Abstract
The slender rice1 mutant (slr1) shows a constitutive gibberellin (GA) response phenotype. To investigate the mode of action of SLR1, we generated transgenic rice expressing a fusion protein consisting of SLR1 and green fluorescent protein (SLR1-GFP) and analyzed the phenotype of the transformants and the subcellular localization of GFP in vivo. SLR1-GFP worked in nuclei to repress the GA signaling pathway; its overproduction caused a dwarf phenotype. Application of GA3 to SLR1-GFP overproducers induced GA actions such as shoot elongation, downregulation of GA 20-oxidase expression, and upregulation of SLR1 expression linked with the disappearance of the nuclear SLR1-GFP protein. We also performed domain analyses of SLR1 using transgenic plants overproducing different kinds of truncated SLR1 proteins. The analyses revealed that the SLR1 protein can be divided into four parts: a GA signal perception domain located at the N terminus, a regulatory domain for its repression activity, a dimer formation domain essential for signal perception and repression activity, and a repression domain at the C terminus. We conclude that GA signal transduction is regulated by the appearance or disappearance of the nuclear SLR1 protein, which is controlled by the upstream GA signal.
INTRODUCTION
Gibberellins (GAs) are growth factors with a tetracyclic diterpenoid structure that are essential regulators of diverse growth and developmental processes of plants (Davies, 1995). A series of genes encoding the enzymes involved in the GA biosynthetic pathway has been cloned from a variety of species (reviewed by Hedden and Phillips, 2000). Expression analysis has revealed that the developmental regulation of the expression of these genes plays an important role in controlling the many aspects of GA-regulated plant growth, such as stem elongation, flower development, and seed germination (Silverstone et al., 1997a; Yamaguchi et al., 1998; Itoh et al., 1999; Rebers et al., 1999). In contrast to the rapid progress in the study of GA biosynthesis, much less is known about how plants perceive GA and how the GA signal is transmitted to cause GA-regulated plant growth.
Ikeda et al. (1999a) isolated a constitutive GA-responsive mutant of rice, slender rice1 (slr1), which shows a slender phenotype with elongated stem, leaf sheath, and blade similar to that of rice plants treated exogenously with GA3. Through phenotypic analysis of slr1, SLR1 is thought to encode a negative regulator for the GA signal transduction pathway (Ikeda et al., 1999b). Recently, we cloned SLR1, and its sequencing analysis has revealed that it encodes a putative transcriptional regulator with a structure similar to those of Arabidopsis Gibberellin Insensitive (GAI) and Repressor of ga1-3 (RGA), wheat Reduced height (Rht), and maize dwarf8 (d8) (Ikeda et al., 2001). Dominant alleles at the Arabidopsis GAI, wheat Rht-B1/Rht-D1, and maize D8 loci confer GA-insensitive mutants with dwarf phenotype (Koornneef et al., 1985; Harberd and Freeling, 1989; Winkler and Freeling, 1994; Peng et al., 1997, 1999), and molecular cloning of Arabidopsis GAI has demonstrated that the in-frame deletion of its N terminus domain occurs in the gai mutant (Peng et al., 1997). According to the dominant phenotype caused by the gai mutant protein, Peng et al. (1997) suggested that the native GAI product represses the action of GA and that its repression can be released by GA. They also suggested that the internal deletion of the GAI protein in the gai mutant is resistant to the GA signal. According to this speculation, mutants with the loss of function of this product should show a constitutive GA response with the slender phenotype regardless of the presence or absence of GA. However, plants with the loss-of-function alleles of gai show only a slight reduction in GA dependence (Peng et al., 1997).
The absence of a clear phenotype of GAI knockout plants has been suggested to be caused by the presence of genes redundant with GAI. Indeed, the RGA gene has a structure highly similar to that of GAI, and its loss of function does not show a typical constitutive GA response phenotype but partially suppresses the dwarf phenotype conferred by the GA deficiency mutation ga1-3 (Silverstone et al., 1997b, 1998). In contrast to the Arabidopsis genome, the rice genome has only one gene encoding a protein orthologous to GAI/RGA/Rht/d8 (Ogawa et al., 2000; H. Itoh and M. Matsuoka, unpublished results); consequently, rice plants with a loss-of-function allele of SLR1 show the constitutive GA-responsive phenotype (Ikeda et al., 2001). Such nonredundancy in rice should provide an advantage in studying the function of SLR1/GAI/RGA/Rht/d8 members.
Using transgenic plants overproducing a fusion protein consisting of SLR1 and green fluorescent protein (SLR1-GFP), we have demonstrated that SLR1 acts in the nucleus to repress GA action and that a GA signal causes the level of SLR1 protein in nuclei to decrease, resulting in the induction of stem elongation. To gain further insight into the function of SLR1, we also performed domain analysis. The analysis revealed that the SLR1 protein can be divided into four domains with distinct functions.
RESULTS
Transgenic Rice Plants Overproducing SLR1-GFP Show the Dwarf Phenotype
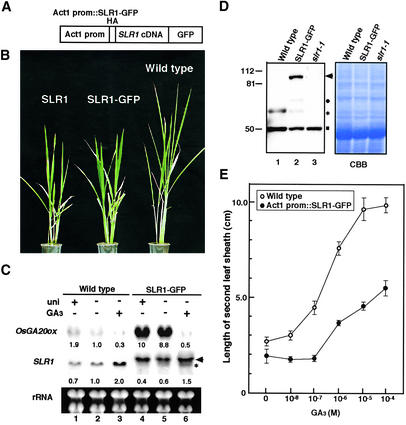
To confirm the repressive function of the SLR1 protein in rice, we generated transgenic rice plants that highly and constitutively produced hemagglutinin (HA)-tagged SLR1-GFP fusion protein under the control of the rice Actin1 promoter (Figure 1A; McElroy et al., 1990). Approximately 60% of the overexpressor plants were 60 to 80% (mild dwarf) as tall as the wild-type plants (Figure 1B). The phenotype with dwarf also was induced by the introduction of Act1 prom::SLR1 cDNA, indicating that fusion with GFP does not affect the severity of the transformants (Figure 1B). These dwarf plants were elongated by treatment with 100 μM GA3 (see below). The construct also caused the same phenomena in the slr1-1 mutant and thus complemented the mutant phenotype (data not shown). These results demonstrate that the SLR1-GFP fusion protein is functional in rice.
Figure 1.
Phenotypic Comparison between Wild-Type and SLR1-GFP–Overexpressing Rice Plants.
(A) Scheme of the chimeric construct consisting of the SLR1 cDNA fused with GFP at the 3′ side and HA at the 5′ side in an in-frame manner under the control of the rice Actin1 promoter (Act1 prom).
(B) Gross morphologies of 45-day-old wild-type (right) and transgenic plants transformed with Act1 prom::SLR1 (left) or Act1 prom::SLR1-GFP (center).
(C) Expression of two GA-regulated genes, OsGA20ox and SLR1, in wild-type and SLR1-GFP plants. RNA gel blot analysis was performed using total RNA from wild-type and SLR1-GFP seedlings grown in water with (+) or without (−) 1 μM uniconazol (uni) or 100 μM GA3. Ten micrograms of total RNA was loaded per lane and stained with ethidium bromide (rRNA). The arrowhead and asterisk (middle) indicate the transcript bands corresponding to SLR1-GFP and the endogenous wild-type SLR1, respectively. The values at the bottom of the OsGA20ox and SLR1 panels indicate the relative levels of OsGA20ox and endogenous SLR1 transcript. Each transcript was normalized by rRNA level after quantification using NIH Image software version 1.61. The transcript level in the wild-type plant without any treatments (−GA3, −uni) was set at 1.0.
(D) Protein gel blot analysis of the endogenous SLR1 protein and the SLR1-GFP fusion protein in wild-type (lane 1), SLR1-GFP (lane 2), and slr1-1 (lane 3) seedlings. Ten micrograms of protein extracts was loaded per lane and probed with anti-SLR1 antibody. Molecular mass markers (in kD) are indicated at left. The extract from slr1-1 was used as a negative control (lane 3). The arrowhead and asterisk indicate the protein bands corresponding to SLR1-GFP and the endogenous wild-type SLR1, respectively. The circle shows the degraded protein derived from the SLR1-GFP protein, because this protein also was recognized by the anti-HA antibody. The antibody also recognized a 50-kD protein (square), which is present in slr1-1 and therefore is not related to SLR1. As a loading control, the Coomassie brilliant blue (CBB) staining profile is shown.
(E) Elongation of the second leaf sheath in response to GA3 treatment in wild-type (open circles) and SLR1-GFP (closed circles) plants. Error bars represent standard deviation from the mean (n = 6).
The dwarf phenotype of the SLR1 overproducers suggests that high-level expression of SLR1-GFP suppresses the action of GA. To elucidate the action of GA at the gene expression level in the overproducers, we examined the expression of GA 20-oxidase (OsGA20ox). Toyomasu et al. (1997) demonstrated that the OsGA20ox transcript level was controlled in a negative feedback manner by the level of bioactive GA. We found that the transcript level in the wild-type plants was increased by treatment with a GA biosynthetic inhibitor, uniconazol, and decreased by GA3 treatment (Figure 1C, top, lanes 1 to 3). In the SLR1-GFP plants, the OsGA20ox transcript level was increased approximately nine times more than that in the wild-type plants under normal growth conditions (Figure 1C, top, lane 2 versus lane 5). The increased level was suppressed to a level similar to that in the GA-treated wild-type plants by the application of 100 μM GA3 (Figure 1C, top, lane 3 versus lane 6). Treatment of the SLR1-GFP plants with uniconazol did not affect the transcript level as much, probably because the expression of OsGA20ox was saturated in the transgenic plants (Figure 1C, top, lane 4 versus lane 5).
In contrast to the expression of OsGA20ox, the expression of SLR1 was regulated positively by the application of GA3 (Ogawa et al., 2000). Indeed, we found that the SLR1 transcript level was upregulated approximately two times by GA3 treatment in the wild-type plants (Figure 1C, middle, asterisked band in lanes 2 and 3). In SLR1-GFP plants, the endogenous SLR1 transcript also was upregulated in the GA-treated plants (Figure 1C, middle, asterisked band in lanes 5 and 6), although the high amount of SLR1-GFP mRNA (Figure 1C, middle, arrowhead) made the level of the endogenous transcript unclear. Thus, we also performed protein gel blot analysis with anti-SLR1 antibody (Figure 1D). The anti-SLR1 antibody recognized 65- and 50-kD proteins in the wild-type plant and four proteins with molecular masses of 95, 70, 65, and 50 kD in the SLR1-GFP plants, whereas it recognized a 50-kD protein in the slr1-1 mutant (Figure 1D, lane 3). According to the calculated molecular mass of SLR1 and HA–SLR1-GFP chimeric proteins, the 95- and 65-kD proteins correspond to the intact HA–SLR1-GFP chimeric protein (arrowhead) and the endogenous SLR1 protein (asterisk), respectively, whereas the 50-kD protein (square) is unrelated to SLR1 because this protein also was recognized in the slr1-1 mutants. The 70-kD protein (circle) is considered a degraded product from the intact HA–SLR1-GFP protein, because it was detected only in the transgenic plants and was recognized by the anti-HA antibody (data not shown).
The blot clearly showed the downregulation of the intact SLR1 protein in the SLR1-GFP–overexpressing plants relative to the level in the wild-type plants (asterisked bands). These observations demonstrate that a large amount of SLR1-GFP represses the action of GA, stunting plant height and upregulating the expression of OsGA20ox or downregulating the expression of SLR1. However, strong repression of the action of GA in the overproducers does not mean that the plants lose their responses to GA, because exogenous GA3 treatment can release the GA-repressive action. This finding suggests that a greater amount of the SLR1 protein increases the critical level of GA at which the GA signal is transduced. To elucidate this possibility, we tested the GA responsiveness of the wild-type and overproducing plants by treatment with various concentrations of GA3 (Figure 1E). The wild-type plants responded to GA3 greater than 10−8 M and started to elongate, but the overproducers did not respond to GA3 until a higher concentration (>10−7 M) was reached. This finding confirms that a much higher concentration of GA is necessary for induction of the action of GA in the overproducers than in the wild-type plants.
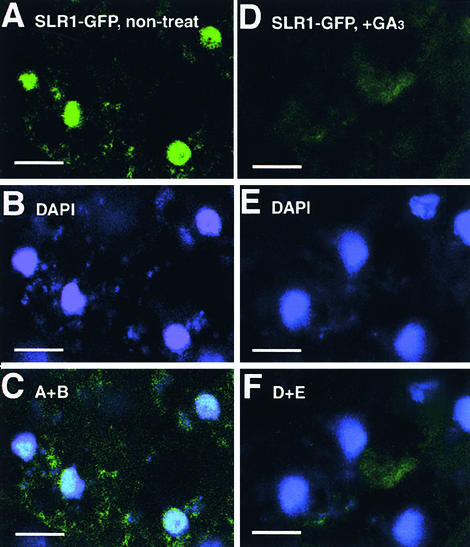
SLR1-GFP Was Localized in Nuclei and Disappeared in Response to Exogenous GA3
We examined the intercellular localization of the SLR1-GFP fusion protein based on the fluorescent localization of GFP. Leaf sheaths of transgenic rice plants were cut with a razor blade, and the small leaf pieces were observed with a confocal microscope. Under normal growth conditions, bright green spots of GFP fluorescence were localized in the nuclei (Figure 2A), as confirmed by specific staining with 4′,6-diamidino-2-phenylindole dihydrochloride n-hydrate (DAPI; Figure 2B). The merged image confirms the overlapping localization of GFP and DAPI fluorescence (Figure 2C). When transgenic rice plants were grown in water containing 100 μM GA3, however, no fluorescence in nuclei was observed (Figures 2D to 2F). These results suggest that SLR1 functions as a repressor in the nucleus and that the disappearance of SLR1 from nuclei caused by GA treatment releases the suppression and allows the plants to transmit the GA signal downstream.
Figure 2.
Effect of GA3 on the Subcellular Localization of SLR1-GFP.
(A) and (D) Confocal microscopic images of GFP fluorescence in young leaf sections from SLR1-GFP overexpressor lines under the control of the rice Actin1 promoter.
(B) and (E) Nuclei in the same cells as in (A) and (D) were stained with DAPI.
(C) and (F) Merged images of GFP and DAPI fluorescence.
Plants were grown with (+GA3, [D] to [F]) or without (non-treat, [A] to [C]) 100 μM GA3 for several days before GFP fluorescence analysis. Bars = 10 μm.
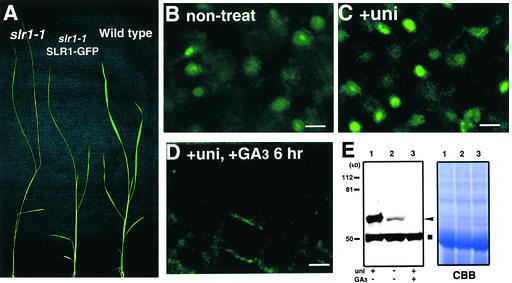
Because the transformant carrying Act1 prom::SLR1-GFP showed dwarfism, it was possible that the disappearance of GFP fluorescence was an artificial event in the plants that overproduced SLR1. To confirm the nuclear SLR1-GFP disappearance under more natural conditions, we generated SLR1-GFP transgenic plants under the control of the SLR1 promoter containing a 1.5-kb fragment of the 5′ flanking sequence. This construct complemented the slr1 mutant phenotype when it was introduced into the slr1-1 plant (Figure 3A), whereas the wild-type plants transformed with the construct showed no detectable phenotype. This finding suggests that SLR1 prom::SLR1-GFP works in a more natural manner in transgenic plants. Under these conditions, nuclear GFP fluorescence was very weak (Figure 3B). These results suggest that SLR1 may accumulate in nuclei at a low level. When the plants were grown with 1 μM uniconazol, the SLR1-GFP fluorescence became a clear spot in nuclei (Figure 3C). This fluorescence disappeared again after 6 hr of treatment with 100 μM GA3 (Figure 3D). Shoot elongation occurred by ∼48 hr after the application of GA3 (data not shown), which suggests that the disappearance of SLR1 is a much earlier event than shoot elongation.
Figure 3.
Effect of GA3 on the Amount of SLR1.
(A) Complementation of the slr1 phenotype with SLR1 prom::SLR1-GFP. Introduction of SLR1 prom::SLR1-GFP rescued the slender phenotype (middle plant). slr1-1 (left) and wild-type (right) plants are shown as control plants.
(B) to (D) Confocal microscopic images of GFP fluorescence in young leaf sections from SLR1 prom::SLR1-GFP transgenic lines. To block GA biosynthesis, the transgenic rice seedlings were pretreated with 1 μM uniconazol ([C] and [D], +uni) and then treated with 100 μM GA3 for 6 hr ([D], +uni, +GA3 6 hr). non-treat indicates normal growth conditions without any treatment (B). Bars = 10 μm.
(E) Protein gel blot analysis of the SLR1 protein. Rice seedlings were grown for 1 week under normal conditions (lane 2) or with 1 μM uniconazol (lane 1; uni). For the GA treatment, the seedlings treated with uniconazol then were sprayed with 100 μM GA3 and collected after 6 hr (lane 3). The arrowhead and square indicate the protein bands corresponding to endogenous SLR1 and SLR1 nonrelated protein, respectively. Each lane contains 10 μg of total protein. As a loading control, the Coomassie brilliant blue (CBB) staining profile is shown.
We also examined the disappearance of the SLR1 protein in response to GA3 by protein gel blot analysis. Crude proteins extracted from 1-week-old seedlings grown under various conditions were electrophoresed and then probed with anti-SLR1 antibody (Figure 3E). A weak immunoreactive band was observed in the crude extract from plants grown under normal conditions (Figure 3E, lane 2, arrowhead), whereas a strong band was seen in plants grown with 1 μM uniconazol (Figure 3E, lane 1). The strong SLR1 band was completely eliminated after 6 hr of treatment with 100 μM GA3 (Figure 3E, lane 3), whereas a 50-kD protein (square) always reacted with the antibody at a similar level in any crude extract (Figure 3E, lanes 1 to 3). All of these observations strongly suggest that the appearance or disappearance of SLR1 in nuclei depends strictly on the endogenous GA level, that SLR1 accumulation in nuclei occurs only without the GA signal, and that the disappearance of SLR1 occurs rapidly when the GA signal is present.
Functional Domain Analyses of the SLR1 Protein
The results described above indicate that the GA signal should be transmitted downstream through the derepression of SLR1 caused by the disappearance of SLR1 in nuclei. This consideration led us to speculate that the SLR1 protein possesses at least two domains for the expression or regulation of its function. One is a repression domain against the action of GA. The other functions in GA signal perception and may be involved in the protein's nuclear localization or disappearance by the GA signal.
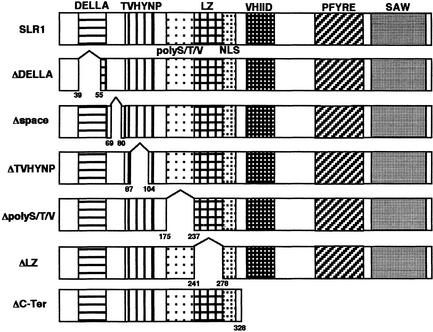
SLR1 has been characterized as a member of the GRAS family based on sequence similarity. Sequences can be subdivided into four distinct sequence motifs (Figure 4): a leucine heptad repeat (LZ), the VHIID motif, the PFYRE motif, and the SAW motif (Pysh et al., 1999). Comparison of the amino acid sequences between SLR1 and one specific subfamily of the GRAS family (GAI, RGA, Rht, and d8) reveals other conserved domains located at the N-terminal halves of this family that are not shared with other GRAS family members: a homopolymeric region rich in serine/threonine (an 11–amino acid stretch) and valine (polyS/T/V), a DELLA box, and a TVHYNP region (Figure 4). To clarify the function of each domain of SLR1, we generated six kinds of deleted SLR1 cDNAs fused with the GFP sequence and introduced them into rice under the control of the Actin1 promoter.
Figure 4.
Diagram of the Deleted Constructs for Domain Analysis of SLR1.
Each domain—DELLA, TVHYNP, Ser/Thr/Val-rich domain (polyS/T/V), LZ, nuclear localization signal (NLS), VHIID, PFYRE motif, and SAW motif—is indicated by different shading. The deleted SLR1 mutants were fused with the GFP coding sequence to generate overproducers for phenotypic analysis (see Figure 5) and subcellular localization studies (see Figure 6). The deletion points in each mutated SLR1 are shown below each box.
Based on plant height, the transgenic plants were categorized into five phenotypes (Table 1). Figure 5 shows the representative phenotypes of 10-day-old plants. The differences in plant height among the phenotypes became greater as the plants grew. In fact, severely dwarf plants reached only 5 to 30 cm even at 3 months and never headed (data not shown). As described previously, plants overproducing intact SLR1 showed a mild dwarf phenotype with GA sensitivity (Table 1, Figures 5B and 5J), whereas the overproducers with ΔDELLA, Δspace, and ΔTVHYNP showed a severe dwarf phenotype with GA insensitivity (Table 1, Figures 5C to 5E and 5K to 5M). These results confirm the importance of the DELLA and TVHYNP regions for GA signaling, as described previously for GAI/Rht/d8 (Peng et al., 1997, 1999). Similarly, the GA-insensitive dwarf phenotype of the Δspace plants demonstrated that the nonconserved spacer region between DELLA and TVHYNP also is important for GA signaling in rice.
Table 1.
Summary of Phenotypic Analysis of Transgenic Rice with SLR1 Domain Deletions
| Phenotype
|
|||||||
|---|---|---|---|---|---|---|---|
| slr1 Mutant-Like | Tall | Normal | Mild Dwarf | Severe Dwarf | |||
| Relative heighta
|
|||||||
| Construct | >150 | >120 | 100% | 60 to 80% | <50% | Totalb | GA Responsec |
| Intact | 0 (0)d | 0 (0) | 9 (25) | 21 (58) | 6 (17) | 36 | Yese |
| ΔDELLA | 0 (0) | 0 (0) | 2 (5) | 7 (19) | 28 (76) | 37 | No |
| Δspace | 0 (0) | 0 (0) | 1 (7) | 3 (21) | 10 (71) | 14 | No |
| ΔTVHYNP | 0 (0) | 0 (0) | 3 (6) | 10 (21) | 34 (72) | 47 | No |
| ΔpolyS/T/V | 0 (0) | 0 (0) | 5 (7) | 20 (29) | 44 (64) | 69 | Yes |
| ΔLZ | 0 (0) | 0 (0) | 21 (88) | 3 (12) | 0 (0) | 24 | Yes |
| ΔC-Ter | 10 (37) | 9 (33) | 8 (30) | 0 (0) | 0 (0) | 27 | Yesf |
Relative height indicates the dwarfism of each transgenic plant compared with wild type as 100%.
Total number of independent T1 transgenic plants examined.
The GA response was determined whether or not application of 100 μM GA3 caused shoot elongation.
Numbers in parentheses indicate the percentages of T1 transgenic plants in each line exhibiting each phenotype.
Yes or no indicates whether or not shoot elongation occurred.
In ΔC-Ter plants, the tall or normal phenotype plants were used to determine the GA response.
Figure 5.
Gross Morphologies of 10-Day-Old Wild-Type and Transgenic Seedlings Overproducing the Truncated SLR1-GFP Proteins with GA3 Treatment ([I] to [P]) or Nontreatment ([A] to [H]).
Plants were grown for 6 days under normal conditions and then treated with or without 100 μM GA3 for another 4 days. Because ΔDELLA, Δspace, ΔTVHYNP, and ΔpolyS/T/V transgenic plants showed a severe dwarf phenotype and never produced any fertile flowers, we used T1 generation plants for the analyses. Asterisks in (I), (J), (N), (O), and (P) show the top of the elongated fourth leaf sheath. Bars = 1 cm.
Overproducers with the SLR1 protein missing the homopolymeric serine/threonine and valine region (ΔpolyS/T/V) also displayed a severe dwarf phenotype (Table 1, Figure 5F). However, the dwarfism of the ΔpolyS/T/V plants was recovered by the application of GA3 (Figure 5N). Most plants overproducing the ΔLZ protein did not show any detectable phenotype; some exceptions showed a very mild dwarf phenotype (Table 1, Figure 5G). We confirmed by protein gel blot analysis that all transgenic lines with this construct expressed the mutated SLR1 protein at a high level (data not shown). These plants showed GA responsiveness and were elongated by GA3 application (Figure 5O).
Interestingly, the overproducers of the ΔC-Ter protein, which contained the N-terminal half but not the C-terminal half from the VHIID domain, showed a slender phenotype similar to that of the slr1 loss-of-function phenotype (Figure 5H). As with the slr1 mutation, this mutation was sterile. We could not examine the GA response of the plants because they showed a constitutive GA response phenotype. However, some plants with a mild slender phenotype (tall) or a normal phenotype were elongated by GA3 treatment (Figure 5P), which indicates that these plants retained GA responsiveness.
Nuclear Localization of the Mutated SLR1 Proteins
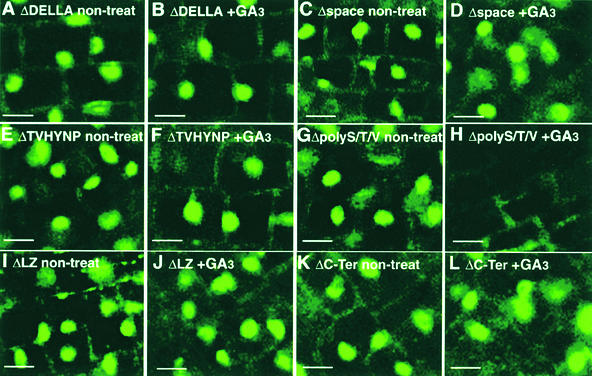
The nuclear localization of each mutated SLR1 protein also was examined with or without the application of GA3. In ΔDELLA, Δspace, and ΔTVHYNP overexpressors, the GFP fluorescence was localized in the nuclei even when GA3 was applied (Figures 6A to 6F), but the intact SLR1-GFP disappeared under the same conditions (Figure 2D). The ΔpolyS/T/V-GFP protein was localized in the nucleus without the GA treatment (Figure 6G) and disappeared in the presence of GA3 (Figure 6H), as did the intact SLR1-GFP. Shoot elongation caused by GA treatment was correlated with the disappearance of the ΔpolyS/T/V-GFP protein.
Figure 6.
Nuclear GFP Fluorescence Pattern in Young Leaves of Transgenic Plants.
The same plants shown in Figure 5 were used for analysis of the nuclear localization of SLR1-GFP. To confirm the nuclear localization of the GFP fluorescence, the positions of nuclei were always tested by DAPI staining. non-treat, nontreatment. Bars = 10 μm.
In the ΔLZ-GFP transgenic plants, the GFP fluorescence was localized in nuclei with or without the application of GA3 (Figures 6I and 6J), but the transgenic plants expressing this construct normally responded to the exogenous GA3 and elongated (Figures 5G and 5O). These observations suggest that the ΔLZ protein lost both functions of SLR1, namely, the repression of GA action and GA sensitivity. Similarly, the ΔC-Ter protein was localized in the nucleus regardless of GA3 treatment (Figures 6K and 6L). This constitutive nuclear localization of ΔC-Ter suggests that the protein lost the function of GA-dependent disappearance.
The ΔpolyS/T/V Protein Has Stronger Repression Activity Than the Intact SLR1
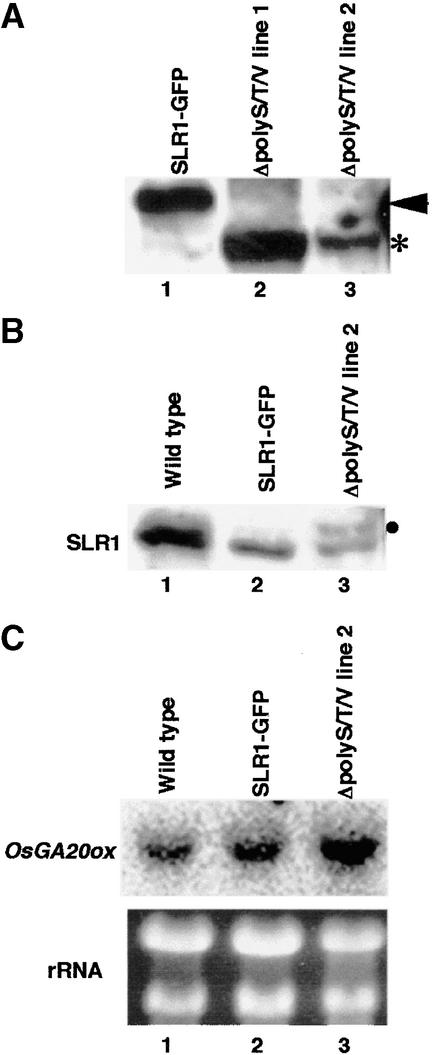
As shown in Table 1, the ΔpolyS/T/V protein induced a more severe dwarf phenotype than the intact SLR1 protein, although both transgenic plants were induced to elongate by the GA treatment. To elucidate the reason why the ΔpolyS/T/V protein induced the severe dwarf phenotype, we compared the level of protein in the transgenic plants directly (Figure 7A). The level of the ΔpolyS/T/V protein (asterisk) in the two independent severe dwarf lines (Figure 7A, lanes 2 and 3) was similar to or less than that of the intact protein (arrowhead) in the mild dwarf plant (Figure 7A, lane 1). This result shows that the severity of the dwarfism in the ΔpolyS/T/V plants is not caused by higher amounts of protein accumulation but by the characteristics of the ΔpolyS/T/V protein itself.
Figure 7.
Effect of PolyS/T/V Deletion on the Regulation of GA Action.
(A) Protein gel blot analysis of the fusion proteins in an SLR1-GFP–overproducing plant (lane 1) and two independent lines of ΔpolyS/T/V-GFP–overproducing plants (lanes 2 and 3). Crude extracts were extracted from the shoot apices of each plant. Twenty micrograms of total protein was subjected to SDS-PAGE, electroblotted, and probed with an anti-HA antibody. The arrowhead and asterisk indicate the positions of intact SLR1-GFP and ΔpolyS/T/V-GFP proteins, respectively.
(B) Endogenous SLR1 protein level in a wild-type plant (lane 1), a SLR1-GFP plant (lane 2), and the ΔpolyS/T/V-GFP line 2 plant used in (A) (lane 3). Protein extracts were subjected to SDS-PAGE and probed with anti-SLR1 antibody. Each lane contains 20 μg of total protein. The circle shows the degraded protein derived from the ΔpolyS/T/V-GFP protein.
(C) Level of OsGA20ox transcript. RNA gel blot analysis was performed with total RNA (10 μg) isolated from the shoot apices of the same plants used in (B).
All of these samples were prepared from adult plants grown for 30 days.
We also compared the steady state levels of endogenous SLR1 and the OsGA20ox transcript in wild-type, SLR1-GFP, and ΔpolyS/T/V transgenic plants. The ΔpolyS/T/V plants showed less SLR1 protein and more OsGA20ox transcript than the wild-type or the intact SLR1-GFP plants (Figures 7B and 7C). An extra minor immunoreactive band was seen in ΔpolyS/T/V transgenic line 2 (Figure 7B, circle), which corresponds to the degraded version of ΔpolyS/T/V protein because this extra band also was recognized against anti-HA antibody (data not shown). These observations confirm that GA action is more strongly suppressed by the ΔpolyS/T/V protein than by the intact SLR1 protein. Consequently, the polyS/T/V region may be involved in the regulatory mechanism of the SLR1 suppressive function (see Discussion).
SLR1 Forms a Homodimer through the Interaction of the LZ Region
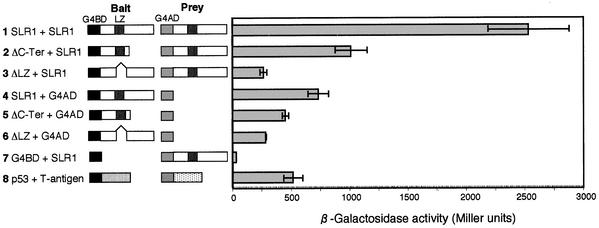
As shown in Figure 5H, ΔC-Ter overexpressors showed a slender phenotype similar to that of the slr1 loss-of-function mutant. This result indicates that the ΔC-Ter protein may act in a dominant negative fashion to interfere with the function of the endogenous SLR1 protein. On the other hand, transgenic plants expressing ΔLZ did not exhibit any significant phenotype. These results suggest that SLR1 functions as a dimer through the LZ domain and that the dominant negative effect of ΔC-Ter can be explained by the interaction between ΔC-Ter containing the LZ domain and the intact SLR1 protein to inhibit the function of SLR1. To examine the dimer formation of SLR1, we tested the homodimerization of the SLR1 protein directly using the yeast two-hybrid system. When the intact SLR1 cDNA was fused with the GAL4 DNA binding domain (G4BD) on the bait plasmid, high lacZ activity was induced, whereas low lacZ activity was induced when the intact SLR1 was fused with the GAL4 activation domain (G4AD) on the prey construct (Figure 8, line 4 versus line 7). This result indicates that the SLR1 protein has autoactivation activity probably caused by its transactivation domain, as reported previously by Ogawa et al. (2000). The combination of G4BD-SLR1 and G4AD-SLR1 induced a high activity of lacZ relative to the SLR1 autoactivation activity (Figure 8, line 1 versus line 4), indicating that SLR1 was able to interact with itself to form a homodimer. The lacZ activity was decreased to a level similar to that of the negative control by deletion of the LZ region (Figure 8, line 3 versus line 6). This result indicates that the LZ region is a critical domain for SLR1 homodimerization. In contrast to ΔLZ, ΔC-Ter interacted with the intact SLR1 protein, and it had less interaction activity than that between intact proteins (Figure 8B, line 1 versus line 2) but still much more activity than the ΔLZ protein and the positive control for this assay system (Figure 8, line 2 versus lines 3 and 8). These results strongly suggest that the dominant negative phenotype in the ΔC-Ter overproducers was caused by the interference of the ΔC-Ter protein with the dimer formation of SLR1.
Figure 8.
Dimer Formation of SLR1 through the LZ Domain in the Yeast Two-Hybrid Assay.
For bait constructs (Bait), full-length SLR1, ΔLZ, or ΔC-Ter (see Figure 4) was fused with the G4BD. For the prey construct (Prey), the full-length SLR1 was fused with the G4AD. The relative lacZ activity of various combinations is presented. p53 and T-antigen (simian virus 40 T-antigen) were used as positive controls to evaluate relative binding affinity. For each pairwise combination, five individual transformants were used to measure relative lacZ activity. Error bars represent standard deviations.
DISCUSSION
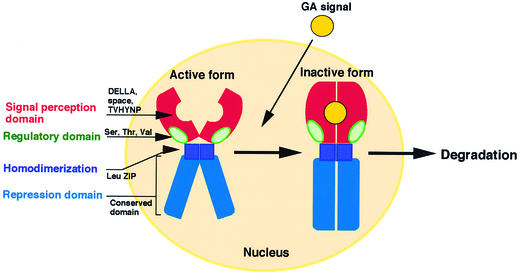
We have demonstrated that the SLR1 protein functions as a negative regulator of GA action in the nucleus and that the GA signal is transmitted downstream through the disappearance of the nucleus-localized SLR1 protein. To confirm the suppressive function of SLR1 and the fact that it is regulated by the GA signal, we also performed functional analyses of several characteristic domains of SLR1. We have summarized the function of each domain in Figure 9. The three parts at the N-terminal end (DELLA, space, and TVHYNP, colored red) act as the GA signal perception domain, which is essential for the GA signal–triggered disappearance of SLR1 in the nucleus. The homopolymeric Ser, Thr, and Val (polyS/T/V) domain behind the N-terminal domain (green) may function as a regulatory domain for SLR1's repressor activity. The LZ region (dark blue) is essential for dimer formation. The C-terminal region (blue), containing the conserved VHIID domain shared with other GRAS family genes, represses the action of GA.
Figure 9.
Scheme of the Functional Domains of SLR1 for the GA Signaling Pathway.
The GA signal (yellow circle) is received by the signal perception domain, which consists of the conserved DELLA and TVHYNP regions and the nonconserved spacer region (red). The SLR1 received with the GA signal is degraded rapidly and disappears in the nuclei. The leucine zipper domain (dark blue) is essential for dimer formation by SLR1. The C-terminal half of SLR1 (blue), which is shared with other GRAS family genes, functions as a repression domain to prevent the action of GA. The Ser/Thr/Val-rich region (green) may work as a regulatory domain through the target sites of O-GlcNAcylation–phosphorylation regulation (see text for details).
Derepression of the Action of GA Can Be Achieved by the Disappearance of the Nuclear SLR1 Protein
Previous genetic analyses of loss-of function mutants of SLR1 in rice (Ikeda et al., 2001) and a gain-of-function mutant of GAI in Arabidopsis (Peng et al., 1997) indicate that SLR1/GAI functions as a repressor for the action of GA and that active GA derepresses the function of these proteins. In this study, we have provided some biochemical evidence for this GA-dependent derepression mechanism of SLR1. The SLR1-GFP fusion protein, which functions in vivo as a repressor in the same way that the intact SLR1 protein does, was localized in the nucleus under normal conditions, and GA application induced the disappearance of the nucleus-localized SLR1-GFP (Figures 2 and 3). These observations demonstrate that SLR1 works as a repressor in nuclei and that the GA signal from upstream causes the nuclear SLR1 to disappear, resulting in the release of the repressive state of the action of GA to the signal-transducing state.
The steady state level of the SLR1 protein was regulated by the level of GA, and the application of GA3 or the inhibition of endogenous GA biosynthesis by uniconazol caused the downregulation or upregulation, respectively, of the SLR1 protein level (Figure 3E). Furthermore, overexpression analysis of SLR1-GFP also showed that GA-regulated responses, such as shoot elongation and OsGA20ox expression, were regulated quantitatively by the level of SLR1 protein in nuclei (Figures 1, 3E, 5B, and 5J). These observations suggest that the level of nucleus-localized SLR1 is regulated quantitatively by the upstream GA signal and that the downstream GA response(s) also is controlled quantitatively by the level of SLR1 protein in nuclei.
On the other hand, ΔDELLA-GFP, which has the same internal deletion as the gai mutant protein (Peng et al., 1997), and ΔTVHYNP-GFP, which has a similar internal deletion as the maize D8-2023 mutant protein (Peng et al., 1999), were localized constitutively in nuclei to induce the GA-insensitive severe dwarf phenotype (Figures 5 and 6). These results support the hypothesis that the internal deletions or truncations in the N-terminal domain of SLR1/GAI members lock the proteins into a conformation that can no longer respond to the GA signal (Silverstone and Sun, 2000). The results also indicate that such a conformational change in SLR1/GAI proteins causes their constitutive nuclear localization even in the presence of GA.
Four Distinct Domains for Expressing SLR1 Function
GA Signal Perception Domain
The description of SLR1 function above indicates that SLR1 should contain at least two distinguishable functions, GA signal perception and GA repression. As reported for gai/Rht-B1/Rht-D1/D8 (Peng et al., 1997, 1999), the N-terminal region containing the DELLA and TVHYNP regions is important for GA signaling. In the deletion analyses of SLR1, the deleted proteins ΔDELLA and ΔTVHYNP were localized constitutively in the nucleus with or without GA3 treatment, and their overproducers showed a GA-insensitive severe dwarf phenotype (Figures 5 and 6). These observations confirm the previous results for gai/Rht-B1/Rht-D1/D8 and indicate that these N-terminal regions in SLR1 work as a GA signal perception domain and that the regions are important for the GA-triggered disappearance of SLR1. In addition to the importance of the conserved DELLA and TVHYNP regions, our results show that the nonconserved spacer region between DELLA and TVHYNP also plays an important role in interacting with the GA signal. This finding suggests that the spacing between the conserved DELLA and TVHYNP is necessary for regular signal perception. It is possible that the tertiary arrangement of the DELLA and TVHYNP regions is important for the perception of a GA signal molecule and that the deletion of the spacer region inhibits the correct conformation of this N-terminal region.
Repression Domain
The suppressive function of SLR1 depends on the C-terminal region, which contains the VHIID, PFYRE, and SAW domains (Figure 4). This fact is supported by the observations that the deleted proteins missing the N-terminal regions (ΔDELLA, Δspace, and ΔTVHYNP) had a constitutive suppressive function, as mentioned above (Figure 5), and that the null alleles of slr1 often contained nucleotide substitutions or deletions in the C-terminal region. One allele (slr1-4) with a nucleotide substitution located just 16 nucleotides upstream from the stop codon shows the slender phenotype (Ikeda et al., 2001), which indicates that several amino acid residues at the C-terminal end are essential for the SLR1 suppressive function. Another allele with one amino acid exchange in the SAW domain (Thr-606 to Pro) also showed the mutant phenotype (H. Itoh and M. Matsuoka, unpublished results), which indicates that the SAW domain is important for the suppressive function. The importance of the C-terminal half to the suppression activity also has been described for gai/RGA (Peng and Harberd, 1993; Peng et al., 1997; Silverstone et al., 1998). Analyses of the Arabidopsis gai intragenic suppressor mutant have demonstrated that all of these suppressor alleles possess additional mutations in their C-terminal halves that disrupt the GAI open reading frame. Moreover, one of the Arabidopsis rga strong alleles, rga-2, has a missense mutation in the PFYRE domain of its C-terminal half.
Dimer Formation
The rice SLR1 protein contains the LZ domain, which is conserved among plant GRAS family proteins. As with other transacting factors, the LZ domain promotes dimer formation, and a protein missing this region failed to interact with the intact protein in yeast cells (Figure 8). The lack of a specific phenotype of the overproducer with the ΔLZ protein suggests that dimer formation by SLR1 is essential for the SLR1 suppressive function. The role of the LZ domain in dimer formation is consistent with the dominant negative phenotype of the plants overproducing the ΔC-Ter protein, which contains the N-terminal half with LZ but not the repression domain. The presence of the dominant negative phenotype caused by the ΔC-Ter protein strongly suggests that the protein interacts with the intact protein produced by the endogenous gene and that the heterodimer does not retain the repression function. Interestingly, the nuclear localization of the ΔLZ protein was observed in the GA-treated plants, even though ΔLZ contains the GA signal perception domain (Figures 4 and 6). This observation indicates that dimer formation also may be necessary for the GA-dependent disappearance of SLR1 in nuclei (Figure 6).
All of the results described above indicate the importance of dimer formation for the transmission of the GA signal and for the disappearance of SLR1 in nuclei. However, this does not mean that dimer formation is sufficient to remove SLR1 from nuclei. In fact, the ΔC-Ter protein, which contains the GA signal perception and LZ domains and can form the homodimer, did not disappear with GA treatment (Figure 6). This result indicates that the C-terminal region also is essential for the signal-dependent disappearance. It is possible that the perception of the GA signal at the N-terminal region may change the C-terminal conformation and cause the SLR1 protein to be targeted by proteinase(s) for degradation, similar to the situation in the auxin and light signal transduction pathways (Gray et al., 1999; Osterlund et al., 2000).
Regulatory Domain
Overexpression of the ΔpolyS/T/V protein caused the severe dwarf phenotype (Table 1, Figure 5), which indicates that the S/T/V-rich domain may work negatively against the function of SLR1. The application of GA3 restored the dwarf phenotype and elongated the overproducers of the ΔpolyS/T/V protein, which indicates that the protein can receive the GA signal to disappear in the nuclei (Figure 6). Based on the characteristic structure of polyS/T/V, this region has been suggested to be a target site for O-linked GlcNAc (O-GlcNAc) modification (Silverstone et al., 1998). According to the proposed model of the functional regulation of GAI/RGA, these proteins may be modified through O-GlcNAcylation at this region by the O-GlcNAc transferase activity of the SPINDLY protein, and this modification may cause their repressive function to increase (Silverstone et al., 1998; reviewed by Thornton et al., 1999). On the other hand, our observation of the increased repressive activity of ΔpolyS/T/V suggests that the polyS/T/V domain has a decreasing effect on its repressive function. This discrepancy may be caused by the difference in the regulatory mechanism between rice and Arabidopsis. For example, the length of the S/T sequence of rice SLR1 (11 amino acids) is much longer than that of Arabidopsis GAI (3 amino acids) or RGA (5 amino acids and 8 amino acids). Another possibility is that this polyS/T/V region is targeted for phosphorylation and that the phosphorylated SLR1 has lower repression activity. Indeed, some proteins are modified competitively or reciprocally with glucosylation and phosphorylation at the same sites (regions) in animal systems (Kelly et al., 1993; Chakraborty et al., 1994; Chou et al., 1995). In addition, studies related to these protein modifications have revealed that phosphorylation is a key step in the targeted protein degradation pathway by proteasomes (Aberle et al., 1997; Skowyra et al., 1997; Winstone et al., 1999). In this situation, the deletion of the polyS/T region causes a defect in the phosphorylation of SLR1, increasing its half-life. We are now elucidating the precise function of this region and the regulatory mechanism of the repressive function of SLR1 through O-GlcN-Acylation and phosphorylation.
METHODS
Construction of SLR1-GFP and Its Derivatives
For the construction of the chimera consisting of the SLR1 cDNA fused to green fluorescent protein (GFP) at the 3′ side and with hemagglutinin (HA) at the 5′ side, the SLR1 cDNA sequence was amplified by polymerase chain reaction (PCR) using primers 5′-CCCCCGGGGAAATGAAGCGCGAGTACCAA-3′ (5′ side, with the underlined SmaI site as a linker) and 5′-CGTCTAGACGCCGCGGCG-ACGCGCCA-3′ (3′ side, with the underlined XbaI site as a linker). The resulting PCR product possessed one additional Glu at the front of the first Met, and the stop codon was replaced with the XbaI site for in-frame fusion with GFP at its C-terminal end. The PCR product and a short synthetic DNA fragment (5′-CCGGGCCCCCATGGA-GTACGACGTACCAGATTACGCTCCCGGGCC-3′; ApaI and SmaI sites underlined, start codon in boldface), which encodes the amino acids of the HA epitope (Met-Asp-Tyr-Asp-Val-Ser-Val-Tyr-Ala), were ligated simultaneously at the ApaI–XbaI site of pBluescript SK+ (pBsSK+; Stratagene, La Jolla, CA). This clone was sequenced to confirm that there was no nucleotide substitution. The clone was digested with ApaI and XbaI to obtain the HA-SLR1 fragment, which was ligated into the same site of the cassette vector containing the Act1 prom::GFP-NOS terminator, modified from the CaMV35S-GFP-NOS cassette vector (Chiu et al., 1996). The cassette vector possesses the KpnI site at the front of the Actin1 promoter and SpeI behind the GFP stop codon. This clone was digested with KpnI and blunted and then redigested with SpeI. Finally, the fragment (Act1 prom::HA-SLR1-GFP) was ligated into the binary vector pBI101-Hm2 (Ohta et al., 1990) at the blunted HindIII and XbaI sites.
To construct ΔC-Ter, we amplified the SLR1 cDNA by PCR using the 5′ primer 5′-CCCCCGGGGAAATGAAGCGCGAGTACCAA-3′ and the 3′ primer 5′-CGTCTAGAGTGGGCGAACTTGAGGTAG-GGGC-3′. The resulting PCR product was inserted into the SmaI–XbaI site of pBsSK+. To create the internal deletion constructs, the PCR-amplified N-terminal fragment with SmaI (5′ side) and BamHI (3′ side) and the C-terminal fragment with BamHI (5′ side) and XbaI (3′ side) were ligated simultaneously into the SmaI (5′)–XbaI (3′) site of pBsSK+. All of the deleted SLR1 cDNAs were sequenced to confirm the fact that no nucleotide substitution had occurred during amplification. These fragments were ligated into the cassette vector containing Act1 prom::GFP-NOS, and the whole inserts were moved into the binary vector as described above.
Antibody Production
A DNA fragment encoding the gibberellin (GA) signal perception domain (Met-1 to Val-133) of SLR1 cDNA was amplified by PCR using specific primers and cloned into a pET32a vector (Novagen, Madison, WI). The cloned DNA fragment was verified without nucleotide substitutions during PCR by sequencing. The resulting recombinant N-terminal SLR1 protein was overexpressed in Escherichia coli. The overproduced recombinant protein was purified using Talon Metal Affinity Resin (Clontech, Palo Alto, CA) from an insoluble fraction under denaturing conditions, according to the manufacturer's instructions. Finally, the recombinant protein was separated by SDS-PAGE, and the gel was used directly for the production of rabbit polyclonal antibodies.
Protein Gel Blot Analysis
Protein was extracted by grinding the seedlings with an equal volume of 2 × sample buffer (1 × sample buffer is 67.5 mM Tris-HCl, pH 6.8, 2% [w/v] SDS, 10% [w/v] glycerol, 0.01% [w/v] bromphenol blue, and 0.1 M [w/v] DTT) and boiling for 5 min. Protein samples were separated by 8% SDS-PAGE and transferred to a Hybond enhanced chemiluminescence membrane (Amersham Pharmacia Biotech, Little Chalfont, UK) by semidry blotting. The blots were incubated with anti-SLR1 or anti-HA (Medical Biological Labs, Nagoya, Japan) antiserum raised in rabbit and then with goat anti–rabbit IgG horseradish peroxidase–conjugated secondary antibody. Detection of the peroxidase activity was performed according to the instruction manual from Pierce (Rockford, IL).
RNA Isolation and RNA Gel Blot Analysis
Total RNA was isolated from seedlings by the method described by Chomczynski and Sacchi (1987). Ten micrograms was electrophoresed on a 1% agarose gel and then transferred to a Hybond N+ membrane (Amersham Pharmacia Biotech). Hybridization was performed at 65°C in 6 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate), 5 × Denhardt's solution (1 × Denhardt's solution is 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% BSA), 0.5% SDS, 10% dextran sulfate, and 0.1 mg·mL−1 denatured salmon sperm DNA. DNA probes were labeled with α-32P-dCTP. Filters were washed twice with 2 × SSC and 0.1% SDS at 65°C for 30 min and once with 0.2 × SSC and 0.1% SDS at 65°C for 10 min.
Microscopic Observation
Young leaves of transgenic rice (Oryza sativa) plants were sectioned by razor blade, and the sections were placed on glass slides. Samples were soaked in 2 μg·μL−1 4′,6-diamidino-2-phenylindole dihydrochloride n-hydrate (DAPI; Dojindo, Kumamoto, Japan) solution for visualization of the nucleus in analyses of the nuclear localization of SLR1-GFP derivatives. The stained samples were observed through a confocal microscanning laser microscope (FV500; Olympus, Tokyo, Japan). The laser scan images were obtained with a combination of 488-nm laser excitation and 505- to 525-nm emission filters. The images obtained were recorded automatically.
Yeast Two-Hybrid Assay
The Matchmaker Two-Hybrid System (Clontech) was used. The intact SLR1 cDNA was inserted into the yeast expression vector pACT2, and the SLR1 derivatives were ligated into pGBT9. To determine the interaction affinity, we used yeast strain Y187 (MATα, ura3-52, his3-200, ade2-101, trp1-901, leu2-3, 112, gal4Δ, met−, gal80Δ, URA3::GAL1UAS-GAL1TATA-lacZ). The β-galactosidase liquid assay was performed according to the Clontech manual.
Plant Transformation, Growth Conditions, and Chemical Treatment
Binary vectors were introduced into Agrobacterium tumefaciens strain EHA101 (Hood et al., 1986) by electroporation. Rice transformation was performed as described by Hiei et al. (1994). Wild-type rice plants (cv T-65) and slr1-1 mutants were used for the analyses. Transgenic plants were selected on medium containing 50 mg·L−1 hygromycin. Hygromycin-resistant plants were transplanted to soil and grown at 30°C in a 16-hr-light/8-hr-dark cycle.
For the analyses of GA response, the seedlings of each transgenic plant (SLR1-GFP) were grown in water. Several days before analysis, the plants were transferred to water containing 100 μM GA3.
GA Induction of Shoot Elongation
To investigate the role of GA in the elongation of the second leaf sheath, 10 rice seed from wild type and SLR1-GFP overproducers were sterilized and allowed to imbibe at 30°C for 1 day. The seed were placed on agar containing various concentrations of GA3 and incubated at 30°C under continuous light. After 6 days of incubation, the lengths of the second leaf sheaths were measured. The SLR1-GFP overproducers were used in this experiment, which has the same genotype confirmed by genomic DNA gel blot analysis.
Acknowledgments
We are grateful to Dr. Yuji Kamiya (RIKEN, Saitama, Japan) for providing the plasmid for OsGA20ox and Dr. Yasuo Niwa (Shizuoka Prefectural University, Shizuoka, Japan) for providing the CaMV35S-GFP-NOS cassette vector. We also thank Dr. Kunio Yasuda and Dr. Takashi Hashimoto (Nara Institute of Science and Technology, Nara, Japan) for their excellent help with confocal microscopy. This work was supported in part by a Grant-in-Aid from the Program for the Promotion of Basic Research Activities for Innovative Biosciences (M.M.), Grant-in-Aid for Center of Excellence (M.M.), the Special Coordination Fund of the Science and Technology Agency (M.M.), and a research fellowship from the Japan Society for the Promotion of Science (H.I.).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010319.
References
- Aberle, H., Bauer, A., Stappert, J., Kispert, A., and Kemler, R. (1997). β-Catenin is a target for the ubiquitin-proteasome. EMBO J. 16, 3797–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty, A., Saha, D., Bose, A., Chatterjee, M., and Gupta, N.K. (1994). Regulation of elF-2 α-subunit phosphorylation in reticulocyte lysate. Biochemistry 33, 6700–6706. [DOI] [PubMed] [Google Scholar]
- Chiu, W.-L., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6, 325–330. [DOI] [PubMed] [Google Scholar]
- Chomczynski, P., and Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 162, 156–159. [DOI] [PubMed] [Google Scholar]
- Chou, T.Y., Gerald, W.H., and Dang, C.V. (1995). c-Myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas. J. Biol. Chem. 270, 18961–18965. [DOI] [PubMed] [Google Scholar]
- Davies, P.J. (1995). Plant Hormones: Physiology, Biochemistry and Molecular Biology. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Gray, W.M., Carlos del Pozo, J., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., Ma, H., and Estelle, M. (1999). Identification of an SCF ubiquitin–ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harberd, N.P., and Freeling, M. (1989). Genetics of dominant gibberellin-insensitive dwarfism in maize. Genetics 121, 827–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden, P., and Phillips, A.L. (2000). Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 12, 523–530. [DOI] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Hood, E.E., Helmer, G.L., Fraley, R.T., and Chilton, M.-D. (1986). The hypervirulence of Agrobacterium tumefaciens A281 is en-coded in a region of pTiBo542 outside of T-DNA. J. Bacteriol. 168, 1291–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda, A., Tanaka, S., Yamaguchi, J., and Futsuhara, Y. (1999. a). Character expression and mode of inheritance of a slender mutant with constitutive gibberellin-response in rice [Japanese]. Sci. Rep. Fac. Agric. Meijo Univ. 35, 7–13. [Google Scholar]
- Ikeda, A., Tanaka, S., Yamaguchi, J., and Futsuhara, Y. (1999. b). Characterization of gibberellin signal transduction pathway in the slender mutant with constitutive gibberellin-response in rice [Japanese]. Sci. Rep. Fac. Agric. Meijo Univ. 35, 15–22. [Google Scholar]
- Ikeda, A., Ueguchi-Tanaka, M., Sonoda, Y., Kitano, H., Koshioka, M., Futsuhara, Y., Matsuoka, M., and Yamaguchi, J. (2001). slender rice, a constitutive response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Kawaide, H., Chen, X., Kamiya, Y., and Matsuoka, M. (1999). The gene encoding tobacco gibberellin 3β-hydroxylase is expressed at the site of GA action during stem elongation and flower development. Plant J. 20, 15–24. [DOI] [PubMed] [Google Scholar]
- Kelly, W.G., Dahmus, M.E., and Hart, W.G. (1993). RNA polymerase II is a glycoprotein. J. Biol. Chem. 268, 10416–10424. [PubMed] [Google Scholar]
- Koornneef, M., Elgersma, A., Hanhart, C.J., von Loenen-Martinet, E.P., van Rign, L., and Zeevaart, J.A.D. (1985). A gibberellin insensitive mutant of Arabidopsis thaliana. Physiol. Plant. 65, 33–39. [Google Scholar]
- McElroy, D., Zhang, W., Cao, J., and Wu, R. (1990). Isolation of an efficient promoter for use in rice transformation. Plant Cell 2, 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, M., Kusano, T., Katsumi, M., and Sano, H. (2000). Rice gibberellin-insensitive gene homologue, OsGAI, encodes a nuclear- localized protein capable of gene activation at the transcriptional level. Gene 245, 21–29. [DOI] [PubMed] [Google Scholar]
- Ohta, S., Mita, S., Hattori, T., and Nakamura, K. (1990). Construction and expression in tobacco of β-glucuronidase (GUS) reporter gene containing an intron within the coding sequence. Plant Cell Physiol. 31, 805–813. [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. [DOI] [PubMed] [Google Scholar]
- Peng, J., and Harberd, N.P. (1993). Derivative alleles of the Arabidopsis gibberellin-insensitive (gai) mutation confer a wild-type phenotype. Plant Cell 5, 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., Carol, P., Richards, D.E., King, K.E., Cowling, R.J., Murphy, G.P., and Harberd, N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11, 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., et al. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261. [DOI] [PubMed] [Google Scholar]
- Pysh, L.D., Wysocka-Diller, J.W., Camilleri, C., Bouchez, D., and Benfey, P.N. (1999). The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18, 111–119. [DOI] [PubMed] [Google Scholar]
- Rebers, M., Kaneta, T., Kawaide, H., Yamaguchi, S., Yang, Y.Y., Imai, R., Sekimoto, H., and Kamiya, Y. (1999). Regulation of gibberellin biosynthesis genes during flower and early fruit development of tomato. Plant J. 17, 245–250. [DOI] [PubMed] [Google Scholar]
- Silverstone, A.L., and Sun, T.P. (2000). Gibberellins and the green revolution. Trends Plant Sci. 5, 1–2. [DOI] [PubMed] [Google Scholar]
- Silverstone, A.L., Chang, C., Krol, E., and Sun, T.P. (1997. a). Developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant J. 12, 9–19. [DOI] [PubMed] [Google Scholar]
- Silverstone, A.L., Mak, P.Y.A., Martinez, E.C., and Sun, T.P. (1997. b). The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146, 1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Ciampaglio, C.N., and Sun, T.P. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10, 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra, D., Craig, K.L., Tyers, M., Elledge, S.J., and Harper, J.W. (1997). F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91, 209–219. [DOI] [PubMed] [Google Scholar]
- Thornton, T.M., Swain, S.M., and Olszewski, N.E. (1999). Gibberellin signal transduction presents … the SPY who O-GlcNAc'd me. Trends Plant Sci. 4, 424–428. [DOI] [PubMed] [Google Scholar]
- Toyomasu, T., Kawaide, H., Sekimoto, H., von Numers, C., Phillips, A.L., Hedden, P., and Kamiya, Y. (1997). Cloning and characterization of a cDNA encoding gibberellin 20-oxidase from rice (Oryza sativa) seedlings. Physiol. Plant. 99, 111–118. [Google Scholar]
- Winkler, R.G., and Freeling, M. (1994). Physiological genetics of the dominant gibberellin-nonresponsive maize dwarf, Dwarf8 and Dwarf9. Planta 193, 341–348. [Google Scholar]
- Winstone, J.T., Strack, P., Beer-Romero, P., Chu, C.Y., Elledge, S.J., and Harper, J.W. (1999). The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulated IκBα ubiquitination in vitro. Genes Dev. 13, 270–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, S., Smith, M.W., Brown, R.G., Kamiya, Y., and Sun, T.P. (1998). Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10, 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]