is likely that the rpn12a-1 mutation causes a more subtle change in proteasome activity. This possibility was supported by immunological measurements with anti-ubiquitin antibodies. Whereas other ubiquitin/26S proteasome pathway mutants often increase the steady state levels of ubiquitinated proteins (Kominami et al., 1995; Bailly and Reed, 1999; Girod et al., 1999; Doelling et al., 2001), these levels remained indistinguishable from wild-type levels in homozygous rpn12a-1 plants (Figure 2D). This lack of effect suggested that the mutation does not perturb the function of the Arabidopsis 26S proteasome globally and thus may affect RPN12a-related functions more specifically.
Development of rpn12a-1 Seedlings
Phenotypic analysis of homozygous rpn12a-1 seedlings indicated that the T-DNA insertion altered Arabidopsis seedling development substantially. In all cases, the mutation behaved as a recessive trait, consistent with the presence of the RPN12a-NPTII fusion protein only in homozygous rpn12a-1 plants. Young rpn12a-1 seedlings grown under continuous white light accumulated 2.5 times more anthocyanins in both hypocotyls and petioles (Figure 4A) (A530 − A650 for 10 seedlings was 0.032 ± 0.001 for the wild type versus 0.083 ± 0.003 for rpn12a-1). An increased percentage of rpn12a-1 seedlings developed with a single cotyledon compared with wild type (2.4% versus <0.05%) (Figure 4B). This higher percentage was observed in all rpn12a-1 lines created after backcrossing to wild type, indicating a linkage to the mutation. The rpn12a-1 plants showed a slower rate of rosette leaf emergence (Figure 4C) and a delay in flowering time (33.3 ± 3.1 days for rpn12a-1 versus 25.2 ± 2.1 days for wild type). During the first week of growth, rpn12a-1 roots grew more rapidly than wild-type roots. Thereafter, they elongated more slowly, such that after 14 days rpn12a-1 roots were significantly shorter than wild-type roots (Figure 4D). Dark-grown rpn12a-1 seedlings showed a delay in skotomorphogenesis, with a reduced rate of cotyledon opening and petiole elongation (Figure 4E).
Figure 4.
Developmental Changes Induced by the rpn12a-1 Mutation.
(A) Increased accumulation of anthocyanins in hypocotyls of 1-week- old rpn12a-1 seedlings. WT, wild type.
(B) Example of a monocotyledonous seedling in an rpn12a-1 population. Left, normal seedling; right, seedling with one cotyledon.
(C) Delayed leaf emergence of rpn12a-1 plants after 14 days of growth. Both wild-type and rpn12a-1 seedlings germinated at the same time.
(D) Reduced root elongation of an rpn12a-1 seedling after 14 days of growth. Red arrowheads indicate the root tips.
(E) Reduced cotyledon opening and petiole growth of an etiolated rpn12a-1 seedling after 10 days of growth in darkness.
rpn12a-1 Plants Have Altered Growth Responses to Exogenous Cytokinins
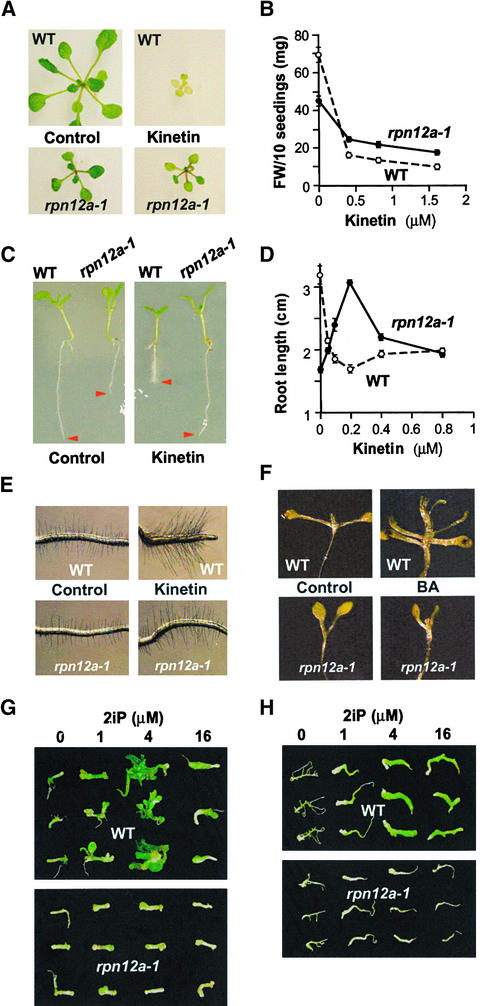
Some of the phenotypes associated with the rpn12a-1 mutation were suggestive of an attenuated cytokinin response (e.g., delayed shoot development and skotomorphogen-esis, increased percentage of monocotyledonous seedlings [Chaudhury et al., 1993; Chory et al., 1994; Mok and Mok, 1994]). Therefore, we tested the effect of exogenous cytokinins on rpn12a-1 seedlings. As can be seen in Figure 5A, wild-type seedlings were strongly inhibited in their growth and eventually became chlorotic when germinated on medium containing 0.1 μM kinetin. In contrast, rpn12a-1 seedlings were much less affected. This difference in growth inhibition was not restricted to germinating seedlings. Shoot growth of rpn12a-1 seedlings germinated first in the absence of cytokinins was less sensitive to kinetin when the seedlings were transferred subsequently to medium containing the hormone (Figure 5B).
Figure 5.
The rpn12a-1 Mutant Has Decreased Sensitivity to the Cytokinins Kinetin, BA, and 2iP.
(A) Shoot growth inhibition and chlorosis induced by kinetin is stronger in wild-type (WT) than in rpn12a-1 seedlings. Plants were germinated and maintained for 14 days on 0.1 μM kinetin.
(B) Inhibition of shoot growth of wild-type and rpn12a-1 seedlings by a range of kinetin concentrations. Seedlings were germinated in the absence of kinetin and then transferred to medium containing the indicated kinetin concentrations. After 21 days, the mean fresh weights of rosettes from 10 seedlings were determined (±sd). FW, fresh weight.
(C) The rpn12a-1 mutation alters the effect of kinetin on root elongation. Seeds were germinated on growth medium, and after 4 days seedlings were transferred to plates containing 0.2 μM kinetin. The red arrowheads indicate root tips after an additional 4 days of growth.
(D) Effect of a range of kinetin concentrations on root elongation. Wild-type and rpn12a-1 seeds were germinated on growth medium, and after 4 days seedlings were transferred to plates containing the indicated kinetin concentrations. After an additional 14 days of growth, the mean root lengths of 10 plants were determined (±sd).
(E) The promotion of root hair formation and elongation by kinetin is reduced by the rpn12a-1 mutation. Seeds were germinated on growth medium. After 4 days, seedlings were transferred to medium containing 0.2 μM kinetin and grown for an additional 4 days.
(F) Promotion of deetiolation by BA is delayed by the rpn12a-1 mutation. Wild-type and rpn12a-1 seedlings were grown for 28 days in the dark in the presence or absence of 20 μM BA.
(G) The induction of greening and shoot formation by 2iP is decreased in rpn12a-1 hypocotyls. Hypocotyl segments of 5-day-old seedlings were incubated for 2 weeks in the presence of 1 μM indole-3-acetic acid supplemented with a range of 2iP concentrations.
(H) The induction of greening by 2iP is decreased in rpn12a-1 roots. Root segments of 5-day-old wild-type and rpn12a-1 seedlings were incubated for 2 weeks in the presence of 1 μM indole-3-acetic acid supplemented with a range of 2iP concentrations.
In both (G) and (H), representative segments from separate 2iP treatments were aligned before photography.
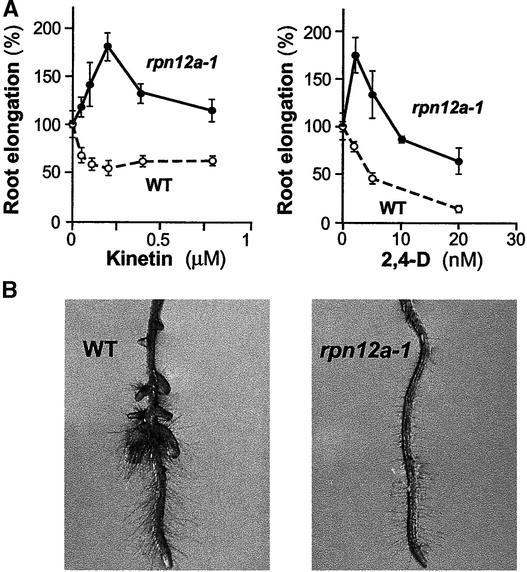
Cytokinins are inhibitors of root elongation growth (Cary et al., 1995). When wild-type plants germinated in the absence of cytokinin were transferred to plates containing 200 nM or more kinetin, a dramatic reduction in root elongation was evident (Figures 5C and 5D). Surprisingly, the rpn12a-1 mutant displayed the opposite effect. Whereas rpn12a-1 roots were significantly shorter than wild-type roots without kinetin treatment, the addition of low concentrations increased root growth substantially, with 0.2 μM kinetin restoring elongation to nearly the level seen for wild-type plants without hormone (Figures 5C and 5D). Higher concentrations became inhibitory, such that rpn12a-1 roots treated with 0.8 μM kinetin were as short as wild-type roots treated with the same concentration. Another root response to cytokinin treatment is the production and elongation of root hairs, probably via the induction of ethylene biosynthesis (Cary et al., 1995). Whereas the roots of the wild type became densely covered with longer hairs after treatment with 0.2 μM kinetin, rpn12a-1 roots did not show such a dramatic effect (Figure 5E).
Cytokinin application is known to induce deetiolation in dark-grown seedlings, characterized by accelerated opening of the cotyledons, elongation of the petioles, and development of true leaves (Chory et al., 1994). Whereas wild-type Arabidopsis seedlings showed these effects when treated with 20 μM concentrations of the cytokinin benzyladenine (BA), this response was delayed severely in rpn12a-1 seedlings (Figure 5F). However, there was no difference between mutant and wild type for the ability of cytokinins to inhibit hypocotyl elongation of etiolated seedlings (data not shown).
Although most of the growth response assays suggested a decrease in cytokinin sensitivity for rpn12a-1 seedlings, root elongation was more complicated to interpret. To further analyze the cytokinin insensitivity, we tested the ability of 2-isopentenyladenine (2iP) to induce greening and shoot formation in hypocotyls and root segments (Mok and Mok, 1994). Wild-type hypocotyls and root segments responded to the 2iP by becoming green, which was reduced noticeably in the rpn12a-1 mutant (Figures 5G and 5H). Whereas 1 to 4 μM 2iP also strongly stimulated shoot formation in wild-type hypocotyls, the mutant hypocotyls failed to develop shoots at any 2iP concentration tested (Figure 5G). Thus, it appears that both roots and shoots from rpn12a-1 have decreased sensitivity to cytokinins.
Cytokinin-Inducible Genes CYCD3 and NIA1 Are Upregulated in the rpn12a-1 Mutant
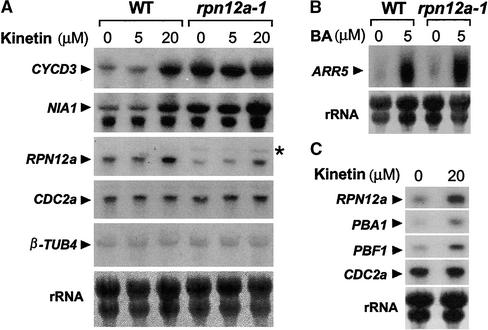
To analyze the cytokinin response at the molecular level, we measured the abundance of the cytokinin-inducible transcripts CYCD3, NIA1, and ARR5. Based on the reduced cytokinin sensitivity of the rpn12a-1 seedlings, we predicted that the cytokinin-induced accumulation of all three mRNAs would be attenuated as well. CYCD3 and NIA1 encode a D-type cyclin and nitrate reductase, respectively, and both show prolonged transcriptional upregulation by cytokinins (Yu et al., 1998; Riou-Khamlichi et al., 1999). Figure 6A confirmed this effect in wild-type plants. Compared with con-trol mRNAs for CDC2a and β-TUB4, 20 μM kinetin increased the mRNA levels substantially for CYCD3 and NIA1 in seedlings after a 4-hr treatment. Surprisingly, when the rpn12a-1 seedlings were tested, the levels of CYCD3 and NIA1 mRNAs were upregulated even in the absence of hormone (Figure 6A). In fact, the addition of 20 μM kinetin did not further increase the level of the CYCD3 mRNA and only increased the level of NIA1 mRNA modestly compared with that in untreated rpn12a-1 plants.
Figure 6.
Expression of Specific Cytokinin-Inducible Genes Is Altered by the rpn12a-1 Mutation.
Wild-type (WT) and rpn12a-1 seedlings were grown in liquid medium for 14 days and then treated with the cytokinin kinetin or BA. After the treatments, total RNA was extracted and subjected to RNA gel blot analysis. Probes included sequences for the 26S proteasome genes RPN12a, PBA1, and PBF1, the cytokinin-inducible genes NIA1, ARR5, and CYCD3, and the cytokinin-noninducible genes CDC2a and β-TUB4. Equal loading of RNA was verified by methylene blue detection of rRNA. Autoradiographic exposure times were varied to best reflect the changes in expression patterns. For each panel, representative blots from at least three independent experiments are shown.
(A) and (C) Seedlings treated for 6 hr with various concentrations of kinetin. The asterisk indicates the position of the RPN12a-NPTII fusion transcript.
(B) Seedlings treated for 15 min with 5 μM BA.
Analysis of the ARR5 mRNA showed that this constitutive upregulation does not occur for all cytokinin-regulated transcripts. ARR5 encodes a response regulator presumably associated with the cytokinin sensory transduction chain and displays a strong and rapid, but transient, increase in mRNA accumulation after exposure to the cytokinin BA (Brandstatter and Kieber, 1998). For this mRNA, rpn12a-1 plants responded like the wild type. Both contained low levels of the ARR5 mRNA without hormone and showed a strong increase after a 15-min exposure to 5 μM BA (Figure 6B).
RNA gel blot analysis revealed that the level of the RPN12a transcript also was upregulated modestly (approximately twofold) by kinetin (Figure 6A). To determine whether this effect was specific for RPN12a or included other subunits of the 26S proteasome, we measured the expression level of the PBA1 and PBF1 genes, which encode β-subunits of the CP (Figure 6C). Levels of both mRNAs, like that of RPN12a, were increased by the treatment of seedlings with 20 μM kinetin.
rpn12a-1 Roots Have Decreased Auxin Sensitivity
Several plant hormone response mutants have altered sen-sitivity to more than one hormone, indicating a close interplay among the associated signaling pathways (Coenen and Lomax, 1997). Consequently, we tested the response of rpn12a-1 seedlings to other hormones, including 1 to 20 nM of the synthetic auxin 2,4-D, 0.01 to 10 μM abscisic acid, 0.01 to 1 μM of the ethylene precursor 1-aminocyclopropane-1-carboxylic acid, 0.5 to 10 μM methyl jasmonate, and 0.2 to 5 μM 22(S),23(S)-homobrassinolide. By using the inhibition of root growth as a parameter, no differences were observed between wild-type and rpn12a-1 seedlings for all concentrations of all hormones except 2,4-D (data not shown). For 2,4-D, the response was remarkably similar to that observed with cytokinins; rpn12a-1 roots actually grew better on low concentrations of 2,4-D rather than being inhibited (Figure 7A). Auxins also are known to stimulate lateral root growth. As can be seen in Figure 7B, 20 nM 2,4-D enhanced lateral root formation in the wild type, but this effect was delayed in rpn12a-1 seedlings. In contrast to the effects with cytokinins, the decreased auxin sensitivity of rpn12a-1 seedlings appeared restricted to roots, because the responses of other tissues to exogenous 2,4-D (e.g., shoot growth, hypocotyl elongation) were like those of the wild type (data not shown).
Figure 7.
Roots of rpn12a-1 Seedlings Are Less Sensitive to the Auxin 2,4-D.
(A) Effect of the rpn12a-1 mutation on the inhibition of root elongation by kinetin (left) and 2,4-D (right). Wild-type (WT) and rpn12a-1 seedlings were grown at a range of concentrations of kinetin or 2,4-D. After 14 days, the mean root lengths of 10 plants were determined and are expressed as percentages of the lengths of the respective untreated controls (±sd).
(B) Effect of the rpn12a-1 mutation on 2,4-D–stimulated lateral root formation. Seedlings were grown for 10 days on 20 nM 2,4-D.
Complementation of rpn12a-1 by Overexpressing the RPN12a cDNA
To confirm that the rpn12a-1 phenotype was caused by the disruption of the RPN12a gene, we attempted to complement the mutation by ectopic expression of the wild-type RPN12a cDNA under the direction of the 35S promoter of Cauliflower mosaic virus. When the transformed rpn12a-1 plants were examined phenotypically, they regained their wild-type morphology and sensitivity to cytokinins and 2,4-D (Figures 8A and 8B; data not shown). All 21 35S-RPN12a transgenic lines containing high levels of the RPN12a protein looked indistinguishable from the wild type, indicating that overexpression of the RPN12a protein was not deleterious to Arabidopsis development. Like the results with hemizygous rpn12a-1 plants (Figure 2C), overexpression of the wild-type protein resulted in the loss of the RPN12a-NPTII fusion protein (Figure 8C), presumably by competitively blocking the association of the fusion protein with the 26S proteasome. From this finding, we conclude that the observed phenotypes for the rpn12a-1 seedlings were the direct result of the T-DNA insertion affecting the RPN12a gene and not another gene located nearby.
Figure 8.

Complementation of the rpn12a-1 Mutation with Wild-Type RPN12a.
The rpn12a-1 mutant was transformed with a cDNA containing the entire RPN12a coding region expressed under the control of the 35S promoter of Cauliflower mosaic virus.
(A) Growth of wild type (WT), the homozygous mutant (rpn12a-1), or the homozygous rpn12a-1 mutant harboring either the empty pGSVE9 vector T-DNA or the 35S-RPN12a transgene. Seedlings were grown for 14 days without (top) or with 0.2 μM kinetin (bottom). Arrowheads identify the tips of the longest roots.
(B) Levels of the RPN12a and RPN12a-NPTII proteins in total crude extracts from 7-day-old seedlings from wild type, rpn12a-1, or the complementation line rpn12a-1::35S-RPN12a. Immunoblots were probed with antibodies against RPN12a or UBC1.
DISCUSSION
Recent studies have implicated the ubiquitin/26S proteasome proteolytic pathway in many environmental and developmental responses in higher plants (Callis and Vierstra, 2000). Here, we extend its importance to cytokinin regulation with the demonstration that an Arabidopsis mutation affecting the synthesis of the RPN12a subunit of the 26S proteasome alters development consistent with a reduction in cytokinin responses. Previous data implicated the ubiquitin/26S proteasome system in the response to hormones, auxins, jasmonate, and abscisic acid (Xie et al., 1998; Gray and Estelle, 2000; Lopez-Molina et al., 2001). Given the complexity of this pathway in plants (in Arabidopsis, it includes more than 1100 genes), it is likely that most if not all hormone responses in plants are regulated at some level by such proteolysis.
In agreement with data from other eukaryotes, RPN12a is a component of the Arabidopsis 26S proteasome. At present, its biochemical function is unknown. RPN12a does not have any recognizable sequence motifs present in other RPN subunits that could provide hints of a function (Glickman et al., 1998; Fu et al., 1999b). It also is distinct from other RP subunits in that it displays the greatest dissimilarity compared with orthologs from other species and by being one of the few Arabidopsis RPN subunits that fails to complement a corresponding yeast deletion (Fu et al., 1999b; H. Fu and R.D. Vierstra, unpublished data). Yeast RPN12 maps to the lid subcomplex of the RP, where it possibly interacts with RPN10 (Kominami et al., 1995; Glickman et al., 1998). Given the predicted role of the RP in substrate selection, RPN12 orthologs could have a unique role in recruiting specific targets.
In addition to RPN12a, we also detected a second Arabidopsis gene (RPN12b) encoding a truncated version of the protein. Although the expression of RPN12b has not yet been demonstrated, three lines of evidence suggest that it is a functional gene. First, the high nucleotide sequence identity between RPN12a and RPN12b exists only within the exons of the coding region, indicating selective evolutionary pressure for the maintenance of the correct coding sequence. Moreover, the predicted beginning of the RPN12b protein coincides with the beginning of a highly conserved stretch (GLNLLXLL) within all RPN12 orthologs from different animal and fungal species (Figure 1B). Second, when the RPN12b genomic sequence was expressed ectopically in Arabidopsis using the 35S promoter of Cauliflower mosaic virus, reverse transcriptase–mediated PCR analysis of the resulting mRNA indicated that the transcript was spliced correctly at the predicted exon/intron junctions to produce a mRNA encoding this 159–amino acid protein (data not shown). However, we were unable to detect immunologically a RPN12b protein of the expected size (18 kD) in these transgenic seedlings using anti-RPN12a antibodies. Third, searches of various databases revealed a Dictyostelium discoideum gene (designated DdM4) encoding a truncated form of RPN12 beginning at a similar position as RPN12b (Kimmel and Firtel, 1985). It was identified as a mRNA of unknown function bearing a short interspersed repeat appended to its 5′ end. The predicted DdM4 protein starts six amino acids downstream of AtRPN12b and terminates 66 residues before AtRPN12b, generating a protein of only 88 residues (Figure 1B). The possibility that the RPN12b locus also encodes a functional subunit raises the intriguing possibility that the RPN12a and RPN12b subunits are interchangeable, thus creating 26S proteasome populations with distinct properties.
The exon-trap T-DNA mutant rpn12a-1 was created by a fortuitous insertion of the ARP-NPTII gene downstream of RPN12a. Transcription, processing, and translation of the rpn12a-1 locus directed the simultaneous synthesis of two proteins, one indistinguishable from wild-type RPN12a and another organized as a RPN12a-NPTII fusion protein. Like its counterparts in budding and fission yeast (Nisogi et al., 1992; Gordon et al., 1996), we expect that null alleles of RPN12a will be lethal in Arabidopsis. Given the relatively subtle phenotypes elicited by the rpn12a-1 mutation and the presence of both the RPN12a and RPN12a-NPTII fusion proteins in homozygous plants, it is reasonable to conclude that rpn12a-1 represents a weak allele. In fact, biochemical fractionation of the 26S proteasome detected complexes containing either the wild-type or fusion protein, indicating that only part of the RP pool is compromised. Additional results support the possibility that rpn12a-1 confers a relatively subtle effect. First, unlike strong mutants affecting other 26S proteasome subunits, including yeast RPN12 (Kominami et al., 1995; Bailly and Reed, 1999; Girod et al., 1999), increased levels of ubiquitinated proteins, characteristic of a backup in protein breakdown, were not observed in rpn12a-1 plants (Figure 2). Second, preliminary biochemical analysis of 26S proteasomes containing RPN12a-NPTII indicate that they are similar in size to those bearing wild-type RPN12a, suggesting that integration of the fusion protein does not radically destabilize the complex (Figure 3). Third, Kominami and Toh-e (1994) demonstrated that RPN12-LacZ fusion proteins can complement a yeast rpn12 mutant, showing that this protein can tolerate large C-terminal additions.
Although the expression of fusion proteins often confers a dominant phenotype, the rpn12a-1 mutation resulting in the RPN12a-NPTII fusion protein behaved as a recessive trait. This can be explained by the ability of the wild-type protein to outcompete the RPN12a-NPTII fusion protein for binding to the 26S proteasome. Accordingly, when sufficient levels of wild-type protein are available (as is the presumed case of hemizygous rpn12a-1 plants or homozygous rpn12a-1 plants expressing a 35S-RPN12a transgene), the fusion protein is blocked from binding and few mutant 26S proteasomes are assembled. Our failure to detect RPN12a-NPTII in both situations implies that the free protein is degraded rapidly. However, for the homozygous rpn12a-1 plants, the wild-type protein created by alternative splicing of the chimeric mRNA is limiting, allowing the RPN12a-NPTII protein to assemble and thus generate a pool of altered 26S proteasomes. The phenotype of the rpn12a-1 plants would be indicative of a specific reduction in RPN12a function and not in the activity of the whole 26S proteasome complex.
The phenotype of rpn12a-1 plants is consistent with a decreased sensitivity of both roots and shoots to cytokinins. Although it is possible that the mutant also is impaired in cytokinin biosynthesis, the fact that rpn12a-1 plants cannot be rescued by exogenous cytokinins suggests otherwise. rpn12a-1 plants also are affected in their response to auxin, indicating that auxin signaling is affected as well. Interestingly, several of the auxin-resistant mutants also are less sensitive to cytokinins, suggesting that these hormone signaling pathways share one or more components (Coenen and Lomax, 1997). The rpn12a-1 mutation appears less severe than the strong cytokinin-resistant mutant cyr1 and may reflect the mild biochemical effect on the RPN12a protein. Thus, it is possible that a stronger defect in RPN12a will further reduce cytokinin sensitivity, leading to a stronger morphological phenotype, as was observed for the cyr1 mutant. The rpn12a-1 phenotype also is different from that of amp1, which is characterized by an increase in endogenous cytokinin levels and by a developmental phenotype consistent with a constitutive cytokinin response (Chaudhury et al., 1993). Instead of a delay in leaf formation in the light and a delayed opening of cotyledons in the dark, amp1 seedlings have accelerated leaf formation and constitutive photomorphogenesis, mimicking wild-type seedlings treated with cytokinin (Chin-Atkins et al., 1996). Although amp1 increases the likelihood of seedlings with more than two cotyledons (Chin-Atkins et al., 1996), we found that rpn12a-1 increased the percentage of monocotyledonous seedlings.
The ability of low cytokinin concentrations to stimulate the elongation of rpn12a-1 roots was unexpected, because exogenous cytokinins are known to inhibit wild-type root growth (Cary et al., 1995). Because untreated rpn12a-1 roots were much shorter than wild-type roots, one possible explanation is that endogenous cytokinins normally promote root growth at low concentrations but become inhibitory at high concentrations. The altered sensitivity of the rpn12a-1 plants could reflect a shift in the dose–response curve toward hormone concentrations higher than those found naturally in the plant. Another apparent contradiction is the enhanced accumulation of anthocyanins in young rpn12a-1 seedlings. Because anthocyanin accumulation is augmented by exogenous cytokinins, a decrease in hormone sensitivity was expected to decrease anthocyanin accumulation, as was observed for the cyr1 mutant (Deikman and Ulrich, 1995). However, decreased cytokinin sensitivity combined with increased anthocyanin accumulation also has been observed in the fus6 mutant (Castle and Meinke, 1994). Thus, it is possible that cyr1 on one side and rpn12a-1 and fus6 on the other side affect different branches of a cytokinin signaling pathway. Interestingly, the FUS6 gene encodes a subunit of the COP9/signalosome complex; this complex is related to the RP and appears to modulate RP subunit composition (Peng et al., 2001). Recent data indicate that the COP9/signalosome and the 26S proteasome work in concert to affect auxin signaling in addition to cytokinins (Schwechheimer et al., 2001).
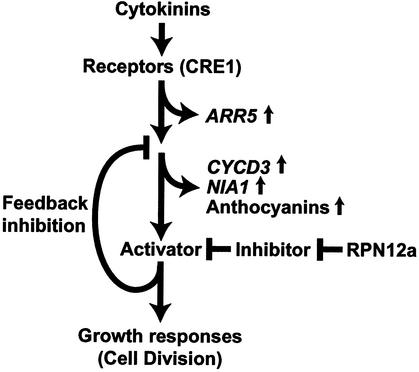
Although the developmental phenotype of rpn12a-1 plants indicated a decreased cytokinin response, transcript levels of the cytokinin-inducible genes CYCD3 and NIA1 implied an accentuated cytokinin response. Both of these transcripts were upregulated even in the absence of hormone treatment. CYCD3 overexpression was shown previously to lead to an increase in the rate of leaf formation (Riou-Khamlichi et al., 1999). Given the fact that a decreased rate of leaf formation was evident in the rpn12a-1 mutant, this increased expression of CYCD3 appeared ineffective in the rpn12a-1 background. How can this be explained? One possible model suggests the participation of an inhibitory feedback loop requiring RPN12a (Figure 9). Such feedback loops are used by many signaling systems to desensitize organisms to the input stimulus and thus adapt. In fact, negative feedback control of gene expression has been shown to be part of both the cytokinin and auxin response pathways (Dominov et al., 1992) and to be involved in the desensitization of plant cell cultures to cytokinins (Mok and Mok, 1994). For cytokinins, it is possible that the pathway that leads to the final responses also activates this negative feedback loop to repress continued activation. Mutations that attenuate both the output response and the feedback pathway could enhance the response of intermediate steps. The increased levels of CYCD3 and NIA1 transcripts in rpn12a-1 seedlings could indicate that both steps are upstream of the event(s) controlled by RPN12a, a possibility also suggested by the absence of any CYCD3 overexpression phenotype in rpn12a-1 plants (Figure 9).
Figure 9.
Possible Role for RPN12a in Cytokinin Signaling.
Cytokinins interact with receptors such as CRE1, resulting in the early induction of ARR5 transcription and subsequent increases in anthocyanin accumulation and CYCD3 and NIA1 expression. The proteasome subunit RPN12a degrades an inhibitor of the pathway immediately upstream of steps that activate cell division, thus allowing cytokinin-induced growth responses to proceed. Destabilization of this inhibitor also activates a negative feedback loop repressing upstream components such as CYCD3 and NIA1 expression. The rpn12a-1 mutation decreases the activity of RPN12a, stabilizing the inhibitor and attenuating both growth responses and feedback inhibition. As a result, CYCD3 and NIA1 expression and anthocyanin synthesis are upregulated but growth responses are downregulated.
In contrast to NIA1 and CYCD3, the expression level of the early response gene ARR5 remained unaffected by the rpn12a-1 mutation in the absence of as well as after a short (15-min) cytokinin treatment (Figure 6B). This lack of effect could imply that RPN12a does not control the cytokinin response leading to early ARR5 induction (Figure 9). Alternatively, we may have used too much BA for the analysis of ARR5. Although Brandstatter and Kieber (1998) reported previously that 5 μM BA was sufficient to induce expression, it is possible that much lower concentrations are effective as well.
How could RPN12a participate in regulating cytokinin growth responses? RPN12a appears to be involved specifically, because T-DNA mutations in other Arabidopsis subunit genes encoding 26S proteasome lid subunits did not lead to a similar phenotype (J. Smalle and R.D. Vierstra, unpublished data). Based on the function of the 26S proteasome, RPN12a likely helps degrade one or more factors that repress cytokinin signaling; stabilization of these regulatory proteins in rpn12a-1 plants would decrease hormone sensitivity. Given the role of cytokinins in promoting cell division (den Boer and Murray, 2000), possible targets include inhibitors of the cell cycle. In yeast, RPN12 has a specific role in the G1/S and G2/M transitions of the cell cycle (Kominami et al., 1995). Although its participation in the G2/M transition is unknown, one function in the G1/S phase is to degrade the cyclin-dependent kinase inhibitor SIC1 (Bailly and Reed, 1999). Disruption of yeast RPN12 leads to the stabilization of SIC1; SIC1 then prevents entry into the cell cycle by associating with and inhibiting the Cln/Cdc28 kinase complex. Interestingly, the Arabidopsis functional homolog of Cdc28 (Cdc2a) interacts with cyclin D3, a mediator of cytokinin-activated cell division and possibly of other cytokinin responses, such as chloroplast development and senescence (Riou-Khamlichi et al., 1999; Healy et al., 2000). Assuming a similar role for Arabidopsis RPN12a, we can expect that it also participates in the breakdown of SIC1-like proteins. Degradation of SIC1 analogs triggered by cytokinins then would activate the CYCD3/CDC2a complex and increase cell division and growth rate. Conversely, their stabilization, resulting from the loss of RPN12a function, would attenuate CYCD3/CDC2a activity and subsequently decrease cytokinin-related growth responses. As yet, no sequence relatives of yeast SIC1 are apparent in the nearly complete Arabidopsis genome. However, several unrelated Arabidopsis cyclin-dependent kinase inhibitors have been identified that could function as inhibitors of the CYCD3/CDC2a complex (Wang et al., 1998; Lui et al., 2000). Whether these are stabilized in the rpn12a-1 mutant awaits testing.
It has been accepted generally that the specificity of ubiquitin/26S proteasome-dependent degradation is controlled to a large extent by the enzymes that conjugate ubiquitin. However, evidence is accumulating that 26S proteasome subunits also have roles in target recognition. Specific RP subunits may function to recognize substrates without previous ubiquitination, may serve as scaffolds to recruit specific E3s, or may identify directly specific ubiquitinated proteins (Murakami et al., 1992; van Nocker et al., 1996b; Sheaff et al., 2000; Xie and Varshavsky, 2000). In budding yeast, for example, the loss of RPN12 stabilizes SIC1, whereas the loss of RPN3 stabilizes the G1 cyclin Cln2 and the anaphase inhibitor PDS1 (Bailly and Reed, 1999). Therefore, instead of a single entry point for all substrates, multiple routes to the CP may exist, each involving different RP subunits. The effects of the rpn12a-1 mutation on Arabidopsis support such a possibility. Instead of conferring a general defect in proteolysis, alteration of the protein led to a specific defect in cytokinin regulation. Certainly, continued investigation of other RPN subunits will reveal how selective each is to the recognition and degradation of the various ubiquitin/26S proteasome targets.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana C24 ecotype, obtained from Dr. M. Jacobs (Vrije Universiteit, Brussels, Belgium), was used as wild type for all phenotypic assays of rpn12a-1. For sterile conditions, plants were grown on Gamborg's B-5 medium (growth medium) (Gibco BRL, Gaithersburg, MD). Unless noted otherwise, plants were irradiated with continuous white fluorescent light. The hormones kinetin, benzyladenine (BA), 2-isopentenyladenine (2iP), indole-3-acetic acid, 2,4-D, abscisic acid, 1-aminocyclopropane-1-carboxylic acid, methyl jasmonate, and 22(S),23(S)-homobrassinolide were obtained from Sigma (St. Louis, MO). For root elongation assays, seedlings were transferred 4 days after germination onto solid medium containing hormones and placed vertically. Root elongation was measured after 9 days on a minimum of 10 seedlings per treatment. For fresh weight analyses, seedlings were germinated on growth medium, transferred after 4 days to medium containing different kinetin concentrations, and grown for an additional 21 days. For the shoot induction experiments, root and hypocotyl segments were dissected from 5-day-old light-grown seedlings, transferred to medium containing 1 μM auxin without or with a range of (2iP) concentrations, and incubated for another 2 weeks in continuous light. Anthocyanin content was measured in hypocotyls of 7-day-old seedlings according to Kubasek et al. (1992) after extraction in methanol containing 1% (v/v) HCl.
Analysis of the Arabidopsis RPN12 Genes and Proteins
RPN12a and RPN12b were located on the lower arms of chromosomes I and V and were sequenced as part of bacterial artificial chromosomes F1N19 and MJC20, respectively. Exon/intron junctions for RPN12a were identified by alignment of the genomic sequence with that for a full-length cDNA (obtained from the Arabidopsis Biological Resource Center, Ohio State University, Columbus). DNA sequencing was performed using dye terminator chemistry (Perkin-Elmer, Foster City, CA). Amino acid sequence comparisons were performed using ClustalW (http://www2.ebi.ac.uk/clustalw) and displayed using Mac BoxShade (Institute of Animal Health, Pirbright, UK).
Proteasome Purification and Activity Assays
The 26S proteasome was partially purified from wild-type or rpn12a-1 seedlings grown in a 16-hr-light/8-hr-dark photoperiod. Plants were frozen to liquid nitrogen temperatures, pulverized, and ground in 4 volumes of buffer A (50 mM Na phosphate, pH 7.5, 2 mM MgCl2, 5% glycerol, 1 mM 2-mercaptoethanol, 1 mM ATP, and 5% polyvinylpyrrolidone-360). After filtering through cheesecloth, the extract was clarified for 15 min at 3000g and polyethylene glycol 8000 was added to a final concentration of 2% (w/v). The extract was reclarified for 15 min at 3000g, and the supernatant was adjusted to 10% polyethylene glycol 8000. The precipitate was collected by centrifugation for 15 min at 3000g, and the pellet was resuspended in buffer B (buffer A with 20% glycerol but without polyvinylpyrrolidone). Proteins were separated by fast protein liquid chromatography (FPLC) using a 1- × 20-cm Sephacryl S-400 column equilibrated with buffer B. Fractions containing the 26S proteasome were identified by peptidase activity using the substrate N-succinyl-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin (Sigma) and by immunoblot analysis with appropriate antibodies.
Antibody Production and Immunoblot Analysis
The RPN12a coding region was amplified by polymerase chain reaction (PCR) from the full-length cDNA using the primers 5′-AAAACC-ATAGAACATATGGATCCGCAGCTA-3′ and 5′-CAGTGAGAAAAG-TCGACTCGAGCACGATACGCTCCA-3′ designed to contain 5′ and 3′ flanking NdeI and XhoI sites, respectively. Upon NdeI–XhoI digestion, this product was cloned into pET28b (Novagen, Madison, WI) for expression in Escherichia coli strain BL21 (DE3). After 3 hr of induction with 0.4 mM isopropyl 1-thio-β-d-galactopyranoside, the RPN12a protein was collected in the inclusion body fraction by centrifugation. The pellet was dissolved in SDS-PAGE sample buffer and subjected to preparative SDS-PAGE. The portion of the gel containing the 31-kD RPN12a protein (visualized by Coomassie Brilliant Blue R-250) was excised, emulsified, and injected directly into rabbits (Polyclonal Antibody Service, Madison, WI). Anti-RPN12a antibodies were affinity purified from serum by adsorption to nitrocellulose coated with recombinant protein (Sambrook et al., 1989). Antibodies against RPN5a, RPT1a, RPT2a, PAC1, and PBA1 were prepared similarly except that recombinant PBA1 protein remained soluble and was purified using nickel–nitrilotriacetic acid agarose chromatography under nondenaturing conditions, as recommended (Qiagen, Valencia, CA). Anti-RPN10, anti-ubiquitin, and anti-UBC1 antibodies were as described previously (Sullivan et al., 1994; van Nocker et al., 1996a).
For immunoblot analyses, proteins subjected to SDS-PAGE were transferred to nitrocellulose membranes (Immobilon NC; Millipore, Bedford, MA) and the membranes were blocked using 10% milk powder (Sambrook et al., 1989). Membranes probed with anti-ubiquitin antibodies were autoclaved before blocking. Both primary and secondary antibodies were diluted 1:1000 in PBS containing 0.2% Tween 20 and 1% BSA. Secondary antibodies used were alkaline phosphatase– or peroxidase-labeled goat anti–rabbit immunoglobulins (Kirkegaard and Perry Laboratories, Gaithersburg, MD).
RNA Isolation, 5′ Rapid Amplification of cDNA Ends PCR, and RNA Gel Blot Analysis
Total RNA was extracted using Trizol reagent (Gibco BRL) from 14-day-old seedlings grown in liquid growth medium. 5′ rapid amplification of cDNA ends PCR was conducted as described by Babiychuk et al. (1997). For RNA gel blot analysis, 20 μg of total RNA was electrophoresed on 1% agarose-formaldehyde gels and transferred to Zeta-Probe GT genomic membranes (Bio-Rad, Hercules, CA). Bound RNA was stained using methylene blue to verify loading and transfer. Probe hybridization, washing, and autoradiography were performed as described previously (Kurepa et al., 1997). 32P-labeled riboprobes were synthesized with T7 or SP6 RNA polymerase using the appropriate linearized plasmids and the Riboprobe Gemini II core system (Promega). The CYCD3 template used the pJSCYCD3 plasmid linearized with SalI–SpeI. This plasmid was created by PCR amplification of the CYCD3 cDNA from a cDNA library using primers 5′-GATCTTATCTTTCTTCTCATTCTTGAGTTT-3′ and 5′-AACAGTAAA-TATCATATAAACGAAATCGAG-3′ and insertion of the product into pGEM-T (Promega). The NIA1 template used the pJSNIA1 plasmid linearized with SphI–AatII. This plasmid was created by PCR amplification of the NIA1 cDNA using primers 5′-GACCTCCGTCGATAA-CCGCCATTATCCCAC-3′ and 5′-ACGATGATTCTCTTTAACCAT-TTAACCATC-3′ and insertion of the product into pGEM-T. The ARR5, RPN12a, and CDC2a templates used expressed sequence tag clones 103N10T7, 198J17T7, and 205H2T7, respectively, linearized with KpnI–PstI. The β-TUB4, PBA1, and PBF1 probes were described previously (Fu et al., 1998b). RNA gel blot signal intensities were estimated using the NIH Image program (National Institutes of Health, Bethesda, MD).
rpn12a-1 Complementation
The RPN12a cDNA was amplified by PCR using primers 5′-AGA-AGAGAAAAAACTCTAGAAGCGATGGAT-3′ and 5′-TGAACACTT-AAACTCTAGAATATAACGTAA-3′ designed to contain an XbaI site at both ends. The resulting product was digested with XbaI and inserted into the binary T-DNA vector pGSVE9 that was digested similarly (E. Babiychuk and S. Kushnir, unpublished data). The resulting construction allowed for expression of the RPN12a coding sequence driven by the 35S promoter. The vector was introduced into Agrobacterium tumefaciens strain AtC58RifR (pMP90), and the strain was used to transform homozygous rpn12a-1 seedlings by the floral dip method (Clough and Bent, 1998). Transformants were identified by hygromycin resistance. Initial transformants (T1) were selfed, and the progeny (T2) were tested for hygromycin resistance. These hygromycin-resistant plants were again selfed, and the progeny (T3) were tested for both kanamycin and hygromycin resistance. Lines that were homozygous for both kanamycin resistance (i.e., containing the rpn12a-1 T-DNA) and hygromycin resistance (i.e., containing the 35S-RPN12a transgene) were analyzed for phenotypic complementation.
Accession Numbers
The accession numbers for the sequences described in this article are AC009579 (RPN12a) and AB017067 (RPN12b). The accession numbers for the bacterial artificial chromosomes described are AC009519 (F1N19) and AB017067 (MJC20), and the accession numbers for the sequences shown in Figure 1 are AE003527 (DmRPN12), AC005783 (Hsp31), P02889 (DdM4), Q23449 (CeS14), X92682 (SpMTS3), CAA15600 (PfS14), and D10515 (ScRPN12).
Acknowledgments
We thank Hongyong Fu for help with the antibody production, Jed Doelling and Joseph Walker for technical assistance, Sarah Patterson for critical reading of the manuscript, and the Arabidopsis Biological Resource Center for providing expressed sequence tag clones. This work was supported by grants from the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (Grant Nos. 97-35301-4218 and 00-35301-9040) and the Research Division of the University of Wisconsin College of Agriculture and Life Sciences (Hatch 142-E443) to R.D.V. and a North Atlantic Treaty Or-ganization research fellowship to J.S.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010381.
References
- Babiychuk, E., Fuangthong, M., Van Montagu, M., Inzé, D., and Kushnir, S. (1997). Efficient gene tagging in Arabidopsis thaliana using a gene trap approach. Proc. Natl. Acad. Sci. USA 94, 12722–12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair, A., Becker, F., Masterson, R.V., and Schell, J. (1990). Perturbation of the ubiquitin system causes leaf curling, vascular tissue alterations and necrotic lesions in a higher plant. EMBO J. 9, 4543–4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly, E., and Reed, S.I. (1999). Functional characterization of rpn3 uncovers a distinct 19S proteasomal subunit requirement for ubiquitin-dependent proteolysis of cell cycle regulatory proteins in budding yeast. Mol. Cell. Biol. 19, 6872–6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, J., Kempf, R., Jeblick, W., and Kauss, H. (2000). Induction of competence for elicitation of defense responses in cucumber hypocotyls requires proteasome activity. Plant J. 21, 311–316. [DOI] [PubMed] [Google Scholar]
- Brandstatter, I., and Kieber, J.J. (1998). Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 10, 1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis, J., and Vierstra, R.D. (2000). Protein degradation in signaling. Curr. Opin. Plant Biol. 3, 381–386. [DOI] [PubMed] [Google Scholar]
- Cary, A.J., Liu, W., and Howell, S.H. (1995). Cytokinin action is coupled to ethylene in its effects on the inhibition of root and hypocotyl elongation in Arabidopsis thaliana seedlings. Plant Physiol. 107, 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle, L.A., and Meinke, D.W. (1994). A FUSCA gene of Arabidopsis encodes a novel protein essential for plant development. Plant Cell 6, 25–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury, A.M., Letham, S., Craig, S., and Dennis, E.S. (1993). amp1, a mutant with high cytokinin levels and altered embryonic pattern, faster vegetative growth, constitutive photomorphogenesis and precocious flowering. Plant J. 4, 907–916. [Google Scholar]
- Chin-Atkins, A.N., Craig, S., Hocart, C.H., Dennis, E.S., and Chaudhury, A.M. (1996). Increased endogenous cytokinin in the Arabidopsis amp1 mutant corresponds with de-etiolation responses. Planta 198, 549–556. [DOI] [PubMed] [Google Scholar]
- Chory, J., Reinecke, D., Sim, S., Washburn, T., and Brenner, M. (1994). A role for cytokinins in de-etiolation in Arabidopsis. Plant Physiol. 104, 339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, R.C., Jordan-Beebe, E.T., Lohman, K.N., Marita, J.M., Walker, J.M., Gatz, C., and Vierstra, R.D. (1999). Sequences within both the N- and C-terminal domains of phytochrome A are required for PFR ubiquitination and degradation. Plant J. 17, 155–167. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Coenen, C., and Lomax, T.L. (1997). Auxin-cytokinin interactions in higher plants: Old problems and new tools. Trends Plant Sci. 2, 351–356. [DOI] [PubMed] [Google Scholar]
- Deikman, J., and Ulrich, M. (1995). A novel cytokinin-resistant mutant of Arabidopsis with abbreviated shoot development. Planta 195, 440–449. [DOI] [PubMed] [Google Scholar]
- den Boer, B.G., and Murray, J.A. (2000). Triggering the cell cycle in plants. Trends Cell Biol. 10, 245–250. [DOI] [PubMed] [Google Scholar]
- Dieterle, M., Zhou, Y.-C., Schafer, E., Funk, M., and Kretsch, T. (2001). EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev. 15, 939–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling, J.H., Yan, N., Kurepa, J., Walker, J., and Vierstra, R.D. (2001). The ubiquitin-specific protease UBP14 is essential for early embryo development in Arabidopsis thaliana. Plant J. 27, 393–405. [DOI] [PubMed] [Google Scholar]
- Dominov, J.A., Stenzler, L., Lee, S., Schwarz, J.J., Leisner, S., and Howell, S.H. (1992). Cytokinins and auxins control the expression of a gene in Nicotiana plumbaginifolia cells by feedback regulation. Plant Cell 4, 451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley, D., et al. (1998). Unified nomenclature for subunits of the Saccharomyces cerevisiae proteasome regulatory particle. Trends Biochem. Sci. 23, 244–245. [DOI] [PubMed] [Google Scholar]
- Fu, H., Sadis, S., Rubin, D.M., Glickman, M., van Nocker, S., Finley, D., and Vierstra, R.D. (1998. a). Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26 S proteasome subunit Mcb1. J. Biol. Chem. 273, 1970–1981. [DOI] [PubMed] [Google Scholar]
- Fu, H., Doelling, J.H., Arendt, C.S., Hochstrasser, M., and Vierstra, R.D. (1998. b). Molecular organization of the 20S proteasome gene family from Arabidopsis thaliana. Genetics 149, 677–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H., Doelling, J.H., Rubin, D.M., and Vierstra, R.D. (1999. a). Structural and functional analysis of the six regulatory particle triple-A ATPase subunits from the Arabidopsis 26S proteasome. Plant J. 18, 529–539. [DOI] [PubMed] [Google Scholar]
- Fu, H., Girod, P.A., Doelling, J.H., van Nocker, S., Hochstrasser, M., Finley, D., and Vierstra, R.D. (1999. b). Structure and functional analysis of the 26S proteasome subunits from plants. Mol. Biol. Rep. 26, 137–146. [DOI] [PubMed] [Google Scholar]
- Fu, H., Reis, N., Lee, Y., Glickman, M.H., and Vierstra, R.D. (2001). Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 20, 7096–7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik, P., Criqui, M.C., Parmentier, Y., Derevier, A., and Fleck, J. (1998). Cell cycle-dependent proteolysis in plants. Identification of the destruction box pathway and metaphase arrest produced by the proteasome inhibitor MG 132. Plant Cell 10, 2063–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girod, P.-A., Fu, H., Zryd, J.-P., and Vierstra, R.D. (1999). Multiubiquitin chain binding subunit MCB1 (RPN10) of the 26S proteasome is essential for developmental progression in Physcomitrella patens. Plant Cell 11, 1457–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman, M.H., Rubin, D.M., Coux, O., Wefes, I., Pfeifer, G., Cjeka, Z., Baumeister, W., Fried, V.A., and Finley, D. (1998). A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94, 615–623. [DOI] [PubMed] [Google Scholar]
- Gordon, C., McGurk, G., Wallace, M., and Hastie, N.D. (1996). A conditional lethal mutant in the fission yeast 26S protease subunit mts3+ is defective in metaphase to anaphase transition. J. Biol. Chem. 271, 5704–5711. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., and Estelle, I. (2000). Function of the ubiquitin-proteasome pathway in auxin response. Trends Biochem. Sci. 25, 133–138. [DOI] [PubMed] [Google Scholar]
- Healy, J.M.S., Menges, M., Doonan, J.H., and Murray, J.A.H. (2000). The Arabidopsis D-type cyclins CycD2 and CycD3 both interact in vivo with the PSTAIRE cyclin-dependent kinase Cdc2a but are differentially controlled. J. Biol. Chem. 276, 7041–7047. [DOI] [PubMed] [Google Scholar]
- Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Inoue, T., Higuchi, M., Hashimoto, Y., Seki, M., Kobayashi, M., Kato, T., Tabata, S., Shinozaki, K., and Kakimoto, T. (2001). Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409, 1060–1063. [DOI] [PubMed] [Google Scholar]
- Kimmel, A.R., and Firtel, R.A. (1985). Sequence organization and developmental expression of an interspersed, repetitive element and associated single-copy DNA sequences in Dictyostelium discoideum. Mol. Cell. Biol. 5, 2123–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominami, K., and Toh-e, A. (1994). Characterization of the function of the NIN1 gene product of Saccharomyces cerevisiae. Exp. Cell Res. 211, 203–211. [DOI] [PubMed] [Google Scholar]
- Kominami, K., DeMartino, G.N., Moomaw, C.R., Slaughter, C.A., Shimbara, N., Fujimuro, M., Yokosawa, H., Hisamatsu, H., Tanahashi, N., and Shimizu, Y. (1995). Nin1p, a regulatory subunit of the 26S proteasome, is necessary for activation of Cdc28p kinase of Saccharomyces cerevisiae. EMBO J. 14, 3105–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubasek, W.L., Shirley, B.W., Mckillop, A., Goodman, H.M., Briggs, W., and Ausubel, F.M. (1992). Regulation of flavonoid biosynthetic genes in germinating Arabidopsis seedlings. Plant Cell 4, 1229–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa, J., Herouart, D., Van Montagu, M., and Inzé, D. (1997). Differential expression of CuZn- and Fe-superoxide dismutase genes of tobacco during development, oxidative stress, and hormonal treatments. Plant Cell Physiol. 38, 463–470. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina, L., Mongrand, S., and Chua, N.-H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98, 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui, H., Wang, H., Delong, C., Fowke, L.C., Crosby, W.L., and Fobert, P.R. (2000). The Arabidopsis Cdc2a-interacting protein ICK2 is structurally related to ICK1 and is a potent inhibitor of cyclin-dependent kinase activity in vitro. Plant J. 21, 379–385. [DOI] [PubMed] [Google Scholar]
- Mironov, V., De Veylder, L., Van Montagu, M., and Inzé, D. (1999). Cyclin-dependent kinases and cell division in plants: The nexus. Plant Cell 11, 509–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok, D.W.S., and Mok, M.C. (1994). Cytokinins: Chemistry, Activity and Function. (Boca Raton, FL: CRC Press).
- Murakami, Y., Matsufuji, S., Kameji, T., Hayashi, S., Igarashi, K., Tamura, T., Tanaka, K., and Ichihara, A. (1992). Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature 360, 597–599. [DOI] [PubMed] [Google Scholar]
- Nisogi, H., Kominami, K., Tanaka, K., and Toh-e, A. (1992). A new essential gene of Saccharomyces cerevisiae, a defect in it may result in instability of nucleus. Exp. Cell Res. 200, 48–57. [DOI] [PubMed] [Google Scholar]
- Peng, Z., Staub, J.M., Serino, G., Kwok, S.F., Kurepa, J., Bruce, B.D., Vierstra, R.D., Wei, N., and Deng, X.-W. (2001). The cellular level of pr500, a protein complex related to the 19S regulatory particle of the proteasome, is regulated in response to stresses in plants. Mol. Biol. Cell 12, 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou-Khamlichi, C., Huntley, R., Jacqmard, A., and Murray, J.A. (1999). Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283, 1541–1544. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schwechheimer, C., Serino, G., Callis, J., Crosby, W.L., Lyapina, S., Deshaies, R.J., Gray, W.M., Estelle, M., and Deng, X.-W. (2001). Interactions of the COP9 signalosome with the E3 ligase SCRTIR1 in mediating auxin response. Science 292, 1379–1382. [DOI] [PubMed] [Google Scholar]
- Sheaff, R.J., Singer, J.D., Swanger, J., Smitherman, M., Roberts, J.M., and Clurman, B.E. (2000). Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol. Cell 5, 403–410. [DOI] [PubMed] [Google Scholar]
- Sullivan, M.L., Carpenter, T.B., and Vierstra, R.D. (1994). Homologues of wheat ubiquitin-conjugating enzymes: TaUBC1 and TaUBC4 are encoded by small multigene families in Arabidopsis thaliana. Plant Mol. Biol. 24, 651–661. [DOI] [PubMed] [Google Scholar]
- Taniguchi, M., Kiba, T., Sakakibara, H., Ueguchi, C., Mizuno, T., and Sugiyama, T. (1998). Expression of Arabidopsis response regulator homologs is induced by cytokinins and nitrate. FEBS Lett. 429, 259–262. [DOI] [PubMed] [Google Scholar]
- van Nocker, S., Deveraux, Q., Rechsteiner, M., and Vierstra, R.D. (1996. a). Arabidopsis MBP1 gene encodes a conserved ubiquitin recognition component of the 26S proteasome. Proc. Natl. Acad. Sci. USA 93, 856–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nocker, S., Saddis, S., Rubin, D.M., Glickman, M., Fu, H., Coux, O., Wefes, I., Finley, D., and Vierstra, R.D. (1996. b). The multiubiquitin chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol. Cell. Biol. 16, 6020–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges, D., Zwickl, P., and Baumeister, W. (1999). The 26S proteasome: A molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68, 1015–1068. [DOI] [PubMed] [Google Scholar]
- Wang, H., Qi, Q., Schorr, P., Cutler, A.J., Crosby, W.L., and Fowke, L.C. (1998). ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana, interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 15, 501–510. [DOI] [PubMed] [Google Scholar]
- Xie, D.X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xie, Y., and Varshavsky, A. (2000). Physical association of ubiquitin ligases and the 26S proteasome. Proc. Natl. Acad. Sci. USA 97, 2497–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X., Sukumaran, S., and Marton, L. (1998). Differential expression of the Arabidopsis Nia1 and Nia2 genes: Cytokinin-induced nitrate reductase activity is correlated with increased Nia1 transcription and mRNA levels. Plant Physiol. 116, 1091–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]