Abstract
An Arabidopsis cDNA (AtCAP1) that encodes a predicted protein of 476 amino acids highly homologous with the yeast cyclase-associated protein (CAP) was isolated. Expression of AtCAP1 in the budding yeast CAP mutant was able to rescue defects such as abnormal cell morphology and random budding pattern. The C-terminal domain, 158 amino acids of AtCAP1 possessing in vitro actin binding activity, was needed for the regulation of cytoskeleton-related defects of yeast. Transgenic plants overexpressing AtCAP1 under the regulation of a glucocorticoid-inducible promoter showed different levels of AtCAP1 accumulation related to the extent of growth abnormalities, in particular size reduction of leaves as well as petioles. Morphological alterations in leaves were attributable to decreased cell size and cell number in both epidermal and mesophyll cells. Tobacco suspension-cultured cells (Bright Yellow 2) overexpressing AtCAP1 exhibited defects in actin filaments and were unable to undergo mitosis. Furthermore, an immunoprecipitation experiment suggested that AtCAP1 interacted with actin in vivo. Therefore, AtCAP1 may play a functional role in actin cytoskeleton networking that is essential for proper cell elongation and division.
INTRODUCTION
The plant cytoskeleton has crucial functions in a number of cellular processes that are essential for cell morphogenesis, organogenesis, and development. The presence of diverse microfilament configurations has been established in a majority of plant cell types (Pathsarathy et al., 1985; Meagher and Williamson, 1994; Kost et al., 1998; Mathur et al., 1999). Microfilament arrays govern important processes, including the determination of division plane during cytokinesis, cell elongation, and cell wall deposition, which are critical to plant development and cell differentiation (Kost et al., 1999). Plant cells contain a dynamic network of cytoskeletal elements that remodel cytoplasmic architecture in response to external and internal stimuli (Kadota and Wada, 1992). The reorganization of the actin cytoskeleton is thought to be mediated by actin binding proteins within minutes to several hours depending on the stimulus and plant tissue involved. Although several actin binding proteins are known, the precise mechanism regulating their role in cell elongation and division is not understood fully. In this context, Ramachandran et al. (2000) noted the role of profilin in cell elongation by analyzing transgenic Arabidopsis plants in which profilin level was downregulated. However, the overall role of proteins associated with actin was not well demonstrated in the elongation and division of plant cells.
Adenylyl cyclase of Saccharomyces cerevisiae has been shown to consist of at least two subunits, a 200-kD catalytic subunit, the product of the cyr1 gene (Kataoka et al., 1985), which is involved in the generation of cAMP from ATP (Toda et al., 1985), and a 70-kD subunit of adenylyl cyclase–associated protein (CAP) (Field et al., 1990). Deletion studies of the CAP gene have shown that the adenylyl cyclase binding site and the cAMP-related function were mapped to the N-terminal region of CAP, whereas the cytoskeleton phenotypes were attributed to the C-terminal part of CAP (Gerst et al., 1991; Mintzer and Field, 1994). In fact, the C-terminal domain of CAP was sufficient to induce the depolymerization of F-actin (Zelicof et al., 1996). CAP was found to localize to actin patches in yeast cells and to be associated with stress fibers in mammalian cells (Lila and Drubin, 1997; Freeman and Field, 2000).
In Drosophila, Act up (a CAP homolog) was shown to be required to prevent excessive accumulation of actin filaments in the eye disc, preventing premature Hedgehog-induced photoreceptor differentiation ahead of the morphogenetic furrow (Benlali et al., 2000). Kawamukai et al. (1992) revealed that CAP of Schizosaccharomyces pombe also has two functions related to cAMP level and the cytoskeleton. In the middle region, CAP possesses two proline-rich segments, P1 and P2, resembling a consensus sequence for binding to Src homology 3 (SH3) domains of several proteins, including the actin-associated protein Abp1p (Freeman et al., 1996; Lambrechts et al., 1997). The SH3 domain of yeast Abp1p mediates the association of CAP with the cortical actin cytoskeleton (Lila and Drubin, 1997); thus, the middle region is a third functional domain.
Genes encoding CAP have been isolated from human (Matviw et al., 1992; Yu et al., 1994), rat (Zelicof et al., 1993; Swiston et al., 1995), mouse (Vojtek and Cooper, 1993), hydra (Fenger et al., 1994), Dictyostelium discoideum (Gottwald et al., 1996), Lentinus edodes (Zhou et al., 1998), and Drosophila (Baum et al., 2000; Benlali et al., 2000). None of the CAP homologs could restore the hyperactivation of adenylyl cyclase by RAS2val19 in CAP-deficient yeast cells. In contrast, all of them suppressed the C-terminal domain defects (Gerst et al., 1991), indicating a high conservation of the cytoskeleton-related function during the evolution of CAP. Nonetheless, a common feature of CAP proteins is the presence of a CAP motif in their N-terminal domains. The motif is characterized by RLE repeats, which have the potential to form an amphipathic helix that mediates protein–protein interactions (Cohen and Parry, 1986; Bush and Sassone-Corsi, 1990). In budding yeast, the RLE repeats are known to be involved in the binding to adenylyl cyclase (Gerst et al., 1991; Vojtek and Cooper, 1993). Therefore, the N-terminal function also may be conserved among various organisms.
Recently, we isolated a cotton cDNA clone encoding a putative CAP homolog (GhCAP) (Kawai et al., 1998). In this study, using a GhCAP cDNA, we isolated an Arabidopsis CAP gene (AtCAP1). Overexpression of AtCAP1 complemented the yeast CAP mutant that had the defect in the C-terminal region. Transgenic plants overexpressing AtCAP1 exhibited morphological alterations that were associated with decreases in cell number as well as cell size in leaf tissues. Similar defects in cell proliferation were confirmed in suspension-cultured tobacco cells (Bright Yellow 2 [BY-2]), in which overproduction of AtCAP1 correlated with the loss of actin filaments and resulted in the arrest of cell division. These results suggest that AtCAP1 may play a crucial role in regulating actin filaments in plant cells.
RESULTS
Isolation of an Arabidopsis cDNA Encoding a CAP Homolog
The cDNA encoding a possible adenylyl CAP from cotton (GhCAP) (Kawai et al., 1998) was used as a probe to screen a cDNA library of Arabidopsis. After the screening of 6 × 105 plaques, 12 clones were isolated. The sequencing analysis revealed that all of the clones were partial because no ATG start codon was found upstream of the CAP motif, the conserved amino acid sequence at the N terminus of the CAP protein family (Freeman et al., 1996). To isolate the full-length cDNA, 5′ rapid amplification of cDNA ends was conducted to amplify a unique 500-bp fragment from the Arabidopsis cDNA mixture (data not shown). The sequencing analysis showed that an ATG start codon was located 12 bp upstream of the conserved CAP motif in all of the independent clones analyzed. Therefore, we concluded that the amplified fragment contained the 5′ end of the full-length cDNA.
The coding region of the cDNA encodes a protein of 476 amino acids that shares an overall identity of 28% with the budding yeast CAP (Figure 1). The similarity was much higher (76%) with the cotton GhCAP. Therefore, the isolated cDNA was designated AtCAP1 (Arabidopsis thaliana CAP homolog). The C-terminal domain (the last 158 amino acids of AtCAP1) showed a higher identity (34%) than the N-terminal domain (the first 224 amino acids of AtCAP1) (22%) compared with the S. cerevisiae CAP. The N-terminal CAP motif including the RLE repeats was highly conserved in AtCAP1 (Figure 1). The proline-rich stretch from amino acids 227 to 237 (Figure 1) may have the potential to bind to SH3 proteins because the conserved prolines at positions 7 and 10 match the consensus PXXP sequence (where X is any amino acid) that is involved in interaction with the SH3 domain. CAPs from other organisms also have one or two proline-rich sequences in the middle region, which is shown as an SH3 protein binding motif (Freeman et al., 1996).
Figure 1.
Alignment of Deduced Amino Acid Sequences of CAP Homologs.
Identical amino acids are highlighted. Dashes represent gaps introduced to give maximal identity. The CAP motif is denoted by a box, and its RLE repeats are underlined. A double line indicates the putative SH3 binding site of AtCAP1. GhCAP, cotton (Kawai et al., 1998); ScCAP, S. cerevisiae (Fedor-Chaiken et al., 1990; Field et al., 1990); SpCAP, S. pombe (Kawamukai et al., 1992); AtCAP1, Arabidopsis (this study).
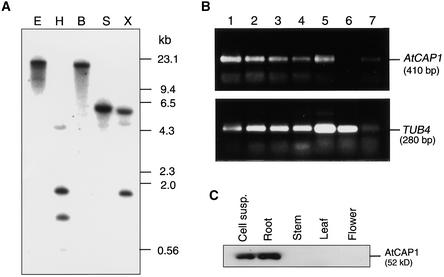
Arabidopsis genomic DNA was digested with several restriction enzymes and subjected to DNA gel blot analysis using a 1.5-kb EcoRI–XhoI cDNA fragment (missing the first 30 bp of the coding region) as a probe. A single band of 6 kb was detected in the SacI digest, whereas XbaI and HindIII, which cleaved the 5′ end of the cDNA probe, produced two bands (Figure 2A). In the BamHI and EcoRI digest, which did not cleave the AtCAP1 cDNA, a single band was observed. When the membrane was washed under low-stringency conditions, no additional bands were observed (data not shown), suggesting that the AtCAP1 gene is present at a single locus in the Arabidopsis genome. In fact, a single genomic sequence of AtCAP1 was reported to localize to chromosome IV of Arabidopsis.
Figure 2.
DNA Gel Blot and Protein Gel Blot Analyses.
(A) Genomic DNA gel blot of AtCAP1. Total DNA (20 μg) digested with EcoRI (E), HindIII (H), BamHI (B), SacI (S), or XbaI (X) was subjected to gel electrophoresis followed by hybridization with a 32P-labeled 1.5-kb cDNA fragment derived from the partial clone pBK-AtCAP16-1.
(B) Analysis of transcript levels of AtCAP1 in Arabidopsis tissues. Total RNAs isolated from suspension-cultured cells (lane 1), roots of seedlings (lane 2), cotyledons (lane 3), rosette leaves (lane 4), shoots of seedlings (lane 5), green siliques (lane 6), and flowers (lane 7) were subjected to RT-PCR with primers directed to AtCAP1 or TUB4 (Marks et al., 1987) genes.
(C) Total soluble protein (20 μg) isolated from suspension-cultured cells (Cell susp.), young roots, stems, rosette leaves, and flowers were subjected to protein gel blot analysis with a polyclonal anti-AtCAP1 antibody directed to the C-terminal domain.
Total RNA samples isolated from different tissues of Arabidopsis, such as cotyledons, leaves, shoots, flowers, green siliques, and suspension cells, were subjected to RNA gel blot analysis to investigate the expression pattern of AtCAP1; however, no signal could be detected. Then we performed reverse transcriptase–mediated polymerase chain reaction (RT-PCR) as described in Methods to determine the level of AtCAP1 in distinct Arabidopsis tissues. After RT-PCR, a specific 410-bp fragment was detected in all tissues analyzed except siliques (Figure 2B). The expression level of AtCAP1 was similar in roots, cotyledons, leaves, shoots, flowers, and suspension cells. The control TUB4 gene was expressed at a similar level in all tissues analyzed (Figure 2B).
To characterize the level of accumulation of the AtCAP1 protein in Arabidopsis tissues, total proteins from suspension-cultured cells, roots, stems, leaves, and flowers were extracted and subjected to protein gel blot analysis as described in Methods. The AtCAP1 antibody specifically recognized a 52-kD protein, which is the predicted molecular mass of the AtCAP1 protein. A detectable level of AtCAP1 protein was found in suspension-cultured cells and roots (Figure 2C).
Overexpression of AtCAP1 Suppressed Cytoskeleton-Related Defects of Budding Yeast CAP-Deficient Cells
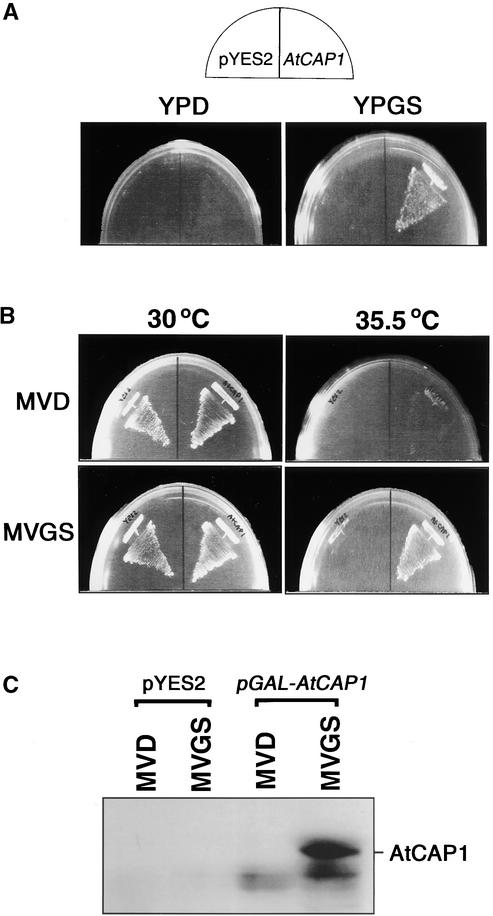
In S. cerevisiae, loss of the C-terminal function of CAP results in abnormal cellular morphology, growth inhibition at high temperature or on rich medium, abnormal actin cytoskeleton distribution, and random budding (Fedor-Chaiken et al., 1990; Field et al., 1990). To determine whether the expression of AtCAP1 is able to complement such defects associated with the loss of the C-terminal domain, the AtCAP1 cDNA was placed under the control of the GAL1 promoter in the yeast multicopy expression vector pYES2. This expression plasmid (pGAL-AtCAP1) then was introduced into the SKN32 yeast strain, which has the CAP gene disrupted with a HIS3 auxotrophic marker (Field et al., 1990).
In the analysis of the suppression of nutritional sensitivity, SKN32 cells carrying either pYES2 empty vector or pGAL-AtCAP1 plasmid were plated on YPD (glucose-containing rich medium) or YPGS (galactose-containing rich medium) and incubated at 30°C for 2 days. SKN32 cells transformed with pYES2 failed to grow on either YPD or YPGS (Figure 3A), suggesting the rich medium sensitivity of this strain. In contrast, SKN32 cells carrying pGAL-AtCAP1 could grow on YPGS but not on YPD (Figure 3A). This result indicates that the expression of AtCAP1 on YPGS suppressed the rich medium sensitivity of SKN32 cells.
Figure 3.
Expression of AtCAP1 Suppressed Nutritional and Temperature Sensitivity of Budding Yeast CAP-Deficient Cells.
(A) Suppression of the rich medium sensitivity of SKN32 cells by overexpression of AtCAP1. SKN32 was transformed with pYES2 or pGAL-AtCAP1 (AtCAP1), plated onto YPD or YPGS, and incubated at 30°C for 2 days.
(B) Suppression of the temperature sensitivity of SKN32 cells. SKN32 was transformed with pYES2 or pGAL-AtCAP1 (AtCAP1), plated onto MVD or MVGS, and incubated at 30 or 35.5°C for 3 days.
(C) AtCAP1 is expressed specifically in SKN32 cells harboring pGAL-AtCAP1 and grown in galactose medium. Twenty micrograms of total protein of SKN32 cells harboring either pYES2 or pGAL-AtCAP1 was subjected to protein gel blot analysis using anti-AtCAP1 antibody. The AtCAP1 protein was detected in SKN32 cells grown in MVGS.
To analyze the temperature sensitivity of SKN32 cells transformed with either pYES2 or pGAL-AtCAP1, transformants were grown on MVD (glucose-containing minimal medium) or MVGS (galactose-containing minimal medium) at 30 or 35.5°C for 3 days. SKN32 cells transformed with an empty pYES2 plasmid grew on both media at 30°C but failed to grow at 35.5°C (Figure 3B). Cells transformed with pGAL-AtCAP1 plasmid were able to grow on both MVD and MVGS media at 30°C, but at 35.5°C, only cells expressing AtCAP1 on MVGS could grow (Figure 3B), indicating that the temperature sensitivity of SKN32 cells was rescued by the overexpression of AtCAP1. The suppression of defects of CAP-deficient budding yeast cells correlated with the accumulation of the AtCAP1 protein (Figure 3C).
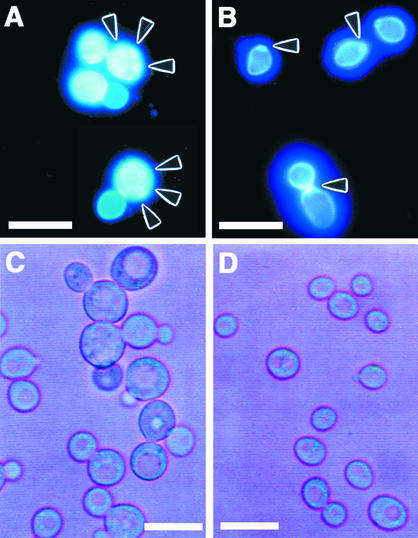
In a wild-type budding yeast strain, buds emerge adjacent to one another, as indicated by the clustered bud scars, and locate at one pole of the cell. In contrast, CAP-deficient yeast cells display random budding (Vojtek et al., 1991). SKN32 cells transformed with pYES2 showed a random deposition of bud scars (Figure 4A), whereas the expression of AtCAP1 in MVGS resulted in one polar budding pattern (Figure 4B), suggesting restoration of the CAP-deficient phenotype.
Figure 4.
Effects of AtCAP1 Expression on Budding Pattern and Cellular Morphology of SKN32 Cells.
(A) and (B) Suppression of the random budding pattern of SKN32 cells by overexpression of AtCAP1. SKN32 was transformed with pYES2 (A) or pGAL-AtCAP1 (B) and cultured in galactose-containing medium at 30°C. Cells were collected and stained with calcofluor. Arrowheads indicate positions of scars. Bars = 20 μm.
(C) and (D) Effects of AtCAP1 expression on the cellular morphology of SKN32 cells. SKN32 was transformed with pYES2 (C) or pGAL-AtCAP1 (D) and cultured in galactose-containing liquid medium at 30°C. Bars = 20 μm.
To characterize the cell morphology, we cultured SKN32 cells transformed with either pYES2 or pGAL-AtCAP1 in liquid MVGS at 30°C. The abnormal morphology with round and enlarged shapes was observed in cells carrying the pYES2 vector (Figure 4C), whereas the expression of AtCAP1 made the cells smaller (Figure 4D). Prolonged incubation of SKN32 cells with an empty pYES2 plasmid resulted in a more severe phenotype that was characterized by the presence of large vacuoles (Gerst et al., 1991).
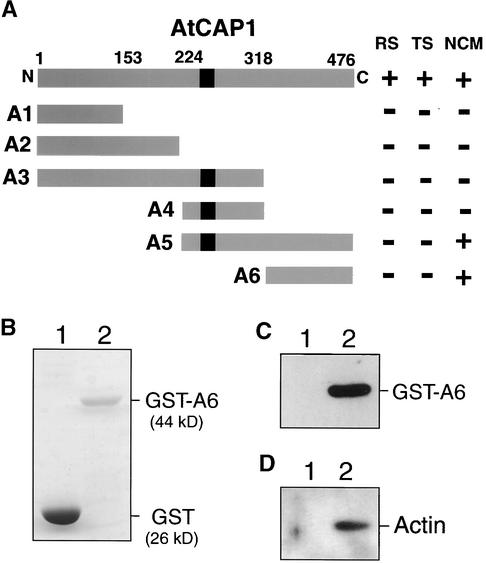
Functional Domains of AtCAP1
To analyze the functional domains of AtCAP1, we created six types of AtCAP1 deletion mutants with 5′ and/or 3′ truncation (Figure 5A, A1 t, A1). SKN32 cells were transformed with each derivative of AtCAP1 on pYES2 vector. The complementation test showed that the abnormal cell morphology was suppressed partially by expression of the deletion derivatives A5 and A6, which encode the C-terminal domain of AtCAP1 (Figure 5A), suggesting that the C-terminal region of AtCAP1 appears to be sufficient to restore the normal cell shape of budding yeast. In contrast, expression of the N-terminal region of AtCAP1, even when including the middle proline-rich region (Figure 5A, A1 t, A1), failed to restore the wild-type morphology.
Figure 5.
Deletion Analysis of AtCAP1 and in Vitro Binding of GST-A6 to Actin.
(A) The putative SH3 binding site is indicated by a black box. Numbers indicate amino acids corresponding to the ends of each deletion mutant (A1 to A6). The results of complementation tests are represented as follows: +, regaining of wild-type phenotype; −, failure to restore wild-type phenotype. The phenotypes of SKN32 cells tested for suppression are as follows: RS, ability to grow on rich media; TS, ability to grow at 35.5°C; NCM, ability to restore normal cell morphology.
(B) Coomassie blue staining of GST (lane 1) and GST-A6 (158 amino acids of the C-terminal domain of AtCAP1) (lane 2) proteins on a gel before binding assay to actin.
(C) Protein gel blot of GST (lane 1) or GST-A6 (lane 2) treated with anti-AtCAP1 antibody.
(D) In vitro binding of the C-terminal domain of AtCAP1 to actin. GST (lane 1) and GST-A6 (lane 2) were subjected to in vitro binding assay to actin and then analyzed by protein gel blotting using anti-actin antibody.
CAP proteins were reported to have actin binding activity in several organisms (Freeman et al., 1995; Lila and Drubin, 1997; Zhou et al., 1998; Freeman and Field, 2000), and its C-terminal domain was sufficient to sequester actin monomers (Freeman et al., 1995). Therefore, we cloned a deletion derivative of AtCAP1 encoding the C-terminal domain (A6; the last 158 amino acids) into the EcoRI site of the glutathione S-transferase (GST) gene fusion vector pGEX2T. The resulting fusion protein (GST-A6) was purified (Figure 5B) as described in Methods. An antibody raised against the C-terminal peptide of AtCAP1 (ETTPVSHSGA) detected a 44-kD protein encoding the GST-A6 fusion protein (Figure 5C). The GST-A6 protein mixed with bovine muscle actin was subjected to in vitro binding assay followed by protein gel blot analysis. As shown in Figure 5D, the GST-A6 bound to actin in vitro (lane 2), whereas the control GST did not (lane 1).
Expression of AtCAP1 in Transgenic Plants
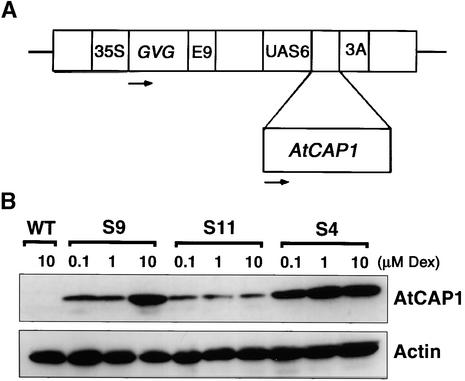
We obtained several transgenic lines possessing a chimeric AtCAP1 gene under a GAL4 promoter, which is recognized by the GVG transcription factor upon activation with dexamethasone (Dex) (Aoyama and Chua, 1997) (Figure 6A). Three independent transgenic plants, S4, S9, and S11, each with a single copy of AtCAP1 (data not shown) and with different expression levels of the AtCAP1 protein (Figure 6B), were chosen for further analysis.
Figure 6.
Analysis of Expression of AtCAP1 in Transgenic Lines upon Dex Treatment.
(A) Scheme of the glucocorticoid-inducible construct. 35S, 35S promoter of Cauliflower mosaic virus; GVG, the chimeric GVG transcription factor; E9, pea rbcS-E9 poly(A) addition sequence; UAS6, six copies of the DNA binding sites for GAL4; AtCAP1, AtCAP1 coding sequence; 3A, pea rbc-3A poly(A) addition sequence. Arrows indicate the direction of transcription.
(B) Protein gel blot detection of AtCAP1 (52 kD) and actin (46 kD). Plants were grown on MS plates for 7 days and then treated with different concentrations of Dex for 2 days. Protein samples were extracted from shoot tissues, and 20 μg of total protein per line was loaded in duplicate gels. Results from Arabidopsis wild-type plants (WT) and transgenic plants overexpressing AtCAP1 (S9, S11, and S4) are shown.
The quantitative estimation of the amount of protein accumulation indicated that although the actin level was maintained constant in all plants, the AtCAP1 level increased as the amount of Dex was increased in transgenic plants (Figure 6B). S4 plants accumulated AtCAP1 protein at high levels, whereas S11 plants accumulated AtCAP1 at low levels.
Overexpression of AtCAP1 Caused Organ Size Reduction and Affected Both the Number and Size of Leaf Cells
To characterize the morphological effects of AtCAP1 overexpression in Arabidopsis plants, we used young Arabidopsis seedlings that had produced and partially expanded the first two leaves before the third rosette leaf primordia were established (defined as the appearance of a leaf 1 mm long; Kim et al., 1998) to induce the accumulation of AtCAP1 protein by Dex treatment (1 μM) for 7 days. All anatomical comparisons were performed in third rosette leaves, except as noted otherwise. As shown in Figure 7A, a typical transgenic plant overexpressing AtCAP1 showed cotyledons and rosette leaves of reduced size but a main root of normal length compared with a wild-type plant. When grown on Dex-free medium, overexpression lines showed no phenotypic differences from wild-type plants (data not shown), suggesting that leaf organ size reduction occurs only when AtCAP1 is expressed. The severity of the organ size reduction correlated with the level of accumulation of the AtCAP1 transgene (Figure 6, Table 1). All transgenic plants showed reductions of leaf blade width and length as well as of petiole length (Figure 7B, Table 1[A]). The S4 plant overexpressing AtCAP1 at high levels showed the most severe phenotype, with 31.9 and 37% reductions of leaf blade width and length, respectively. The petiole lengths in such plants were reduced severely (73.8%) compared with those of wild-type plants (Table 1[A](a)). A second experiment with S4 transgenic plants (Table 1) showed a similar leaf organ size reduction upon induction of AtCAP1 accumulation (Table 1[A](b)), suggesting that overexpression of AtCAP1 in Arabidopsis plants correlates with a reduction of leaf organ size.
Figure 7.
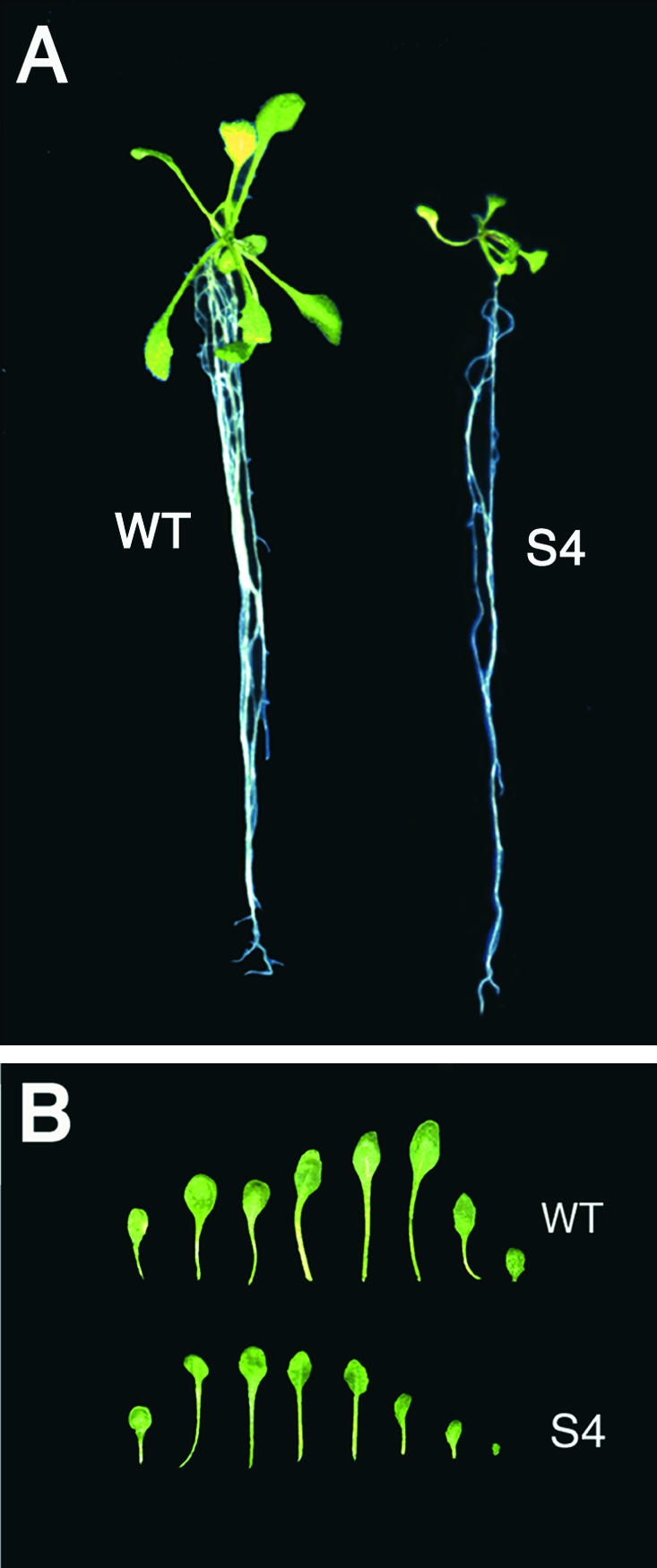
Anatomic Comparison of Transgenic Arabidopsis Plants Overexpressing AtCAP1.
(A) Plants grown on Dex-free medium for 7 days were transferred to Dex medium (1 μM) and grown for 7 days. WT, wild type.
(B) Leaves of the wild-type plants and S4 transgenic plants. The leaves in each row are, from left, cotyledon and rosette leaves.
Table 1.
Comparison of Organ Size [A], Cell Area [B], and Cell Number [C] of Third Leaf Dimensions of Arabidopsis Plants
| [A]
|
|
|
|
|
|
|
|---|---|---|---|---|---|---|
| Plant Line | Width of Leaf Blade (mm) |
Relative Width (%) | Length of Leaf Blade (mm) |
Relative Length (%) | Length of Leaf Petiole (mm) |
Relative Length (%) |
| (a) | ||||||
| WT (n = 9) | 3.0 ± 0.4 | 100.0 | 3.9 ± 0.5 | 100.0 | 5.6 ± 1.0 | 100.0 |
| S11 (n = 12) | 2.6 ± 0.4 | 86.2 | 3.2 ± 0.5 | 82.2 | 3.7 ± 0.7 | 66.7 |
| S9 (n = 9) | 2.2 ± 0.4 | 74.0 | 3.3 ± 0.6 | 83.0 | 3.1 ± 0.8 | 55.8 |
| S4 (n = 8) | 2.1 ± 0.2 | 68.1 | 2.5 ± 0.4 | 63.0 | 1.5 ± 0.3 | 26.2 |
| (b) | ||||||
| WT (n = 4) | 4.5 ± 0.7 | 100.0 | 6.5 ± 0.9 | 100.0 | 14.5 ± 2.1 | 100.0 |
| S4 (n = 4) | 2.3 ± 0.3 | 52.2 | 3.2 ± 0.4 | 50.2 | 5.2 ± 0.9 | 36.2 |
| [B] | ||||||
| Plant Line
|
Abaxial Epidermal Cell Area (μm2)
|
Relative Area (%)
|
Palisade Cell Area (μm2)
|
Relative Area (%)
|
||
| WT | 2301.4 ± 822.6 | 100.0 | 1095.5 ± 201.9 | 100.0 | ||
| S4 | 1805.1 ± 760.4 | 78.4 | 702.0 ± 227.0 | 64.1 | ||
| [C] | ||||||
| Plant Line
|
Total No. of Epidermal Cellsa (%)
|
Total No. of Palisade Cellsb (%)
|
||||
| WT | 142.5 ± 4.9 (100.0) | 142 ± 2.8 (100.0) | ||||
| S4 | 104.5 ± 6.4 (73.3) | 84 ± 1.4 (59.1) | ||||
Arabidopsis wild-type (WT) and transgenic plants overexpressing AtCAP1 (S11, S9, and S4) were used. Observations were made 7 days after Dex treatment (1 μM) of 4-day-old {[A](a)} or 7-day-old {[A](b) and [B]} plants. Data are means (±sd) of more than 128 cells per leaf [B].
a Cell number of a single cell layer in a cross-section of the adaxial surface.
b Cell number of the first layer of palisade cells.
To determine whether the leaf organ size reduction in S4 transgenic plants was attributable to a change in cell expansion or a change in cell number, we performed an anatomical analysis of S4 plant leaves, comparing them with the wild type (Table 1[B]). The epidermal cells of S4 transgenic plants were smaller than those of wild-type plants. The cell areas of abaxial epidermal cells and palisade cells in S4 transgenic plants were 78.4 and 64.1% of the wild-type areas, respectively (Table 1[B]). The number of palisade cells in the third leaves of S4 plants in the leaf width direction was ∼59% of the wild-type cell number (Table 1[C]). These anatomical results suggest that a decrease in cell size and cell number directly affected the leaf morphology in S4 plants.
Depolymerization of Actin Filaments in BY-2 Suspension Cells Overexpressing AtCAP1
The cytoskeleton is known to be required for a number of cellular processes, including cell division and elongation. We postulated that the actin network might be affected by the overexpression of AtCAP1 in transgenic plants. However, we were unable to obtain clear results with the whole plant system; therefore, we moved to a tobacco BY-2 cell system that offers the advantage of low background when analyzing the cytoskeleton.
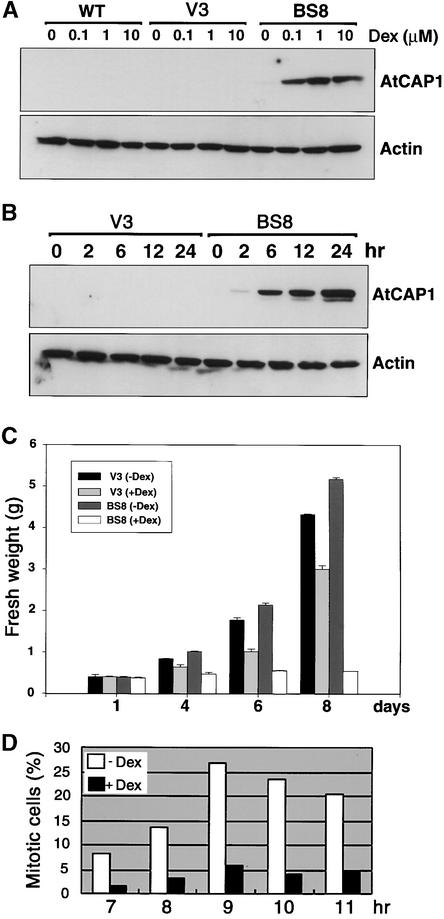
The pTA-AtCAP1 construct or the pTA7002 empty vector was transformed into tobacco BY-2 cells using Agrobacterium tumefaciens–mediated transformation. Twelve independent lines were isolated for each construct. Line BS8, which stably expressed AtCAP1, and line V3 (empty vector) (Figure 8) were chosen for further analysis. Transgenic BY-2 cells cultured for 7 days were transferred to fresh Murashige and Skoog (1962) (MS) liquid medium containing 1 μM Dex and grown at 27°C for 24 hr. Quantitative estimation of the amount of protein accumulation indicated that although the actin level was maintained constant in all BY-2 lines, the AtCAP1 level was increased as the amount of Dex was increased in the BS8 line (Figure 8A). Furthermore, accumulation of AtCAP1 was detected after 2 hr of Dex treatment (1 μM) (Figure 8B).
Figure 8.
Effect of Dex on the Expression of AtCAP1 and Actin in Tobacco BY-2 Cells, and Frequency Distribution of Mitotic Cells in BY-2 Cells Overexpressing AtCAP1.
(A) Protein gel blot detection of AtCAP1 (52 kD) and actin (46 kD) protein levels in BY-2 suspension-cultured cells at different Dex concentrations. The wild-type line (WT), an empty vector cell line (V3), and the AtCAP1 transgenic cell line (BS8) were grown in liquid medium for 7 days and then treated with different concentrations of Dex for 24 hr.
(B) Time course analysis of AtCAP1 and actin proteins in BY-2 transgenic suspension-cultured cells. BY-2 transgenic suspension cell lines V3 and BS8 were grown for 7 days in liquid medium and then treated with 1 μM Dex for the times indicated.
(C) Growth comparison of BY-2 suspension-cultured cells. Transgenic BY-2 cells cultured for 7 days were transferred to fresh MS liquid medium with or without Dex (1 μM) and cultured at 27°C. Samples were taken at the days indicated, and fresh weight was measured. Data represent averages of two independent experiments. TA, pTA7002 glucocorticoid-inducible vector.
(D) Transgenic BY-2 cells cultured for 7 days were subcultured in medium containing aphidicolin (5 mg/L) for 24 hr. After extensive washing, cells were resuspended in fresh medium with or without Dex (1 μM). Samples were removed at the times indicated, and mitotic cells were recorded as indicated in Methods. At least 500 cells were counted per treatment.
To determine whether the effect of the overexpression of AtCAP1 in BY-2 cells is similar to that in transgenic Arabidopsis plants, we examined the proliferation of BY-2 cells and found that AtCAP1 level was correlated with the arrest of cell proliferation (Figure 8C). In contrast, although the V3 BY-2 cell line showed growth retardation in the presence of Dex, cell proliferation was not inhibited as much as in the BS8 line. Tobacco BY-2 cells are well known for the study of mitotic events caused by high levels of synchronization after aphidicolin treatment (Nagata and Kumagai, 1999). Thus, we compared mitotic cells of the AtCAP1 transgenic BS8 line in the presence or absence of Dex in the early stage of cell culture. The results indicated that the mitotic activity of BY-2 cells overexpressing AtCAP1 was not sustained (Figure 8D), which coincided with the expression time of the AtCAP1 protein (6 hr). These result clearly demonstrate that the cell division process is inhibited when AtCAP1 is overexpressed in BY-2 cells, which is in a good agreement with our previous observations.
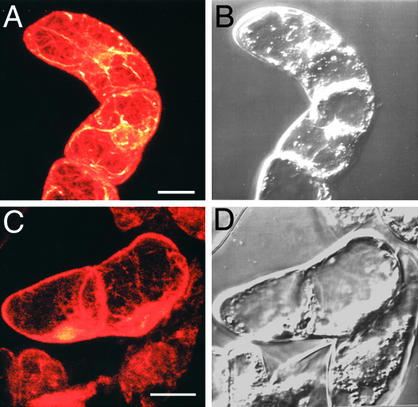
Next, we were prompted to investigate the integrity of the actin cytoskeleton in BY-2 cells. BY-2 cells (cultured for 7 days) were transferred to fresh MS liquid medium containing 1 μM Dex and incubated at 27°C for 24 hr. Then, BY-2 cells were fixed and stained with rhodamine phalloidin as described in Methods. Normal actin cytoskeleton arrays were observed in the wild type (Figure 9A), whereas BY-2 cells overexpressing AtCAP1 showed loss of actin filaments (Figure 9C). These results suggest that the abnormalities observed in both transgenic Arabidopsis plants and tobacco BY-2 cells overexpressing AtCAP1 are the result of depolymerization of the actin cytoskeleton.
Figure 9.
Microscopic Analysis of Actin Filament Arrays in Tobacco BY-2 Cells.
BY-2 cells cultured for 7 days were transferred to a Dex-containing medium (1 μM Dex) and cultured for 24 hr at 27°C. Then, BY-2 cells were fixed and stained with rhodamine phalloidin as described in Methods. Fluorescent (A) and (C) and corresponding bright-field (B) and (D) images of wild-type BY-2 cells (A) and (B) and AtCAP1 transgenic BY-2 cells (C) and (D) are shown. Bars = 20 μm.
Interaction of AtCAP1 with Actin in BY-2 Cells
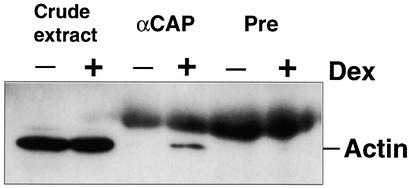
CAP proteins have been suggested to have actin binding activity in several organisms (Freeman et al., 1995; Lila and Drubin, 1997; Zhou et al., 1998; Freeman and Field, 2000). Therefore, we were interested to determine whether AtCAP1 also might bind to actin in our BY-2 cell system. BY-2 cells cultured for 7 days (BS8) were transferred to Dex-free or Dex-containing (1 μM) medium and cultured for 24 hr. Proteins were extracted as described in Methods. Total protein (150 μg) extracts were immunoprecipitated with either AtCAP1 antibody or preimmune serum and then subjected to immunoblot analysis using an actin antibody. As shown in Figure 10, actin protein was immunoprecipitated by the AtCAP1 antibody but not the preimmune serum, indicating that AtCAP1 has a conserved function to bind directly or indirectly to actin through another cell component. The large smear that can be observed in Figure 10 (lanes 3 to 6) corresponds to a low cross-reactivity reaction of anti-mouse IgG with rabbit immunoglobulin added to the immunoprecipitation.
Figure 10.
Binding of AtCAP1 to Actin.
BY-2 cells (cultured for 7 days) overexpressing AtCAP1 under glucocorticoid-inducible promoter were transferred to Dex-free (−) or Dex-containing (+) medium (1 μM Dex) and cultured for 24 hr at 27°C. Total protein extracts (150 μg) were immunoprecipitated with either anti-AtCAP1 antibody (αCAP) or preimmune serum (Pre) and subjected to protein gel blot analysis using an anti-actin antibody. Twenty micrograms of total protein (crude extract) was loaded for each sample.
DISCUSSION
Molecular Characterization of AtCAP1 and Complementation of Yeast Cells
In this study, we isolated an Arabidopsis cDNA (AtCAP1) that encodes a protein of 476 amino acids that is highly homologous with the adenylyl CAP of S. cerevisiae. The CAP is a multifunctional protein; its N-terminal domain is required for the cAMP-mediated signaling pathway by interaction with adenylyl cyclase, and the C-terminal domain is needed for nutritional response and cytoskeleton organization in yeast cells (Fedor-Chaiken et al., 1990; Field et al., 1990; Gerst et al., 1991). Budding yeast cells lacking the C-terminal domain of CAP have abnormal responses to nutrient deprivation and excess, altered cell shape and size, random budding pattern, and sensitivity to increased temperatures (Field et al., 1990; Gerst et al., 1991). In this study, we demonstrate that overexpression of AtCAP1 could suppress the phenotypes associated with the defect of the C-terminal domain by restoring the wild-type phenotype that includes resistance to rich medium, ability to grow at high temperatures, and normal budding pattern and cell morphology. The defects associated with the C-terminal domain also have been rescued by the expression of other CAP homologs (Kawamukai et al., 1992; Matviw et al., 1992; Vojtek and Cooper, 1993; Zelicof et al., 1993; Yu et al., 1994) and profilin (Vojtek et al., 1991). Therefore, AtCAP1 functionally complements both the lack of proper cytoskeletal function and the inability to respond to the nutritional stress of budding yeast CAP-deficient cells.
A deletion analysis of AtCAP1 revealed that expression of a C-terminal region as short as 158 amino acids partially complemented the abnormal cell morphology of budding yeast CAP-deficient cells but failed to rescue the nutritional defects of mutant cells. Vojtek et al. (1991) suggested that CAP alters the formation of second messengers resulting from phosphoinositide metabolism and that these second messengers are important in regulating cellular morphology and cellular adaptation to a variety of nutritional environments. Recently, Gottwald et al. (1996) reported that binding to phosphatidylinositol-4-monophosphate (PIP2) by the N-terminal domain of the D. discoideum CAP regulates its G-actin sequestering activity. Moreover, two point mutants of profilin with substitution at either Arg-72 or Arg-81 that prevents them from binding PIP2 bind normally to actin and complement profilin-deficient yeast cells, but they fail to complement the loss of the C-terminal domain of CAP (Vojtek et al., 1991). Thus, binding to PIP2 by the N-terminal domain of CAP might be required to respond properly to nutritional stresses, which may explain the inability of the C-terminal domain of AtCAP1 to complement the rich medium sensitivity of CAP-deficient cells.
CAPs have been shown to inhibit actin polymerization in vitro by sequestering monomeric actin (Freeman et al., 1995; Gottwald et al., 1996). This actin binding activity has been mapped to the C-terminal region of CAP (Gerst et al., 1991; Freeman et al., 1995). Recently, Baum et al. (2000) reported a “verprolin homology”–related domain in all CAPs, just C-terminal of the polyproline-rich domain, and suggested that this region of the protein may be used to facilitate actin binding. To test the binding of the C-terminal domain of AtCAP1 to actin, we fused the deletion derivative A6, encoding the last 158 amino acids of AtCAP1, to GST to generate a GST-A6 fusion protein. The in vitro binding assay suggested that the C-terminal domain of AtCAP1 has a conserved function to bind actin like other CAP proteins (Freeman et al., 1995; Gottwald et al., 1996; Freeman and Field, 2000).
Overexpression of AtCAP1 in Transgenic Arabidopsis Plants Causes Morphological Defects
The CAP proteins were shown to have actin binding activity (Freeman et al., 1995; Lila and Drubin, 1997; Zhou et al., 1998), and knockout mutants for the CAP gene in Drosophila displayed an increased actin filament cytoskeleton and disruption in cell polarity, suggesting that CAP function prevents the excessive actin filament polymerization that is required for normal eye disc development and proper oocyte polarity (Baum et al., 2000; Benlali et al., 2000). However, the effect of increased CAP activity on cell development has not been demonstrated. Therefore, we generated sense Arabidopsis transgenic plants overexpressing a chimeric AtCAP1 gene under the control of a glucocorticoid-inducible promoter. Transgenic plants expressed AtCAP1 at different levels upon Dex induction, and the extent of AtCAP1 protein accumulation was found to be associated directly with the degree of growth abnormalities, in particular, size reduction of leaves and petioles. Our anatomic results on third rosette leaves showed that the observed leaf size reduction was caused by a decrease in both the size and number of epidermal and palisade cells. Kim et al. (1998) reported that four days after the appearance of the fifth leaf primordium, the division of the leaf palisade cells ceased in wild-type plants. Under the conditions used in this study, the overexpression of AtCAP1 in third leaves of transgenic plants was induced since the appearance of the leaf primordia before cell division was completed. Therefore, overexpression of AtCAP1 not only may have affected the normal elongation of leaf cells but also may have altered the normal cell division.
Recently, Dong et al. (2001) reported that the overexpression of actin-depolymerizing factor (AtADF1) in Arabidopsis resulted in the disappearance of thick actin cables in different cell types, causing irregular cellular and tissue morphogenesis and reducing the growth of cells and organs. Moreover, Arabidopsis plants germinated and grown in a latriculin B–containing medium developed morphologically normal seedlings, but as a result of the absence of cell elongation, these were stunted, resembling either genetic dwarfs or environmental bonsai plants (Baluska et al., 2001). Thus, to determine whether loss of the F-actin cytoskeleton was responsible for the abnormalities in Arabidopsis plants overexpressing AtCAP1, we examined the actin cytoskeleton. Nevertheless, we could not obtain clear results with our plant system; therefore, we moved to the analysis of BY-2 tobacco suspension cells overexpressing AtCAP1 (as discussed below). Further studies of the effect of the overexpression of AtCAP1 in other plant tissues such as flowers and siliques may help us determine the role of the actin cytoskeleton in the development of reproductive organs.
Loss of Actin Filaments in Tobacco BY-2 Cells Overexpressing AtCAP1
BY-2 cells overexpressing the AtCAP1 protein inhibited cell proliferation and showed a severe decrease in mitotic cells, indicating that the cell division process in BY-2 cells was arrested. Furthermore, our unpublished results indicate that overexpression of AtCAP1 in transgenic tobacco plants resulted in morphological defects similar to those observed in Arabidopsis plants.
Microscopic visualization of the actin cytoskeleton in BY-2 cells revealed that the overexpression of AtCAP1 correlated with the loss of F-actin; such actin filament depolymerization was not observed in either wild-type or empty vector BY-2 lines. The anti-AtCAP1 antibody was raised against the last 10 amino acids of the C-terminal domain of AtCAP1. Thus, the antibody specifically recognizes a single band in crude extracts of Arabidopsis tissues. No cross-reactivity was detected with the endogenous CAP of tobacco. Therefore, in the coimmunoprecipitation assay, the endogenous tobacco CAP was not immunoprecipitated along the AtCAP1 protein. Under such circumstances, AtCAP1 bound directly or indirectly to tobacco actin, resulting in the loss of actin filaments in vivo. It has been demonstrated that the CAP protein regulates the actin cytoskeleton primarily through an actin monomer binding activity in several classes of organisms, including yeast, fungi, and mammals (Freeman et al., 1995; Zelicof et al., 1996; Zhou et al., 1998; Freeman and Field, 2000). Consequently, AtCAP1-expressing cells appear as if the actin cytoplasmic strands have collapsed completely, and the nucleus and cytoplasm are at the cell periphery. This is exactly what would be expected to happen if the actin filaments had mostly depolymerized, because the actin cables in these strands are known to be necessary to stabilize them. For example, in Tradescantia stamen hair cells injected with high concentrations of profilin, actin is virtually depolymerized, leading to localization of the cytoplasm and nucleus at the cell periphery (Staiger et al., 1994). Loss of actin as a result of the Dex-induced expression of AtCAP1 would seem to fit with findings discussed for Drosophila in which CAP loss-of-function mutants showed excessive accumulation of actin filaments. Furthermore, studies with actin-depolymerizing drugs (latriculin B) or transgenic plants overexpressing AtADF1 have shown a reduction in cell expansion resulting from the loss of actin filaments (Baluska et al., 2001; Dong et al., 2001), which fits well with the results observed in this study. It is not apparent why the loss of actin filaments resulting from AtCAP1 overexpression would arrest cell division in BY-2 cells; perhaps it is the result of a failure to complete cell plates at cytokinesis, which is known to be a consequence of actin depolymerization after profilin microinjection in dividing and interphase stamen hair cells of Tradescantia (Valster et al., 1997).
Actin is a fundamental component of the cytoskeleton in all eukaryotes and directs the spatial organization of many essential subcellular processes, including cell expansion and division. Interestingly, in contrast to what is known of other eukaryotic organisms, cell divisions apparently occur in plant cells devoid of all actin filaments (Baluska et al., 2001). Therefore, we cannot exclude the possibility that the defects in cell division observed in Arabidopsis plants overexpressing AtCAP1 were attributable to defects in other processes in which AtCAP1 might be involved. CAP proteins were suggested to participate in a signal transduction pathway through their N-terminal domain binding to adenylyl cyclase (Fedor-Chaiken et al., 1990; Field et al., 1990; Kawamukai et al., 1992) or to interact with SH3 proteins by their middle proline-rich sequences (Freeman et al., 1996; Lambrechts et al., 1997). Alterations of the activity of other interacting partners when AtCAP1 is overexpressed remain to be investigated.
The evidence presented here that the overexpression of AtCAP1 inhibited both cell division and cell elongation by alteration of the actin cytoskeleton suggests that AtCAP1 regulates cytoskeleton organization, leading to proper cell proliferation in higher plants.
METHODS
cDNA Library Screening
An Arabidopsis thaliana cDNA library prepared from suspension cells was provided by Dr. Csaba Koncz (Max-Planck-Institute, Cologne, Germany). A 1.5-kb BamHI–KpnI fragment corresponding to the coding region of the cotton CAP (GhCAP) (Kawai et al., 1998) was labeled with α-32P-dCTP (167 TBq/mmol; ICN Biomedicals, Costa Mesa, CA) and used to screen an Arabidopsis cDNA library. After hybridization, membranes were washed in 2 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS two times at room temperature for 10 min and then in 1 × SSC and 0.1% SDS two times at 65°C for 30 min and exposed with a Fujix Bas 1000 Image Analyzer (Fuji Photo Film, Tokyo, Japan). The isolated cDNA containing the partial sequence of AtCAP1 was subcloned into pBluescript II SK− (Stratagene) to create pBK-AtCAP16-1. Analysis of the phylogenetic relationship of CAP homologs was performed using CLUSTAL V (Higgins et al., 1992).
Rapid Amplification of cDNA Ends of the 5′ End of AtCAP1
The Marathon cDNA Amplification Kit (Clontech, Palo Alto, CA) was used to construct an Arabidopsis rapid amplification of cDNA ends adaptor-ligated cDNA library according to the manufacturer's instructions. Two oligonucleotide primers hybridizing to AtCAP1, one located 350 bp (5′-CGCATGCAGCCTTCAAGTGATTG-3′) and the other located 210 bp (5′-CGAACAAGCAGCTCCTTTTGGGA-3′) downstream of the CAP motif, and two nested primers (AP1 and AP2) directed to the Marathon cDNA adaptor primer (Clontech) were used to amplify the 5′ end of AtCAP1. A 100-ng rapid amplification of cDNA ends cDNA library was used as a template to perform the polymerase chain reaction (PCR): 1 cycle at 94°C for 1.5 min, 25 cycles at 94°C for 1 min, and 68°C for 3 min using the Marathon cDNA Amplification Kit (Clontech) and Advantage Klen Taq polymerase (Clontech). Then the amplified products were cloned into pCR2.1 (Invitrogen, Carlsbad, CA) to create plasmid pCR5′-AtCAP1. All clones encoded the same sequence, suggesting that no mutation was introduced during PCR.
Plasmid Construction
The plasmid pBK-AtCAP16-1 was digested with XbaI to remove the 5′ end of the partial clone and ligated with the XbaI fragment derived from the 5′ end of the cDNA on pCR5′-AtCAP1 to create the plasmid pBK-AtCAP1, which contained the full-length cDNA of AtCAP1. The AtCAP1 coding region (open reading frame) was amplified by PCR using oligonucleotide primers directed to the 5′ and 3′ ends of the open reading frame that contain the EcoRI recognition sequence. The PCR fragment was digested with EcoRI and subcloned into a pBluescript II SK− EcoRI site to create the plasmid pBS-AtCAP1. The yeast expression vector pYES2 contains the URA3 gene and 2-μm sequence; it also contains the GAL1 promoter and terminator sequence flanking an EcoRI site. The coding region of AtCAP1 derived from pBS-AtCAP1 was inserted into the EcoRI site of pYES2 to produce the pGAL-AtCAP1 plasmid.
AtCAP1 deletion mutants with 5′ and/or 3′ truncations (A1 to A6) were created by amplification of specific segments of AtCAP1 cDNA by PCR. Six types of deletion mutants were created: A1 (amino acids 1 to 153), A2 (amino acids 1 to 223), A3 (amino acids 1 to 318), A4 (amino acids 224 to 318), A5 (amino acids 224 to 476), and A6 (amino acids 319 to 476). Oligonucleotide primers used for PCR contained mismatches to create the initiation and termination codons and also included an EcoRI recognition site to clone the PCR products into the EcoRI site of pBluescript II SK− (Stratagene). After confirming the sequences, each deletion derivative was subcloned into the EcoRI site of pYES2.
The binary transformation vector pTA7002 containing the two-component glucocorticoid system (Aoyama and Chua, 1997) was digested with XhoI and SpeI (Takara, Tokyo, Japan). The AtCAP1 coding sequence was isolated from pBS-AtCAP1 after digestion with XhoI and SpeI and cloned into pTA7002 to give rise to pTA-AtCAP1.
DNA and RNA Analysis
Genomic DNA was prepared from Arabidopsis plants (ecotype Columbia), digested with EcoRI, HindIII, BamHI, or SacI, size-fractionated by electrophoresis on 0.7% agarose gels, and transferred to Hybond-N+ membranes (Amersham Pharmacia) according to the manufacturer's instructions. Probes used for hybridization were labeled with α-32P-dCTP (167 TBq/mmol; ICN) using Random Primer Labeling Kit version 2 (Takara) according to the manufacturer's instructions. After hybridization, the membranes were washed under high stringency conditions (2 × SSC and 0.1% SDS two times at room temperature for 10 min, 0.1 × SSC and 0.1% SDS two times at 65°C for 30 min).
Total RNA samples were isolated from suspension-cultured cells, cotyledons, rosette leaves, roots, stems, siliques, and flowers. Two oligonucleotide primers hybridizing to AtCAP1, one homologous with the 5′ end (5′-GGAATTCCGATGGAACTTCAAATGGGTCGC-3′) and a second homologous with the 3′ end (5′-CAGCGCATGTTCCACCCAATCTCCA-3′), were used to amplify a 410-bp fragment. Amplification of a 280-bp fragment encoding the 5′ end of the noncoding sequence of the β-Tubulin 4 gene (TUB4; Marks et al., 1987) was performed with TUB4 sense (5′-CGCGGATCCAGTTATTCC-CAAGAAACCGG-3′) and antisense (5′-CGCGGATCCCTTCTCTGC-TTCCTCTTTGC-3′) primers. Two micrograms of total RNA was used as a template to perform the PCR: 1 cycle at 94°C for 1.5 min and 30 cycles at 94°C for 30 sec, 55°C for 30 sec, and 72°C for 1.5 min using a One-Step RT-PCR Kit (Takara) in a TaKaRa PCR Thermal Cycler.
Protein Analysis
For in vitro actin binding assay, a C-terminal fragment of AtCAP1 (A6) that encodes a putative actin binding domain (Freeman et al., 1995) was cloned in the EcoRI site of glutathione S-transferase gene fusion vector pGEX2T (Amersham Pharmacia) to create the GST-A6 fusion protein. This fusion protein expressed in Escherichia coli BL21 overnight at 27°C in the presence of 0.1 mM isopropyl-β-d-thiogalactoside was purified using glutathione–Sepharose 4B (Amersham Pharmacia) according to the manufacturer's protocol. Proteins thus obtained were mixed with bovine monomeric actin (Sigma) followed by absorption using glutathione–Sepharose 4B beads (Amersham Pharmacia). After several washes of beads, the eluted protein samples were subjected to the protein gel blot analysis described below.
For protein gel blot analysis, SDS-PAGE was performed on 12% acrylamide slab gels. Polypeptides were transferred onto polyvinylidene difluoride membranes (Immobilon P; Millipore, Bedford, MA) and reacted with antibodies. A polyclonal antibody against the C-terminal region (residues 466 to 476) of AtCAP1 raised in rabbit and a mouse anti-actin monoclonal antibody (clone 4; ICN Biomedicals) were used. All antibodies were detected with an enhanced chemiluminescence kit (ECL-PLUS; Amersham Pharmacia) and were captured on x-ray film (Fuji Photo Film).
For the analysis of plant proteins, total proteins from different organs of Arabidopsis plants or BY-2 suspension cell cultures were extracted using a procedure described previously (Magyar et al., 1997).
To analyze the in vivo association of AtCAP1 with actin, total protein (150 μg) extracts were immunoprecipitated with either anti-AtCAP1 antibody or preimmune serum, which then were subjected to protein gel blot analysis using an anti-actin antibody.
Yeast Transformation
Yeast transformation was performed by the lithium acetate method (Ito et al., 1983). Other yeast standard genetic manipulations were performed as described by Sherman et al. (1986). Strains were grown on rich medium YPD (1% Bacto yeast extract [Difco], 2% peptone [Difco], and 2% glucose) or YPGS (1% Bacto yeast extract, 2% peptone, and 5% galactose) or minimal medium MVD (0.7% yeast nitrogen base [Difco] and 2% glucose) or MVGS (0.7% yeast nitrogen base and 2% galactose) supplemented with the required amino acids or nucleotide base. MVD or MVGS medium lacking uracil was used to select transformants. Budding yeast strain SKN32 (MATa leu2 ura3 trp1 ade8 can1 cap::HIS3) was described previously by Field et al. (1990). For morphological observation, yeast transformants were cultured in liquid MVGS at 30°C until log phase, collected by brief centrifugation, and subjected to microscopic observation using light microscopy (Nikon, Tokyo, Japan). Bud scars were visualized by staining with the fluorescent dye calcofluor as described by Pringle et al. (1989).
Generation of Transgenic Plants
pTA-AtCAP1 was introduced into Agrobacterium tumefaciens (EHA105) and then into Arabidopsis (ecotype Columbia) using the in planta transformation method (Bechtold et al., 1993). To confirm that each line contained the AtCAP1 construct, genomic DNA was isolated from the potential lines by homogenizing one young leaf according to Nucleon Phytopure DNA Extraction Kit instructions (Amersham Pharmacia), and PCR was performed with oligonucleotide primers directed to the 5′ and 3′ ends of AtCAP1. T2 and T3 seed were collected from these lines and grown on Murashige and Skoog (1962) (MS) medium for subsequent experiments. All plants were grown under continuous light at 23°C.
Tobacco (Nicotiana tabacum cv Bright Yellow 2 [BY-2]) suspension-cultured cells were transformed with A. tumefaciens harboring pTA-AtCAP1 (Sigee et al., 1982). Transformed cells were selected on a medium containing hygromycin (150 μg/mL).
Maintenance of Cell Cultures and Synchronization
Tobacco cells were maintained in a liquid medium (3% sucrose, 4.3 g/L MS salts, 100 mg/mL inositol, 1 mg/L thiamine, 0.2 mg/mL 2,4-D, and 255 mg/L KH2PO4, pH 5.6) on a rotary shaker at 25°C. Cells were subcultured weekly (1:50) into new medium.
For synchronization experiments, 5 mL of BY-2 cells (cultured for 7 days) was transferred to 95 mL of fresh liquid medium containing 5 mg/L aphidicolin (Sigma). After 24 hr of incubation on a rotary shaker, cells were washed four times with 250 mL of fresh medium and finally resuspended in 95 mL with or without 1 μM dexamethasone (Dex). After release from aphidicolin, samples were fixed in 2% formaldehyde, 1 × PBS, and 1 μg/mL 4′,6-diamidino-2-phenylindole to determine the mitotic index (number of mitotic cells/total number of cells) (Hasezawa et al., 2000).
Morphological Observations
The leaves were numbered from the first rosette leaf that emerged after the cotyledons to the last rosette leaf. The appearance of a leaf (1 mm long) was defined as the initiation of a leaf primordium as defined by Kim et al. (1998). The definitions of directions within each leaf blade were those provided originally by Tsuge et al. (1996).
To make the morphological comparison between Dex-treated transgenic and control plants, seed were subjected to cold treatment (4°C) for 1 week to break dormancy and stimulate the synchronization of germination. Plants thus obtained were grown in Dex-free medium until the first two pairs of rosette leaves expanded partially (2 to 3 days). Then, plants were transferred to new medium containing Dex and grown for 7 days at 22°C under continuous light. Third leaves were removed, and blade as well as petiole dimensions were measured. For observation of epidermal and palisade cells, leaf pieces were cut from the central portions of the leaf blades from both the right and left sides. The samples then were submerged in water followed by vacuum for a few seconds until intercellular air spaces were filled. The samples were placed on a glass slide and photographed under bright-field illumination to obtain paradermal images of the layers of leaf cells. The average epidermal cell area was determined by measuring the total area of epidermal cells on a photograph and then dividing this area by the number of epidermal cells in the photograph.
For anatomic analysis, samples were embedded in Technovit 7100 resin (Kulzer and Co., Wehrheim, Germany) and examined as described previously (Kim et al., 1998). To calculate the number of epidermal and palisade cells, transverse sections were made at the center of the first rosette leaves and the number of cells in a single row from one margin to the other, in the width direction, was counted.
The observation of the presence of actin was performed as follows. BY-2 suspension-cultured cells were treated as described by Wasteneys et al. (1997). After fixation and immunolabeling procedures using rhodamine phalloidin (Molecular Probes, Eugene, OR) diluted to a final concentration of 0.33 μM, images of samples were recorded with either a Laser Scanner Fluoview BX50 microscope (Olympus, Tokyo, Japan) or a confocal scanning microscope (Leica, Wetzlar, Germany) on a 600 × 800 or 512 × 512 pixel frame, respectively.
Accession Number
The accession number for AtCAP1 is AB014759.
Acknowledgments
We thank Drs. Maki Kawai, Makoto Kawamukai, Csaba Koncz, Takashi Aoyama, Nam-Hai Chua, Hirokazu Tsukaya, Fumi Kumagai, and Seiichiro Hasezawa for their help with instrumentation and their gifts of materials. Technical assistance from Hiromi Suzuki also is appreciated. This work was supported by the Research for the Future program of the Japan Society for the Promotion of Science and by a Grant-in-Aid for Scientific Research on Priority Areas (Grant No. 10182102) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010301.
References
- Aoyama, T., and Chua, N.H. (1997). A glucocorticoid-mediated transcription factor induction system in transgenic plants. Plant J. 11, 605–612. [DOI] [PubMed] [Google Scholar]
- Baluska, F., Jasik, J., Edelmann, H.G., Salajová, T., and Volkmann, D. (2001). Latriculin B-induced plant dwarfism: Plant cell elongation is F-actin-dependent. Dev. Biol. 231, 113–124. [DOI] [PubMed] [Google Scholar]
- Baum, B., Li, W., and Perrimon, N. (2000). A cyclase-associated protein regulates actin and cell polarity during Drosophila oogenesis and in yeast. Curr. Biol. 10, 964–973. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316, 1194–1199. [Google Scholar]
- Benlali, A., Draskovic, I., Hazelett, D.J., and Treisman, J.E. (2000). act up controls actin polymerization to alter cell shape and restrict Hedgehog signaling in the Drosophila eye disc. Cell 101, 271–281. [DOI] [PubMed] [Google Scholar]
- Bush, S., and Sassone-Corsi, P. (1990). Dimers, leucine zippers and DNA-binding domains. Trends Genet. 6, 36–40. [DOI] [PubMed] [Google Scholar]
- Cohen, C., and Parry, D. (1986). Alpha-helical coiled-coils: A widespread motif in proteins. Trends Biochem. Sci. 11, 245–248. [Google Scholar]
- Dong, C.H., Xia, G.X., Hong, Y., Ramachandran, S., Kost, B., and Chua, N.H. (2001). ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. Plant Cell 13, 1333–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor-Chaiken, M., Deschenes, R.J., and Broach, V. (1990). SRV2, a gene required for RAS activation of adenylate cyclase in yeast. Cell 61, 329–340. [DOI] [PubMed] [Google Scholar]
- Fenger, U., Hofmannn, M., Galliot, B., and Schaller, H.C. (1994). The role of the cAMP pathway in mediating the effect of head activator on nerve-cell determination and differentiation in hydra. Mech. Dev. 47, 115–125. [DOI] [PubMed] [Google Scholar]
- Field, J., et al. (1990). Cloning and characterization of CAP, the S. cerevisiae gene encoding the 70kd adenylyl cyclase-associated protein. Cell 61, 319–327. [DOI] [PubMed] [Google Scholar]
- Freeman, N.L., and Field, J. (2000). Mammalian homolog of the yeast cyclase associated protein, CAP/Srv2p, regulates actin filament assembly. Cell Motil. Cytoskeleton 45, 106–120. [DOI] [PubMed] [Google Scholar]
- Freeman, N.L., Chen, Z., Horensteins, J., Weber, A., and Field, J. (1995). An actin monomer binding activity localizes to the carboxy-terminal half of the Saccharomyces cerevisiae cyclase-associated protein. J. Biol. Chem. 270, 5680–5685. [DOI] [PubMed] [Google Scholar]
- Freeman, N.L., Lila, T., Mintzer, K.A., Chen, Z., Pahk, A.J., Ren, R., Drubin, D.G., and Field, J. (1996). A conserved proline-rich region of the Saccharomyces cerevisiae cyclase-associated protein binds SH3 domains and modulates cytoskeletal localization. Mol. Cell. Biol. 16, 548–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerst, J.E., Ferguson, K., Vojtek, A., Wingler, M., and Field, J. (1991). CAP is a bifunctional component of the Saccharomyces cerevisiae adenylyl cyclase complex. Mol. Cell. Biol. 11, 1248–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwald, U., Brokamp, R., Karakesisoglou, I., Schleicher, M., and Noegel, A. (1996). Identification of a cyclase-associated protein (CAP) homologue in Dictyostelium discoideum and characterization of its interaction with actin. Mol. Biol. Cell 7, 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasezawa, S., Ueda, K., and Kumagai, F. (2000). Time-sequence observations of microtubule dynamics throughout mitosis in living cell suspensions of stable transgenic Arabidopsis-direct evidence for the origin of cortical microtubules at M/G1 interface. Plant Cell Physiol. 41, 244–250. [DOI] [PubMed] [Google Scholar]
- Higgins, D.G., Bleasby, A.J., and Fuchs, V. (1992). CLUSTAL V: Improved software for multiple sequence alignment. Comput. Appl. Biosci. 2, 189–191. [DOI] [PubMed] [Google Scholar]
- Ito, H., Fukuda, Y., Murata, K., and Kimura, A. (1983). Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota, A., and Wada, M. (1992). Photoinduction of formation of circular structures by microfilaments on chloroplasts during intracellular orientation in protonemal cells of the fern Adiantum capillus-veneris. Protoplasma 167, 97–107. [Google Scholar]
- Kataoka, T., Broek, D., and Wigler, M. (1985). DNA sequence and characterization of the S. cerevisiae gene encoding adenylate cyclase. Cell 43, 493–505. [DOI] [PubMed] [Google Scholar]
- Kawai, M., Aotsuka, S., and Uchimiya, H. (1998). Isolation of a cotton CAP gene: A homologue of adenylyl cyclase-associated protein highly expressed during fiber elongation. Plant Cell Physiol. 39, 1380–1383. [DOI] [PubMed] [Google Scholar]
- Kawamukai, M., Gerst, J., Field, J., Riggs, M., Rodgers, L., Wigler, M., and Young, D. (1992). Genetic and biochemical analysis of the adenylyl cyclase associated protein, cap, in S. pombe. Mol. Cell. Biol. 3, 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, G.T., Tsukaya, H., and Uchimiya, H. (1998). The CURLY LEAF gene controls both division and elongation of cells during the expansion of the leaf blade in Arabidopsis thaliana. Planta 206, 175–183. [DOI] [PubMed] [Google Scholar]
- Kost, B., Spielhofer, P., and Chua, N.H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 6, 393–401. [DOI] [PubMed] [Google Scholar]
- Kost, B., Mathur, J., and Chua, N.H. (1999). Cytoskeleton in plant development. Curr. Opin. Plant Biol. 2, 462–470. [DOI] [PubMed] [Google Scholar]
- Lambrechts, A., Verschelde, J.L., Jonckheere, V., Goethals, M., Vandekerckhove, J., and Ampe, C. (1997). The mammalian profilin isoforms display complementary affinities for PIP2 and proline-rich sequences. EMBO J. 16, 484–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lila, T., and Drubin, D.G. (1997). Evidence for physical and functional interactions among two Saccharomyces cerevisiae SH3 domain proteins, an adenylyl cyclase-associated protein and the actin cytoskeleton. Mol. Biol. Cell 8, 367–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar, Z., et al. (1997). Cell cycle phase specificity of putative cyclin-dependent kinase variants in synchronized alfalfa cells. Plant Cell 9, 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks, M.D., West, J., and Weeks, D.P. (1987). The relatively large beta-tubulin gene family of Arabidopsis contains a member with an unusual transcribed 5′ noncoding sequence. Plant Mol. Biol. 10, 91–104. [DOI] [PubMed] [Google Scholar]
- Mathur, J., Spielhofer, P., Kost, B., and Chua, N.H. (1999). The actin cytoskeleton is required to elaborate and maintain spatial patterning during trichome cell morphogenesis in Arabidopsis thaliana. Development 126, 5559–5568. [DOI] [PubMed] [Google Scholar]
- Matviw, H., Yu, G., and Young, D. (1992). Identification of a human cDNA encoding a protein that is structurally and functionally related to the yeast adenylyl cyclase-associated CAP proteins. Mol. Cell. Biol. 12, 5033–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher, R.B., and Williamson, R.E. (1994). The plant cytoskeleton. In Arabidopsis, E. Meyerowitz and C. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 1049–1084.
- Mintzer, K.A., and Field, J. (1994). Interactions between adenylyl cyclase, CAP, and RAS from Saccharomyces cerevisiae. Cell Signal. 6, 681–694. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nagata, T., and Kumagai, F. (1999). Plant cell biology through the window of the highly synchronized tobacco BY-2 cell line. Methods Cell Sci. 21, 123–127. [DOI] [PubMed] [Google Scholar]
- Pathsarathy, M.V., Perdue, T.D., Witzmun, A., and Alvernaz, J. (1985). Actin network as a normal component of the cytoskeleton in many vascular plant cells. Am. J. Bot. 72, 1318–1323. [Google Scholar]
- Pringle, J., Preston, R., Adams, A., Stearms, T., Drubin, D., Haarer, B., and Jones, E. (1989). Fluorescence microscopy methods for yeast. Methods Cell Biol. 31, 357–435. [DOI] [PubMed] [Google Scholar]
- Ramachandran, S., Christensen, H.E.M., Ishimaru, Y., Dong, C.H., Cho-Ming, W., Cleary, A.L., and Chua, N.H. (2000). Profilin plays a role in cell elongation, cell shape maintenance, and flowering in Arabidopsis. Plant Physiol. 124, 1637–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., Fink, G.R., and Hicks, J.B. (1986). Techniques and protocols. In Laboratory Course Manual for Methods in Yeast Genetics, F. Sherman, G.R. Fink, and J.B. Hicks, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp.117–167.
- Sigee, D.C., Smith, V.A., and Hindley, J. (1982). Passage of bacterial DNA into host cells during in vitro transformation of Nicotiana tabacum by Agrobacterium tumefaciens. Microbios 34, 113–132. [PubMed] [Google Scholar]
- Staiger, C.J., Yaun, M., Valenta, R., Shaw, P.J., Warn, R.M., and Lloyd, C.W. (1994). Microinjected profilin affects cytoplasmic streaming in plant cells by rapidly depolymerizing actin microfilaments. Curr. Biol. 4, 215–219. [DOI] [PubMed] [Google Scholar]
- Swiston, J., Hubbertey, A., Yu, G., and Young, D. (1995). Differential expression of CAP and CAP2 in adult rat tissues. Gene 165, 273–277. [DOI] [PubMed] [Google Scholar]
- Toda, T., Uno, I., Ishikawa, T., Powers, S., Kataoka, T., Broek, D., Cameron, S., Broach, J., Matsumoto, K., and Wingler, M. (1985). In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell 40, 27–36. [DOI] [PubMed] [Google Scholar]
- Tsuge, T., Tsukaya, H., and Uchimiya, H. (1996). Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 122, 1589–1600. [DOI] [PubMed] [Google Scholar]
- Valster, A.H., Pierson, E.S., Valenta, R., Hepler, P.K., and Emons, A.M. (1997). Probing the plant actin cytoskeleton during cytokinesis and interphase by profilin microinjection. Plant Cell 9, 1815–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek, A., Haarer, B., Field, J., Gerst, J., Pollard, T., Brown, S., and Wigler, M. (1991). Evidence for a functional link between profilin and CAP in the yeast S. cerevisiae. Cell 66, 497–505. [DOI] [PubMed] [Google Scholar]
- Vojtek, A.B., and Cooper, J.A. (1993). Identification and characterization of a cDNA encoding a mouse CAP: A homolog of the yeast adenylyl cyclase associated protein. J. Cell Sci. 105, 777–785. [DOI] [PubMed] [Google Scholar]
- Wasteneys, G.O., Willingale-Theune, J., and Menzel, D. (1997). Freeze shattering: A simple and effective method for permeabilizing higher plant cell walls. J. Microsc. 188, 51–61. [DOI] [PubMed] [Google Scholar]
- Yu, G., Swiston, J., and Young, D. (1994). Comparison of human CAP and CAP2, homologs of the yeast adenylyl cyclase-associated proteins. J. Cell Sci. 107, 1671–1678. [DOI] [PubMed] [Google Scholar]
- Zelicof, A., Gatica, J., and Gerst, J.E. (1993). Molecular cloning and characterization of a rat homolog of CAP, the adenylate cyclase-associated protein from Saccharomyces cerevisiae. J. Biol. Chem. 268, 13448–13453. [PubMed] [Google Scholar]
- Zelicof, A., Protopopov, V., David, D., Lin, X.Y., Lustgarten, V., and Gerst, J.E. (1996). Two separate functions are encoded by the carboxyl-terminal domains of the yeast cyclase-associated protein and its mammalian homologs: Dimerization and actin binding. J. Biol. Chem. 271, 18243–18252. [DOI] [PubMed] [Google Scholar]
- Zhou, G.L., Miyazaki, Y., Nakagawa, T., Tanaka, K., Shishido, K., Matsuda, H., and Kawamukai, M. (1998). Identification of a CAP (adenylyl-cyclase-associated protein) homologous gene in Lentinus edodes and its functional complementation of yeast CAP mutants. Microbiology 144, 1085–1093. [DOI] [PubMed] [Google Scholar]