Abstract
The tumor-suppressor p53 undergoes extensive poly(ADP-ribosyl)ation early during apoptosis in human osteosarcoma cells, and degradation of poly(ADP-ribose) (PAR) attached to p53 coincides with poly(-ADP-ribose)polymerase-1, (PARP-1) cleavage, and expression of p53 target genes. The mechanism by which poly(ADP-ribosyl)ation may regulate p53 function has now been investigated. Purified wild-type PARP-1 catalyzed the poly(ADP-ribosyl) of full-length p53 in vitro. In gel supershift assays, poly(ADP-ribosyl)ation suppressed p53 binding to its DNA consensus sequence; however, when p53 remained unmodified in the presence of inactive mutant PARP-1, it retained sequence-specific DNA binding activity. Poly(ADP-ribosyl)ation of p53 by PARP-1 during early apoptosis in osteosarcoma cells also inhibited p53 interaction with its DNA consensus sequence; thus, poly(ADP-ribosyl)ation may represent a novel means for regulating transcriptional activation by p53 in vivo.
Keywords: poly(ADP-ribose) polymerase, p53, DNA consensus sequence, poly(ADP-ribose), DNA binding
Introduction
One of the earliest nuclear events triggered by the DNA strand breakage associated with DNA repair, DNA replication, and apoptosis is the poly(ADP-ribosyl)ation of various DNA-binding proteins [1–6]. Utilizing NAD as a substrate, this posttranslational modification is catalyzed by a poly(ADP-ribose) polymerase (PARP) gene family, which now includes PARP-1, PARP-2, and PARP-3 [7,8]. PARP-1 is activated by binding to DNA ends through its two zinc fingers [9,10] and undergoes extensive autopoly(ADP-ribosyl)ation in a central automodification domain through an ester linkage between poly(ADP-ribose) (PAR) homopolymers and 15 glutamic acid residues in this domain [9,11]. In contrast, PARP-2 and PARP-3 lack DNA binding and automodification domains [7] and account for the residual PARP activity in PARP-1-deficient cells [8,12]. Given that each PARP-1 molecule has 18 to 28 automodification sites [13], PAR chains of up to 200 residues are covalently bound mainly to PARP-1 and, to a lesser extent, to other nuclear acceptor proteins. Restricted mostly to the potential targets located adjacent to DNA breaks [14], poly(ADP-ribosyl)ation of nuclear proteins in response to DNA strand breakage is transient in intact cells. The half-life of PAR is only 1 to 2 min as a result of its rapid degradation by PAR glycohydrolase [15].
The nuclear protein substrates of PARP-1 include histones, DNA topoisomerases I and II [16,17], SV40 large T antigen [18], DNA polymerases α and δ, proliferating cell nuclear antigen, and ∼15 components of the DNA synthesome [17]. Poly(ADP-ribosyl)ation of enzymes, such as DNA polymerases α and δ [19,20] and topoisomerases I and II [16,21,22], modulates their activities; in most instances, such modification inhibits enzyme activity, presumably as a result of a marked decrease in affinity of the proteins for DNA caused by electrostatic repulsion between the negatively charged DNA and PAR. Extensive automodification of PARP-1 also allows it to cycle on and off DNA ends during DNA repair in vitro [23–26]; whereas unmodified PARP-1 binds tightly to DNA ends and is thought to interfere with repair, the subsequent release of PARP-1 triggered by automodification allows access of repair enzymes to the DNA. PARP-1 and/or poly(ADP-ribosyl)ation are implicated in base excision repair (BER) [27–31], and PARP-1 has been shown to interact with components of the BER complex (i.e., XRCC1, DNA pol β, and DNA ligase lll) [30,32].
We have recently shown that a transient poly(ADP-ribosyl)ation of nuclear proteins occurs early during apoptosis, before commitment to cell death, in various cell lines and with different inducers of apoptosis, and that this event is followed by cleavage and inactivation of PARP-1 [6,33,34]. Prevention of this early PARP-1 activation by expression of PARP antisense RNA or by PARP-1 gene knockout blocks progression of apoptosis, thus correlating this early poly(ADP-ribosyl)ation with later events in the cell death cascade [6]. The impairment of apoptosis, as well as a lack of expression of the tumor-suppressor proteins p53 and Rb in cells of PARP-1 knockout mice, were also recently shown by comparative genomic hybridization analysis to be associated with genomic instability [35]. The expression of p53 is induced by a variety of proapoptotic stimuli and is required for apoptosis in many cell types [36]; transcriptional activation of target genes and interaction with other proteins are thought to contribute to p53-dependent apoptosis. A functional association of PARP and p53 has recently been suggested by coimmunoprecipitation of each protein in vitro by antibodies to the other [37,38]. Together with a variety of other nuclear proteins, p53 undergoes extensive poly(ADP-ribosyl)ation early during spontaneous apoptosis in human osteosarcoma cells; this event is accompanied by a marked increase in the intracellular abundance of p53 [39].
Subsequent degradation of PAR covalently attached to p53 occurs simultaneously with the onset of proteolytic processing and activation of caspase-3, caspase-3-mediated cleavage of PARP-1, and internucleosomal DNA fragmentation. The removal of PAR from p53 during apoptosis also coincides with a marked induction of expression of the p53-responsive genes encoding the proapoptotic proteins Bax and Fas [39], suggesting that poly(ADP-ribosyl)ation may regulate p53 function.
It has been shown that p53 is poly(ADP-ribosyl)ated by partially purified PARP-1 in vitro, and that binding of p53 to a specific DNA consensus sequence prevents its covalent modification [40]. We now show that poly(ADP-ribosyl)ation of p53 with PARP-1 in vitro and in vivo inhibits the binding of p53 to its DNA consensus sequence, providing mechanistic insights on how poly(ADP-ribosyl)ation affects p53 functions in vivo. These results suggest that, similar to the mechanism proposed for PARP-1 itself, p53 may cycle on and off its DNA consensus sequences depending on the extent of their poly(ADP-ribosyl)ation. PARP-1 may thus regulate the transcriptional activation of target genes by p53 during p53-dependent apoptosis.
Materials and Methods
Recombinant p53: Expression and Purification of Recombinant Wild-Type and Inactive Mutant PARP-1 Proteins
Purified recombinant p53, an 80-kDa GST fusion protein of full-length human p53 (amino acids 1–393) expressed in Escherichia coli and purified by Ni resin column chromatography, and GST were obtained from Santa Cruz Biotechnology. Recombinant human wild-type PARP-1 and a catalytically inactive PARP-1 mutant were expressed as six histidine-tagged fusion proteins in E. coli, purified to >95% homogeneity by Ni resin column chromatography, and renatured. The catalytically inactive PARP-1 mutant bears a single-point mutation at Lys893, which has been shown to be critical for activity [41], introduced by site-directed mutagenesis of the catalytic domain of human PARP-1.
Immunoprecipitation, Immunodepletion, and Immunoblot Analysis
For immunoblot analysis, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transfer of proteins to nitrocellulose membranes were performed according to standard procedures. The membranes were stained with Ponceau S (0.1%) to confirm equal loading and transfer of proteins, and were then incubated, unless indicated otherwise, with mouse monoclonal antibodies (mAbs) to p53 (1:200 dilution) (PAb421; Calbiochem), to PAR (1:250 dilution) [42], or to glutathione S-transferase (GST) (1:200 dilution) (Santa Cruz Biotechnology), or with polyclonal antibodies (pAb) to PARP-1 (1:1000 dilution) (Pharmingen). Immune complexes were detected with appropriate horseradish peroxidase-conjugated secondary antibodies (1:3000 dilution) and enhanced chemiluminescence (Pierce).
Immunoprecipitation and immunodepletion were performed as described previously [43], with slight modifications. Equal amounts of poly(ADP-ribosyl)ation reaction mixtures or of cell extracts (10 µg protein) were precleared by incubation for 30 minutes at 4°C in 200 µl of EBC buffer [50 mM Tris-HCl (pH 8.0), 120 mM NaCl, 0.5% NP-40, aprotinin (0.1 T IU/ml)] containing control mouse or rabbit (as appropriate) IgG (1 µg) and protein A/G agarose beads (20 µl; Pharmacia). After centrifugation, PARP-1 was immunodepleted from the supernatants by incubation for 1 hour at 4°C with anti-PARP (1 µg; Pharmingen) and protein A/G agarose beads (20 µl; Pharmacia). After centrifugation, the resulting supernatants were subjected to either immunoprecipitation or electrophoretic mobility supershift assays (EMSSA). Immunoprecipitation was performed by incubation of supernatants for 1 hour at 4°C in 500 µl of NET-N buffer [20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 0.5% NP-40] containing either mAb to p53 (Ab-1; Calbiochem) or control IgG (each at a concentration of 2 µg/ml). After addition of protein A/G agarose beads (20 µl), the mixtures were incubated for 1 hour at 4°C. The beads were then separated by centrifugation and washed extensively with NET-N buffer, after which bound proteins were subjected to immunoblot analysis, first with the mAb to PAR and then with pAb to p53 (Calbiochem) or to PARP.
Poly(ADP-ribosyl)ation of p53 In Vitro by Purified Recombinant PARP-1
Purified recombinant p53 (1 µg) was incubated for 30 minutes at 37°C with purified recombinant wild-type or mutant PARP-1 (0.1 µg) in a reaction mixture (50 µl) containing 50 mM Tris-HCl (pH 7.8), 25 mM MgCl2, 1 mM dithiothreitol, 4 µg of nicked calf thymus DNA, and 100 µM NAD. The reaction was terminated by the addition of SDS-PAGE sample buffer and subjected to immunoblot analysis with anti-PAR. In some experiments, the poly(ADP-ribosyl)ation of p53 was assessed by incubation of these proteins for 1, 3, or 5 minutes at 37°C in the same reaction mixture but containing 1 µM [32P]NAD in place of 100 µM NAD; the mixtures were subsequently analyzed by SDS-PAGE and autoradiography.
EMSSA
EMSSA analysis was performed according to standard procedures, with slight modifications. Oligonucleotides containing p53 consensus binding sequences (Santa Cruz Biotechnology) were labeled at their 5′ ends with [γ-32P]ATP by T4 polynucleotide kinase, and were purified by phenol-chloroform extraction and ethanol precipitation. After incubation with excess poly(dI:dC) (10 µg) and bovine serum albumin (10 µg) in DNA binding buffer (Promega), cell extracts (10 µg of protein) or poly(ADP-ribosyl)ation reaction mixtures (20 µl) were immunodepleted of PARP-1 as described above. Given that recombinant p53 has been shown to be essentially unable to bind its DNA consensus sequence in EMSSAs unless binding is first selectively activated by mAb to p53 PAb421 [44], gel supershift assays were performed by incubation of the PARP-depleted supernatants for 1 hour on ice in a final volume of 40 µl with mAb to p53 (1 µg) and then for 15 minutes at room temperature with the 32P-labeled DNA probe (100,000 cpm). The DNA-protein complexes were then analyzed by electrophoresis on native 5% polyacrylamide gels in Tris-borate-EDTA buffer, followed by autoradiography.
Results
Poly(ADP-ribosyl)ation of p53 by Purified Recombinant PARP-1 In Vitro
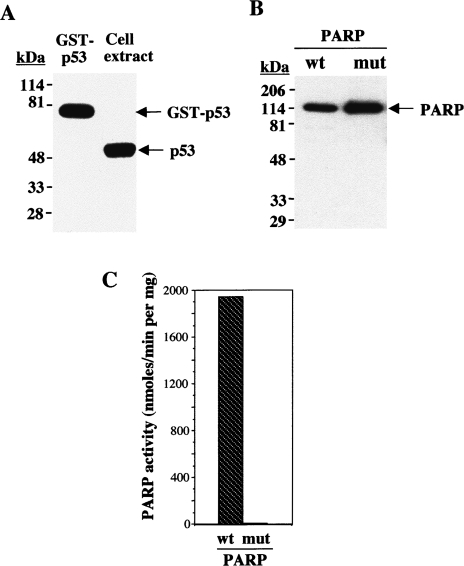
We examined the mechanism by which poly(ADP-ribosyl)ation may regulate p53 function with the use of purified recombinant proteins in vitro. The purified GST fusion protein containing full-length p53 was analyzed by immunoblot analysis with anti-p53. These antibodies specifically recognized endogenous p53 in human osteosarcoma cell extracts and the 80-kDa GST-p53(1–393) fusion protein, consistent with the expected molecular mass of GST fused to full-length human wild-type p53 (Figure 1A). Similarly, immunoblot analysis with anti-PARP of purified recombinant wild-type PARP-1 and a catalytically inactive mutant, both expressed as histidine-tagged fusion proteins, revealed ∼120-kDa immunoreactive proteins (Figure 1B). Measurement of the incorporation of [32P]NAD into acid-insoluble acceptors verified that the wild-type PARP-1 protein was catalytically active (1.9x103 nmol/min per milligram), while the mutant was not (8 nmol/min per milligram) (Figure 1C).
Figure 1.
Immunoblot analysis of recombinant p53 and PARP-1 proteins; catalytic activities of wild-type and mutant PARP-1. (A) Immunoblot analysis with anti-p53 of purified GST-p53(1–393) (0.1 µg) and of an osteosarcoma cell extract (30 µg of protein). (B) Immunoblot analysis with anti-PARP of purified wild-type (wt) and catalytically inactive mutant (mut) PARP-1. The positions of molecular size standards (in kDa) and of the proteins (arrows) are indicated. (C) Relative catalytic activities of wild-type and mutant PARP-1 were determined by incubation of PARP-1 (0.1 µg) for 1 minute at 25°C in a reaction mixture (50 µl) containing 50 mM Tris-HCl (pH 7.8), 25 mM MgCl2, 1 mM dithiothreitol, 4 µg of activated DNA, 100 µM NAD, and 1 µl [32P]NAD (2 mCi/mmol), as described in Materials and Methods section. Data are expressed as nanomoles of [32P]NAD incorporated per minute per milligram of protein, and are means of triplicate determinations from a representative experiment.
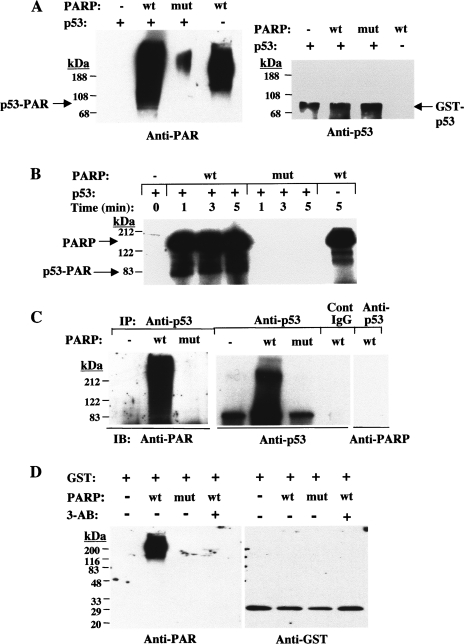
Incubation of GST-p53(1–393) with purified recombinant wild-type PARP-1, but not with the mutant enzyme, in the presence of 100 µM NAD and nicked DNA resulted in poly(ADP-ribosyl)ation of the fusion protein, as revealed by immunoblot analysis with anti-PAR (Figure 2A). One unavoidable technical complication of these experiments with purified proteins is that PARP-1 also undergoes automodification, adding to the complexity of the immunological analyses. The immunoreactive band migrating just below the position corresponding to ∼108 kDa (which was not evident in the reaction mixture lacking p53 fusion protein) likely corresponds to poly(ADP-ribosyl)ated p53; the immunoreactive bands migrating at positions corresponding to 108 to >200 kDa likely include automodified PARP-1 and long-chain, highly modified p53. An ∼80-kDa band in the anti-PAR immunoblot coincided with the GST-p53(1–393) fusion protein as revealed by reprobing the blot with anti-p53. Poly(ADP-ribosyl)ation of GST-p53(1–393) by wild-type PARP-1, but not by the mutant protein, was also demonstrated by incubation in the presence of 1 µM [32P]NAD and autoradiography (Figure 2B). Most of the radioactivities evident at ∼122 kDa and in two lower bands reflected automodified PARP-1 and its degradation products, given that these bands were also present in the lane corresponding to wild-type PARP-1 alone; a band at ∼80 kDa present in the samples containing GST-p53(1–393) and wild-type PARP-1 thus corresponds to the poly(ADP-ribosyl) ated GST-p53 fusion protein. As expected, although there was 10-fold more p53 than PARP-1, PARP-1 was the main acceptor protein in these reactions, with heteromodification of p53 occurring at a lesser extent. This preferential automodification of PARP-1 is attributed to the presence of 28 automodification sites (glutamate residues) for the covalent attachment of PAR homopolymers of up to 200 residues long [9,11,13]. In contrast, binding experiments with free PAR and p53 revealed only two PAR-binding sites in the sequence-specific core DNA binding domain of p53 and a third site in the oligomerization domain [45]. There is no apparent increase in intensity of the bands (PAR covalently bound to PARP or to p53) with time because, under the conditions of the in vitro poly(ADP-ribosyl)ation used in this experiment (with 1 µM 32P-NAD), the reaction is only linear for less than 1 minute; at this NAD concentration, only short polymers are covalently bound to acceptor proteins; thus, the molecular weight of PARP stays around 116 kDa. However, an increase in size of p53, as revealed by a shift in mobility to a slightly higher position in the gel, indicates addition of longer polymer chains with time.
Figure 2.
Poly(ADP-ribosyl)ation of GST-p53(1–393) and GST by wild-type PARP-1 in vitro. (A) Recombinant wild-type or mutant PARP-1 (0.1 µg) was incubated for 30 minutes at 37°C in the presence of 100 µM NAD and in the absence or presence of GST-p53(1–393) (1 µg), as indicated. The reaction mixtures were then subjected to immunoblot analysis with anti-PAR (left panel), after which the blot was stripped of antibodies and reprobed with anti-p53 (right panel). Arrows: p53-PAR, poly(ADP-ribosyl)ated p53 fusion protein; GST-p53, GST-p53(1–393). (B) Recombinant wild-type or mutant PARP-1 (0.1 µg) was incubated for the indicated times at 37°C in the presence of 1 µM [32P]NAD and in the absence or presence of GST-p53(1–393) (1 µg). The reaction mixtures were then subjected to SDS-PAGE and autoradiography. Arrows indicate the positions of poly(ADP-ribosyl)ated PARP-1 and p53 fusion protein. (C) GST-p53(1–393) was incubated in the absence or presence of wild-type or mutant PARP-1 as in (A). The reaction mixtures were immunodepleted of PARP-1 and then subjected to immunoprecipitation with anti-p53 or with control IgG, as indicated. The resulting immunocomplexes were subjected to immunoblot analysis with anti-PAR, after which the blot was stripped of antibodies and reprobed with anti-p53 and then with anti-PARP, as indicated. (D) Purified GST (1 µg) was incubated for 30 minutes at 37°C in the presence of 100 µM NAD and in the absence or presence of wild-type or mutant PARP-1 (0.1 µg) and 1 mM 3-aminobenzamide (3-AB), as indicated. The reaction mixtures were then subjected to immunoblot analysis with anti-PAR (left panel), after which the blot was stripped of antibodies and reprobed with anti-GST (right panel).
To confirm that GST-p53(1–393) is poly(ADP-ribosyl)ated by wild-type PARP-1 in vitro, the reaction mixture containing nonradioactive NAD was immunodepleted of PARP by immunoprecipitation with anti-PARP, and the resulting supernatant was subjected to immunoprecipitation with anti-p53. Immunoblot analysis of the resulting precipitates with anti-PAR revealed extensive poly(ADP-ribosyl)ation of GST-p53(1–393) in the presence of wild-type PARP-1, but not with the mutant enzyme (Figure 2C). Reprobing of the immunoblot with anti-p53 and anti-PARP verified that the modified protein was indeed p53 and not PARP-1 (Figure 2C). When immunoprecipitation of the PARP-1-depleted supernatant was performed with control IgG, immunoblot analysis with anti-p53 failed to detect any specific immunoreactivity (Figure 2C). To verify that it was the p53 portion, and not the GST component, of GST-p53 that was poly(ADP-ribosyl)ated by PARP-1, purified recombinant GST was incubated with wild-type and mutant PARP-1 in the presence of NAD. Immunoblot analysis with anti-PAR and anti-GST revealed that wild-type PARP-1 was the only protein to undergo poly(ADP-ribosyl)ation, a reaction that was inhibited by the specific PARP inhibitor 3-aminobenzamide (Figure 2D).
Poly(ADP-ribosyl)ation of p53 by PARP-1 In Vitro Blocks Binding to its DNA Consensus Sequence
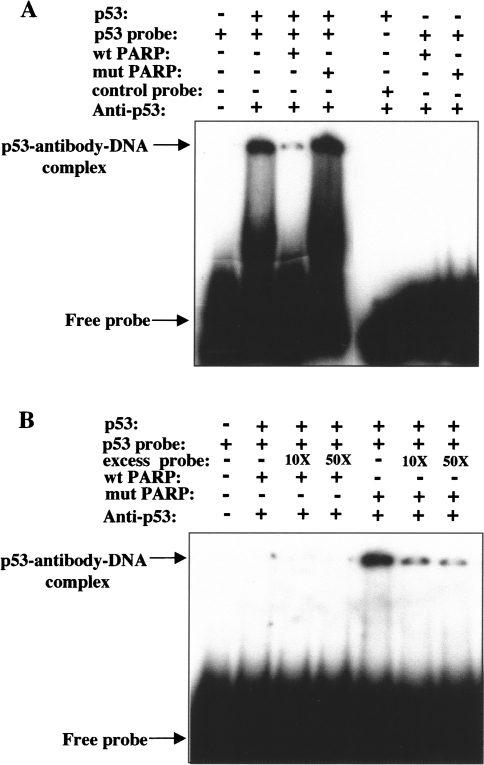
To investigate whether poly(ADP-ribosyl)ation of p53 by PARP-1 affects binding to the corresponding DNA consensus sequence element, we performed gel supershift assays with a 32P-labeled 30-bp oligonucleotide containing the specific p53-binding sequence as the DNA probe. Recombinant p53 has been shown to be essentially inactive in sequence-specific DNA binding in EMSSAs, but binding is strongly and selectively activated by mAb to p53 PAb421 [44]. Thus, in gel supershift assays, poly(ADP-ribosyl)ation reaction mixtures were immunodepleted of PARP-1, incubated first with mAb to p53 and then with the DNA probe, and analyzed by electrophoresis and autoradiography.
Binding of unmodified GST-p53(1–393) to the 32P-labeled probe was apparent by the appearance of a distinct shifted band corresponding to the p53-antibody-DNA complex (Figure 3A). The specificity of the interaction was confirmed by the observations that incubation of GST-p53(1–393) with an unrelated control oligonucleotide (NF-κB consensus sequence) did not result in the formation of a DNA-protein complex (Figure 3A), and that binding of the p53 fusion protein to the p53-binding sequence was inhibited in a concentration-dependent manner by the presence of excess nonradioactive-specific probe (Figure 3B). Poly(ADP-ribosyl)ation of GST-p53(1–393) by wild-type PARP-1 in the presence of NAD markedly inhibited the binding of the p53 fusion protein to the specific DNA probe (Figure 3A). In contrast, preincubation of GST-p53(1–393) with mutant PARP-1 and NAD did not affect the formation of the specific DNA-protein complex. The shifted bands were not due to binding of any remaining PARP-1 in the reaction mixtures to the DNA probe because wild-type or mutant PARP-1, in the absence of p53, did not bind to the p53 consensus sequence. Similar EMSSA analysis has also previously shown that free PAR noncovalently attached to p53 in vitro blocks the sequence-specific interaction of p53 with its consensus binding sequence [45].
Figure 3.
Effect of poly(ADP-ribosyl)ation of GST-p53(1–393) by PARP-1 in vitro on binding of p53 to its DNA consensus sequence. (A) After incubation for 30 minutes at 37°C, poly(ADP-ribosyl)ation reaction mixtures containing NAD and the indicated combinations of GST-p53(1–393) and wild-type or mutant PARP-1 were immunodepleted of PARP-1 and then subjected to EMSSA by incubation first with anti-p53 and then with a 32P-labeled 30-bp oligonucleotide probe corresponding to the consensus p53-binding sequence (or with a control probe corresponding to the NF-κB-binding sequence). (B) EMSSA analysis was performed as in (A), with the exception that the incubation with the 32P-labeled p53-specific probe was performed in the absence or presence of x10 or x50 excesses of the same unlabeled probe. The positions of free DNA probe and the antibody-p53-DNA complex are indicated.
Poly(ADP-ribosyl)ation of p53 at an Early Stage of Apoptosis in Osteosarcoma Cells Inhibits Binding to its DNA Consensus Sequence In Vitro
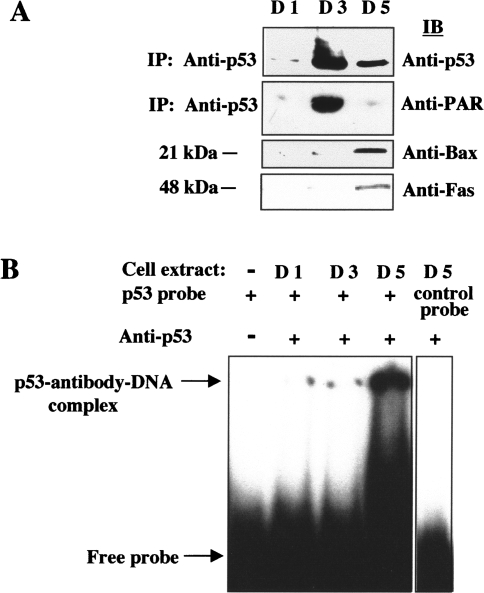
Degradation of PAR attached to p53 coincides with the onset of caspase-3-mediated PARP-1 cleavage, internucleosomal DNA fragmentation, and activation of the p53 target genes encoding Bax and Fas during spontaneous apoptosis in human osteosarcoma cells [39], suggesting that poly(ADP-ribosyl)ation may regulate p53 function. Extracts of osteosarcoma cells prepared before (day 1), after (day 5), and at (day 3) the peak of poly(ADP-ribosyl)ation that occurs at the early reversible stage of apoptosis were subjected to immunoprecipitation with anti-p53 and immunoblot analysis with anti-PAR and anti-p53. Although endogenous levels of p53 protein were significantly increased at days 3 and 5, transient extensive poly(ADP-ribosyl)ation of endogenous p53 was apparent only at day 3 (Figure 4A). Immunoblot analysis of osteosarcoma cell extracts at different stages of apoptosis with antibodies to Bax and Fas further showed that expression of these p53-responsive genes (lower panel) was negligible when p53 was in a poly(ADP-ribosyl)ated state (day 3), but increased at day 5 when p53 was largely unmodified (middle panel). Thus, poly(ADP-ribosyl)ation may modulate p53 function early in apoptosis perhaps by altering its binding to specific DNA sequences in the promoters of p53 target genes such as Bax and Fas. The effect of poly(ADP-ribosyl)ation on the binding of p53 to its DNA consensus sequence was therefore examined by gel supershift assays with cell extracts prepared before (day 1), after (day 5), and at (day 3) the peak of poly(ADP-ribosyl)ation. Consistent with our in vitro results with purified proteins, extensive poly(ADP-ribosyl)ation of endogenous p53 by PARP-1 (at day 3) in intact cells also inhibited p53 binding to its specific DNA consensus sequence (Figure 4B). The specificity of the interaction was confirmed by the incubation of cell extract (D5) with an unrelated control oligonucleotide (NF-κB consensus sequence), which did not result in the formation of a DNA-antibody-protein complex (Figure 4B, right panel). Thus, by modulating p53 binding to its DNA consensus sequence while in a negatively charged modified state, poly(ADP-ribosyl)ation may represent an additional posttranslational modification besides phosphorylation [46] and acetylation [47], by which p53-dependent transcription may be transiently regulated in vivo.
Figure 4.
Effect of poly(ADP-ribosyl)ation of p53 during early apoptosis in osteosarcoma cells on p53 binding to its DNA consensus sequence in vitro. (A) Cell extracts from osteosarcoma cells before (day 1), after (day 5), and at (day 3) the peak of poly(ADP-ribosyl)ation during the early stage of apoptosis were subjected to immunoblot analysis with antibodies to Bax and Fas (lower panel) and to immunoprecipitation with anti-p53. After immunoprecipitation with anti-p53, the resulting immunocomplexes were then subjected to immunoblot analysis with anti-p53 (upper panel) and anti-PAR (middle panel). (B) Osteosarcoma cell extracts prepared before (D1), after (D5), and at the peak of poly(ADP-ribosyl)ation (D3) were subjected to gel supershift assays with anti-p53 and the 32P-labeled 30-bp oligonucleotide containing the consensus p53-binding sequence. Control EMSSAs were also performed in the absence of cell extract, or with an unrelated control oligonucleotide (NF-κB consensus sequence) to confirm specificity of binding.
Discussion
We have previously shown that a transient poly(ADP-ribosyl)ation of nuclear proteins occurs early during apoptosis, before commitment to cell death, in 3T3-L1, HL-60, Jurkat, and osteosarcoma cells and in immortalized PARP-1+/+ fibroblasts with different inducers of apoptosis. This transient PARP-1 activation precedes the proteolytic processing of caspase 3, caspase-mediated cleavage of PARP-1 and the DNA fragmentation factor (DFF45), and internucleosomal DNA fragmentation [6,33,34]. Given that the activity of PARP is dependent on DNA strand breaks, this early nuclear poly(ADP-ribosyl)ation during apoptosis is consistent with the appearance of large (1 Mb) chromatin fragments at this reversible stage [48], as well as a marked decrease in NAD concentration indicative of increased PAR synthesis, and a subsequent recovery in NAD levels before internucleosomal DNA cleavage [49]. PARP activation also occurs during apoptosis induced by various DNA-damaging agents, such as alkylating agents, topoisomerase inhibitors, adriamycin, X-rays, ultraviolet radiation, mitomycin C, and cisplatin [50–54]. Progression of Fas-mediated apoptosis is blocked by prevention of this early PARP-1 activation by expression of PARP antisense RNA or by PARP-1 gene knockout, thus correlating this early poly(ADP-ribosyl)ation with later events in the cell death cascade [6].
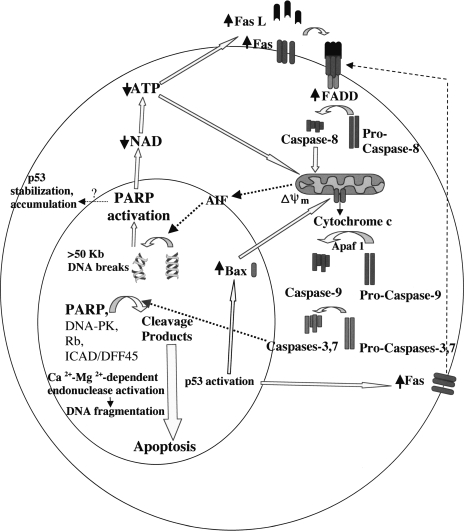
We recently proposed a model, summarized in Figure 5, whereby PARP-1 and poly(ADP-ribosyl)ation may play a role in an “amplification loop” towards caspase activation and downstream apoptotic events [34]. Stimulation of the Fas receptor induces the gradual release of mitochondrial factors, such as the apoptosis-inducing factor (AIF), that translocates from the mitochondrial intermembrane space to the nucleus and induces low levels of caspase-independent cleavage of chromatin into large 50-kb fragments [55]. These large DNA breaks can stimulate PARP activity, rapidly decreasing NAD and ATP levels, which contributes to both receptor and mitochondrial pathways of apoptosis. Partial depletion of ATP (∼10–65% of the control) has been shown to upregulate Fas, Fas ligand, and Fas-associating protein with the death domain (FADD), resulting in induction of caspase-8 and caspase-3 activity [56]. Binding of Fas to Fas ligand recruits FADD via shared protein motifs (death domains), resulting in subsequent activation or amplification of the caspase cascade leading to apoptosis. Further depletion of ATP below a threshold level inhibits later events in apoptosis [57]; thus, subsequent degradation of PARP by caspase-3-like proteases may prevent depletion of NAD and ATP below this critical level as well as release certain nuclear proteins, such as endonucleases [58,59] or p53, from poly(ADP-ribosyl)ation-induced inhibition.
Figure 5.
PARP-1 activation and cleavage in an “amplification loop” toward caspase activation and downstream apoptotic events. Stimulation of the Fas receptor induces the release of mitochondrial factors, such as AIF, that translocates to the nucleus and induces caspase-independent chromatin cleavage into large 50-kb fragments. These large DNA breaks activate PARP, rapidly decreasing NAD and ATP levels, which contributes to both receptor and mitochondrial pathways of apoptosis. Partial depletion of ATP further upregulates Fas, Fas ligand, and FADD; binding of Fas to Fas ligand recruits FADD, resulting in subsequent activation or amplification of the caspase cascade leading to apoptosis. Subsequent cleavage of PARP by caspase-3 prevents depletion of NAD and ATP below a critical level as well as releases certain nuclear proteins, such as Ca2+ -Mg2+-dependent endonucleases and p53, from poly(ADP-ribosyl)ation-induced inhibition.
Apoptosis in osteosarcoma cells is associated with accumulation of p53 early in the death program, presumably due to induced expression of the protein or stabilization by inhibition of p53 degradation via modification of the protein [39]. Immunoprecipitation experiments further revealed that p53 undergoes extensive poly(ADP-ribosyl)ation during the transient burst of PAR synthesis at the early stages of apoptosis, and subsequent degradation of PAR covalently attached to p53 coincides with caspase-3-mediated cleavage of PARP-1 and expression of the p53-responsive genes encoding Bax and Fas [39]. Induction of Bax expression may influence the decision to commit to apoptosis since homodimerization of Bax promotes cell death and heterodimerization of Bax with Bcl2 inhibits the antiapoptotic function of Bcl2 [60]. Wild-type p53 also upregulates Fas expression during chemotherapy-induced apoptosis, and p53-responsive elements were recently identified within the first intron and the promoter of the Fas gene [61]. Furthermore, p53 activation transiently increases surface Fas expression by enhancing intracellular transport from the Golgi complex, induces Fas-FADD binding, and transiently sensitizes cells to Fas-induced apoptosis [62]. The simultaneous removal of PAR from modified p53 and upregulation of Bax and Fas expression suggested that poly(ADP-ribosyl)ation may transiently inhibit the transactivation activity of p53 early during apoptosis; caspase-mediated PARP-1 cleavage may subsequently release p53 from such inhibition when the cells become irreversibly committed to death. Additionally, a polymer binding site in p53 localized near a proteolytic cleavage site [45] suggests that PAR binding could protect this sequence from proteolysis, given that similar protection has been noted after binding of antibodies adjacent to this region [63]. Thus, the poly(ADP-ribosyl)ation of p53 early in apoptosis could also play a role in p53 accumulation by protecting the protein from proteolytic degradation.
It is also possible that poly(ADP-ribosyl)ation contributes indirectly to the regulation of p53, given that such modification by PARP-1 of DNA-dependent protein kinase stimulates the activity of this enzyme [64], which in turn regulates p53 function by phosphorylation. With the use of purified proteins in vitro, we have now shown that poly(ADP-ribosyl)ation of p53 by PARP-1 inhibits the interaction of p53 with its DNA consensus sequence. Furthermore, poly(ADP-ribosyl)ation of endogenous p53 by PARP-1 during the early stage of apoptosis in human osteosarcoma cells also inhibited the binding of p53 to its consensus binding sequence. These results provide mechanistic insight on how poly(ADP-ribosyl)ation may affect p53 function in vivo. The PARP-1-mediated covalent attachment of PAR to p53 presumably occurs on glutamic acid residues within p53. Consistent with these results, free PAR polymer non-covalently bound to p53 in vitro blocks its specific binding to the cognate DNA consensus sequence, and binding sites for free PAR have been mapped to two sites in the sequence-specific DNA-binding domain of p53 [45]. Our data thus indicate that transient poly(ADP-ribosyl)ation of p53 by PARP-1 may contribute to regulation of p53 function by inhibiting the binding of p53 to its target sequence in the promoters of p53-responsive genes, presumably as a result of the negative charge conferred by the PAR modification, and that it thereby transiently prevents activation of these genes by p53. Although the suppression of p53 binding to its DNA consensus sequence following extensive poly(ADP-ribosylribosyl)ation in vitro is not surprising, it is interesting to note that p53 modified by PARP-1 in vivo does not bind its DNA consensus sequence, consequently blocking expression of p53-responsive genes Bax and Fas, which implies that most of the p53 are poly(ADP-ribosyl)ated at this early reversible stage of apoptosis.
The roles of PARP-1 in gene transcription may be mediated, in part, by its effects on chromatin structure, given that PARP-1 depletion by expression of antisense RNA markedly alters the organization of chromatin [29]. PARP-1 has been implicated to play a dual role in the regulation of transcription. PARP-1, in the absence of NAD, is suggested to enhance activator-dependent transcription by interacting with RNA polymerase II-associated factors [65]. Similarly, PARP-1 binds to transcription enhancer factor 1 (TEF1), and thereby promotes muscle-specific gene transcription [66], as well as to the transcription factor AP-2, and thereby acts as a coactivator in AP-2-mediated transcription [67]. PARP-1 also functions as a coactivator of the retroviral transcriptional activator Tax [68]. Similarly, we recently demonstrated that PARP-1 plays a role in the induction of expression of the transcription factor E2F-1 by increasing the promoter activity of the gene during reentry into S-phase [20,69].
Although, as described above, PARP-1 acts as a positive cofactor for transcription in the absence of NAD, its activation in the presence of NAD inhibits RNA polymerase II-dependent transcription [65]. PARP-1-dependent inhibition of transcription is mediated by the poly(ADP-ribosyl)ation of various transcription factors, which prevents the formation of active transcription complexes [70]. The basal transcription factor TFIIF as well as TEF1 and the transcription factors YY1, SP-1, and CREB are substrates for poly(ADP-ribosyl)ation [66,70,71]. Similar to the effects of this modification on p53 demonstrated in the present study, poly(ADP-ribosyl)ation of these proteins prevents their binding to the respective DNA consensus sequences [70]. These various transcription factors are specific substrates for poly(ADP-ribosyl)ation, given that other proteins, including TBP, TFIIB, c-Jun, and AP-2, have been shown to be ineffective PARP-1 substrates under the same conditions [70,71]. In a manner similar to that of the cycling of PARP-1 on and off DNA ends during DNA repair in vitro [23–26], it is thus possible that p53 may cycle on and off its corresponding DNA consensus sequence depending on the extent of its poly(ADP-ribosyl)ation. Such a mechanism would thus constitute a novel means for regulating transcriptional activation of target genes by p53 in vivo.
Acknowledgements
We thank D. Nicholson for the human osteosarcoma cell line, M. Miwa and T. Sugimura for the mAb to PAR, and Marianne Augustine for invaluable help with some of the experiments.
Abbreviations
- anti
antibodies to
- EMSSA
electrophoretic mobility supershift assay
- GST
glutathione S-transferase
- IgG
immunoglobulin G
- NAD
nicotinamide adenine dinucleotide
- mAb
monoclonal antibody
- PAGE
polyacrylamide gel electrophoresis
- PARP-1
poly(ADP-ribose) polymerase-1
- PAR
poly(ADP-ribose)
Footnotes
This work was supported, in part, by OXIGENE Europe (grant 97A108 to M. S.), by the National Cancer Institute (grants CA25344 to M. S. and 1P01 CA74175 to M. S., M. J., and A. D.), by the US Air Force Office of Scientific Research (grant AFOSR-89-0053 to M. E. S.), and by the US Army Medical Research and Development Command (contracts DAMD17-90-C-0053 to M. S. and DAMD 17-96-C-6065 to D. R.).
References
- 1.Thraves PJ, Smulson ME. Acceptors for the poly ADP-ribosylation modification of chromatin structure are altered by carcinogen-induced DNA damage. Carcinogenesis. 1982;3:1143–1148. doi: 10.1093/carcin/3.10.1143. [DOI] [PubMed] [Google Scholar]
- 2.Malik N, Smulson ME. A relationship between nuclear poly(ADP-ribosyl)ation and acetylation posttranslational modifications: I. Nucleosome studies. Biochemistry. 1984;23:3721–3725. doi: 10.1021/bi00311a023. [DOI] [PubMed] [Google Scholar]
- 3.Berger NA, Sims JL, Catino DM, Berger SJ. Poly(ADP-Ribose)Polymerase Mediates the Suicide Response to Massive DNA Damage: Studies in Normal and DNA Repair-Defective Cells. In: Miwa M, Hayaishi O, Shall S, Smulson M, Sugimura T, editors. ADP Ribosylation, DNA Repair and Cancer. Tokyo: Japan Sci. Soc. Press; 1983. pp. 219–226. [PubMed] [Google Scholar]
- 4.Berger N, Whitacre C, Hashimoto H, Berger S, Chaterjee S. NAD and poly(ADP-ribose) regulation of proteins involved in response to cellular stress and DNA damage. Biochimie. 1995;77:364–367. doi: 10.1016/0300-9084(96)88147-8. [DOI] [PubMed] [Google Scholar]
- 5.Smulson ME, Kang VH, Ntambi JM, Rosenthal DS, Ding R, Simbulan CMG. Requirement for the expression of poly(ADP-ribose) polymerase during the early stages of differentiation of 3T3-L1 preadipocytes, as studied by antisense RNA induction. J Biol Chem. 1995;270:119–127. doi: 10.1074/jbc.270.1.119. [DOI] [PubMed] [Google Scholar]
- 6.Simbulan-Rosenthal CM, Rosenthal DS, Iyer S, Boulares AH, Smulson ME. Transient poly(ADP-ribosyl)ation of nuclear proteins and role for poly(ADP-ribose) polymerase in the early stages of apoptosis. J Biol Chem. 1998;273:13703–13712. doi: 10.1074/jbc.273.22.13703. [DOI] [PubMed] [Google Scholar]
- 7.Johansson M. A human poly(ADP-ribose) polymerase gene family (ADPRTL): cDNA cloning of two novel poly(ADP-ribose) polymerase homologues. Genomics. 1999;57:442–445. doi: 10.1006/geno.1999.5799. [DOI] [PubMed] [Google Scholar]
- 8.Ame J, Rolli V, Schreiber V, Niedergang C, Apiou, Decker P, Muller S, Hoger T, de Murcia J, de Murcia G. PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem. 1999;274:17860–17868. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- 9.Cherney BW, McBride OW, Chen DF, Alkhatib H, Bhatia K, Hensley P, Smulson ME. cDNA sequence, protein structure, and chromosomal location of the human gene for poly(ADP-ribose) polymerase. Proc Natl Acad Sci USA. 1987;84:8370–8374. doi: 10.1073/pnas.84.23.8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menissier-de Murcia J, Molinete M, Gradwohl G, de Murcia G. Zinc-binding domain of poly(ADP-ribose)polymerase participates in the recognition of single-strand breaks on DNA. J Mol Biol. 1989;210:229–233. doi: 10.1016/0022-2836(89)90302-1. [DOI] [PubMed] [Google Scholar]
- 11.Kawaichi M, Ueda K, Hayaishi O. Multiple auto(ADP-ribosyl)ation of rat liver poly(ADP-ribose) synthetase. J Biol Chem. 1981;256:9483–9489. [PubMed] [Google Scholar]
- 12.Shieh WM, Ame JC, Wilson M, Wang ZQ, Koh D, Jacobson M, Jacobson E. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J Biol Chem. 1998;273:30069–30072. doi: 10.1074/jbc.273.46.30069. [DOI] [PubMed] [Google Scholar]
- 13.Desmarais Y, Menard L, Lagueux J, Poirier G. Enzymological properties of poly(ADP-ribose) polymerase: characterization of automodification sites and NADase activity. Biochim Biophys Acta. 1991;1078:179–186. doi: 10.1016/0167-4838(91)99007-f. [DOI] [PubMed] [Google Scholar]
- 14.Thraves PJ, Kasid U, Smulson ME. Selective isolation of domains of chromatin proximal to both carcinogen-induced DNA damage and polyadenosine diphosphate ribosylation. Cancer Res. 1985;45:386–391. [PubMed] [Google Scholar]
- 15.Lin W, Ame J, Aboul-Ela N, Jacobson E, Jacobson M. Isolation and characterization of the cDNA encoding bovine poly(ADP-ribose) glycohydrolase. J Biol Chem. 1997;272:11895–11901. doi: 10.1074/jbc.272.18.11895. [DOI] [PubMed] [Google Scholar]
- 16.Kasid UN, Halligan B, Liu LF, Dritschilo A, Smulson M. Poly(ADP-ribose)-mediated posttranslational modification of chromatin-associated human topoisomerase: I. Inhibitory effects on catalytic activity. J Biol Chem. 1989;264:18687–18692. [PubMed] [Google Scholar]
- 17.Simbulan-Rosenthal CM, Rosenthal DS, Hilz H, Hickey R, Malkas L, Applegren N, Wu Y, Bers G, Smulson M. The expression of poly(ADP-ribose) polymerase during differentiation-linked DNA replication reveals that this enzyme is a component of the multiprotein DNA replication complex. Biochemistry. 1996;35:11622–11633. doi: 10.1021/bi953010z. [DOI] [PubMed] [Google Scholar]
- 18.Baksi K, Alkhatib H, Smulson ME. In vivo characterization of the poly ADP-ribosylation of SV40 chromatin and large T antigen by immunofractionation. Exp Cell Res. 1987;172:110–123. doi: 10.1016/0014-4827(87)90098-x. [DOI] [PubMed] [Google Scholar]
- 19.Yoshihara K, Itaya A, Tanaka Y, Ohashi Y, Ito K, Teraoka H, Tsukada K, Matsukage A, Kamiya T. Inhibition of DNA polymerase α, DNA polymerase β, terminal nucleotidyltransferase and DNA ligase II by poly (ADP-ribosyl)ation reaction in vitro. Biochem Biophys Res Commun. 1985;128:61–67. doi: 10.1016/0006-291x(85)91644-4. [DOI] [PubMed] [Google Scholar]
- 20.Simbulan-Rosenthal CM, Rosenthal DS, Boulares AH, Hickey RJ, Malkas LH, Coll JM, Smulson ME. Regulation of the expression or recruitment of components of the DNA synthesome by poly(ADP-ribose) polymerase. Biochemistry. 1998;37:9363–9370. doi: 10.1021/bi9731089. [DOI] [PubMed] [Google Scholar]
- 21.Ferro AM, Olivera BM. Poly (ADP ribosylation) of DNA topoisomerase I from calf thymus. J Biol Chem. 1984;259:547–554. [PubMed] [Google Scholar]
- 22.Darby MK, Schmitt B, Jongstra BJ, Vosberg HP. Inhibition of calf thymus type II DNA topoisomerase by poly(ADP ribosylation) EMBO J. 1985;4:2129–2134. doi: 10.1002/j.1460-2075.1985.tb03903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Satoh MS, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 24.Satoh MS, Poirier GG, Lindahl T. NAD+-dependent repair of damaged DNA by human cell extracts. J Biol Chem. 1993;268:5480–5487. [PubMed] [Google Scholar]
- 25.Smulson M, Istock N, Ding R, Cherney B. Deletion mutants of poly(ADP-ribose) polymerase support a model of cyclic association and dissociation of enzyme from DNA ends during DNA repair. Biochemistry. 1994;33:6186–6191. doi: 10.1021/bi00186a018. [DOI] [PubMed] [Google Scholar]
- 26.Smulson ME, Pang D, Jung M, Dimtchev A, Chasovskikh S, Spoonde A, Simbulan-Rosenthal C, Rosenthal D, Yakovlev A, Dritschilo A. Irreversible binding of poly(ADP-ribose) polymerase cleavage product to DNA ends revealed by atomic force microscopy: possible role in apoptosis. Cancer Res. 1998;58:3495–3498. [PubMed] [Google Scholar]
- 27.Molinete M, Vermeulen W, Burkle A, de Murcia JM, Kupper JH, Hoeijmakers JH, de Murcia G. Overproduction of the poly(ADP-ribose) polymerase DNA-binding domain blocks alkylation-induced DNA repair synthesis in mammalian cells. EMBO J. 1993;12:2109–2117. doi: 10.1002/j.1460-2075.1993.tb05859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schreiber V, Hunting D, Trucco C, Gowans B, Grunwald P, de Murcia G, de Murcia J. A dominant negative mutant of human PARP affects cell recovery, apoptosis, and sister chromatid exchange following DNA damage. Proc Natl Acad Sci USA. 1995;92:4753–4757. doi: 10.1073/pnas.92.11.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding R, Smulson M. Depletion of nuclear poly(ADP-ribose) polymerase by antisense RNA expression: influences on genomic stability, chromatin organization and carcinogen cytotoxicity. Cancer Res. 1994;54:4627–4634. [PubMed] [Google Scholar]
- 30.Dantzer F, Rubia G, Menissier-de Murcia J, Hostomsky Z, de Murcia G, Schreiber V. Base excision repair is impaired in mammalian cells lacking poly(ADP-ribose) polymerase. Biochemistry. 2000;39:7559–7569. doi: 10.1021/bi0003442. [DOI] [PubMed] [Google Scholar]
- 31.Vodenicharov M, Sallmann F, Satoh M, Poirier G. Base excision repair is efficient in cells lacking poly(ADP-ribose) polymerase 1. Nucleic Acids Res. 2000;28:3887–3896. doi: 10.1093/nar/28.20.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simbulan-Rosenthal CM, Rosenthal DS, Smulson ME. Pleiotropic Roles of Poly(ADP-Ribosyl)ation of DNA- Binding Proteins. In: Czabo C, editor. Cell Death: The Role of PARP. Boca Raton: CRC Press; 2000. pp. 251–277. [Google Scholar]
- 34.Rosenthal DS, Simbulan-Rosenthal CM, Smith W, Benton B, Ray R, Smulson M. Poly(ADP-Ribose) Polymerase is an Active Participant in Programmed Cell Death and Maintenance of Genomic Stability. In: Szabo C, editor. Cell Death: The Role of PARP. Boca Raton: CRC Press; 2000. pp. 227–250. [Google Scholar]
- 35.Simbulan-Rosenthal C, Haddad B, Rosenthal D, Weaver Z, Coleman A, Luo R, Young H, Wang ZQ, Ried T, Smulson M. Chromosomal aberrations in PARP-/- mice: genome stabilization in immortalized cells by reintroduction of poly(ADP-ribose) polymerase cDNA. Proc Natl Acad Sci USA. 1999;96:13191–13196. doi: 10.1073/pnas.96.23.13191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fisher D. Apoptosis in cancer therapy: crossing the threshold. Cell. 1994;78:539–542. doi: 10.1016/0092-8674(94)90518-5. [DOI] [PubMed] [Google Scholar]
- 37.Vaziri H, West M, Allsop R, Davison T, Wu Y, Arrowsmith C, Poirier G, Benchimol S. ATM-dependent telomere loss in aging human diploid fibroblasts and DNA damage lead to the posttranslational activation of p53 protein involving poly(ADP-ribose) polymerase. EMBO J. 1997;16:6018–6033. doi: 10.1093/emboj/16.19.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wesierska-Gadek J, Bugajska-Schretter A, Cerni C. ADP ribosylation of p53 tumor-suppressor protein: mutant but not wild-type p53 is modified. J Cell Biochem. 1996;62:90–101. doi: 10.1002/(sici)1097-4644(199607)62:1<90::aid-jcb10>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 39.Simbulan-Rosenthal CM, Rosenthal DS, Luo R, Smulson ME. Poly(ADP-ribosyl)ation of p53 during apoptosis in human osteosarcoma cells. Cancer Res. 1999;59:2190–2194. [PubMed] [Google Scholar]
- 40.Wesierska-Gadek J, Schmid G, Cerni C. ADP ribosylation of wild-type p53 in vitro: binding of p53 protein to specific p53 consensus sequence prevents its modification. Biochem Biophys Res Commun. 1996;224:96–102. doi: 10.1006/bbrc.1996.0990. [DOI] [PubMed] [Google Scholar]
- 41.Kim H, Jacobson M, Rolli V, Menissier-de Murcia J, Reinbolt J, Simonin F, Ruf A, Schulz G, de Murcia G. Photo-affinity labeling of human poly(ADP-ribose) polymerase catalytic domain. Biochem J. 1997;322:469–475. doi: 10.1042/bj3220469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawamitsu H, Hoshino H, Okada H, Miwa M, Momoi H, Sugimura T. Monoclonal antibodies to poly(adenosine diphosphate ribose) recognize different structures. Biochemistry. 1984;23:3771–3777. doi: 10.1021/bi00311a032. [DOI] [PubMed] [Google Scholar]
- 43.Simbulan C, Suzuki M, Izuta S, Sakurai T, Savoysky E, Kojima K, Miyahara K, Shizuta Y, Yoshida S. Poly (ADP-ribose) polymerase stimulates DNA polymerase alpha. J Biol Chem. 1993;268:93–99. [PubMed] [Google Scholar]
- 44.Hupp T, Meek D, Midgley C, Lane D. Regulation of specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 45.Malanga M, Pleschke J, Kleczkowska H, Althaus F. Poly(ADP-ribose) binds to specific domains of p53 and alters its DNA binding functions. J Biol Chem. 1998;273:11839–11843. doi: 10.1074/jbc.273.19.11839. [DOI] [PubMed] [Google Scholar]
- 46.Ko L, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 47.Gu W, Roeder R. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 48.Neamati N, Fernandez A, Wright S, Kiefer J, McConkey DJ. Degradation of lamin B1 precedes oligonucleosomal DNA fragmentation in apoptotic thymocytes and isolated thymocyte nuclei. J Immunol. 1995;154:3788–3795. [PubMed] [Google Scholar]
- 49.Nosseri C, Coppola S, Ghibelli L. Possible involvement of poly(ADP-ribosyl) polymerase in triggering stress-induced apoptosis. Exp Cell Res. 1994;212:367–373. doi: 10.1006/excr.1994.1156. [DOI] [PubMed] [Google Scholar]
- 50.Marks D, Fox R. DNA damage, poly(ADP-ribosyl)ation and apoptotic cell death as a potential common pathway of cytotoxic drug action. Biochem Pharmacol. 1991;42:1859–1867. doi: 10.1016/0006-2952(91)90582-p. [DOI] [PubMed] [Google Scholar]
- 51.Tanizawa A, Kubota M, Hashimoto H, Shimizu T, Takimoto T, Kitoh T, Akiyama Y, Mikawa H. VP-16-induced nucleotide pool changes and poly(ADP-ribose) synthesis: the role of VP-16 in interphase death. Exp Cell Res. 1989;185:237–246. doi: 10.1016/0014-4827(89)90052-9. [DOI] [PubMed] [Google Scholar]
- 52.Tanizawa A, Kubota M, Takimoto T, Akiyama Y, Seto S, Kiriyama Y, Mikawa H. Prevention of adriamycin-induced interphase death by 3-aminobenzamide and nicotinamide in a human promyelocytic leukemia cell line. Biochem Biophys Res Commun. 1987;144:1031–1036. doi: 10.1016/s0006-291x(87)80067-0. [DOI] [PubMed] [Google Scholar]
- 53.Manome Y, Datta R, Taneja N, Shafman T, Bump E, Hass R, Weichselbaum R, Kufe D. Coinduction of c-jun gene expression and internucleosomal DNA fragmentation by ionizing radiation. Biochemistry. 1993;32:10607–10613. doi: 10.1021/bi00091a010. [DOI] [PubMed] [Google Scholar]
- 54.Yoon Y, Kim J, Kang K, Kim Y, Choi K, Joe C. Poly(ADP-ribosyl)ation of histone H1 correlates with internucleosomal DNA fragmentation during apoptosis. J Biol Chem. 1996;271:9129–9134. doi: 10.1074/jbc.271.15.9129. [DOI] [PubMed] [Google Scholar]
- 55.Lorenzo H, Susin S, Penninger J, Kroemer G. Apoptosis-inducing factor (AIF): a phylogenetically old, caspase-independent effector of cell death. Cell Death Differ. 1999;6:516–524. doi: 10.1038/sj.cdd.4400527. [DOI] [PubMed] [Google Scholar]
- 56.Feldenberg L, Thevananther S, del Rio M, de Leon M, Devarajan P. Partial ATP depletion induces Fas- and caspase-mediated apoptosis in MDCK cells. Am J Physiol. 1999;276:837–846. doi: 10.1152/ajprenal.1999.276.6.F837. [DOI] [PubMed] [Google Scholar]
- 57.Eguchi Y, Srinivasan A, Tomaselli K, Shimizu S, Tsujimoto Y. ATP-dependent steps in apoptotic signal transduction. Cancer Res. 1999;59:2174–2181. [PubMed] [Google Scholar]
- 58.Yakovlev A, Wang G, Stoica B, Simbulan-Rosenthal C, Yoshihara K, Smulson M. Role of DNAS1L3 in Ca2+- and Mg2+-dependent cleavage of DNA into oligonucleosomal and high molecular mass fragments. Nucleic Acids Res. 1999;27:1999–2005. doi: 10.1093/nar/27.9.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yakovlev A, Wang G, Stoica B, Boulares H, Spoonde A, Yoshihara K, Smulson M. A role of the Ca2+/Mg2+-dependent endonuclease in apoptosis and its inhibition by poly(ADP-ribose) polymerase. J Biol Chem. 2000;275:21302–21308. doi: 10.1074/jbc.M001087200. [DOI] [PubMed] [Google Scholar]
- 60.Chinnaiyan A, Orth K, O'Rourke K, Duan H, Poirier G, Dixit V. Molecular ordering of the cell death pathway. Bcl-2 and Bcl-xL function upstream of the CED-3-like apoptotic proteases. J Biol Chem. 1996;271:4573–4576. doi: 10.1074/jbc.271.9.4573. [DOI] [PubMed] [Google Scholar]
- 61.Muller M, Wilder S, Bannasch D, Israeli D, Lelbach K, Li-Weber M, Friedman S, Galle P, Stremmel W, Oren M, Krammer P. p53 activates the CD95 (APO-1/Fas) gene in response to DNA damage by anticancer agents. J Exp Med. 1998;188:2033–2045. doi: 10.1084/jem.188.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bennett M, MacDonald K, Chan S, Luzio J, Simari R, Weissberg P. Cell surface trafficking of Fas: a rapid mechanism of p53-mediated apoptosis. Science. 1998;282:290–293. doi: 10.1126/science.282.5387.290. [DOI] [PubMed] [Google Scholar]
- 63.Li X, Coffino P. Identification of a region of p53 that confers lability. J Biol Chem. 1996;271:4447–4451. doi: 10.1074/jbc.271.8.4447. [DOI] [PubMed] [Google Scholar]
- 64.Ruscetti T, Lehnerr B, Halbrook J, LeTrong H, Hoekstra M, Chen D, Peterson S. Stimulation of DNA PK by poly(ADP-ribose) polymerase. J Biol Chem. 1998;273:14461–14467. doi: 10.1074/jbc.273.23.14461. [DOI] [PubMed] [Google Scholar]
- 65.Meisterernst M, Stelzer G, Roeder R. Poly(ADP-ribose) polymerase enhances activator-dependent transcription in vitro. Proc Natl Acad Sci USA. 1997;94:2261–2265. doi: 10.1073/pnas.94.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Butler A, Ordahl C. Poly(ADP-ribose) polymerase binds with transcription factor 1 to MCAT1 elements to regulate muscle-specific transcription. Mol Cell Biol. 1999;19:296–306. doi: 10.1128/mcb.19.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kannan P, Yu Y, Wankhade S, Tainsky M. Poly(ADP-ribose) polymerase is a coactivator for AP-2-mediated transcriptional activation. Nucleic Acids Res. 1999;27:866–874. doi: 10.1093/nar/27.3.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anderson M, Scoggin K, Simbulan-Rosenthal C, Steadman J. Identification of poly(ADP-ribose) polymerase as a transcriptional coactivator of human T-cell leukemia virus type 1 Tax protein. J Virol. 2000;74:2169–2177. doi: 10.1128/jvi.74.5.2169-2177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simbulan-Rosenthal CM, Rosenthal DS, Luo R, Smulson ME. Poly(ADP-ribose) polymerase upregulates E2F-1 promoter activity and DNA pol α expression during early S-phase. Oncogene. 1999;18:5015–5023. doi: 10.1038/sj.onc.1202900. [DOI] [PubMed] [Google Scholar]
- 70.Oei S, Griesenbeck J, Schweiger M, Ziegler M. Regulation of RNA polymerase II-dependent transcription by poly(ADP-ribosyl)ation of transcription factors. J Biol Chem. 1998;273:31644–31647. doi: 10.1074/jbc.273.48.31644. [DOI] [PubMed] [Google Scholar]
- 71.Rawling J, Alvarez-Gonzalez R. TFllF, a basal eukaryotic transcription factor, is a substrate for poly(ADP-ribosyl)ation. Biochem J. 1997;324:249–253. doi: 10.1042/bj3240249. [DOI] [PMC free article] [PubMed] [Google Scholar]