Abstract
Potassium ions (K+) are the most abundant cations in plants and are necessary for cell growth. Arabidopsis shy3-1 mutant plants have a short hypocotyl, small leaves, and a short flowering stem, and these defects result from decreased cell expansion. The semidominant shy3-1 mutation changes an amino acid in KT2/KUP2, a K+ transporter related to the Escherichia coli Kup protein. Second mutations in the KT2/KUP2/SHY3 gene, including presumed null mutations, suppress the shy3-1 phenotypes. Plants with these intragenic suppressor mutations appear similar to wild-type plants, suggesting that KT2/KUP2/SHY3 acts redundantly with other genes. Expression of the shy3-1 mutant version of KT2/KUP2/SHY3 in wild-type plants confers shy3-1–like phenotypes, indicating that shy3-1 probably either causes a gain of function or creates an interfering protein. The shy3-1 mutation does not eliminate the ability of the KT2/KUP2 cDNA to rescue the growth of a potassium transport-deficient E. coli mutant. A PSHY3::GUS fusion is expressed in growing portions of the plant. These results suggest that KT2/KUP2/SHY3 mediates K+-dependent cell expansion in growing tissues.
INTRODUCTION
Potassium ions (K+) mediate many physiological responses in plants, including stomatal opening and closing, leaf movements, and regulation of membrane polarization. Perhaps the most fundamental role of K+ in plants is in cell growth. Plant cells grow by loosening their cell walls and taking up water (Cosgrove, 1993). K+ provide the necessary osmotic potential for water uptake (Keller and Van Volkenburgh, 1996; Claussen et al., 1997), and intracellular turgor pressure drives cell expansion. Only a subset of cells in a plant grow at any one time, and proper delivery of K+ to growing regions is essential for correct growth and morphology.
Multiple transporters mediate the uptake and movement of K+ in plants (Kochian and Lucas, 1988; Maathuis and Sanders, 1996; Maathuis et al., 1997; Fox and Guerinot, 1998; De Boer, 1999; Rodríguez-Navarro, 2000). K+ transporters in root epidermal and cortical cells take up K+ from soil. Plasmodesmata connect the cytoplasms of root epidermal, cortical, and endodermal cells and allow K+ to traverse the endodermis symplastically. Outward-rectifying K+ channels allow ions to exit stelar cells and enter the apoplast, the extracellular space that is contiguous with the xylem (Wegner and Raschke, 1994; Roberts and Tester, 1995; Gaymard et al., 1998). Ions then can move to the aerial portions of the plant through the xylem, and unidentified transporters import K+ into shoot meristem, stem, and leaf cells. Plasmodesmata connect many of these shoot cells, and K+ may reach some of them symplastically. A similar internal transport stream may supply roots, in which the most active growth occurs in regions where immature cells may have low uptake capacity.
Current studies aim to understand how different plant K+ transporters mediate K+ nutrition and K+-dependent growth. Genes encoding plant K+ transporters have been isolated by sequence similarity to K+ transporters from other organisms and by complementing Saccharomyces cerevisiae potassium uptake mutants (De Boer, 1999; Maathuis et al., 1997; Fox and Guerinot, 1998; Rodríguez-Navarro, 2000). Plants have several genes encoding members of the Shaker superfamily of potassium channels. Shaker proteins have six transmembrane domains, and the superfamily includes both inward- and outward-rectifying potassium channels.
Characterization of Arabidopsis plants with mutations in two genes encoding members of the Shaker superfamily has revealed the physiological roles of the corresponding channels. Mutant akt1 plants grew poorly on medium containing micromolar concentrations of potassium in the presence of ammonium ions, indicating that AKT1 mediates high-affinity K+ uptake into roots (Hirsch et al., 1998). The skor mutant is defective in a stelar potassium (K) outwardly rectifying channel (Gaymard et al., 1998). The SKOR gene is expressed in stelar cells surrounding the vasculature, and skor1 mutant plants contain 50% less potassium in their leaves than wild-type plants (Gaymard et al., 1998). Thus, SKOR is thought to load potassium into the xylem for transport to the shoot (Gaymard et al., 1998). Neither of these mutants has obvious morphological defects when grown with an ample potassium supply, suggesting that other ion transporters can compensate for the absence of either AKT1 or SKOR.
A second family of K+ transporters, called HAK, KT, or KUP, has multiple members in most plants examined (Quintero and Blatt, 1997; Santa-María et al., 1997; Fu and Luan, 1998; Kim et al., 1998; Rubio et al., 2000; Maser et al., 2001; Rigas et al., 2001). Arabidopsis has 13 genes encoding members of this family. These proteins have 12 or 14 predicted transmembrane domains, and they are similar to the Escherichia coli Kup transporter (Schleyer and Bakker, 1993) as well as to potassium transporters of the fungi Schwan-niomyces occidentalis and Neurospora crassa (Bañuelos et al., 1995; Haro et al., 1999). HvHAK1, AtKT1/AtKUP1, and AtHAK5 can mediate high-affinity K+ or rubidium ion (Rb+) uptake into yeast or Arabidopsis cells (Santa-María et al., 1997; Fu and Luan, 1998; Kim et al., 1998; Rubio et al., 2000).
The tiny root hairs (trh1-1) mutation is a T-DNA insertion in the KT3/KUP4 gene encoding a member of this family (Rigas et al., 2001). Plants carrying this mutation have small root hairs that do not elongate properly, indicating that the TRH1/KT3/KUP4 transporter promotes expansion of root hairs, presumably by mediating potassium uptake into these cells.
The Arabidopsis shy3-1 mutation is semidominant and causes a short hypocotyl (Reed et al., 1998). Here we report that a missense mutation in the KT2/KUP2 gene decreases hypocotyl and leaf cell expansion in the shy3-1 mutant. Null mutations in the gene do not have such dramatic phenotypes, suggesting that the shy3-1 allele may alter regulatory properties of the protein or encode an interfering allele.
RESULTS
shy3-1 Mutation Decreases Cell Expansion in the Shoot
We isolated the shy3-1 mutant on the basis of its short hypocotyl phenotype (Reed et al., 1998). shy3-1 plants also had smaller leaves and shorter inflorescence stems than wild-type plants (Figure 1A; see also Figures 4A and 5D below). At a stage at which plants had six true leaves, the youngest leaves (numbers 5 and 6) of mutant plants were 30% smaller than corresponding wild-type leaves (Figure 1A). Moreover, the overall rate of development was similar. For example, new leaves were initiated at approximately the same rate in wild-type and mutant plants, and in short days shy3-1 plants actually flowered slightly earlier than wild-type plants (Reed et al., 1998). In contrast to stems and leaves, in most experiments roots of wild-type and shy3-1 seedlings had similar lengths (Reed et al., 1998; data not shown).
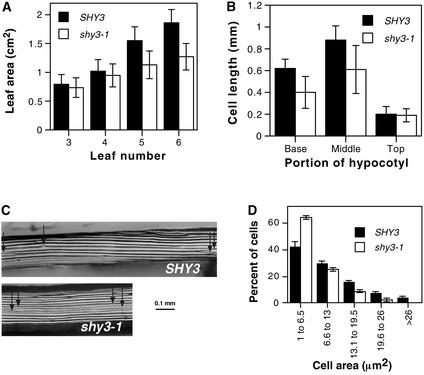
Figure 1.
Phenotypes of shy3-1 Mutant Plants.
(A) Leaf areas of 4-week-old wild-type and shy3-1 plants ±sd (n = 12). Sizes of leaves 3 and 4 were not significantly different between SHY3 and shy3-1 (P > 0.2). Sizes of leaves 5 and 6 were significantly different between SHY3 and shy3-1 (P < 0.005 for leaf 5 and P < 0.001 for leaf 6).
(B) Hypocotyl epidermal cell lengths of different portions of hypocotyls from shy3-1 mutant and wild-type seedlings after 7 days of dark growth. Base, 2 to 5 cells from the root; Middle, 6 to 15 cells from the root; Top, 16 to 20 cells from the root. Data are means of 25 cells from six different seedlings ±sd. Cells in the base and middle of hypocotyls were significantly different in length between wild-type and shy3-1 mutant (P < 0.001), whereas those at the top were not significantly different (P > 0.5).
(C) Superglue imprints of corresponding portions from the middle of seven-day-old dark-grown wild-type (SHY3) and shy3-1 hypocotyl epidermis showing differences in epidermal cell lengths. Arrows indicate ends of representative cells.
(D) Epidermal cell area distribution of the sixth leaf of four-week-old wild-type and shy3-1 plants. Data are means of distributions from three different leaves ±sd (n = 75 cells/leaf).
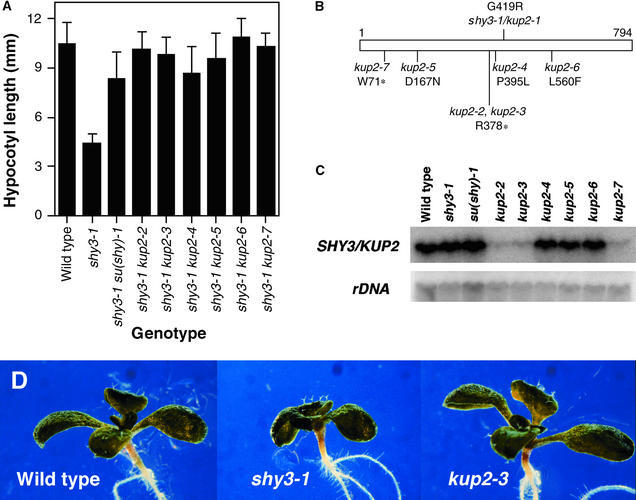
Figure 4.
Recapitulation of the shy3-1/kup2-1 Phenotype in Transgenic Plants Overexpressing shy3-1/kup2-1.
(A) Adult wild-type, shy3-1, and 35S::shy3-1/kup2-1 plants.
(B) RNA gel blot hybridizations of RNA from pooled tall or short segregant progeny of one transgenic 35S::shy3-1 line, hybridized with SHY3/KUP2 or β-tubulin probes. Different lanes contain RNA from different pooled seedlings. The pooled short segregants overexpressed shy3-1/kup2-1, whereas the pooled tall segregants did not. s, short segregants; t, tall segregants; wt, wild type.
Figure 5.
Suppressors of shy3-1/kup2-1.
(A) Hypocotyl lengths of wild-type, shy3-1/kup2-1, and shy3-1/kup2-1 su(shy)/kup2 mutants grown for 5 days in darkness. Data are means of hypocotyl lengths of 19 to 25 seedlings ±sd. All of the mutants were significantly taller than the shy3-1/kup2-1 single mutant (P < 0.001).
(B) Positions of mutations in the SHY3/KUP2 protein. Asterisks indicate stop codons.
(C) RNA gel blot hybridization of RNA from homozygous suppressor lines with SHY3/KUP2 and rDNA probes.
(D) Appearance of wild-type, shy3-1/kup2-1, and kup2-3 null mutant seedlings grown in white light for 9 days. Cotyledons and leaves of the kup2-3 seedling are larger than those of the shy3-1/kup2-1 single mutant seedling and approximately the same size as those of the wild-type seedling.
The shorter hypocotyls and smaller leaves of shy3-1 mutant plants reflected decreased cell enlargement. After one week of growth in the dark, epidermal cells in the lower and middle portions of the hypocotyl of wild-type seedlings had elongated substantially, whereas cells at the top of the hypocotyl remained short. In shy3-1 seedlings, epidermal cells of the lower and middle parts of the hypocotyl elongated just 60% as much as the corresponding wild-type cells (Figures 1B and 1C). Wild-type and mutant seedlings each had an average of 20 epidermal cells in a longitudinal file in the hypocotyl. Light-grown wild-type and shy3-1 leaf epidermal cells had a broad size distribution, but shy3-1 leaves had more small cells and fewer large cells than wild-type leaves in all parts of the leaf (Figure 1D; data not shown). Thus, the primary developmental effect of the shy3-1 mutation is to decrease cell expansion in shoot tissues.
shy3-1 Mutation Affects the KT2/KUP2 Potassium Transporter
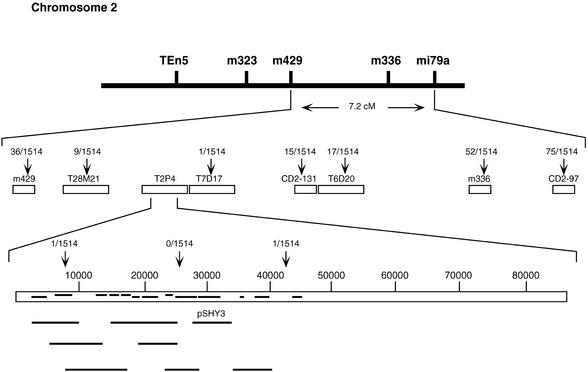
Figure 2 shows details of the cloning of SHY3. We mapped SHY3 to a 34-kb region on the bacterial artificial chromosome clone T2P4 on chromosome 2 using several new cleaved-amplified polymorphic sequence (CAPS) markers. To determine which of the 10 predicted open reading frames in this region encodes SHY3, we constructed eight different plasmid subclones of T2P4 containing each of the candidate genes with their presumed promoters in a plant transformation vector (Figure 2) and transformed these into phyB-1 shy3-1 mutant plants. As shown in Figure 3, dark-grown progeny of transformants homozygous for one subclone, pSHY3, had longer hypocotyls than phyB-1 shy3-1 mutant seedlings. This plasmid carried a single gene called AtKT2 or AtKUP2 that encodes a potassium transporter (Quintero and Blatt, 1997; Kim et al., 1998). We sequenced genomic DNA from the AtKT2/AtKUP2 gene from shy3-1 mutant plants and found a single base change from G to A, which is predicted to change the 419th amino acid from glycine to arginine.
Figure 2.
Map-Based Cloning of SHY3.
Open boxes indicate cosmid and bacterial artificial chromosome clones from which new CAPS markers were made. Vertical arrows indicate CAPS markers, with the number of recombinant chromosomes corresponding to the marker shown. Horizontal bars on bacterial artificial chromosome T2P4 indicate predicted open reading frames (http://www.tigr.org/tdb/at/at.html), with upper bars reading to the right and lower bars reading to the left. Solid lines beneath the T2P4 depiction represent the genomic clones used for the transformation of phyB-1 shy3-1/kup2-1 plants. Only clone pSHY3 partially complemented mutant phenotypes of phyB-1 shy3-1/kup2-1. cM, centimorgan.
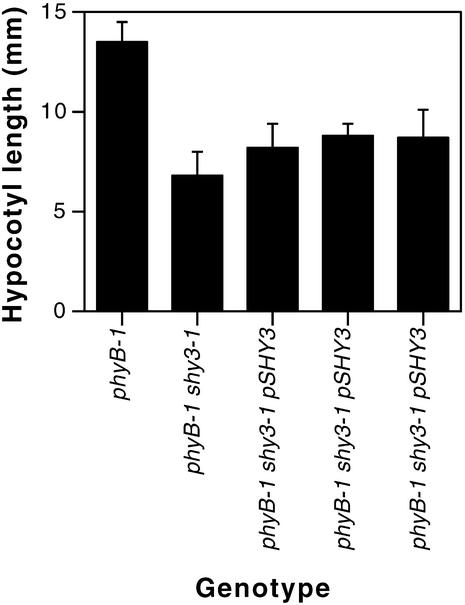
Figure 3.
Partial Rescue of phyB-1 shy3-1/kup2-1 Hypocotyl Length by a Genomic Clone Carrying SHY3/KUP2.
Seedlings were grown for seven days in darkness. Each measurement is the mean of 13 to 16 hypocotyl lengths ±sd. Data shown are for homozygous progeny of three different transformants. Hypocotyls of transformed lines were significantly longer than phyB-1 shy3-1/kup2-1 hypocotyls by t test (P < 0.005). In the dark, the phyB-1 photoreceptor mutation present in these lines has almost no effect on hypocotyl length.
To determine whether this mutation in AtKT2/AtKUP2 could confer dominant inhibition of hypocotyl and leaf growth and therefore cause the shy3-1 phenotypes, we placed the mutant open reading frame behind the strong 35S promoter and introduced this construct into wild-type Landsberg erecta plants. Three of five such 35S::shy3-1 transformants had progeny with short hypocotyls in the dark. As adults, these short hypocotyl seedlings had smaller leaves and shorter inflorescence stems than wild-type plants (Figure 4A). After self-fertilization for two more generations, we did not obtain lines that gave 100% small progeny, suggesting that either homozygosity of the transgene was lethal or the transgene was frequently silenced. However, the short hypocotyl phenotype correlated with the expression level of AtKT2/AtKUP2. As shown in Figure 4B, among pooled progeny of one transformant, those with short hypocotyls in the dark overexpressed the AtKT2/AtKUP2 gene, whereas those with long hypocotyls did not. Among adult progeny of two transformants tested, plants with small leaves and short inflorescence stems overexpressed the transgene, whereas plants of normal size did not (data not shown), indicating that the transgene likely caused both short hypocotyl and small adult shoot phenotypes.
Together with the isolation of intragenic suppressors of shy3-1 described below, these data demonstrate that the AtKT2/AtKUP2 gene is the same as SHY3 and that the mutation in AtKT2/AtKUP2 is the same as shy3-1. For simplicity, we refer to the gene as SHY3/KUP2 and to the mutation as shy3-1/kup2-1.
Intragenic Suppressors of the shy3-1/kup2-1 Mutation Restore Normal Hypocotyl Elongation
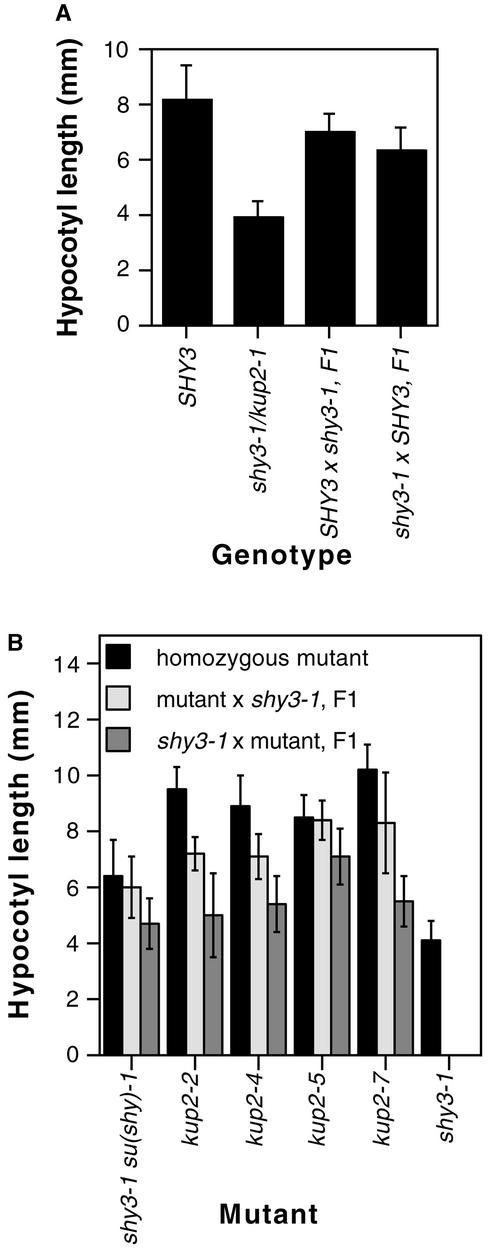
The partial dominance of shy3-1/kup2-1 (Reed et al., 1998) and the phenotypes of 35S::shy3-1 plants suggested that shy3-1/kup2-1 is not a null allele. To obtain null mutations in the SHY3/KUP2 gene, we screened M2 self-progeny of ethyl methanesulfonate–mutagenized shy3-1/kup2-1 seed for individuals having a long hypocotyl in the dark (see Methods). We found seven such su(shy) (suppressor of shy) mutants that retained the original shy3-1/kup2-1 mutation and had long hypocotyls (Figure 5A). Allelism tests (see Methods) showed that six of these mutations [su(shy)-2, su(shy)-3, su(shy)-4, su(shy)-5, su(shy)-6, and su(shy)-7] were alleles of one locus. We outcrossed each of these six shy3-1/kup2-1 su(shy) double mutants to wild type and found that all of the self-progeny of these outcrosses had long hypocotyls, indicating that these six mutations were closely linked to shy3-1/kup2-1.
We sequenced the SHY3/KUP2 gene from these six mutants and found that each had a new mutation in addition to shy3-1/kup2-1. These mutations, therefore, are intragenic suppressors of shy3-1/kup2-1, and we have named them kup2-2 through kup2-7. Figure 5B diagrams the predicted effects of each of these mutations on the SHY3/KUP2 protein. kup2-2 and kup2-3 had identical nucleotide substitution mutations that introduce a stop codon at amino acid 378, and kup2-7 had a nucleotide substitution that introduced a stop codon at amino acid 71. As shown in Figure 5C, these three mutants have very low levels of SHY3/KUP2 transcript; therefore, they are likely to be null mutations. (The residual hybridization signal in these mutants may reflect a low steady state transcript level or cross-hybridization to another KUP gene.) In contrast, kup2-4, kup2-5, and kup2-6 had missense mutations at amino acids 395, 167, and 560, respectively, and had roughly wild-type levels of SHY3/KUP2 transcript (Figure 5C).
In white light, the kup2 mutants appeared very similar to wild-type seedlings at both the seedling and adult stages (Figure 5D; data not shown). Roots of seedlings of kup2-3, a presumed null mutant, also were the same lengths as those of wild-type seedlings (data not shown).
The seventh suppressor of shy3-1/kup2-1, su(shy)-1, was not allelic to the kup2 mutations and did not map to SHY3/KUP2, indicating that it affects a distinct locus. shy3-1/kup2-1 su(shy)-1 plants had a normal level of the SHY3/KUP2 transcript (Figure 5C), suggesting that this mutation does not suppress shy3-1/kup2-1 by changing the regulation of the SHY3/KUP2 gene. The mutant had epinastic leaves that were larger than those of wild-type plants, it senesced more slowly than wild-type plants, and mutant seedlings were resistant to the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (data not shown). These phenotypes indicated that the su(shy)-1 mutant had decreased ethylene responses. We have not characterized this mutant further.
Maternal Effects of kup2 Mutations
We previously found that shy3-1/kup2-1 is partially dominant (Reed et al., 1998). In backcrossing shy3-1/kup2-1, we noticed that the hypocotyl length of heterozygotes depended in part on the polarity of the cross. Figure 6A shows that shy3-1/SHY3 heterozygous plants in which the wild-type allele was from the maternal parent had slightly longer hypocotyls than heterozygotes in which the wild-type allele was derived from the pollen. Older heterozygous plants were not distinguishable, suggesting that this effect was limited to early seedling phenotypes.
Figure 6.
Maternal Effects of the shy3-1/kup2-1 Mutation on Hypocotyl Length.
(A) Hypocotyl lengths of SHY3 × shy3-1/kup2-1 F1 seedlings arising from opposite polarities of the cross. In each cross, the genotype of the maternal parent is listed first. Data are means of hypocotyl lengths of 22 to 29 seedlings ±sd. Values for each reciprocal cross were different from each other by t test (P < 0.005).
(B) Hypocotyl lengths of kup2-x × shy3-1/kup2-1 F1 seedlings arising from opposite polarities of the cross. Data are means of hypocotyl lengths of 13 to 20 seedlings ±sd. Values for each reciprocal cross were different from each other by t test (P < 0.005).
The suppressing kup2 mutations showed a similar (and slightly larger) maternal effect in backcrosses with shy3-1/kup2-1 (Figure 6B). Thus, when we fertilized kup2-x mutant pistils with shy3-1/kup2-1 pollen, hypocotyls of the resulting F1 plants were longer than those of shy3-1/kup2-1 mutant plants and similar to those of the corresponding kup2-x mutant plants. Conversely, when we fertilized shy3-1/kup2-1 pistils with kup2-x pollen, hypocotyls of the resulting F1 plants were shorter than those of the corresponding kup2-x plants and similar to those of shy3-1/kup2-1 plants. That is, for this phenotype, the kup2 mutations were largely recessive to shy3-1/kup2-1 as the paternal allele but largely dominant as the maternal allele. For leaf size, F1 plants from the two polarities of the cross appeared similar to each other.
shy3-1/kup2-1 Mutation Does Not Eliminate K+ Transport
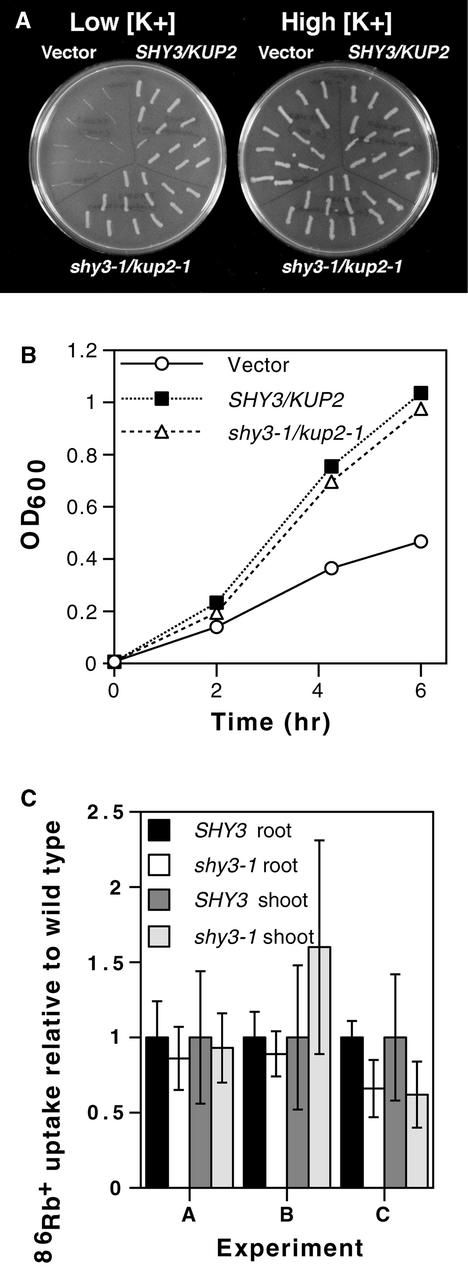
Our data suggest that shy3-1/kup2-1 is either a gain-of-function or a dominant negative allele. As shown in Figure 5C, the SHY3/KUP2 transcript was equally abundant in wild-type and shy3-1/kup2-1 mutant plants. Thus, the shy3-1/ kup2-1 mutation probably changes properties of the transporter protein rather than expression of the gene. The shy3-1/kup2-1 mutation introduces a positively charged amino acid near the external face of the 10th predicted transmembrane helix of SHY3/KUP2. To evaluate the effect of shy3-1/kup2-1 on SHY3/KUP2 K+ uptake activity, we compared the ability of wild-type and mutant cDNAs to rescue the growth of E. coli strain TK2463 cells in low-potassium medium. E. coli strain TK2463 carries mutations in trkD (=kup) and kdp K+ uptake systems and grows poorly in medium containing low potassium (Epstein et al., 1993). The SHY3/KUP2 cDNA clone was found previously to rescue the growth of this strain (Kim et al., 1998). Figures 7A and 7B show that TK2463 cells transformed with wild-type or shy3-1/kup2-1 mutant cDNAs grew at similar rates on low potassium medium, whereas control cells transformed with the vector grew more slowly. These bacterial strains carrying wild-type or shy3-1/kup2-1 mutant versions of the cDNA also grew at similar rates on plates containing a range of different K+ concentrations (5 to 80 mM) at several different pH values (6.6 to 8.1) and on low potassium medium in the presence of increasing amounts of Na+ (5 to 40 mM) (data not shown). These results indicate that SHY3/KUP2 protein carrying the shy3-1/kup2-1 mutation retains potassium transport activity.
Figure 7.
Effect of the shy3-1/kup2-1 Mutation on Ion Transport.
(A) E. coli TK2463 cells transformed with wild-type AtKUP2 cDNA, shy3-1/kup2-1 mutant AtKUP2 cDNA, and vector control streaked on medium containing low (∼2 mM) or high (∼120 mM) potassium.
(B) Growth of E. coli TK2463 cells transformed with wild-type AtKUP2 cDNA, shy3-1/kup2-1 mutant cDNA, and vector control in a medium containing 2 mM potassium. Data are OD600 of the cell cultures. Counts of colony-forming units gave similar results. Longer incubation of these strains led to renewed growth, possibly arising from selection of bacteria with suppressing mutations that allowed increased K+ uptake.
(C) 86Rb+ uptake by wild-type and shy3-1/kup2-1 seedlings. Eight- to 9-day-old light-grown seedlings were fed 86Rb+ through the roots, and radioactivity in roots and shoots was measured after 20 min. Data from three experiments are shown and represent mean uptake per seedling of six measurements ±sd, relative to wild-type uptake, with six seedlings per measurement. Uptake in roots (but not shoots) was significantly less in shy3-1 than in wild type in experiment C (P < 0.005), but in all other experiments, wild-type and mutant uptake rates were not significantly different (P > 0.1).
Similarly, we were unable to measure a significant defect in radioactive Rb+ uptake activity in shy3-1/kup2-1 mutant plants. Rb+ is transported by both high- and low-affinity K+ uptake systems with similar affinity as for K+. We fed 86RbCl to roots of eight- and nine-day-old light-grown seedlings and followed the movement of 86Rb+ into root and shoot. Figure 7C shows that wild-type and mutant plants accumulated 86Rb+ to similar levels in both roots and shoots. Although in some instances we saw a difference between wild-type and mutant 86Rb+ uptake rates, this was not statistically significant in most experiments. Time course experiments also showed similar uptake rates (data not shown).
To assess the effect of the shy3-1/kup2-1 mutation on shoot ionic composition, we measured relative compositions of major elements in adult shoot tissue of wild-type and shy3-1/kup2-1 mutant plants. As shown in Table 1, mutant and wild-type plants had only small differences when values were normalized to dry weight. In particular, K+ contents were just 10% lower in mutant than in wild-type plants. However, shy3-1/kup2-1 mutant plants were smaller than wild-type plants, and these small differences in relative composition of different elements mask a larger absolute difference in growth per plant. Thus, the shy3-1/kup2-1 mutation had a larger effect on the growth of the shoot than on its chemical composition.
Table 1.
Elemental Analysis of SHY3 and shy3-1/kup2-1 Adult Shoot Tissue
| Composition (%)a
|
|||
|---|---|---|---|
| Element | SHY3 | shy3-1/kup2-1 | Relative Difference (%) |
| C | 37.68 ± 0.19 | 38.37 ± 0.01 | +2 |
| N | 3.43 ± 0.04 | 3.93 ± 0.01 | +15 |
| P | 0.62 ± 0 | 0.68 ± 0.01 | +10 |
| K | 3.88 ± 0.02 | 3.50 ± 0.03 | −10 |
| Ca | 2.49 ± 0.02 | 2.56 ± 0.01 | +3 |
| Mg | 0.35 ± 0.01 | 0.32 ± 0 | −9 |
Composition is given as percent dry weight ±sd of two measurements.
SHY3/KUP2 Is Expressed in Growing Tissues
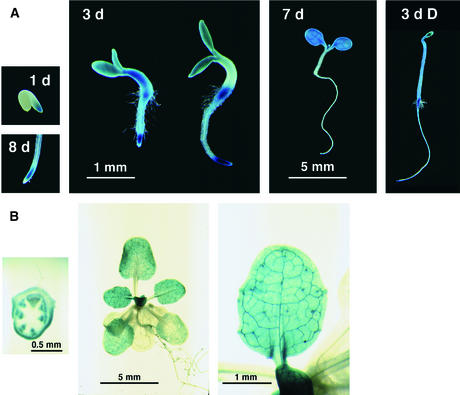
Previous RNA hybridization data showed that SHY3/KUP2 is expressed in stem, leaf, flower, and root tissue (Kim et al., 1998). To determine with finer resolution which tissues express the gene, we fused 2.9 kb of DNA upstream of the start codon of AtKUP2/AtKT2 to the β-glucuronidase (GUS) gene encoding β-glucuronidase, introduced the resulting construct into transgenic plants, and analyzed the expression pattern at different stages of development. Fifteen transformant lines were analyzed for 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (X-gluc) staining after six days of germination. All of these lines showed a similar pattern of staining, and we selected one line with a single insertion locus for further analysis. All growing regions of the plant stained with X-gluc. Throughout the life cycle, we observed staining in the growing region of the root tip (Figure 8). Seedlings three days old or younger stained in all tissues (Figure 8A). At about this stage, staining became more localized to cotyledons, the upper part of the hypocotyl, and the root–hypocotyl junction (Figure 8A). Dark-grown seedlings also stained in the hypocotyl, the cotyledons, the root tip, and at the root–hypocotyl junction. As plants grew, staining disappeared from the hypocotyl and cotyledons and appeared in new leaves. Four-week-old plants showed strong staining in the inflorescence stem, young leaves, and root tips (Figure 8B). In most of these tissues, staining was strongest in or around vascular tissues. Older parts of the plant such as fully expanded leaves and older parts of the root lacked X-gluc staining.
Figure 8.
X-Gluc Staining of PSHY3/KUP2::GUS Transgenic Plants.
(A) Seedling expression patterns. Numbers in panels indicate seedling ages in days. The two three-day-old seedlings shown illustrate the variation in seedling morphology and staining observed. The 1 d and 8 d photographs are at the same scale as the 3 d photograph, and the 3 d D (dark-grown) photograph is at the same scale as the 7 d photograph.
(B) Adult expression patterns. Shown are a cross-section of a flowering stem with the base of a cauline leaf visible, a rosette, and a single leaf. Staining is prominent around the vasculature.
DISCUSSION
Arabidopsis has 13 HAK/KT/KUP genes, and the absence of obvious phenotypes of the kup2 null mutants may reflect redundancy of function between SHY3/KUP2 and other members of this transporter family. Alternatively, feedback regulation of other ion transport pathways may compensate for the loss of SHY3/KUP2. Low external K+ concentration upregulates several genes encoding K+ transporters, including some HAK/KT/KUP genes (Maathuis and Sanders, 1995; Santa-María et al., 1997; Kim et al., 1998; Wang et al., 1998). The skor potassium channel mutant also lacks an overt morphological phenotype, apparently because of alternative ion transport pathways (Gaymard et al., 1998).
Considering that other transporters probably compensate for the absence of SHY3/KUP2 in the kup2 null mutants, at present the phenotypes of the shy3-1/kup2-1 mutant give the best indication of the normal function of SHY3/KUP2. The small cells of mutant tissues suggest that SHY3/KUP2 participates in developmentally regulated cell enlargement. The expression of SHY3/KUP2 in growing tissues also suggests that the gene acts in expanding cells. An attractive model is that the transporter imports K+ to growing cells, thereby ensuring that intracellular osmotic potential is high enough to drive water uptake and cell expansion. The trh1-1 null mutation in another member of this gene family also causes a defect in cell enlargement, specifically in root hairs (Rigas et al., 2001). Together, the shy3-1/kup2-1 and trh1-1 mutant phenotypes suggest that HAK/KT/KUP proteins mediate cell expansion in multiple tissues. Because these proteins are thought to transport K+ actively against an electrochemical gradient (Rodríguez-Navarro, 2000), localized activity of these proteins may determine which cells or tissues of a plant expand. These proteins, therefore, might be targets of the regulatory controls that determine relative growth rates of different organs.
In addition to transporters of the HAK/KT/KUP family, channels of the Shaker superfamily participate in potassium nutrition and growth. Mutant Arabidopsis plants deficient in the AKT1 potassium channel have a defect in K+ uptake into roots at low external K+ concentrations in the presence of ammonium ions (Hirsch et al., 1998). The AKT1 gene is expressed predominantly in root epidermal, cortical, and endodermal cells (Cao et al., 1995; Lagarde et al., 1996), indicating that AKT1 acts specifically to take up K+ in roots. When wild-type or akt1-1 mutant seedlings are starved for K+ (which occurs at higher K+ concentration for akt1-1 than wild type), they appear small, and their actual size reflects the amount of K+ provided (Hirsch et al., 1998). Thus, like the trh1-1 and shy3-1/kup2-1 mutants, the akt1-1 mutant has a defect in cell growth, although the defect in akt1-1 affects the entire plant rather than just the shoot and is apparent at low external K+ concentration rather than high external K+ concentration, as in shy3-1/kup2-1.
The SKOR protein also is thought to be important for feeding K+ to the shoot (Gaymard et al., 1998). Although skor plants have normal shoot growth (J.W. Reed, unpublished observation), they have 50% less potassium in the shoot than wild-type plants and compensating surfeits of other ions such as Ca2+ and Na+ (Gaymard et al., 1998). Furthermore, a PSKOR::GUS fusion was expressed only in root stelar cells and not in the shoot (Gaymard et al., 1998). Thus, skor mutant plants may compensate for a block of potassium movement out of root stelar cells by transporting other cations into the xylem, which then move up to the shoot and are taken up by shoot cells. The differences among transport and morphological phenotypes of the akt1-1, skor, trh1-1, and shy3-1/kup2-1 mutants underscore the multiplicity of distinct localized transport activities that may be required to move ions from the soil to growing sink tissues.
Much of the volume increase of expanding plant cells occurs in a large central vacuole, which may have a different K+ concentration from the cytoplasm (Walker et al., 1996). Because KT1/KUP1, SHY3/KUP2, and HAK1 can mediate K+ or Rb+ uptake by plant, yeast, or E. coli cells, KUP proteins may localize to the plasma membrane. However, it is also possible that they localize to the vacuolar membrane (tonoplast), which has several K+ transporters (Allen and Sanders, 1997). In that case, they could affect cellular K+ uptake indirectly by regulating K+ partitioning between the vacuole and the cytoplasm.
Our finding that shy3-1/kup2-1 and other kup2 mutations have a maternal effect on hypocotyl elongation suggests that SHY3/KUP2 may be expressed in the female gametophyte or that expression may be higher from the maternal than the paternal allele early in embryo or seedling development. Expression of SHY3/KUP2 at this early stage may regulate hypocotyl elongation. Several other genes have been found to have a maternal effect caused by preferential expression of the maternal allele during the first few days after fertilization (Vielle-Calzada et al., 2000). The maternal effect of kup2 alleles is not absolute, because the paternal allele does affect hypocotyl length to some degree (Figure 5). Moreover, adult phenotypes did not show maternal effects.
Our data do not reveal how the shy3-1/kup2-1 mutation affects SHY3/KUP2 protein activity. shy3-1/kup2-1 is not a null allele, and the mutant gene supports potassium transport activity in E. coli. The partial dominance of the shy3-1/kup2-1 mutation (Reed et al., 1998), and the ability of the shy3-1/kup2-1 mutant transgene to confer a short hypocotyl and small leaves to wild-type plants, suggest that the mutation causes a gain of function or creates a mutant protein that interferes with some normal function. Proteins of the HAK/KT/KUP family have 12 predicted transmembrane helices (Fu and Luan, 1998; Kim et al., 1998), and the shy3-1/kup2-1 mutation adds a positive charge to the outer end of the 10th predicted transmembrane helix of SHY3/KUP2. If this helix forms part of the transport pore, the mutation might affect the kinetics or ion specificity of SHY3/KUP2 transport activity in ways that we have not detected in our E. coli assays. Alternatively, the mutation may alter the regulation of SHY3/KUP2 by some other protein not present in E. coli. Finally, the mutant protein may interfere with the function of some other interacting protein whose activity regulates cell growth. More detailed studies of the effect of the shy3-1/kup2-1 mutation on transport, and characterization of additional mutant alleles, will provide a more complete picture of the structural requirements for K+ transport by this protein.
METHODS
Plant Material and Growth Conditions
The Arabidopsis thaliana shy3-1 mutation was isolated as a suppressor of phyB-1 and is in the Landsberg erecta background (Reed et al., 1998). shy3-1 was separated from the phyB-1 mutation by outcrossing to wild type, and the shy3-1 single mutant was used for most of the experiments described in this work. Seed were surface-sterilized and plated on MS/agar/sucrose plates (1 × Murashige and Skoog [1962] salts, 0.8% phytagar [Gibco BRL], 1 × Gamborg's B5 vitamin mix, and 2% sucrose). The potassium concentration of these plates is ∼20 mM.
Hypocotyl and Leaf Measurements
Hypocotyl imprints were obtained from seven-day-old dark-grown seedlings, and leaf imprints were made from the sixth leaf of four-week-old light-grown plants. Seedlings or leaves were placed on a thin layer of Quicktite super glue spread on a microscope slide, and after 2 min the seedlings were peeled off, leaving an imprint of the epidermal cells. Imprints were photographed using Nomarski optics, and NIH Image software (http://rsb.info.nih.gov/nih-image) was used to measure cell areas. Hypocotyl cell lengths were measured using a stage micrometer. Entire leaves from 4-week-old plants were imaged using a slide scanner (Neff and Chory, 1998), and NIH Image software was used to measure the leaf areas. Data were tested for significance using t tests.
Mapping and Cloning
To map the SHY3 gene, we crossed the phyB-1 shy3-1 double mutant (in ecotype Landsberg erecta) with the phyB-9 single mutant (in ecotype Columbia). F2 seed were grown on MS/sucrose/agar plates for 7 days under low fluence red light, and DNA from tall (presumed SHY3+/+) F2 individuals was assayed for Landsberg/Columbia cleaved-amplified polymorphic sequence markers (Konieczny and Ausubel, 1993). We developed several new cleaved-amplified polymorphic sequence markers from cosmids and bacterial artificial chromosome (BAC) clones, as indicated in Figure 2. Details of these markers have been submitted to the Arabidopsis database (http://genome-www.stanford.edu/Arabidopsis). Genomic DNA from BAC clone T2P4 was subcloned into plant transformation vectors derived from pBI121 (but lacking the 35S promoter) (Jefferson et al., 1987). These subclones were transformed into shy3-1 mutant plants by vacuum infiltration (Bechtold et al., 1993). Clone pSHY3, which partially rescued the shy3-1 mutant phenotype, carried a 6.5-kb EcoRI fragment that included 2.4 kb upstream of the start codon and 0.9 kb downstream of the stop codon of AtKT2/AtKUP2. Mutant alleles were sequenced using pooled products from 10 independent polymerase chain reaction (PCR) procedures as templates by the University of North Carolina at Chapel Hill DNA sequencing facility. The shy3-1 mutation created a DdeI restriction site, and the kup2-2/kup2-3 and kup2-4 mutations eliminated XhoI and HpaII restriction enzyme recognition sites. These changes were confirmed by the digestion of PCR products spanning these sites.
The shy3-1/kup2-1 mutant version of SHY3/KUP2 genomic DNA was amplified by PCR and then cloned behind the double 35S promoter between EcoRI and BamHI sites in pRTL2 (Restrepo et al., 1990). The 35S::shy3-1 construct then was excised by partial digestion with PstI and cloned into the plant transformation vector pPZP211 (Hajdukiewicz et al., 1994). Transformants were obtained after vacuum infiltration of wild-type plants (Bechtold et al., 1993).
RNA Gel Blot Hybridization
Total RNA was isolated from seedlings grown in liquid MS/sucrose medium for eight days in light. RNA gel blot hybridization experiments were performed with 30 μg of total RNA, as described previously (Nagpal et al., 2000).
Isolation and Genetic Analysis of shy3-1/kup2-1 Suppressor Mutations
We mutagenized ∼9,000 shy3-1/kup2-1 seed with ethyl methanesulfonate as described previously (Reed et al., 1998) and screened ∼63,000 M2 seed for mutants having long hypocotyls in darkness. Candidate su(shy) (suppressor of shy) mutants were checked for the presence of the starting shy3-1/kup2-1 mutation indicated by the presence of a DdeI restriction site created by shy3-1/kup2-1. For allelism tests, we crossed each of the shy3-1/kup2-1 su(shy) double mutants to each other and found that hypocotyls of F1 plants from all of the crosses were longer than those from the shy3-1/kup2-1 single mutant parent and similar in length to those from the corresponding double mutant parents. To determine whether these results reflected a failure to complement (implying allelism) or the maternal dominance seen in backcrosses, we assessed hypocotyl lengths of F2 self-progeny of each of these F1 plants. For crosses of any combination of mutants among kup2-2, kup2-3, kup2-4, kup2-5, kup2-6, or kup2-7, all of the F2 progeny had long hypocotyls, indicating that all of these mutations were linked closely to each other and therefore probably allelic. For all crosses involving mutation su(shy)-1, the F2 populations segregated seedlings with both long hypocotyls and short hypocotyls, indicating that mutation su(shy)-1 is not linked closely, and therefore is not allelic, to the other mutations. To test linkage to shy3-1/kup2-1, we outcrossed each of the shy3-1/kup2-1 su(shy) double mutants to wild-type plants and examined hypocotyl lengths of F2 self-progeny of these F1 plants. For mutants kup2-2, kup2-3, kup2-4, kup2-5, kup2-6, and kup2-7, all of these F2 seedlings had long hypocotyls in the dark, indicating that these mutations were linked tightly to shy3-1/kup2-1. In contrast, the outcross with su(shy)-1 gave seedlings with short hypocotyls among the F2 progeny, indicating that it is not linked closely to shy3-1/kup2-1.
Escherichia coli Complementation Assay
Escherichia coli strain TK2463 (F− thi lacZ amx82 rha Δ[trkA] trkD1 Δ[Kdp-FAB]5 endA) (Epstein et al., 1993) and the SHY3/KUP2 cDNA clone were kindly provided by J.I. Schroeder (University of California, San Diego). The trkD gene was renamed kup (Schleyer and Bakker, 1993). A 1.2-kb internal fragment of shy3-1/kup2-1 cDNA that included the shy3-1/kup2-1 mutation was amplified from shy3-1/kup2-1 RNA by reverse transcriptase–mediated PCR, cut with BglII and NheI, and cloned between BglII and NheI sites of the SHY3/KUP2 cDNA. E. coli transformants were selected on KML medium (10 g of tryptone, 5 g of yeast extract, and 10 g [∼120 mM] of KCl per liter) (Kim et al., 1998) and tested for their ability to grow in medium containing low (∼2 mM) potassium (10 g of tryptone, 2 g of yeast extract, and 100 mM mannitol per liter, pH 7.0) at 37°C. High- and low-potassium agar plates had the same media plus 1.6% Bacto-agar (Difco). Carbenicillin (100 μg/mL) was used to maintain plasmids in growth experiments.
Rubidium Uptake
Eight- to nine-day-old seedlings were picked from MS/sucrose plates with forceps and placed along the short edge of a glass microscope slide with their roots hanging off the edge. A thin layer of 0.5% agarose was allowed to solidify at the edge of the slide before placing the seedlings to ensure that they would adhere to the slide edge and prevent the shoots from contacting directly the liquid MS/sucrose containing 86Rb+. We placed six seedlings on each slide and used six slides for each genotype. The slides with seedlings were placed in six-well microtiter dishes, each well containing 250 μL of liquid MS/sucrose, for 10 to 30 min. Uptake was started by transferring the slides to similar wells containing 230 μL of liquid MS/sucrose with 86RbCl (Amersham, Piscataway, NJ) so that only roots were in contact with the solution. In the three experiments shown, the 86Rb+ concentrations were 35, 186, and 78 μM. After 86Rb+ uptake for 20 min, the seedlings were washed four times in cold MS/sucrose, briefly blotted dry with Whatman 3MM filter paper, and then cut at the base of the hypocotyl to separate shoots from roots. Time course experiments showed that this 20-min time point was in the linear part of the uptake curve (data not shown). Roots and shoots then were transferred to separate scintillation vials, and radioactivity was measured using an LKB scintillation counter (Bromma, Sweden).
Compositional Analysis
Plants were grown on soil for four weeks under short day conditions, and the shoots were harvested, frozen, and lyophilized. Compositional analyses were performed at the analysis facility of the North Carolina State University Soil Science Department by inductively coupled plasmon emission. For P, K, Ca, and Mg, samples were heated overnight at 500°C in a dry ash muffle furnace, dissolved in 0.5 n HCl, and analyzed on a Perkin-Elmer Plasma 2000 machine (Perkin-Elmer, Norwalk, CT). For C and N, samples were analyzed in a Perkin-Elmer 2400 CHN analyzer.
Promoter::β-Glucuronidase Fusion
A 2.9-kb fragment including the presumed promoter region and the first 14 codons of the SHY3/KUP2 gene was amplified from BAC clone T2P4 by PCR. The PCR product was cloned between BamHI and EcoRI sites of the β-glucuronidase reporter gene fusion vector CAMBIA1381Xa (CAMBIA, Canberra, Australia). The resulting construct was transformed into Landsberg erecta plants by vacuum infiltration (Bechtold et al., 1993), transformants were selected on MS/sucrose/agar plates containing 25 μg/mL hygromycin, and T2 seedlings were stained with 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (Research Products International, Mount Prospect, IL) as described (Jefferson, 1987).
Acknowledgments
We thank Gene Kim and Julian Schroeder for providing the SHY3/KUP2 cDNA and E. coli strain TK2463, Beverly Taylor for performing the compositional analysis, the Arabidopsis Genetic Stock Center for genomic DNA clones, and anonymous reviewers for helpful comments. P.N. worked on this project without pay. This work was supported by National Institutes of Health Grant No. R29-GM52456 and National Science Foundation Grant No. IBN-9983046 to J.W.R.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010322.
References
- Allen, G.J., and Sanders, D. (1997). Vacuolar ion channels of higher plants. Adv. Bot. Res. 25, 217–252. [Google Scholar]
- Bañuelos, M.A., Klein, R.D., Alexander-Bowman, S.J., and Rodríguez-Navarro, A. (1995). A potassium transporter of the yeast Schwanniomyces occidentalis homologous to the Kup system of Escherichia coli has a high concentrative capacity. EMBO J. 14, 3021–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316, 1194–1199. [Google Scholar]
- Cao, Y., Ward, J.M., Kelly, W.B., Ichida, A.M., Gaber, R.F., Anderson, J.A., Uozumi, N., Schroeder, J.I., and Crawford, N.M. (1995). Multiple genes, tissue specificity, and expression-dependent modulation contribute to the functional diversity of potassium channels in Arabidopsis thaliana. Plant Physiol. 109, 1093–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussen, M., Lüthen, H., Blatt, M., and Böttger, M. (1997). Auxin-induced growth and its linkage to potassium channels. Planta 201, 227–234. [Google Scholar]
- Cosgrove, D.J. (1993). How do plant cell walls extend? Plant Physiol. 102, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer, A.H. (1999). Potassium translocation into the root xylem. Plant Biol. 1, 36–45. [Google Scholar]
- Epstein, W., Buurman, E., McLaggan, D., and Naprstek, J. (1993). Multiple mechanisms, roles, and controls of K+ transport in Escherichia coli K-12. Biochem. Soc. Trans. 21, 1006–1010. [DOI] [PubMed] [Google Scholar]
- Fox, T.C., and Guerinot, M.L. (1998). Molecular biology of cation transport in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 669–696. [DOI] [PubMed] [Google Scholar]
- Fu, H.-H., and Luan, S. (1998). AtKUP1: A dual-affinity K+ transporter from Arabidopsis. Plant Cell 10, 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaymard, F., Pilot, G., Lacombe, B., Bouchez, D., Bruneau, D., Boucherez, J., Michaux-Ferrière, N., Thibaud, J.-B., and Sentenac, H. (1998). Identification and disruption of a plant Shaker-like outward channel involved in K+ release into the xylem sap. Cell 94, 647–655. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Haro, R., Sainz, L., Rubio, F., and Rodríguez-Navarro, A. (1999). Cloning of two genes encoding potassium transporters in Neurospora crassa and expression of the corresponding cDNAs in Saccharomyces cerevisiae. Mol. Microbiol. 31, 511–520. [DOI] [PubMed] [Google Scholar]
- Hirsch, R.E., Lewis, B.D., Spalding, E.P., and Sussman, M.R. (1998). A role for the AKT1 potassium channel in plant nutrition. Science 280, 918–921. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. [Google Scholar]
- Jefferson, R.A., Kavanaugh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, C.P., and Van Volkenburgh, E. (1996). Osmoregulation by oat coleoptile protoplasts: Effect of auxin. Plant Physiol. 110, 1007–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, E.J., Kwak, J.M., Uozumi, N., and Schroeder, J.I. (1998). AtKUP1: An Arabidopsis gene encoding high-affinity potassium transport activity. Plant Cell 10, 51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochian, L.V., and Lucas, W.J. (1988). Potassium transport in roots. Adv. Bot. Res. 15, 93–178. [Google Scholar]
- Konieczny, A., and Ausubel, F. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Lagarde, D., Basset, M., Lepetit, M., Conejero, G., Gaymard, F., Astruc, S., and Grignon, C. (1996). Tissue-specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J. 9, 195–203. [DOI] [PubMed] [Google Scholar]
- Maathuis, F.J.M., and Sanders, D. (1995). Contrasting roles in ion transport of two K+-channel types in root cells of Arabidopsis thaliana. Planta 197, 456–464. [DOI] [PubMed] [Google Scholar]
- Maathuis, F.J.M., and Sanders, D. (1996). Mechanisms of potassium absorption by higher plant roots. Physiol. Plant. 96, 158–168. [Google Scholar]
- Maathuis, F.J.M., Ichida, A.M., Sanders, D., and Schroeder, J.I. (1997). Roles of higher plant K+ channels. Plant Physiol. 114, 1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser, P., et al. (2001). Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 126, 1646–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nagpal, P., Walker, L., Young, J., Sonawala, A., Timpte, C., Estelle, M., and Reed, J.W. (2000). AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 123, 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M.M., and Chory, J. (1998). Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero, F.J., and Blatt, M.R. (1997). A new family of K+ transporters from Arabidopsis that are conserved across phyla. FEBS Lett. 415, 206–211. [DOI] [PubMed] [Google Scholar]
- Reed, J.W., Elumalai, R.P., and Chory, J. (1998). Suppressors of an Arabidopsis thaliana phyB mutation identify genes that control light signalling and hypocotyl elongation. Genetics 148, 1295–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo, M.A., Freed, D.D., and Carrington, J.C. (1990). Nuclear transport of plant potyviral proteins. Plant Cell 2, 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigas, S., Desbrosses, G., Haralampidis, K., Vicente-Agullo, F., Feldmann, K.A., Grabov, A., Dolan, L., and Hatzopoulos, P. (2001). TRH1 encodes a potasium transporter required for tip growth in Arabidopsis root hairs. Plant Cell 13, 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, S., and Tester, M. (1995). Inward and outward K+-selective currents in the plasma membrane of protoplasts from maize root cortex and stele. Plant J. 8, 811–825. [Google Scholar]
- Rodríguez-Navarro, A. (2000). Potassium transport in fungi and plants. Biochim. Biophys. Acta 1469, 1–30. [DOI] [PubMed] [Google Scholar]
- Rubio, F., Santa-María, G.E., and Rodríguez-Navarro, A. (2000). Cloning of Arabidopsis and barley cDNAs encoding HAK potassium transporters in root and shoot cells. Physiol. Plant. 109, 34–43. [Google Scholar]
- Santa-María, G.E., Rubio, F., Dubcovsky, J., and Rodríguez-Navarro, A. (1997). The HAK1 gene of barley is a member of a large gene family and encodes a high-affinity potassium transporter. Plant Cell 9, 2281–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleyer, M., and Bakker, E.P. (1993). Nucleotide sequence and 3′-end deletion studies indicate that the K+-uptake protein Kup from Escherichia coli is composed of a hydrophobic core linked to a large and partially essential hydrophilic C terminus. J. Bacteriol. 175, 6925–6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calzada, J.P., Baskar, R., and Grossniklaus, U. (2000). Delayed activation of the paternal genome during seed development. Nature 404, 91–94. [DOI] [PubMed] [Google Scholar]
- Walker, D.J., Leigh, R.A., and Miller, A.J. (1996). Potassium homeostasis in vacuolate plant cells. Proc. Natl. Acad. Sci. USA 93, 10510–10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T.-B., Gassmann, W., Rubio, F., Schroeder, J.I., and Glass, A.D.M. (1998). Rapid up-regulation of HKT1, a high-affinity potassium transporter gene, in roots of barley and wheat following withdrawal of potassium. Plant Physiol. 118, 651–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner, L.H., and Raschke, K. (1994). Ion channels in the xylem parenchyma of barley roots: A procedure to isolate protoplasts from this tissue and a patch-clamp exploration of salt passageways into xylem vessels. Plant Physiol. 105, 799–813. [DOI] [PMC free article] [PubMed] [Google Scholar]