Abstract
Testicular germ cell tumor (TGCT) is the most common tumor type among adolescent and young adult males. Familial clustering and bilateral disease are suggestive of a genetic predisposition among a subgroup of these patients, but susceptibility genes for testicular cancer have not yet been identified. However, suggestive linkage between disease and genetic markers has been reported at loci on chromosome arms 3q, 5q, 12q, 18q, and Xq. We have analyzed primary familial/bilateral (n=20) and sporadic (n=27) TGCTs, including 28 seminomas and 19 nonseminomas, for allelic imbalance (AI) within the autosomal regions. DNA from all tumors were analyzed by fluorescent polymerase chain reaction of 22 polymorphic loci at 3q27-ter, 5q13-35.1, 12q21-ter, and 18q12ter. All tumor genotypes were evaluated against their corresponding constitutional genotypes. The percentages of TGCTs with genetic changes at 3q, 5q, 12q, and 18q, were 79%, 36%, 53% and 43%, respectively. The frequencies at 3q and 12q in nonseminomas were significantly higher than in seminomas (P=.003 and P=.004). In order to evaluate changes at hemizygous Xq loci, five loci were analyzed by co-amplification with an autosomal reference marker known to reveal retained heterozygosity in the tumor DNA. Gain of Xq sequences was seen in more than 50% of the tumors. The degree of amplification varied among the loci in each of five tumors, and based on these breakpoints, a common region of overlapping gains was found at Xq28. No significant differences were found between the frequencies of genetic changes in familial/bilateral versus sporadic tumors, an observation speaking in disfavor of the existence of a single susceptibility gene for TGCT in any of the analyzed regions. Our data suggest that gain of genetic material at distal Xq and losses at 5q and 18q contribute to establishment of seminomas, whereas imbalances at 3q as well as gain at distal part of 12q are associated with further progression into nonseminomas.
Keywords: allelic imbalance, familial cancer, loss of heterozygosity, susceptibility gene, testicular germ cell tumor
Introduction
Testicular germ cell tumor (TGCT) is the most common malignancy among young white males, and the incidence is increasing rapidly [1–3]. TGCTs are subdivided into two main histological entities: the undifferentiated seminomas, and the nonseminomas, composed of embryonic neoplastic germ cells, which mimic the histogenesis of an early embryo. Seminomas are believed to arise from a carcinoma in situ stage, and may develop into nonseminomas [4,5]. TGCTs are characterized by overrepresentation of chromosome arm 12p, often through the presence of isochromosome 12p [6,7], and nonrandom losses and gains of certain chromosomes [5,8–11]. TGCTs are nearly always hyperdiploid, and are frequently in the triploid range [5,12].
The cause of TGCT remains unknown. Increased incidence over time and correlation with socioeconomic class point toward influence of environmental factors. The observed familial clustering of TGCT, particularly among brothers, may be due to their exposure to similar environments, in utero, or as children [13–16]. However, the four-fold increased risk for fatherson transmission indicates a genetic predisposition [14]. Men with GCT in one testis are at increased risk of developing a contralateral malignancy [17]. The presence of bilateral neoplastic changes supports a genetic susceptibility for TGCT, but is also consistent with exposure to environmental carcinogens. Statistical analyses by Nicholson and Harland [18] suggest that patients with bilateral disease carry the same genetic predisposition as familial cases, and that approximately one third of all men with TGCT is genetically predisposed to the disease.
The International Testicular Cancer Linkage Consortium (ITCLC) analyzed 220 polymorphic microsatellite loci throughout the autosomal genome in selected families with two or more cases of testicular cancer. None of the markers showed conclusive evidence of a close map position to a TGCT predisposing gene, but loci on chromosome arms 3q, 5q, 12q, and 18q showed suggestive linkage to the disease [19]. Recently, Rapley et al. [20] found significant linkage between markers at Xq27 and TGCT within a subset of TGCT families (hLOD=4.7).
In the present study, series of familial/bilateral and sporadic TGCTs, comparable according to histology and percentage of tumor cells, were analyzed for somatic alterations at polymorphic microsatellite loci, within and near the five candidate regions.
Materials and Methods
Samples from the TGCT Patients
Primary tumor biopsies and corresponding peripheral blood samples were obtained from 47 Norwegian TGCT patients. The patients were grouped into cases of familial and/or bilateral TGCT (n=20) and cases of sporadic cancer (n=27). Among the 20 familial/bilateral TGCTs, 13 were bilateral, 11 had affected family members, and thus, 4 had both bilateral tumors and familial occurrence of the disease. Four of the familial/bilateral TGCTs were from patients with history of cryptorchidism. Median age at diagnosis was 29 years for the familial/bilateral group and 30 for the sporadic.
Three 5 µm sections were taken from different parts of each frozen tumor sample prior to DNA isolation. The sections were stained with hematoxylin and eosin and visually evaluated by light microscopy. The various tumor components were described according to the WHO's recommendations [21], and percentage of intact neoplastic cells was estimated for each section. An average of the three sections per tumor sample was calculated. Among all tumors, an average of 75% tumor cells was found (range 30–100%). The familial/bilateral and sporadic tumor groups were comparable according to histology and estimated percentage of tumor cells. A total of 28 seminomas included 13 familial/bilateral and 15 sporadic tumors, and among the 19 nonseminomas, 7 were familial/bilateral and 12 sporadic TGCTs.
DNA was isolated from blood and tumor tissues by applying the phenol/chloroform extraction principle [22].
Microsatellite Analyses
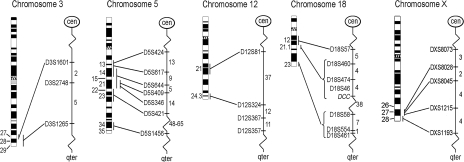
Throughout the five candidate regions suspected to carry a TGCT susceptibility gene [19,20,23], we investigated markers at 27 microsatellite loci (Figure 1). Primer sequences and allele diversities were obtained from the Human Genome Database [24] and the Généthon human linkage map [25].
Figure 1.
Map positions of the analyzed microsatellite markers. All markers have (CA)n dinucleotide repeats, except D5S1456 that has a (GATA)n tetranucleotide repeat. Numbers to the left of each ideogram indicate the chromosome bands. Numbers to the right of each autosomal genetic map indicate the sex-averaged map distance between the markers in centi Morgan (cM). For the X chromosome, this value represents the female recombination (fcM) value, based on the Généthon human linkage map [25].
3q27-ter Five members of a cancer-prone Canadian kindred who all developed TGCT [26] shared a common haplotype for three markers in the 3q telomeric region. We analyzed the same three markers, D3S1601, D3S2748, and D3S1265, which are all located in the 3q27-ter candidate region [19].
5q13-35.1 The candidate region at 5q suggested by ITCLC lies between the markers D5S428 (maps together with the more informative marker D5S617) and D5S421. Leahy et al. [23] suggested a target region between D5S428 and D5S409. The marker D5S346 is closely located to adenomatous polyposis coli (APC) [27], a candidate tumor-suppressor gene on 5q21 [28,29]. Additional three markers were included to flank and refine this candidate region.
12q21-ter The ITCLC results showed increasing linkage evidence along the long arm of chromosome 12, as the markers became more distal [19]. We therefore analyzed three markers in the q telomeric region (12q24.3) as well as one more proximal marker.
18q12-ter The suggestive linkage evidence at 18q spanned several chromosome bands. D18S554 at 18q23 was found to be the marker with the overall highest linkage score (nonparametric linkage=1.6) in the ITCLC study [19]. This and two flanking markers, D18S58 and D18S461, were included in the present study. Five additional markers mapping to 18q12-21 were also analyzed due to the clustering of putative tumor-suppressor genes (e.g. DCC, SMAD2, and DPC4) in this region.
Xq27-ter Five markers were chosen to cover and flank the Xq27 region defined by Rapley et al. [20].
Polymerase chain reaction (PCR) conditions The 10 µl reaction volume consisted of 1x GeneAmp PCR buffer with 1.5 mM MgCl2 (Applied Biosystems, Foster City, CA, USA), 2 to 5 pmol of each primer (DNA Technology, Aarhus, Denmark), 200 µM each of the four dNTPs (Amersham Pharmacia Biotech., London, UK), 0.4 units AmpliTaq DNA Polymerase (Applied Biosystems, Foster City, CA, USA), and 50 ng DNA template. The forward primers were 5′-labeled with HEX, TET, or 6-FAM fluorochromes. Three primer pairs were multiplexed in each PCR.
The PCR was carried out in a 96-well format using an MJ PTC-200 thermocycler (MJ Research, Watertown, MA, USA). Two minutes of denaturation at 94°C was followed by 27 cycles of 30 seconds denaturation at 94°C, 75 seconds annealing at 55°C, and 15 seconds elongation at 72°C, before 6 minutes final extension at 72°C.
Detection of PCR products PCR products from two multiplex reactions (2x0.8 µl) were pooled to allow capillary electrophoresis of six loci simultaneously. This was further mixed with 0.5 µl GeneScan-350 [TAMRA] Size Standard (Applied Biosystems, Foster City, CA, USA) in 12 µl deionized formamide, CH3NO (Kodak Eastman Chemical, New Haven, CT, USA), followed by capillary electrophoresis on an ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). The samples were electrokinetically injected for 1 to 20 seconds into a 47x50 µm capillary, and electrophoresed at 15 kV for 23 minutes. The resulting electropherograms represented relative intensities of four different fluorescent dyes with respect to electrophoresis time (i.e., sizes of DNA fragments). The softwares GeneScan 3.1 and GenoTyper 2.1 (Applied Biosystems, Foster City, CA, USA) were used to analyze the electropherograms, before the allele peak heights were further exported to Microsoft Excel.
Determination of allelic imbalance (AI) and loss of heterozygosity (LOH) A semiquantitative expression of AI, QLOH, was calculated as the ratio of the allele intensity ratios in tumor and blood (constitutional) DNA, as in [tumor allele 1/tumor allele 2]/[blood allele 1/blood allele 2]. When this value was greater than one, QLOH was set to be the inverse. For designation of AI at a locus, we required two independent amplifications of the specific marker where both showed QLOH values less than or equal to 0.84 [30]. The mean QLOH value was used further. The 0.84 cut-off value was determined due to the standard deviation of QLOH among samples with retained heterozygosity (SD=0.083). This gives a probability of 99.75% that a scored AI is real, and not due to technical error, given independence between the errors of repeated PCRs [30].
The TGCTs comprise a heterogeneous group of neoplasms, both with respect to different tumor components, and the varying presence of normal cells in the tumor biopsies. These factors must be considered when scoring LOH. In the present study, LOH was scored when QLOH was less than or equal to the estimated fraction of normal cells in the tumor biopsy. The latter is of course somewhat subjective, but still, this way of LOH scoring is safer than the usual practice, designating all tumors as LOH if their QLOH values are below a certain fixed threshold value. However, no matter how low the QLOH value is, it is still possible that it reflects gain of one allele, and not loss of the other. Therefore, we obtained additional information on the nature of our AIs by comparing our results to those of a separate study, analyzing 33 of the same tumors by comparative genomic hybridization (CGH) [31].
Determination of the results for the X chromosome markers An AI approach is not possible for investigation of X chromosome markers in male tumors because of their constitutional hemizygosity. Together with the X markers, we therefore co-amplified an autosomal reference marker with QLOH value known to be close to 1.00. We then compared the peak heights of the X markers with the peak heights of the reference, in both blood and tumor DNA, to see whether the X markers were over- or underrepresented in tumor DNA, compared to the reference. The results were always confirmed by a second independent PCR.
Results
Analysis of AI and LOH at Autosomal Loci
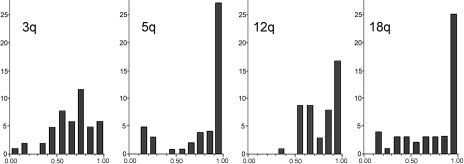
Forty-seven TGCTs were analyzed for AI and LOH at 22 autosomal polymorphic loci covering four autosomal candidate regions for TGCT susceptibility. The distributions of the tumors' average QLOH values are shown in Figure 2. The frequencies of tumors showing alterations (i.e., confirmed QLOH≤0.84) at one or more loci at 3q, 5q, 12q, or 18q were 79%, 36%, 53%, and 43%, respectively (Table 1). The frequency of changes in the 3q region was significantly higher than for each of the 5q, 12q, and 18q regions (P<.001, P=.009, and P<.001, respectively).
Figure 2.
The distribution of QLOH (x-axis) for each of the analyzed autosomal regions. For all tumors, an average QLOH value was found along each of the four investigated autosomal regions, and the distributions are shown in the histograms above, e.g., if a tumor showed 0.39, 0.43, and 0.41 for the three loci at chromosome arm 3q, the average value of 0.41 contributed to the bar representing QLOH values from 0.4 to 0.5 in the 3q histogram. The y-axis shows the number of tumors in each histogram group. The figure illustrates the infrequent LOH at chromosome arm 12q, where only one average QLOH value is less than 0.5. For the chromosome arms 3q, 5q, and 18q, there were 10, 9, and 11 tumors with average QLOH values less than 0.5, respectively. Few tumors have average QLOH value near 1.0 at 3q loci, in contrast to the other regions.
Table 1.
Frequencies of Tumors Showing AI, LOH, and the Total Frequency of Change (AI+LOH).
| All tumors (n=47) (%) | Familial/bilateral (n=20) (%) | Sporadic (n=27) (%) | Seminomas (n=28) (%) | Nonseminomas (n=19) (%) | ||
| 3q | AI | 47 | 30 | 60 | 36 | 63 |
| LOH | 32 | 40 | 26 | 29 | 37 | |
| Total | 79 | 70 | 85 | 64 | 100 | |
| 5q | AI | 15 | 20 | 11 | 18 | 11 |
| LOH | 21 | 25 | 19 | 18 | 26 | |
| Total | 36 | 45 | 30 | 36 | 37 | |
| 12q | AI | 45 | 40 | 48 | 29 | 68 |
| LOH | 9 | 5 | 11 | 7 | 11 | |
| Total | 53 | 45 | 59 | 36 | 79 | |
| 18q | AI | 15 | 15 | 15 | 14 | 16 |
| LOH | 28 | 25 | 30 | 21 | 37 | |
| Total | 43 | 40 | 44 | 36 | 53 | |
LOH was found in 32%, 21%, 9%, and 28% of the tumors at the 3q, 5q, 12q, and 18q loci, respectively (Table 1). The LOH frequency at the 12q loci was significantly lower than for each of the 3q, 5q, and 18q regions (P=.005, .05, and .02, respectively).
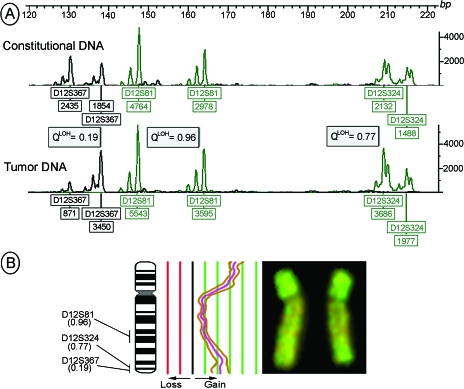
Breakpoints in the AI/LOH pattern within the investigated regions were seen in six tumors. At the 5q region, two tumors showed retained heterozygosity at D5S644, but AI at the more distal markers. At 12q, one tumor revealed retained heterozygosity at D12S81, but increasingly stronger AI toward the telomere (Figure 3A). Another tumor showed AI at D12S324, but retained heterozygosity for the flanking markers. At 18q, two tumors showed either AI or retained heterozygosity at D18S57, and LOH or AI at the more distal markers, respectively.
Figure 3.
Chromosome 12 alterations in a mixed TGCT. (A) The electropherograms of three markers amplified in blood (constitutional) and tumor DNA show the allele intensities in relative fluorescence units (y-axis and peak heights in boxes below the alleles). The tumor showed gradually stronger AI (decreasing QLOH) toward the distal 12q loci. A second PCR of the same markers and templates confirmed the results, and showed QLOH values of 0.20, 0.97, and 0.77. The fourth investigated 12q marker, D12S357 (not shown), was constitutionally homozygous, and thus not informative. (B) CGH of the tumor showed gain of the whole chromosome with additional amplification of two regions. The central curve shows the average fluorescence ratio of 14 chromosomes between tumor and reference DNA, whereas the two flanking curves represent the 95% confidence interval. The gain of the short arm might reflect the isochromosome 12p, a frequent and characteristic aberration in germ cell tumors, but interestingly, the distal part of the long arm is also amplified. A QLOH value of 0.19 (as for D12S367) will almost exclusively, in any AI/LOH study, be interpreted as LOH. However, upon comparison with CGH data, we see that the AI in this tumor is most likely caused by amplification of genetic material (complete CGH — copy number karyotypes — for all tumors will be published elsewhere; Ref. [31]).
Familial/Bilateral versus Sporadic
The overall frequencies of tumors showing AI or LOH in all investigated regions were 51% among the familial/bilateral and 55% among the sporadic tumors. No significant differences were seen comparing the familial/bilateral and the sporadic tumors for genetic changes within the individual regions (Table 1). For 3q and 12q, AI/LOH was found in 85% and 59% of the sporadic tumors, whereas 70% and 45%, respectively, among the familial/bilateral (P=.21 and P=.33).
Seminomas versus Nonseminomas
The overall number of changes was significantly higher among the nonseminomas than for the seminomas (P<.001). The frequencies of genetic changes at 3q and 12q in nonseminomas (100% and 79%, respectively) were significantly higher than in seminomas (64% and 36%; P=.003 and P=.004, respectively).
Analysis of X Chromosome Loci
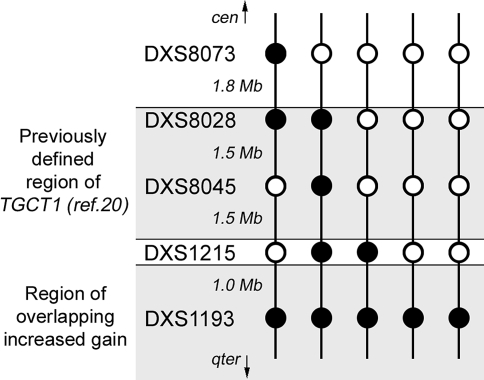
Thirty-eight of the 47 pairs of blood/tumor DNA were analyzed at five loci on the X chromosome. In general, the peak heights showed increased values from blood to tumor, relative to their co-amplified autosomal reference markers. Though heterozygous (QLOH>0.84), the reference markers may still have altered copy numbers in tumor, and thus, the X markers' status as gained or lost is not definite by this approach. More interesting are the observed breakpoints between the peak heights of neighboring X chromosome markers, relative to their common reference marker. Five tumors with such breakpoints were seen, and altogether, they showed increased gain toward the more distal markers (Figure 4).
Figure 4.
Closing in on TGCT1. This panel shows the pattern of the five tumors with breakpoints in their X marker peak heights, compared to their reference peak heights. The filled circles indicate markers with increased gain compared to their neighboring markers (open circles). DXS1193 was the only marker showing increased gain in all these tumors.
Discussion
AI in TGCTs
AI studies of TGCT are complicated by tumor heterogeneity, where both tumor tissues of different histologic types and also normal cells often are intermingled. Furthermore, the tumor can be genetically heterogeneous within a morphologically homogeneous component. Thus, AI can be the result of LOH masked by both normal cells and by other subclones of the tumor with retained heterozygosity. Qualitative and semiquantitative histological examinations of tumor cross sections ensure better interpretation of AI studies. In this study, the percentage of intact tumor tissue was estimated in all biopsies used for DNA isolation, and this was taken into account when scoring LOH among the AI cases. However, detection of AI can also reflect gain of one of the alleles. For better interpretation of the AIs in our study, we therefore compared our data with the results of a separate study [31], where 33 of the same tumors were analyzed by CGH. That study showed net loss at chromosome arms 5q and 18q, in 48% and 52% of the tumors, respectively, and none of the tumors revealed gain. At distal 12q and 3q, 60% and 12% of the tumors showed gain, and none showed loss. When comparing these results with the present study, one should bear in mind the different resolutions inherent in the two methods. Furthermore, skewed intensities between the homologues, but unchanged overall copy number (as is the case with uniparental disomy), are only detected by the AI approach, whereas simultaneous gain of both homologues is only revealed by CGH.
We reported at the 2000 AACR Annual Meeting [32] the high frequencies of AI at 3q, 5q, 12q, and 18q. This was recently confirmed by Faulkner et al. [33] who reported frequencies of LOH comparable to our frequencies of AI. However, this study did not take into account that the imbalances also might reflect gain of genetic material.
Frequent Changes at 3q are Due to Both Loss and Trisomy
The overall frequency of AI was significantly higher for the 3q loci than for any other investigated loci. However, only 4 of 33 TGCTs investigated by CGH showed changes (all gain) at distal 3q. This indicates that AI detected at 3q usually reflects trisomy, which will be hidden by a CGH approach by a near triploid background. This is in keeping with previous cytogenetic findings [5]. Furthermore, studies have indicated trisomy 3 to be more frequent in nonseminomas than in seminomas [5,7], which is in agreement with our AI data for the 3q loci. However, the investigated 3q loci also showed the highest frequencies of LOH, indicating that a substantial share of the changes at 3q is not caused by trisomy. In addition, the group of tumors with lowest QLOH values was not correlated with any aberration seen by CGH, indicating the involvement of a relatively small region. Because the seminomas and nonseminomas both show similar and high frequencies of LOH for the 3q markers, the loss of genetic sequences on chromosome 3 is most likely an early event in TGCT development, and the 3q27-ter candidate region may harbor a TGCT suppressor gene.
AI at 5q and 18q is Due to Loss of Genetic Material
Our AI data, together with the corresponding CGH results, give evidence that the frequent imbalances at 5q and 18q result from loss of genetic material. The observed LOH frequencies at these genomic regions are in keeping with previous studies [34–36], and are similar in both seminomas and nonseminomas. Thus, loss of genetic material from these regions appears to be an early event in the TGCT development.
AI at 12q Loci is Due to Gain of Genetic Material
Isochromosome 12p, i(12p), is present in more than 80% of human TGCTs [7]. However, Rodriguez et al. [7] hypothesized that the pathogenetic trigger in TGCT is not the gain of 12p, but the simultaneous loss of a putative tumor-suppressor gene at 12q. LOH has previously been reported in 50% of TGCTs at one or more loci along 12q [37]. In the present study, we show a similar frequency of AI (55%) at 12q loci. However, the frequent gain of 12q sequences seen by CGH, and not a single event of loss, together with significantly lower frequencies of LOH than at all the other investigated regions, suggests that AI scored at 12q loci reflects gain, rather than loss, of genetic material. The significantly higher proportion of AI among nonseminomas than among seminomas indicates that this gain is involved in the progression of seminomas into nonseminomas.
The results of the ITCLC study showed increasing linkage evidence for the 12q markers as they approached the telomere [19]. This correlates with the gain at distal 12q seen by CGH in some of the tumors (Figure 3B). Microsatellites are underrepresented in subtelomeric regions [38], and analyses of more distal markers could reveal even stronger evidence of linkage in the ITCLC study.
AI/LOH at Syntenic Loci
Due to the low number of breakpoints in the AI/LOH pattern, we have not defined any smallest region of overlapping imbalances within the autosomal regions. This might indicate that it is not the loss or gain of one particular gene, but the unison copy number change of several genes along a chromosomal region that is important for TGCT development. The clustering of known tumor-suppressor genes at 5q21 and 18q21 supports this theory.
Closing in on TGCT1?
Our results on the X chromosome are in agreement with molecular cytogenetic studies on TGCT, showing a general overrepresentation of the X chromosome in the tumor DNA [39–41], and thus indicating the existence of one or more genes on the X chromosome which, upon up-regulation, contributes to TGCT development. Recently, Rapley et al. [20] found evidence for a TGCT susceptibility locus, TGCT1, at Xq27, between the markers DXS8028 and FMR1Di (2.5 female cM proximal to DXS1215). However, this region was limited by only one recombination event on each side of the region. Five of our investigated tumors showed breakpoints in the amplification level among the investigated X chromosome markers (Figure 4). Although speculative, one may hypothesize from these somatic changes that TGCT1 may have a more distal map position, or a second target gene is present on Xq, distal of DXS1215 and TGCT1.
Similar Frequencies of Genetic Changes between Familial/Bilateral and Sporadic TGCTs Speak in Disfavor of One Single Susceptibility Gene
A segregation analysis on TGCT families and an analysis based on the frequency of bilateral disease gave evidence for an autosomal recessive inheritance mode [18,42]. Individuals with familial/bilateral TGCT may thus have inherited two inactive alleles of a tumor-suppressor gene with limited penetrance. Those with sporadic TGCT are then thought to be heterozygous for the gene, and somatic mutation, imprinting, and loss are possible second steps in the total inactivation of the tumor-suppressor gene.
The fact that none of the candidate regions showed significantly different frequencies of genetic changes between the familial/bilateral and the sporadic tumor groups speaks in disfavor of the existence of one single TGCT susceptibility gene. However, the high frequencies of genetic changes within the investigated regions suggest their importance in the development of primary TGCTs. One may hypothesize that different genes located within the different candidate regions are responsible for the predisposition in different individuals, or that several genes together give an elevated risk of TGCT.
Based on the model seminomas arise from carcinomas in situ, and may develop into nonseminomas [5], our data suggest that gain of genetic material at distal Xq, and losses at 5q and 18q, contribute to establishment of seminomas, whereas imbalances at 3q and gain at distal part of 12q are associated with further progression into nonseminomas.
Acknowledgements
R. I. S. and S. M. K. are research fellows of the Research Council of Norway and the Norwegian Cancer Society (NCS), respectively.
Abbreviations
- AI
allelic imbalance
- CGH
comparative genomic hybridization
- GCT
germ cell tumor
- ITCLC
international testicular cancer linkage consortium
- LOH
loss of heterozygosity
- TGCT
testicular germ cell tumor
Footnotes
This study was supported by grants from the NCS (R. A. L.).
References
- 1.Devesa SS, Blot WJ, Stone BJ, Miller BA, Tarone RE, Fraumeni JFJ. Recent cancer trends in the United States. J Natl Cancer Inst. 1995;87:175–182. doi: 10.1093/jnci/87.3.175. [DOI] [PubMed] [Google Scholar]
- 2.Ekbom A, Akre O. Increasing incidence of testicular cancer — birth cohort effects. APMIS. 1998;106:225–229. doi: 10.1111/j.1699-0463.1998.tb01340.x. [DOI] [PubMed] [Google Scholar]
- 3.Hoff WE, Tretli S, Fosså SD. Trends in incidence of testicular cancer in Norway 1955–1992. Eur J Cancer. 1995;31A:2044–2048. doi: 10.1016/0959-8049(95)00321-5. [DOI] [PubMed] [Google Scholar]
- 4.Raghavan D, Sullivan AL, Peckham MJ, Neville AM. Elevated serum alpha-fetoprotein and seminoma: clinical evidence for a histologic continuum? Cancer. 1982;50:982–989. doi: 10.1002/1097-0142(19820901)50:5<982::aid-cncr2820500528>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 5.De Jong B, Oosterhuis JW, Castedo SM, Vos A, te Meerman GJ. Pathogenesis of adult testicular germ cell tumors. A cytogenetic model. Cancer Genet Cytogenet. 1990;48:143–167. doi: 10.1016/0165-4608(90)90115-q. [DOI] [PubMed] [Google Scholar]
- 6.Atkin NB, Baker MC. Specific chromosome change, i(12p), in testicular tumours? Lancet. 1982;2:1349. doi: 10.1016/s0140-6736(82)91557-4. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez E, Mathew S, Reuter V, Ilson DH, Bosl GJ, Chaganti RS. Cytogenetic analysis of 124 prospectively ascertained male germ cell tumors. Cancer Res. 1992;52:2285–2291. [PubMed] [Google Scholar]
- 8.Castedo SM, de Jong B, Oosterhuis JW, Seruca R, Idenburg VJ, Dam A, te MG, Koops HS, Sleijfer DT. Chromosomal changes in human primary testicular nonseminomatous germ cell tumors. Cancer Res. 1989;49:5696–5701. [PubMed] [Google Scholar]
- 9.Castedo SM, de Jong B, Oosterhuis JW, Seruca R, te MG, Dam A, Schraffordt KH. Cytogenetic analysis of ten human seminomas. Cancer Res. 1989;49:439–443. [PubMed] [Google Scholar]
- 10.Lothe RA, Peltomäki P, Tommerup N, Fosså SD, Stenwig AE, Børresen AL, Nesland JM. Molecular genetic changes in human male germ cell tumors. Lab Invest. 1995;73:606–614. [PubMed] [Google Scholar]
- 11.Murty VV, Bosl GJ, Houldsworth J, Meyers M, Mukherjee AB, Reuter V, Chaganti RS. Allelic loss and somatic differentiation in human male germ cell tumors. Oncogene. 1994;9:2245–2251. [PubMed] [Google Scholar]
- 12.Chaganti RS, Rodriguez E, Bosl GJ. Cytogenetics of male germ cell tumors. Urol Clin North Am. 1993;20:55–66. [PubMed] [Google Scholar]
- 13.Heimdal K, Olsson H, Tretli S, Flodgren P, Børresen AL, Fosså SD. Familial testicular cancer in Norway and southern Sweden. Br J Cancer. 1996;73:964–969. doi: 10.1038/bjc.1996.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forman D, Oliver RT, Brett AR, Marsh SG, Moses JH, Bodmer JG, Chilvers CE, Pike MC. Familial testicular cancer: a report of the UK family register, estimation of risk and an HLA class 1 sib-pair analysis. Br J Cancer. 1992;65:255–262. doi: 10.1038/bjc.1992.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldgar DE, Easton DF, Cannon-Albright LA, Skolnick MH. Systematic population-based assessment of cancer risk in first-degree relatives of cancer probands. J Natl Cancer Inst. 1994;86:1600–1608. doi: 10.1093/jnci/86.21.1600. [DOI] [PubMed] [Google Scholar]
- 16.Tollerud DJ, Blattner WA, Fraser MC, Brown LM, Pottern L, Shapiro E, Kirkemo A, Shawker TH, Javadpour N, O'Connell K. Familial testicular cancer and urogenital developmental anomalies. Cancer. 1985;55:1849–1854. doi: 10.1002/1097-0142(19850415)55:8<1849::aid-cncr2820550834>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 17.Reinberg Y, Manivel JC, Zhang G, Reddy PK. Synchronous bilateral testicular germ cell tumors of different histologic type. Pathogenetic and practical implications of bilaterality in testicular germ cell tumors. Cancer. 1991;68:1082–1085. doi: 10.1002/1097-0142(19910901)68:5<1082::aid-cncr2820680529>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 18.Nicholson PW, Harland SJ. Inheritance and testicular cancer. Br J Cancer. 1995;71:421–426. doi: 10.1038/bjc.1995.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.International Testicular Cancer Linkage Consortium, author. Candidate regions for testicular cancer susceptibility genes. APMIS. 1998;106:64–70. [PubMed] [Google Scholar]
- 20.Rapley EA, Crockford GP, Teare D, Biggs P, Seal S, Barfoot R, Edwards S, Hamoudi R, Heimdal K, Fosså SD, Tucker K, Donald J, Collins F, Friedlander M, Hogg D, Goss P, Heidenreich A, Ormiston W, Daly PA, Forman D, Oliver TD, Leahy M, Huddart R, Cooper CS, Bodmer JG, Easton DF. Localization to Xq27 of a susceptibility gene for testicular germ cell tumours. Nat Genet. 2000;24:197–200. doi: 10.1038/72877. [DOI] [PubMed] [Google Scholar]
- 21.Sobin LH. Standardization and the histopathology of tumours. Histopathology. 1977;1:87–92. doi: 10.1111/j.1365-2559.1977.tb01647.x. [DOI] [PubMed] [Google Scholar]
- 22.Kunkel LM, Smith KD, Boyer SH, Borgaonkar DS, Wachtel SS, Miller OJ, Breg WR, Jones HWJ, Rary JM. Analysis of human Y-chromosome-specific reiterated DNA in chromosome variants. Proc Natl Acad Sci USA. 1977;74:1245–1249. doi: 10.1073/pnas.74.3.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leahy MG, Tonks S, Moses JH, Brett AR, Huddart R, Forman D, Oliver RT, Bishop DT, Bodmer JG. Candidate regions for a testicular cancer susceptibility gene. Hum Mol Genet. 1995;4:1551–1555. doi: 10.1093/hmg/4.9.1551. [DOI] [PubMed] [Google Scholar]
- 24.GDB (TM), author Human Genome Database (database online) Baltimore, Maryland, USA: Johns Hopkins University; 1990. Updated daily. Available from Internet: URL http://www.gdb.org. [Google Scholar]
- 25.Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J. A comprehensive genetic map of the human genome based on 5264 microsatellites. Nature. 1996;380:152–154. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- 26.Goss PE, Bulbul MA. Familial testicular cancer in five members of a cancer-prone kindred. Cancer. 1990;66:2044–2046. doi: 10.1002/1097-0142(19901101)66:9<2044::aid-cncr2820660933>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 27.Spirio L, Joslyn G, Nelson L, Leppert M, White R. A CA repeat 30–70 kB downstream from the adenomatous polyposis coli (APC) gene. Nucleic Acids Res. 1991;19:6348. doi: 10.1093/nar/19.22.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leppert M, Dobbs M, Scambler P, O'Connell P, Nakamura Y, Stauffer D, Woodward S, Burt R, Hughes J, Gardner E. The gene for familial polyposis coli maps to the long arm of chromosome 5. Science. 1987;238:1411–1413. doi: 10.1126/science.3479843. [DOI] [PubMed] [Google Scholar]
- 29.Bodmer WF, Bailey CJ, Bodmer J, Bussey HJ, Ellis A, Gorman P, Lucibello FC, Murday VA, Rider SH, Scambler P. Localization of the gene for familial adenomatous polyposis on chromosome 5. Nature. 1987;328:614–616. doi: 10.1038/328614a0. [DOI] [PubMed] [Google Scholar]
- 30.Skotheim RI, Diep CB, Kraggerud SM, Jakobsen KS, Lothe RA. Evaluation of loss of heterozygosity/allelic imbalance in tumor DNA. Cancer Genet Cytogenet. 2001;127(1):64–70. doi: 10.1016/s0165-4608(00)00433-7. [DOI] [PubMed] [Google Scholar]
- 31.Kraggerud SM, Skotheim RI, Szymanska J, Eknæs M, Fosså SD, Stenwig AE, Peltomäki P, Lothe RA. Familial/bilateral and sporadic testicular germ cell tumors analyzed by comparative genomic hybridization. 2001 Submitted. [Google Scholar]
- 32.Skotheim RI, Kraggerud SM, Stenwig AE, Fosså SD, Lothe RA. Testicular germ cell tumors show frequent genetic changes at 3q, 5q, 12q and 18q by allelic imbalance studies and comparative genomic hybridization. Proc Am Assoc Cancer Res. 2000;41:420. (Abstract) [Google Scholar]
- 33.Faulkner SW, Leigh DA, Oosterhuis JW, Roelofs H, Looijenga LH, Friedlander ML. Allelic losses in carcinoma in situ and testicular germ cell tumours of adolescents and adults: evidence suggestive of the linear progression model. Br J Cancer. 2000;83:729–736. doi: 10.1054/bjoc.2000.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng HQ, Liu L, Goss PE, Bailey D, Hogg D. Chromosomal deletions occur in restricted regions of 5q in testicular germ cell cancer. Oncogene. 1999;18:3277–3283. doi: 10.1038/sj.onc.1202662. [DOI] [PubMed] [Google Scholar]
- 35.Murty VV, Li RG, Houldsworth J, Bronson DL, Reuter VE, Bosl GJ, Chaganti RS. Frequent allelic deletions and loss of expression characterize the DCC gene in male germ cell tumors. Oncogene. 1994;9:3227–3231. [PubMed] [Google Scholar]
- 36.Peng HQ, Bailey D, Bronson D, Goss PE, Hogg D. Loss of heterozygosity of tumor-suppressor genes in testis cancer. Cancer Res. 1995;55:2871–2875. [PubMed] [Google Scholar]
- 37.Murty VV, Houldsworth J, Baldwin S, Reuter V, Hunziker W, Besmer P, Bosl G, Chaganti RS. Allelic deletions in the long arm of chromosome 12 identify sites of candidate tumor-suppressor genes in male germ cell tumors. Proc Natl Acad Sci USA. 1992;89:11006–11010. doi: 10.1073/pnas.89.22.11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weissenbach J. Microsatellite polymorphisms and the genetic linkage map of the human genome. Curr Opin Genet Dev. 1993;3:414–417. doi: 10.1016/0959-437x(93)90114-5. [DOI] [PubMed] [Google Scholar]
- 39.Summersgill B, Goker H, Weber-Hall S, Huddart R, Horwich A, Shipley J. Molecular cytogenetic analysis of adult testicular germ cell tumours and identification of regions of consensus copy number change. Br J Cancer. 1998;77:305–313. doi: 10.1038/bjc.1998.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenberg C, Schut TB, Mostert M, Tanke H, Raap A, Oosterhuis JW, Looijenga L. Chromosomal gains and losses in testicular germ cell tumors of adolescents and adults investigated by a modified comparative genomic hybridization approach. Lab Invest. 1999;79:1447–1451. [PubMed] [Google Scholar]
- 41.Mostert MM, van de Pol M, Olde WD, Suijkerbuijk RF, Geurts vK, van Echten J, Oosterhuis JW, Looijenga LH. Comparative genomic hybridization of germ cell tumors of the adult testis: confirmation of karyotypic findings and identification of a 12p-amplicon. Cancer Genet Cytogenet. 1996;89:146–152. doi: 10.1016/0165-4608(96)00043-x. [DOI] [PubMed] [Google Scholar]
- 42.Heimdal K, Olsson H, Tretli S, Fosså SD, Børresen AL, Bishop DT. A segregation analysis of testicular cancer based on Norwegian and Swedish families. Br J Cancer. 1997;75:1084–1087. doi: 10.1038/bjc.1997.185. [DOI] [PMC free article] [PubMed] [Google Scholar]