Abstract
Experimental proteome analysis was combined with a genome-wide prediction screen to characterize the protein content of the thylakoid lumen of Arabidopsis chloroplasts. Soluble thylakoid proteins were separated by two-dimensional electrophoresis and identified by mass spectrometry. The identities of 81 proteins were established, and N termini were sequenced to validate localization prediction. Gene annotation of the identified proteins was corrected by experimental data, and an interesting case of alternative splicing was discovered. Expression of a surprising number of paralogs was detected. Expression of five isomerases of different classes suggests strong (un)folding activity in the thylakoid lumen. These isomerases possibly are connected to a network of peripheral and lumenal proteins involved in antioxidative response, including peroxiredoxins, m-type thioredoxins, and a lumenal ascorbate peroxidase. Characteristics of the experimentally identified lumenal proteins and their orthologs were used for a genome-wide prediction of the lumenal proteome. Lumenal proteins with a typical twin-arginine translocation motif were predicted with good accuracy and sensitivity and included additional isomerases and proteases. Thus, prime functions of the lumenal proteome include assistance in the folding and proteolysis of thylakoid proteins as well as protection against oxidative stress. Many of the predicted lumenal proteins must be present at concentrations at least 10,000-fold lower than proteins of the photosynthetic apparatus.
INTRODUCTION
Recently, the genome of the dicotyledon Arabidopsis was sequenced completely, and 25,498 genes were annotated (Arabidopsis Genome Initiative, 2000). The availability of the Arabidopsis genome now allows the classification of proteins according to predicted domain(s) and subcellular localization. Importantly, the sequenced genome also makes it possible to take advantage of the dramatic improvements in biological mass spectrometry to study gene expression directly at the protein level and to determine protein localization and post-translational modifications efficiently in a systematic manner. The improved mass accuracy, mass resolution, and sensitivity of the latest generation of mass spectrometers allow the rapid identification of picomole to femtomole amounts of proteins and peptides. Examples of excellent reviews on mass spectrometry and their application in biology include those by Jensen et al. (1998), Pandey and Mann (2000), and Yates (2000) and references in the study by Blackstock and Mann (2000). Combined with rapidly expanding plant genome information, these technical improvements are expected to have a profound effect on plant biology (for discussion, see van Wijk, 2001).
A significant subset of the Arabidopsis proteome is localized in different organelles (e.g., the mitochondria and different types of plastids). Plastids are predicted to contain between 10 to 15% of the nucleus-encoded gene products, corresponding to ∼2500 to 3500 proteins, indicating the importance of this organelle in the plant cell (Arabidopsis Genome Initiative, 2000). This prediction is based on the presence of an N-terminal chloroplast transit peptide (cTP) in the nucleus-encoded chloroplast proteins (except for proteins localized in the chloroplast outer membrane) using the cellular localization program TargetP (Emanuelsson et al., 2000). After synthesis in the cytosol, the cTP is recognized at the chloroplast envelope and the precursor protein is translocated into the chloroplast, followed by processing of the cTP. Proteins then are directed into the inner envelope membrane, into the thylakoid membranes, or remain in the chloroplast stroma. The inner envelope proteins, the peripheral thylakoid proteins located at the stromal site of the thylakoid membrane, and most of the integral thylakoid membrane proteins have no additional N-terminal transit peptides. In contrast, proteins located in the thylakoid lumen are targeted and translocated via a second transit peptide, the lumenal transit peptide (lTP), which is located directly C-terminal of the cTP. A few small thylakoid proteins with a single transmembrane (TM) domain also have an lTP (Thompson et al., 1998, 1999). Lumenal proteins are involved in a number of well-characterized functions, such as water splitting, electron transport, and the violaxanthin cycle. However, the function of most newly discovered lumenal proteins is not clear (Kieselbach et al., 1998; Peltier et al., 2000).
The lTPs show strong similarities to bacterial signal peptides and can be divided into a charged N-terminal domain, a hydrophobic core, and a more polar C-terminal domain that ends with short chain residues at the −3 and −1 positions relative to the terminal cleavage site (von Heijne et al., 1989). Soluble lumenal proteins with lTPs are translocated by at least two different mechanisms involving two different sets of proteins, whereas the single TM proteins with lTPs appear to insert spontaneously (reviewed by Dalbey and Robinson, 1999; Keegstra and Cline, 1999). Proteins translocated via the so-called Sec pathway require ATP and cpSecA/Y (Knott and Robinson, 1994; Settles et al., 1997; Mori et al., 1999) and probably cpSecE (Schuenemann et al., 1999; Froderberg et al., 2001). Proteins translocated via the ΔpH or twin-arginine translocation (TAT) pathway require neither soluble factors nor GTP or ATP, but they do require a transthylakoid proton gradient and several TAT proteins. The TAT substrates also require a specific TAT motif, RRx-h-h, where R is arginine and h is a hydrophobic residue (Walker et al., 1999; Robinson and Bolhuis, 2001; for an exception, see Summer et al., 2000).
In a recent study, we began to systematically identify proteins in thylakoids from pea using two-dimensional (2-D) electrophoresis and mass spectrometry (MS) (Peltier et al., 2000; van Wijk, 2000). Because little genomic or protein sequence information is available for pea, most of the proteins were identified based on homology with proteins in Arabidopsis. The nucleus-encoded proteins identified on the 2-D gels were used to test the prediction for chloroplast localization and the transit peptide by the software programs ChloroP (Emanuelsson et al., 1999) and SignalP (Nielsen et al., 1997, 1999). The program SignalP (http://www.cbs.dtu.dk/services/SignalP/) was developed originally for the prediction of cleavable signal peptides of secretory proteins in bacteria and eukaryotes, but it also predicted lTPs as well as their cleavage sites with good success (Peltier et al., 2000).
With the complete annotated genome of Arabidopsis now available, we set out to identify a larger fraction of lumenal proteins and to extend the set of experimentally determined N termini. We expected that this would provide parameters and thresholds for a genome-wide theoretical prediction of the lumenal proteome using modified versions of the cellular localization programs TargetP (an update of ChloroP) and SignalP as a basis. We also thoroughly verified the gene annotation of all experimentally identified lumenal proteins and demonstrate generic strategies for reannotation. The molar ratio between the most abundant and least abundant identified proteins was 5 × 104, and many of the predicted lumenal proteins are expected to have expression levels that are 105- to 106-fold below those of the most abundant lumenal proteins.
RESULTS
Experimental Identification of Lumenal and Weakly Bound Peripheral Proteins by 2-D Electrophoresis and MS
To purify the lumenal proteome, thylakoids from Arabidopsis were purified carefully using differential centrifugation, linear Percoll gradients, and multiple washing steps. Subsequently, the purified thylakoids were disrupted by Yeda press treatment. The lumenal proteome then was separated by 2-D electrophoresis using immobilized pH gradient (IPG) strips in the first dimension (pI 4 to 7 and 6 to 11) and high-resolution Tricine gradient gels in the second dimension. Both analytical (250 μg of protein) and preparative (1.5 to 2 mg of protein) 2-D gels were generated and were stained with silver or Coomassie blue, respectively. Figures 1A and 1B show the analytical 2-D gels of the purified lumen at the two pI ranges. For identification of the proteins, the strategy used was similar to that described by Peltier et al. (2000). In summary, protein spots were picked from the 2-D gels, washed, digested with the site-specific protease trypsin, and extracted. The masses of the extracted peptides were determined by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) MS and/or sequenced by nano-electrospray ionization tandem mass spectrometry (nano-ESI/MS/MS) and followed by identification using different search engines. N-terminal sequences of ∼30 identified proteins were determined by Edman degradation to validate lTP prediction.
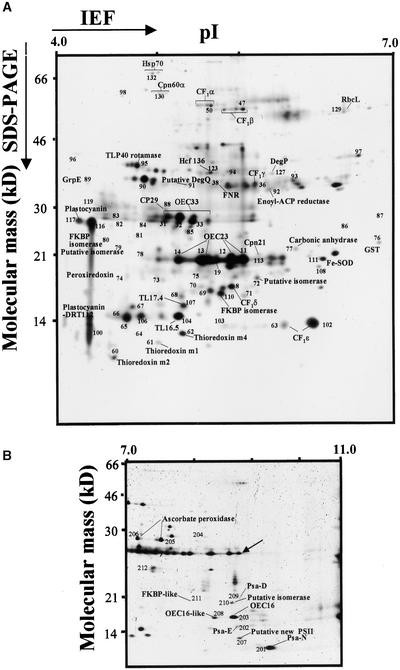
Figure 1.
Silver-Stained 2-D Electrophoresis Maps of Lumenal Proteins of Arabidopsis (ecotype Columbia).
Fractions strongly enriched for thylakoid lumenal proteins isolated from Arabidopsis were separated by 2-D electrophoresis with denaturing isoelectric focusing (IEF) on immobilized pH gradients in the first dimension between pH 4 and 7 (A) or between pH 7 and 11 (B) and Tricine-PAGE in the second dimension ([A] and [B]). Gels were calibrated for molecular mass (in kD) and pI (in pH units) by internal (pH and mass) and external (mass) standards. The protein spot numbers refer to the spot numbers listed in Tables 1 to 4. For a selected number of spots, the identity (in addition to the number) also has been listed on the 2-D maps. The map in (B) also includes the internal standards for the pI, as indicated by the arrow. To more easily compare the Arabidopsis maps with those constructed for pea (as described by Peltier et al., 2000), spot numbers are identical for orthologous pairs.
ACP, acyl carrier protein; FNR, ferredoxin-NADP reductase; GST, glutathione S-transferase; PSII, photosystem II; SOD, superoxide dismutase.
The proteins identified on the 2-D gels and the summarizing data of the identification process are listed in the tables, grouped according to the predicted or most likely cellular localization. For each protein identified, we have listed the percentage of sequence coverage obtained by MALDI-TOF MS (at 50 ppm mass accuracy), one of the several internal sequences determined by tandem MS, and the N-terminal sequence as determined by Edman sequencing. In addition, the localization predictions by TargetP and Predotar, as well as the predictions of lTPs obtained using SignalP in three different search modes (eukaryotic, Gram-positive, and Gram-negative bacteria), are indicated. A prediction of the targeting pathway (TAT or Sec + other) through the thylakoid membrane based on the characteristics of the lTP also is listed. This extensive prediction of location and cleavage sites was performed to set thresholds and parameters for the theoretical genome-wide prediction of the lumenal proteome, as presented in the second half of this article. It is important to stress that we did not attempt to experimentally identify all lumenal proteins (an impossible task); rather, we sought to assemble a set of lumenal proteins that would allow determination of the parameters and thresholds for theoretical prediction. This has the advantage that an overview of the total potential lumenal proteome and function can be obtained, including proteins of very low abundance or proteins that are expressed only under specific conditions or in different types of plastids (see Discussion).
Protein Identities, Function, and Expression of Isoforms and Paralogs
Eighty-one proteins on the 2-D gels were identified with high confidence. Thirty of those are very clearly lumenal proteins, based on characteristic presequences and on very good matches between the experimentally determined N termini and the predicted lTPs (Table 1). Nineteen of those lumenal proteins have signal sequences that are typical for TAT substrates. An additional 12 proteins, listed in Table 2, have fairly clear features of lTPs, and for several of them the experimentally determined N terminus corresponded to the predicted lumenal cleavage site. However, because of an ambiguous Edman sequence tag, because the lTP cleavage site is unusual (see further below), or because of contrasting localization information in the literature, they were not added to Table 1 and their true localization needs to be determined by additional experimentation.
Table 1.
Identification of 30 Lumenal Arabidopsis Proteins from the 2-D Electrophoresis Gels Shown in Figure 1
| Spot No. | Molecular Mass (kD) |
pI | Identitya | Accession No.b |
MALDI Percent Coverage at 50 ppmc |
Localization and Cleavage Site Predictione
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SignalP
|
|||||||||||||
|
MS/MS Sequenced |
TargetingPathway | TargetP | Predotar | Euk | Gram− | Gram+ | N Terminus | ||||||
| 203 | 16.7 | 9.3 | OEC16 | 4583542 (At4g21280) | 44 | TAT | C2/44 | P 1 | 75-VLA-DA | 74-AVL-AD | 77-ADA-IS | 77-ISIKVf | |
| 208 | 16.7 | 9.1 | OEC16-like | 7267278 (At4g05180) |
37 | TAT | C3/48 | P 0.7 | 84-AEA-IP | 84-AEA-IP | 84-IPIKVf | ||
| 11, 12, 13, 14 | 20.5 | 5.7 to 6.0 | OEC23 | 7443217 (At1g06680) |
40 | TAT | C2/31 | P 0.6 | 77-ADA-AY | 77-ADA-AY | 77-ADA-AY | 77-AYGEAf,g,h | |
| 201 | 12.1 | 9.6 | Psa-N | 1709825 (At5g64040) |
35 | TAT | C5/81 | P 0.9 | 84-ASA-NA | 86-ANA-GV | 84-ASA-NA | 84-NAGVIf | |
| 123 | 36.7 | 5.6 | Hcf136 | 6016183 (At5g23120) |
47 | TAT | C2/60 | P 0.9 | 78-ARA-DE | 78-ARA-DE | 78-ARA-DE | 78-DEQLSf,g | |
| 205 to 206 | 30.4 | 7.2 to 7.6 | Putative ascorbate peroxidase |
7267543 (At4g09010) |
32 | TAT | C3/36 | P 1 | 82-AKA-AD | 82-ADLIQf,g | |||
| 207 | 13.2 | 9.3 | Putative new photosystem II protein |
2809245 (At1g03600) |
33 | TAT | C2/67 | P 1 | 68-VSA-AE | 68-VSA-AE | 68-VSA-AE | 68-AEDEEg | |
| 91 | 33.6 | 5.4 | Putative DegQ protease (DegP5) |
2832642 (At4g18370) | 11 | TAT | C3/26 | P 0.5 | 10-AFS-SI | 71-AIA-LE | 71-XEQXXh | ||
| 19 | 19.4 | 5.6 | Putative protein OEC23 related |
2244908 (At4g15510) |
23 | TAT | C1/75 | M 0.7 | 104-AFA-ST | 104-AFA-ST | 54-QSA-KS | 104-STPVFf | |
| 71 | 17.2 | 6.0 | Not annotated OEC23 related |
AC007171 | SLDQFGSPQFVAD (3) | TAT (KR) | C4/34 | P 0.8 | 71-AKS-ME | 26-SHH-KI | 69-NKA-KS | ||
| 204 | 31.4 | 8.5 | Putative protein OEC23 related |
7019666 (At3g55330) |
31 | TAT | C1/26 | Neither | 74-SFA-AE | 74-SFA-AE | 74-SFA-AE | 74-AESKKg | |
| 212 | 26.3 | 7.6 | Putative protein OEC23 related |
7485407 (At2g39470) |
54 | TAT | C3/72 | Neither | 73-LLA-EE | 73-LLA-EE | 73-LLA-EE | ||
| 108 | 19.0 | 6.5 | Putative protein OEC23 related |
2829916 (At1g77090) | 33 | TAT | C4/35 | M 1 | 63-ALA-FP | 63-ALA-FP | 70-VVKQGh | ||
| 111 | 19.5 | 6.5 | Putative protein OEC23 related |
7594543 (At3g56650) |
GSTVVLFVVSATEK (7) | TAT | C1/75 | P 0.9 | 66-ISA-AR | 66-ISA-AR | 67-REVEVg,h | ||
| 210 | 17.3 | 9.3 | Putative isomerase | 6143884 (At3g10060) |
50 | TAT | C2/56 | Both | 73-ASG-IL | 82-AEA-VS | 82-AEA-VS | ||
| 110 | 17.3 | 5.7 | Putative FKBP isomerase |
2289010 (At2g43560) | 50 | TAT | C4/48 | Neither | 59-AAG-LP | 57-AYA-AG | 57-AYA-AG | AGLPPf | |
| 80 | 25.4 | 4.7 | Similar to FKBP isomerase |
7543908 (At5g13410) |
24 | TAT | C1/29 | P 0.6 | 31-VAA-RL | 17-ALA-GT | 89-SQFADf | ||
| 104 | 16.0 | 5.2 | Putative protein | 2262151 (At4g02530) | 23 | TAT | C1/38 | P 0.9 | 58-LVA-IG | 58-LVA-IG | 73-AILEAg | ||
| 70 | 17.8 | 5.5 | Putative protein OEC23 related |
6642664 (At1g76450) | VEAFAETLVSGLDR (3) | TAT | C2/32 | P 0.8 | 80-AFA-ET | 80-AFA-ET | 80-AFA-ET | 80-ETNASf | |
| 117 | 28.9 | 4.2 | Plastocyanin 2 | 130261 (At1g76100) |
23 | Sec + other | C1/32 | P 0.9 | 72-AMA-ME | 72-AMA-ME | 72-AMA-ME | 72-MEVLLf | |
| 90, 100, 116 | 14 to 35 | 4.4 to 4.9 | Plastocyanin 1 DRT112 |
1169201 (At1g20340) |
29 | IEVLLGGGDGSLAFIPNDFSIAK (4) | Sec + other | C1/75 | P 1 | 68-AMA-IE | 68-AMA-IE | 68-IEVLLf,h | |
| 31 to 33 | 28.5 | 5.1 to 5.6 | OEC33 | 3286693 (At5g66570) |
55 | Sec + other | C2/26 | P 0.8 | 85-ASA-EG | 85-ASA-EG | 85-ASA-EG | 85-EGAPKf | |
| OEC33-like | 4835233 (At3g50820) |
37 | Sec + other | C2/28 | P 0.9 | 84-AGA-EG | 84-AGA-EG | 84-AGA-EG | 84-EGAPKf | ||||
| 127 | 36.5 | 6.2 | DegP1 | 2565436 (At3g27925) |
31 | Sec + other | C3/35 | P 1 | 100-VES-AS | 99-AVE-SE | 100-VES-AS | 103-FVVSTf | |
| 95 | 38.5 | 4.8 | TLP40 rotamase | 6016707 (At3g01480) |
53 | Sec + other | C1/75 | P 1 | 82-AHA-VA | 82-AHA-VA | 82-AHA-VA | 92-VLISGf | |
| 211 | 21.2 | 8.5 | FKBP-like protein | 6686798 (At4g39710) |
32 | Sec + other | C3/74 | M 0.6 | 73-ADA-TR | 73-ADA-TR | 73-ADA-TR | ||
| 107 | 16.7 | 5.3 | Putative protein pentapeptide repeat |
6226234 (At5g53490) |
36 | AFVGNTIGQADGVYDKPLDLR (1) | Sec + other | C4/65 | P 0.7 | 77-VIA-AN | 77-VIA-AN | 77-VIA-AN | 77-ANQRLg |
| 103 | 15.0 | 5.7 | Putative protein pentapeptide repeat |
2344892 (At2g44920) | 31 | Sec + other | C1/56 | P 0.8 | 81-ALA-FK | 81-ALA-FK | 81-ALA-FK | 81-FKGGGg | |
| 106 | 16.0 | 4.8 | Putative protein | 8809586 (At5g52970) | VLAQNYPVTPGLAIK (4) | Sec + other | C4/83 | P1 | 75-ADA-KV | 75-ADA-KV | 75-ADA-KV | 75-KVGVNg | |
| 73 | 18.2 | 5.0 | Putative protein | 3776572 (At1g54780) |
44 | Sec + other | C2/53 | P 1 | 84-ALA-SE | 84-ALA-SE | 84-ALA-SE | 84-SEFNIh | |
| 68 | 17.1 | 5.3 | Putative protein | 4455236 (At4g24930) |
FWLEDTPYGR (2) | Sec + other | C4/66 | M 0.5 | 63-ALA-IP | 63-ALA-IP | 63-ALA-IP | 63-IPSLSf | |
Proteins were identified by MALDI-TOF MS and/or nanoESI/MS/MS. N termini of all proteins were determined by Edman sequencing. All accession numbers in italics showed mis-assignments in the database and have been corrected.
Identity given in NCBI completed by domain prediction found by Pfam, Blocks, Prints, Prodom, or Phi-Blast.
Accession numbers in NCBI and in MIPS (in parentheses). All accession numbers in italics showed misassignments in the database and have been corrected.
Percentage of coverage at 50 ppm for the MALDI-TOF peptides.
Sequence tag identified by ESI/MS/MS; additional sequence tags were obtained (numbers listed in parentheses) but are not shown.
Localization, reliability class, and cleavage site prediction by TargetP (C for chloroplast), localization and score by Predotar (P for plastid, M for mitochondria), cleavage site prediction of the lumenal transit peptide by three versions of SignalP developed for eukaryotes (Euk), Gram-negative or Gram-positive bacteria, and in boldface when this prediction fits with the N-terminal Edman sequence.
N-terminal Edman sequence tag found in the literature or in public databases.
N-terminal Edman sequence tag identified in our previous study of pea.
N-terminal Edman sequence tag identified in this study.
Table 2.
Identification of 12 Arabidopsis Proteins from the 2-D Electrophoresis Gels Shown in Figure 1
| Spot No. | Molecular Mass (kD) |
pI | Identitya | Accession No.b |
MALDI Percent Coverage at 50 ppmc |
Localization and Cleavage Site Predictione
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SignalP
|
|||||||||||||
|
MS/MS Sequenced |
TargetingPathway | TargetP | Predotar | Euk | Gram− | Gram+ | N Terminus | ||||||
| 74 | 18.4 | 4.7 | 2-Cys peroxiredoxin | 7242491 (At3g11630) |
APDFEAEAVFDQEFIK (1) | TAT | C2/48 | P 1 | 69-AQA-DD | 69-AQA-DD | |||
| 68 | 17.1 | 5.3 | Peroxiredoxin-like | 7529720 (At3g52960) |
YAILADDGVVK (3) | TAT | C1/56 | P 0.9 | 71-VTI-SI | 73-ASI-SV | 73-SVXXKf | ||
| 79 | 22.2 | 4.7 | Putative isomerase | 7287985 (At3g60370) |
38 | TAT | C5/42 | Neither | 53-SSS-AK | 54-SSA-KT | 54-SSA-KT | ||
| 89 | 34.6 | 4.3 | Chaperone GrpE | 4583546 (At5g11710) |
45 | TAT | C2/64 | Neither | 67-ASG-EA | 68-SGE-AE | 67-ASG-EA | ||
| 69 | 17.6 | 5.6 | Putative protein | 5840783 (At5g58250) |
VHFLVANAK (7) | Sec + other | C1/52 | M 0.8 | 18-AAA-CR | 58-KTA-AT | 58-KTA-AT | ||
| 60 | 12.1 | 4.6 | Thioredoxin m1 | 2809238 (At1g03680) |
35 | Sec + other | C2/48 | Neither | 69-CEA-QD | ||||
| 61 | 13.3 | 5.0 | Thioredoxin m2 | 4206206 (At4g03520) |
34 | Sec + other | C1/56 | Neither | 75-CEA-QE | 73-EAQETf | |||
| 62 | 14.3 | 5.3 | Thioredoxin m4 | 6539614 (At3g15360) |
21 | Sec + other | C1/82 | P 0.9 | 27-SSA-AP | 27-SSA-AP | 69-RIA-RG | 76-EAQDTf | |
| 77 | 21.3 | 6.3 | Carbonic anhydrase | 14343 (At3g01500) |
46 | Sec + other | C2/32 | P 0.8 | 19-SQS-SL | 64-VFA-AP | 64-VFA-AP | 66-APXIAf | |
| 131 | 75.9 | 4.9 | Hsp70 | 7441883 (At4g24280) |
21 | Sec + other | C1/92 | P 0.8 | 93-AVA-AM | ||||
| 130 | 61.8 | 5.0 | Cpn60 | 2506276 (At2g28000) |
24 | Sec + other | C2/45 | P 1 | 14-VLC-SS | 49-ANV-KE | |||
| 113 | 20.6 | 6.2 | Cpn21 | 4127456 (At5g20720) |
23 | Sec + other | C2/50 | P 1 | 19-SLA-SL | 92-AQS-KP | |||
The locations of these proteins are either in the thylakoid lumen or peripherally attached to the stroma side of the thylakoid membrane. All proteins were identified by MALDI-TOF MS and/or nano-ESI/MS/MS. N termini for four proteins were determined by Edman sequencing.
Identity given in NCBI completed by domain prediction found by Pfam, Blocks, Prints, Prodom, or Phi-Blast.
Accession numbers in NCBI and in MIPS (in parentheses).
Percentage of coverage at 50 ppm for the MALDI-TOF peptides.
Sequence tag identified by ESI/MS/MS; additional sequences were identified (numbers in parentheses) but are not shown.
Localization, reliability class, and cleavage site prediction by TargetP (C for chloroplast), localization and score by Predotar (P for plastid, M for mitochondria), cleavage site prediction of the lumenal transit peptide by three versions of SignalP developed for eukaryotes (Euk), Gram-negative or Gram-positive bacteria, and in boldface when this prediction fits with the N-terminal Edman sequence.
N-terminal Edman sequence tag identified in this study.
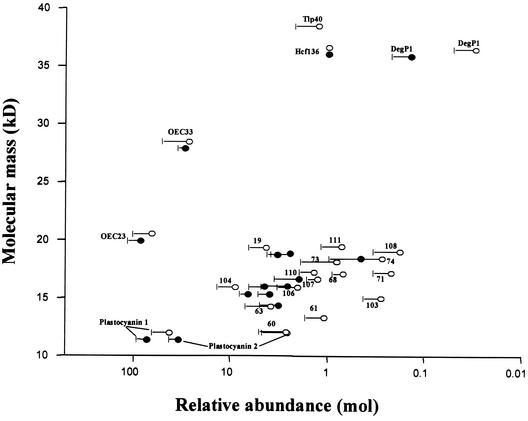
To quantify the expression levels of the proteins in both pea (Figure 2 from Peltier et al., 2000) and Arabidopsis, silver-stained and Coomassie blue–stained gels (pI 4 to 7 maps) were analyzed using the 2-D software package Melanie. All expression levels were calculated on a molar basis and normalized within each gel to the expression of Hcf136, because Hcf136 spots could be well quantified on both the Coomassie blue– and silver-stained maps (Figure 2). Spots are shown only if they were detected in duplicate on both the Coomassie blue– and silver-stained gels (thus, four gels for each spot). The expression levels of the most abundant proteins (the oxygen-evolving complex [OEC] proteins and plastocyanin [PC]) were ∼10,000-fold higher than those of the least abundant proteins (see below for details). In general, the expression levels for pea and Arabidopsis corresponded very well (Figure 2).
Figure 2.
Relative Expression Levels of Thylakoid Proteins in Pea and Arabidopsis.
Quantification of expression levels of lumenal and other thylakoid proteins from Arabidopsis proteins (closed symbols) and pea proteins (open symbols) calculated on a molar basis and normalized to the expression level of Hcf136. Duplicate Coomassie blue–stained and duplo silver-stained 2-D gels of two independent experiments with a pH range of 4 to 7 were analyzed. Standard errors (n = 4) are indicated, and the x axis is in log scale.
Apart from OEC23, seven weakly related OEC23 paralogs were identified on the 2-D gels (Table 1). In our earlier study of pea thylakoids, we also observed expression of an eighth paralog (spot 22). Sequence tags for some of the OEC23 paralogs (spots 19, 108, 111, and 204) also were identified on our previous maps of the pea lumen; however, at the time, we were not always able to find the corresponding genes or the OEC23 domains. The expression levels of the paralogs were 20- to 300-fold lower than that of OEC23. Thus, OEC23 is by far the most strongly expressed protein in this family. Database searching identified a ninth OEC-like protein that was 88% identical to OEC23 (accession number At2g30790). However, expression data could not be found from expressed sequence tags (ESTs) or at the protein level. Sequence analysis of this OEC23-related gene showed a frameshift (as a result of a base pair loss) at the end of the first exon in the lTP, possibly preventing protein expression.
For three other lumenal proteins (OEC16, OEC33, and PC) and several peripheral proteins (fibrillins, RNA binding protein 29, and m-type thioredoxins), one or more very closely related paralogs were identified (Figures 3A and 3B). The difference in expression levels within each group of paralogs was on the order of a factor 5 to 25. Alignment of pairs of these closely related paralogs showed that the presequences were less conserved than were the mature proteins, with an average 63% identity for the presequences and 78% for the mature proteins (Figures 3A and 3B). It is likely that these paralogs originate from gene duplications (see Discussion).
Figure 3.
Sequence Analysis of Pairs of Isoforms/Paralogs Present in the Thylakoid Lumen or Associated with the Thylakoid Membrane of Arabidopsis Chloroplasts.
Alignments of two pairs of paralogs of thylakoid proteins identified on the 2-D gels shown in Figure 1. The cleavage sites of the lTP are indicated by the arrows.
(A) OEC16 isoforms in spots 203 and 208.
(B) Plastocyanin (PC) isoforms in spots 116 and 117.
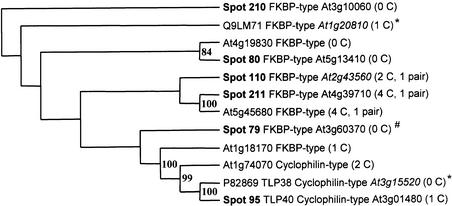
Interestingly, a multigene family of five lumenal isomerases (Table 1) and one potential lumenal isomerases (Table 2) was identified experimentally, of which only TLP40 was identified previously (Fulgosi et al., 1998). The relationship for this isomerase family is shown in Figure 4 in the form of a rooted tree. Two of the isomerases (spots 210 and 211) are basic proteins (pI 8.3 and 9.3) and were expressed at much lower levels than was OEC16 (Figure 1B). The expression levels of TLP40 and spot 110 were quite similar to the expression of Hcf136 (Figure 2), whereas spot 79 was expressed at very low levels (<0.01).
Figure 4.
The Phylogenetic Relationship between Members of the Lumenal Isomerase Family.
The rooted tree of lumenal isomerases in spots 79, 80, 95, 110, 210, and 211, four predicted isomerases, and two isomerases found in Swiss-Prot (marked with *). The tree was built in Phylip using both parsimony and distance methods on protein sequences and reflects consensus trees with branches supported by the highest possible bootstrap values. The rooting was done with an outgroup sequence from the moss Physcomitrella and is an ortholog of spot 210. The location of Spot 79 is ambiguous (#).
Other lumenal or putative lumenal proteins include several known chaperones (GrpE, HSP70, Cpn60, and Cpn21), the assembly factor Hcf136 (Meurer et al., 1998), the protease DegP1 described by Itzhaki et al. (1998), and carbonic anhydrase. In addition, eight lumenal proteins (Table 1) and two more potential lumenal proteins (Table 2) with unknown functions were identified. We identified three m-type thioredoxins (m1, m2, and m4) as well as two 2-Cys peroxiredoxins and sequenced the N termini of m2 and m4 and one of the peroxiredoxins. Recently, these five proteins were implicated directly in antioxidant defense (Baier and Dietz, 1999; Baier et al., 2000; Issakidis-Bourguet et al., 2001), and their localization is not clear but probably is peripheral on the stromal side of the thylakoid membrane (Baier et al., 2000).
In addition to the lumenal proteins, 32 proteins were identified that are either (1) peripheral thylakoid proteins attached to the stromal side and are part of photosynthetic complexes (two Psa proteins and five CF1 proteins) or (2) associated with the thylakoid membranes as part of their function (six ribosomal proteins, four RNA binding proteins, a ribosome recycling factor, six fibrillins, Fe–superoxide dismutase, and ClpS1) and very low levels of the two ribulose-1,5-bisphosphate carboxylase/oxygenase subunits, which are the most abundant stromal proteins in the chloroplast (Table 3). Interestingly, we also identified ROC4, a cyclophilin with peptidyl-prolyl cis-trans isomerase activity, which was determined earlier to be in chloroplasts by protein gel blotting (Lippuner et al., 1994). The copurification with the lumenal proteins indicates that they interact with the thylakoid membrane but that they are released easily by either the Yeda press or the sonication used to open the lumenal compartment. Because we did not observe any significant amount of the very abundant Calvin cycle enzymes, it is clear that this set of 32 proteins interacts with the membrane as part of their function.
Table 3.
Arabidopsis Proteins Copurified with the Thylakoid Membranes and Identified from the 2-D Electrophoresis Gels Shown in Figure 1
| Spot No. | Apparent Molecular Mass (kD) |
pI | Identitya | Accession No.b | MALDI-TOF Percent Coverage at 50 ppmc |
MS/MS Sequenced |
Localization and Cleavage Site Predictione
|
||
|---|---|---|---|---|---|---|---|---|---|
| TargetP | Predotar | SignalP | |||||||
| 75 | 18.7 | 5.5 | ClpS1 | 5123926 (At4g25370) | AIAWAIDEK (1) | C1/63 | P 1 | −73 PIA-QP | |
| 92 | 33.3 | 6.2 | Enoyl-acyl carrier protein reductase |
4006834 (At2g05990) | 35 | C2/74 | M 0.8 | None | |
| 111 | 19.5 | 6.5 | Fe-superoxide dismutase |
1351082 (At4g25100) | 13 | TFMTNLVSWEAVSAR (2) | No chloroplast | Neither | None |
| 72 | 18.3 | 6.1 | ROC4 isomerase | 461899 (At5g13120) | 21 | C2/67 | P 0.8 | −55-HYA-SP +47-GIA-LS |
|
| 81 | 28.5 | 4.7 | Fibrillin homolog | 7484966 (At4g22240) | 42 | C2/59 | P 1 | None | |
| 82 | 28.9 | 4.7 | Probable fibrillin | 7488105 (At4g04020) | 42 | C2/55 | P 1 | None | |
| 83 | 28.2 | 4.9 | Fibrillin CDSP34 homolog 1 + |
6729544 (At3g58010) | FFMISYLDDEILIVR (6) | C1/53 | P 1 | None | |
| Fibrillin CDSP34 homolog 2 |
2673904 (At2g42130) | LKEEYVEGMLETPTVIEEAVPEQLK (4) | C1/48 | P 1 | ±24 ASP-SR | ||||
| 84 | 26.1 | 5.4 | Similar to fibrillin | 11994325 (At3g23400) | LIPVTLGQVFQR (3) | C1/72 | P 0.8 | −18 ALL-SD +104 LVA-SV |
|
| 96 | 40.9 | 4.1 | Putative fibrillin | 3608139 (At2g35490) | 30 | C1/53 | M 0.9 | −45 YRP-KP +50 RFS-KI |
|
| 88 | 29.9 | 5.1 | CP29 A′ RNA-BP+ | 681904 (At3g53460) | 21 | C1/65 | Neither | −12 AFN-PK | |
| Putative RNA-BP | 3608147 (At2g35410) | 13 | C1/74 | P 1 | −45 SNL-SP +75 TSA-DE |
||||
| 119 | 32.1 | 4.1 | CP31 RNA-BP | 681908 (At4g24770) | 40 | C1/93 | P 1 | −85 DWA-EE +16 AMA-DS |
|
| 85 | 24.3 | 4.8 | CP29 B′ RNA-BP | 4056477 (At2g37220) | 35 | C1/47 | P 1 | −37 LSF-KL +107 AQL-AQ |
|
| 113 | 20.6 | 6.2 | Similar to RNA-BP | 4678944 (At3g52150) | 21 | C1/56 | P 0.6 | −42 SLA-GT | |
| 86 | 26.5 | 6.6 | Ribosomal S5 P | 4836870 (At1g78630) | 20 | C1/49 | P 1 | −17 LHT-RT +50 VKA-SS |
|
| 107 | 17.1 | 5.3 | Ribosomal S7 P | 7525079 | 34 | Chloroplast encoded | |||
| 87 | 26.8 | 6.8 | Ribosomal L4 P | 3914666 (At1g07320) | 41 | C2/32 | P 1 | −20 LFL-SS +25 SHQ-IP |
|
| 67 | 16.3 | 4.8 | Ribosomal L12 P | 548655 (At3g27850) | ILVDYLQDK (4) | C1/54 | P 0.8 | −21 TCA-ST +58 VEA-PE |
|
| 78 | 19.5 | 4.9 | Ribosomal L13 P | 1707008 (At2g33800) | 23 | C1/56 | P 1 | −19 VKS-SG +47 IYA-NS |
|
| 65 | 15.9 | 4.7 | Ribosomal L27 P | 9759141 (At5g40950) | 29 | C4/19 | P 1 | −27 SFL-NR | |
| 64 | 15.2 | 4.9 | Putative ribosomal recycling factor |
7523401 (At3g63190) | 31 | C2/18 | P 0.9 | +48 LIA-CS | |
| 209 | 18.9 | 9.3 | Psa-D | 4587564 (At1g03130) | 40 | C1/43 | P 1 | +43 AIR-AE | |
| 202 | 14.2 | 9.3 | Psa-E | 7443149 (At4g28750) | 47 | C2/44 | P 1 | ±22 AGA-SS | |
| 38 | 33.9 | 5.6 | FNR | 5730139 (At5g66190) | 35 | C2/64 | P 0.7 | ±48 VKA-QV | |
| 102 | 15.2 | 6.4 | RbcS (20.2) | 4204274 (At1g67090) | 21 | C3/54 | P 1 | ±18 AQA-TM | |
| 129 | 50.6 | 6.6 | RbcL (47.3) | 1944432 | 13 | Chloroplast encoded | |||
| 48 to 52 | 53.4 | 5.4 to 5.6 | CF1α (55.1) | 5881679 | 41 | Chloroplast encoded | |||
| 43 to 47 | 51.3 | 5.7 to 6.0 | CF1β (53.1) | 5881701 | 57 | Chloroplast encoded | |||
| 34 | 34.5 | 6.1 | CF1γ (53.1) | 461550 (At4g04640) | 22 | C1/42 | P 0.9 | −18 SLS-AD +43 SRA-SS |
|
| 8 | 17.8 | 5.8 | CF1δ (53.1) | 5916447 (At4g09650) | 55 | C4/48 | P 0.8 | −58 AMA-LA | |
| 102 to 53 | 15.2 | 6.3 to 6.4 | CF1ε (53.1) | 7525039 | 53 | Chloroplast encoded | |||
These proteins are located at the stromal side of the thylakoids and were released by Yeda press. Proteins were identified by MALDI-TOF MS and/or nano-ESI/MS/MS.
Identity given in NCBI completed by domain prediction found by Pfam, Blocks, Prints, Prodom, or Phi-Blast.
Accession numbers in NCBI and in MIPS (in parentheses).
Percentage of coverage at 50 ppm for the MALDI-TOF peptides.
Sequence tag obtained identified by ESI/MS/MS; other sequences were obtained (number of sequences identified in parentheses) but are not shown.
Localization, reliability class, and cleavage site prediction by TargetP (C for chloroplast), localization and score by Predotar (P for plastid, M for mitochondria), cleavage site prediction of the lumenal transit peptide by two versions of SignalP developed for Gram-negative (−) or Gram-positive (+) bacteria.
Finally, low levels of six nonchloroplast proteins from different cellular locations were identified (Table 4). All six proteins are known to be very abundant in each of these locations. Several of these proteins were identified exclusively by sequence tags and not by MALDI-TOF MS. Together, these six proteins contribute <0.5% of total mass, indicative of the low level of contamination from nonchloroplast locations.
Table 4.
Small Set of Arabidopsis Proteins from the 2-D Electrophoresis Gel Shown in Figure 1 That Are Highly Abundant in Plant Cells and Are Not Localized in the Chloroplast
| Spot No. | Molecular Mass (kD) |
pI | Identitya | Accession No.b | MALDI Percent Coverage at 50 ppmc |
MS/MS Sequenced |
Localization |
|---|---|---|---|---|---|---|---|
| 66 | 16.1 | 4.7 | GCSH protein | 121075 (At2g35370) | 17 | FFMISYLDDEILIVR (2) | Mitochondria |
| 97 | 40.1 | 6.7 | Formate dehydrogenase | 6625953 (At5g14780) | 31 | Mitochondria | |
| 98 | 59.4 | 4.7 | Similar to PDI | 5263328 (At1g21750) | 42 | TNVEVDQIESWVK (3) | Endoplasmic reticulum |
| 94 | 36.7 | 5.8 | Annexin | 4959106 (At1g35720) | LLVSLVTSYR (2) | Cytosol | |
| 93 | 35.6 | 6.5 | Annexin | 4959108 (At5g65020) | LLLPLVSTFR (2) | Cytosol | |
| 76 | 22.0 | 6.8 | GST | 3201613 (At2g30860) | 47 | Cytosol | |
| 74 | 18.4 | 4.7 | TCTP homolog | 11994618 (At3g16640) | VVDIVDTFR (1) | Cytosol/nucleus |
The cellular location for each protein is listed. Proteins were identified by MALDI-TOF MS and/or nano-ESI/MS/MS.
Identity given in NCBI completed by domain prediction found by Pfam, Blocks, Prints, Prodom, or Phi-Blast.
Accession numbers in NCBI and in MIPS (in parentheses).
Percentage of coverage at 50 ppm for the MALDI-TOF peptides.
Sequence tags identified by ESI/MS/MS and the number of sequences identified (in parentheses).
Correction of Gene Annotations by MS Analysis and Comparison with ESTs
We systematically analyzed the proteins listed in Table 1 and several others for quality of gene annotation in the Arabidopsis database. Significant errors were detected in 30% of the sequences analyzed, varying from incorrect N and C termini prediction to errors in intron/exon boundary prediction and missing exons; the accession numbers for those genes are listed in italics in Table 1. These errors became evident because a number of sequence tags obtained by MS/MS did not match or only partially matched the predicted protein sequences and by cross-correlating theoretical mass and pI values with the experimental values determined from the 2-D gels (see below). By systematic matching of ESTs against the genomic sequences, in many cases we arrived at the correct protein sequence (see also Mann and Pandey, 2001). We have selected several examples to demonstrate some of these annotation problems and reannotation strategies and to discuss implications for identification and localization (Figures 5A to 5D).
Figure 5.
Correction of Gene Annotation and Other Sequence Analysis of Arabidopsis Proteins Identified on the 2-D Gels in Figure 1.
(A) Spot 71, identified as a 17.2-kD protein at pI 6.0 on the 2-D gel, was not annotated in the Arabidopsis genome, and no overlapping EST could be found. However, a homolog could be reconstructed in soybean using three overlapping ESTs. Using the reconstructed cDNA from soybean, the corresponding gene could be identified in the Arabidopsis genome on chromosome II. Three internal sequences determined by nano-ESI/MS/MS matched sequences in the gene and are indicated by boxes. Analysis with the functional domain predictor Pfam indicated that the protein in spot 71 belongs to the OEC23 family. The predicted lTP or cTP cleavage sites are indicated with arrows. Amino acid residues conserved among all three sequences are shown in red, and those conserved between two sequences are shown in blue. RR and KR motifs in the lTP are boxed.
(B) Spot 83 is a fibrillin (on chromosome II) and is an example of a serious misassignment in intron/exon boundaries. The misassignments were corrected by matching of the genomic sequence with a homologous fibrillin (verified entirely by overlapping ESTs) on chromosome III in Arabidopsis (T10K17.220) and verified by matching protein sequences obtained by nano-ESI/MS/MS; these sequence tags are boxed. To arrive at the correct amino acid sequence, amino acids shown in red need to be removed from the annotated sequence, and amino acids shown in blue need to be included. Fifty percent of the protein sequence was changed compared with the predicted sequence.
(C) The annotated genome sequence of a FKBP (accession number 22989010) identified in spot 110 has an incorrectly predicted N terminus (MLLVL…). Orthologs in tomato (1) (AW041520), barley (2) (11193249), alfalfa (3) (11902372), and C. reinhardtii (4) (AV624465) all show typical bipartite presequences with typical lumenal TAT motifs. The N terminus of the protein in spot 110 could be extended with one overlapping EST (T76027); however, no EST was found for the very N-terminal end. Amino acid residues conserved among all four sequences are shown in red, and those conserved between fewer than four sequences are shown in blue. The cleavage site for the lTP is indicated with an arrow.
(D) Alignment of the Arabidopsis protein sequence of spot 104 with its homologs in seven other plant species. Spot 104 has a typical TAT motif, and in vitro analysis has shown that this protein translocated via the TAT pathway (Mant et al., 1999). Interestingly, the first arginine residue of the twin arginines is replaced by a lysine residue in four of the orthologs. Orthologs are from potato (1) (10447846), tomato (2) (5900060), soybean (3) (7795408), alfalfa (4) (11900582), Mesembryanthemum crystallinum (5) (8330419), barley (6) (11198348), and Physcomitrella patens (7) (6102372). Amino acid residues conserved among all seven sequences are shown in red, and those conserved between fewer than seven sequences are shown in blue. The cleavage site for the lTP is indicated with an arrow.
The first example is a protein (spot 71, an OEC23 paralog) that was not annotated at all (Figure 5A). No ESTs for Arabidopsis were found either. We did not identify the protein by MALDI-TOF MS. However, we did identify two nucleotide stretches that matched the same region of chromosome II of the Arabidopsis genome by BLAST searching using two sequence tags determined by nano-ESI/MS/MS. The sequence tags also matched ESTs from soybean, and a protein of 214 amino acid residues could be reconstructed from overlapping EST sequences in soybean and potato. Alignment of these reconstructed genes with the nonannotated Arabidopsis genome sequence helped to identify the gene on chromosome II, and one additional sequence tag could be matched (Figure 5A). Directly N terminal of each sequence tag is a lysine or arginine, which confirms the reconstructed protein, because the protein was digested originally with trypsin, which cleaves C terminal of lysine or arginine. The Arabidopsis protein is predicted to contain a typical N-terminal chloroplast transit peptide and also has a putative lTP.
A second example is a peripheral fibrillin (spot 83) for which ∼50% of the predicted protein was misassigned (Figure 5B). No ESTs are available for this protein either, but several ESTs were found for a closely related paralog. Comparison with this homologous fibrillin in Arabidopsis indicated that 240 bp of the sequence annotated as encoding sequence was likely to consist of introns and should be removed, whereas 309 bp was predicted to consist of introns but in fact consisted of exons. After correction of the fibrillin protein sequence, three sequence tags obtained by MS/MS matched to the sequence (Figure 5B).
The annotated genome sequence of a FK506 binding protein (FKBP) identified in spot 110 has a truncated N terminus (probably 60 to 70 amino acid residues are missing) starting with MLLVL. Orthologs in tomato, barley, alfalfa, and the green alga Chlamydomonas reinhardtii all show bipartite presequences with typical lumenal twin-arginine motifs. The N terminus of the protein in spot 110 could be extended with 37 amino acid residues using one overlapping EST; however, no EST was found for the very N-terminal end. Several other proteins were found in which the N terminus was assigned incorrectly, leading to an incorrect localization prediction (see also Peltier et al., 2001).
More than 50% of the identified lumenal proteins have a typical twin-arginine motif, which is indicative of targeting through the TAT pathway. In many cases, all orthologs in other plant species also contain this TAT motif, indicating that the targeting route is conserved generally from algae to higher plants (although perhaps less conserved from cyanobacteria to plants [e.g., Hcf136] [Hynds et al., 2000]). However, spot 104 (Table 1) is a protein that has been shown experimentally to translocate via the TAT pathway (Mant et al., 1999). Database searching identified orthologs in seven other plant species. Interestingly, in five of these orthologs, the twin arginines were altered to KR, although the other features of a TAT motif were preserved. This finding suggests that in some cases TAT substrates also contain KR. Interestingly, very recently, a similar observation was made for an Escherichia coli protein (Hinsley et al., 2001). Further experimentation is needed to confirm that the orthologs with KR in fact translocate via the TAT pathway.
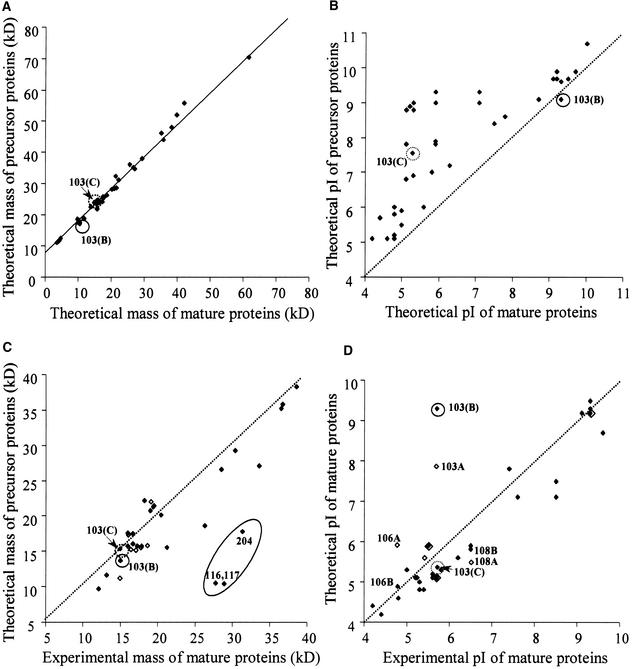
Correlation between Experimental and Theoretical Molecular Mass and pI Values, Interspecies Comparison, and Expression Levels
When searching the database with MS data, it is possible to use the pI and molecular mass values from the 2-D gels to reduce the number of proteins searched in the database. However, all lumenal localized proteins encoded by nuclear genes are synthesized with a bipartite presequence, and these presequences must be taken into account. To evaluate the contribution of the presequences of the lumenal proteins on pI and molecular mass, we cross-correlated the theoretical molecular mass and pI values of the precursors to those of the corresponding mature lumenal proteins (Figures 6A and 6B). The cross-correlation plot for the molecular mass shows a straight line parallel to a perfect correlation, with a shift of ∼9 kD, and thus an average length of the cTP + lTP of ∼81 amino acid residues (Figure 6A). This also indicates that the length of the presequence is independent of the size of the mature protein, which is very important for genome-wide prediction of the lumenal proteome (see below). Cross-correlation between the theoretical pI of the precursor versus the mature protein shows that the precursors are far more basic than are the mature proteins. Processing of the precursor proteins consistently gave a pI shift downward of up to 4 pI units, with generally larger shifts for the smaller proteins.
Figure 6.
Cross-Correlation between Experimental and Theoretical Molecular Masses and pI Values of Precursors and Mature Proteins in Arabidopsis.
Cross-correlation of predicted and experimental molecular mass ([A] and [C]) and pI values ([B] and [D]) of the proteins from Arabidopsis identified in Figure 1 before ([A] and [B]) and after ([C] and [D]) removal of the cTP and lTP. Dotted lines indicate perfect correlations. Protein spot numbers are indicated for strongly deviated points. The circled data points correspond to spot 103 (see Figure 7). Open symbols represent values based on incorrect annotation (e.g., 103A, 106A, and 108A) ([C] and [D]). Three (most likely) monodimers (spots 116, 117, and 204) are indicated in (C).
We also cross-correlated the theoretical sequences (from the original annotation and reannotation) and experimental values for pI and molecular mass of the lumenal proteins (Figures 6C and 6D). We, and many others for nonplant species, have observed that experimental pI and molecular mass generally correspond to the theoretical values. However, if there is a significant mistake in gene assignment, or if there is a large post-translational modification or processing event, these data points will lie outside of the correlation and thus will highlight such modifications or annotation errors. Generally, the correlation between predicted and experimental mass on the Arabidopsis maps were within a range of ±5 kD (Figure 6C). However, three proteins (two PC spots and 204) were completely outside of this range and could correspond to the presence of homodimers. This dimerization could have occurred during the isoelectric focusing when the proteins reached their pI. The dimers apparently could not be solubilized before separation in the second dimension. Most of the pI values observed on the 2-D maps agreed with the corresponding theoretical pI values within a range of ±0.5 pH unit. Four protein spots were in the range ±0.5 to 1 pH unit. This larger difference is caused by post-translational modifications, visible on the gel as trains of spots (e.g., OEC23 spots 11 to 14), and might be caused by carbamylation. There are a number of post-translational modifications described for stromal and thylakoid proteins (glycosylation and palmitoylation; see Peltier et al., 2000); however, it is unknown whether such modifications occur on the lumenal proteins.
One spot (spot 103A) showed a very large difference (>3 pH units) between predicted and experimental pI value (Figure 6D). This immediately suggested a possible problem of gene annotation. Therefore, the annotated genomic sequence was verified by overlapping ESTs. A reannotated gene was constructed, and the predicted protein sequence is shown (Figure 7, sequence B). Complete coverage of the reannotated gene was obtained and showed that exon IV had not been recognized as an exon and that exon V was predicted too short and was frameshifted. The theoretical pI value of sequence B still did not match the experimental value (Figure 6D). Interestingly, one additional EST covering the C-terminal end was found and a second protein sequence could be predicted (Figure 7, sequence C). In this case, exon VI was shorter than in sequence B, and two additional exons (exons VII and VIII) were added, as in the original annotation. The theoretical pI value of the processed protein now matched the experimental pI value (Figure 6D), and 10 matching peptide masses from MALDI-TOF MS were identified. Thus, it is highly likely that the correct protein sequence of spot 103 is sequence C. This is a clear case of alternative splicing, which occurs (in the case of sequence C) in the middle of exon VI. In other cases, reannotation affected the theoretical pI only moderately (spots 106 and 108) or hardly at all (Figure 6D).
Figure 7.
Alternative Splicing of Lumenal Protein Spot 103.
The gene annotation for protein spot 103 is incorrect in the database. The top of the figure shows a scheme of the genomic sequence and the positions of the exons. Based on overlapping ESTs, it is clear that the gene annotation in the database (sequence A) is incorrect. Exon IV was not recognized as an exon, and exon V is too short and was frameshifted (indicated in black). One full-length cDNA and corresponding proteins (sequence B) can be reconstructed from the overlapping ESTs. Sequence B is constructed from six exons (I, II, III, IV, V, V′, and VI) and encodes a protein with pI value of 9.35 for the precursor and 9.14 for the mature protein. Sequence C is constructed from exons I to V′ plus exons VI, VII, and VIII and encodes a protein with a pI value of 7.55 for the precursor and 5.30 for the mature protein. The alternative splice site (in sequence C) occurs in the middle of exon VI. Two ESTs were used to reconstruct the protein in sequence C. The pI value and molecular mass of the processed protein match exactly the experimental coordinates on the 2-D gel (Figure 1A). Ten peptide masses determined by MALDI-TOF MS match this protein (mass accuracy within 50 ppm). Red indicates conservation, and blue or green indicates no conservation of the sequence between the different annotations A, B, and C. Matching Arabidopsis ESTs are indicated. The lumenal processing site is indicated by the arrows.
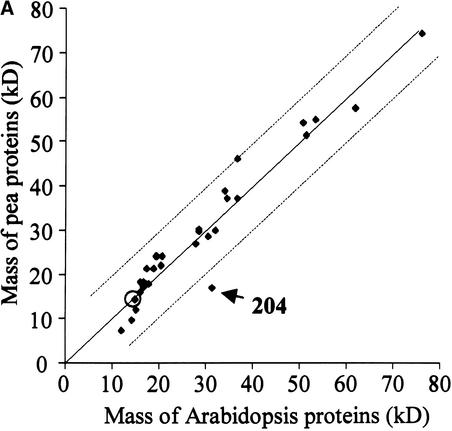
2-D maps of different plant species have been, and will be, generated for different subproteomes. To obtain a better insight into the interspecies correlation of 2-D gel patterns, we compared the maps generated for Arabidopsis with those generated earlier for pea (see Figure 2 in Peltier et al., 2000). Superficially, the pea and Arabidopsis 2-D maps show similar overall patterns, both in terms of expression levels and as coordinates for the different proteins, although the maps cannot be overlaid. To express this more quantitatively, we cross-correlated the molecular mass and pI values of the two species (Figures 8A and 8B). Nearly all pairs of pea/Arabidopsis proteins matched within a range of ±10 kD and ±0.75 pH, with the few exceptions indicated in Figures 8A and 8B. Interestingly, a nearly perfect correlation was found for spot 103, indicating that the acidic form was expressed preferentially in both organisms (Figure 7). All lumenal proteins identified on the pea maps also were identified on the Arabidopsis maps, with the exception of two spots, indicating that the lumenal proteomes of the two species are rather similar and showing the reproducibility of our preparations and 2-D gels. Naturally, more proteins were identified in the current study as a result of the completion of Arabidopsis genome sequencing and because the identification of pea proteins necessarily was based mostly on homology.
Figure 8.
Cross-Correlation of the Experimental Molecular Mass (A) and pI Values (B) of the Lumenal Proteins from Pea and Arabidopsis Identified on the 2-D Maps.
The broken lines indicate a deviation of 10 kD or 0.75 pI units. Protein spot numbers are indicated for strongly deviated points. The circled data points correspond to spot 103 (see Figure 7).
Sequence Analysis of cTP and lTP of the Experimentally Identified Proteins and Their Orthologs
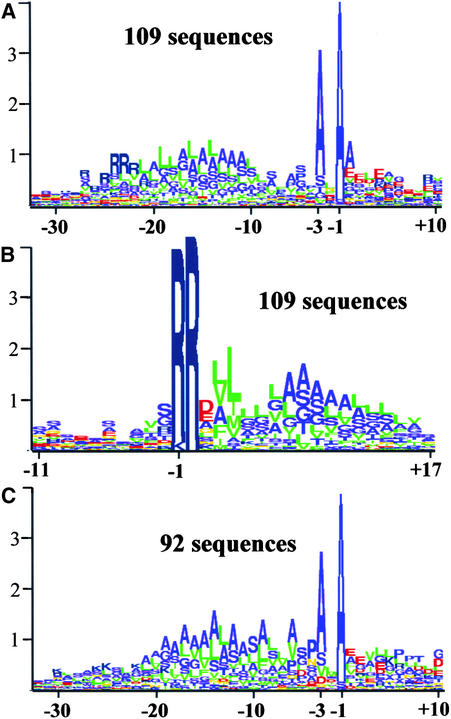
To obtain cutoff parameters and selection criteria for a genome-wide prediction of lumenal proteins (presented below), we analyzed and assembled information concerning the lTPs of all experimentally identified and N-terminally sequenced lumenal proteins, both from our own work and from the literature. The N termini for 41 nonredundant lumenal proteins (including membrane proteins with lTPs, such as PsbY1,2, PsbW, and PsbX) were identified experimentally in Arabidopsis or other plant species. To obtain a more statistically significant set, closely related orthologs of these nonredundant lumenal proteins were identified by BLAST searching of all plant and algal (C. reinhardtii) protein and DNA sequence entries. Thus, a total of 201 lumenal proteins were identified, and 109 of these have a typical TAT motif. Approximately 10% were orthologs in green algae. The cTPs of the algae are not well predicted by TargetP (see Discussion), but their lTPs were very similar to those of higher plants. It is important to note that lTPs show much less sequence conservation than does the mature protein sequence, between both paralogs and orthologs, thus reducing the redundancy in this lTP set. The lTPs then were analyzed using logoplots of the sequences aligned according to the experimentally identified N termini (Figures 9A and 9C) or to the TAT motif (Figure 9B). In the logoplots, the sequence alignment is represented by a sequence of stacked letters in which the total height of the stack at each position shows the amount of conserved information, whereas the relative height of each letter shows the relative abundance of the corresponding amino acid (Schneider and Stephens, 1990).
Figure 9.
Sequence Analysis of cTP and lTP of the Experimentally Identified Proteins in Arabidopsis and Their Homologs/Orthologs in Other Plant Species.
(A) and (B) Logoplot of 109 lumenal proteins with a typical TAT motif identified experimentally in Arabidopsis and their homologs in other plant species aligned according to the experimentally identified N terminus (A) and RR motif (B).
(C) Logoplot of 92 lumenal proteins without a typical TAT motif identified experimentally in Arabidopsis and their orthologs in other plant species aligned according to the experimentally identified N terminus.
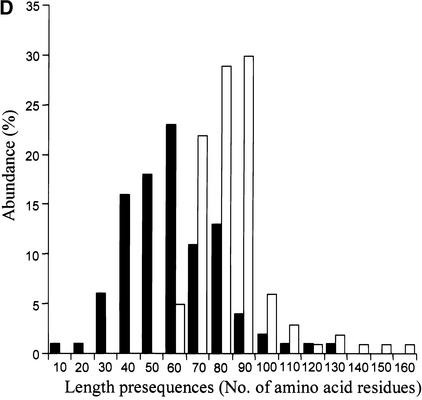
(D) Length distribution of cTP for the experimental training set of TargetP (141 proteins) (closed bars) and the length distribution of cTP + lTP (open bars) for the 174 experimentally identified lumenal proteins. Proteins are divided into classes of 10 amino acid residues.
The comparison between the logoplots in Figures 9A and 9C shows that the N-terminal charged domains contain almost exclusively arginine in the TAT substrates, whereas in the Sec substrates the charged amino acid is preferentially lysine. In the Sec substrates, the hydrophobicity of the core domain is more pronounced and the C-terminal polar region is shorter, with a preference for a proline at position −4. No significant differences were found for positions −1 and −3, and the consensus sequence for the lTP cleavage site remains AxA for both substrates.
The logoplot in Figure 9B shows the alignment of the lumenal TAT substrates according to the TAT motif and shows that the TAT motif in plants is RRxh(h/u) (where x is any residue, h is any hydrophobic residue, and u is any uncharged residue) and is more degenerated than it is in bacteria, which has a consensus motif of S/TRRxFLK (Berks et al., 2000). We did not observe any significant differences between lTPs of Arabidopsis proteins and those of other plants species or between lTPs of monocotyledonous versus dicotyledonous plants.
To define additional cutoff values for a theoretical genome-wide prediction of lumenal proteins, the length distribution of cTPs and lTPs was analyzed (Figure 9D). A total of 141 proteins were used for the length distribution of cTP and are part of the experimental training set of TargetP (Emanuelsson et al., 2000); 174 lumenal proteins, which are part of the set of 201 lumenal proteins described above (some of the 201 proteins missed the very N terminus), were used for the length distribution of the total presequence of the lumenal proteins (cTP + lTP). The two distributions are almost Gaussian, with an average size of 54 amino acids for cTP and 81 amino acids for cTP + lTP and a maximum length of 129 for cTP and 151 for cTP + lTP.
Genome-Wide Localization Predictions and Analysis
With the information summarized in Figures 9A to 9D, we sought to predict the lumenal proteome in Arabidopsis. The set of Arabidopsis sequences for the theoretical analysis was obtained from Munich Information Center for Protein Sequences (MIPS) (see Methods) and contained 25,460 entries, including functional annotations (if any). The general prediction strategy is summarized in Figure 10A, and the prediction process can be followed through Table 5. To predict the potential lumenal proteins, the protein sequences were first processed through the plant version of TargetP (Emanuelsson et al., 2000), which discriminates between proteins targeted to the chloroplast, the mitochondrion, and the secretory pathway (and as a fourth group, “other” destinations). TargetP also predicts the cleavage site of the cTP. A total of 3646 proteins were predicted to be localized to the chloroplast (using all reliability classes) and were processed further through SignalP 2.0 HMM version (SignalP-HMM; Nielsen et al., 1997, 1999), which discriminates between sequences containing a cleavable signal peptide, a noncleavable signal anchor, or neither. SignalP was designed originally to detect signal peptides for secretion in bacteria and eukaryotes, but lTPs show strong similarities to the secretory peptides (von Heijne et al., 1989). Evaluation of lTP prediction by SignalP on the set of experimentally identified lumenal proteins listed in Table 1 shows no bias in successful prediction in either Gram-positive or Gram-negative mode. The performance of SignalP in the eukaryotic mode was less successful (Table 1). Thus, we used the union of the Gram-negative and Gram-positive versions of SignalP-HMM.
Figure 10.
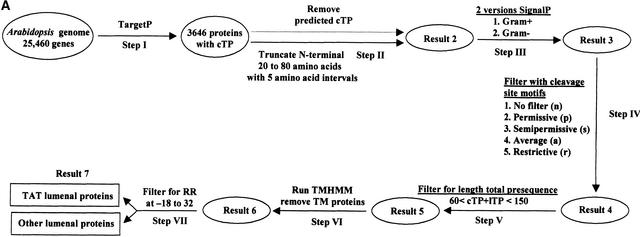
Genome-Wide Prediction of the Lumenal Proteins with a Typical TAT Motif in the Genome of Arabidopsis.
(A) Summarizing scheme of the genome-wide prediction of lumenal proteins with a TAT signal.
Step I. 25,460 Arabidopsis open reading frames (see Methods) were processed through TargetP, resulting in 3646 protein sequences predicted to have a cTP (Result 1).
Step II. For each of the 3646 proteins, 20, 25, … 80 residues were removed from the N terminus to mimic the cleavage of the cTP. (Thus, each protein was present in 13 differently truncated versions.)
Step III. The truncated proteins were processed through SignalP (both Gram-negative and Gram-positive versions) to predict the potential presence of a lTP. If any of the 13 truncated versions of a protein were predicted to contain a lTP by at least one of the two SignalP versions, the protein was kept in Result 3 (1224 proteins).
Step IV. The proteins predicted to have a lTP were checked for the presence of the four versions of the −3,−1 motif (p, s, a, and r; see Results) at the lTP cleavage site.
Step V. Length restrictions were imposed. Proteins that did not meet the length criteria were excluded (Result 5; 596 proteins using the p motif).
Step VI. The remaining proteins were processed through the TM region predictor TMHMM. All proteins that contained one or more TM regions in the predicted mature part (i.e., C terminal of the predicted lTP cleavage site) were removed.
Step VII. Proteins with a TAT pathway motif (twin arginine; RR) in region −32 to −18 relative to the predicted lTP cleavage site were sorted to the TAT lumenal protein set (93 proteins using lTP cleavage site motif p). The rest of the proteins were kept in the “other lumenal proteins” set (380 proteins using the p motif).
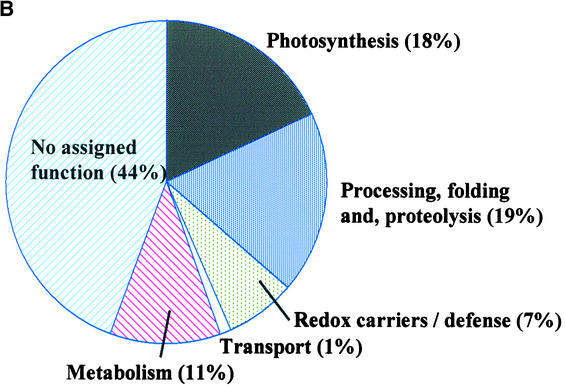
(B) Functional catalog of the 71 predicted lumenal proteins with a TAT motif based on known function or functional domain prediction.
Table 5.
Summary of the Different Result Steps during the Genome-Wide Screening for Lumenal Proteins, following the Strategy Shown in Figure 10A
| Result Steps (see Figure 10A) |
No. of lTPs | False Positive Rate on the Set of 211 Known Stromal Proteins |
True Positive Rate on the Set of Experimentally Identified Lumenal Proteins (Sensitivity) |
|---|---|---|---|
| Result 3 | 1224 | 0.38 | 0.94 |
| Result 4 | |||
| n | 1224 | 0.38 | 0.94 |
| p | 943 | 0.34 | 0.94 |
| s | 630 | 0.29 | 0.88 |
| a | 525 | 0.27 | 0.76 |
| r | 230 | 0.17 | 0.59 |
| Result 5 | |||
| p | 596 | 0.29 | 0.85 |
| s | 411 | 0.18 | 0.82 |
| a | 343 | 0.17 | 0.74 |
| r | 161 | 0.12 | 0.56 |
| Result 7, TAT | |||
| p | 93 | 0.06 | 0.83 |
| s | 62 | 0.05 | 0.83 |
| a | 52 | 0.05 | 0.67 |
| r | 27 | 0.02 | 0.39 |
| Result 7, other | |||
| p | 380 | 0.14 | 0.60 |
| s | 267 | 0.12 | 0.53 |
| a | 221 | 0.11 | 0.47 |
| r | 105 | 0.09 | 0.40 |
To mimic the actual sorting process, we removed the predicted cTP before submitting the sequences to the SignalP-HMM predictor (Figure 10A). Our first idea was to remove the stretch of residues corresponding to the TargetP-predicted cTP cleavage site, but that was abandoned because the TargetP prediction of cTP cleavage sites is quite inaccurate (∼45% correct within two amino acid residues), in contrast to the more reliable chloroplast localization prediction (85% success rate). Instead, we processed the 3646 predicted chloroplast-targeted sequences by removing from 20 to 80 amino acids from their N termini in steps of five residues (90% of the cTPs in the TargetP training set fall into this length interval; Figure 9D) and submitted these sequences to SignalP for lTP prediction. Thus, for each predicted chloroplast protein, we obtained 26 SignalP predictions (in total, 13 processed versions of the sequences [removing 20, 25, 30, … 80 residues]) using both Gram-positive and Gram-negative versions of SignalP-HMM (Figure 10A). The prediction results are based on the union of these predictions and resulted in 1224 proteins with potential lTPs (Table 5, result 3). The sensitivity of the prediction at this stage was 94% (thus, there were 6% false negatives for the known lumenal proteins), but, as expected, there was a strong overprediction on the set of 211 stromal proteins, resulting in a false positive rate of 38%.
Constraints then were added in the form of a demand for the −3,−1 motif to be present at the cleavage site, as shown in the logoplots of Figures 9A and 9C. The residues in the −3,−1 motif are preferentially alanines, but there also can be other small and apolar residues at the −3 position and (with very low frequency) at the −1 position (Figures 9A and 9C). The motif was applied in four versions of various stringency: (1) the restrictive (r) motif A-x-A (present at 75% of lTP cleavage sites in the set of 201 known lumenal proteins); (2) the average (a) motif [ASV]-x-A (present in 87%); (3) the semipermissive (s) motif [ADGLSTV]-x-A (present in 95%); and (4) the permissive (p) motif [ADGLSTV]-x-[AGS] (present in 97%). The p motif included all amino acids that were present more than twice at positions −1 and −3 in the set of 201 lumenal proteins. For each motif, a list of cTP + lTP–containing proteins was generated, and these are summarized in Table 5. This showed that the sensitivity (tested on the known lumenal proteins) decreased from 0.94 to 0.59 with increasing stringency, whereas the false positive rate decreased from 0.38 to 0.17 (tested on the set of known stromal proteins).
Additionally, to be considered a potential lumenal protein, the total length of the predicted cTP + lTP was required to be between 60 and 150 residues (corresponding to experimentally verified lengths in the set of 174 complete lumenal proteins [Figure 9D]), and the processed mature part of the protein was required to be at least 50 amino acid residues. Using the p motif, 774 proteins were left in result 5, whereas applying the r motif reduced the set to 190 proteins (Figures 6A and 10A). At this stage, the sensitivity was 0.85 and 0.56 and the false positive rate was 0.21 and 0.1 for the p and r motifs, respectively.
Next, we verified the sequences for potential TM regions using the predictor program TMHMM (version 2.0) (Krogh et al., 2001). All sequences that were predicted to contain a TM region either overlapping with the lTP cleavage site or downstream of it were discarded, because our aim was to find soluble lumenal proteins. TM domains inside of the lTP regions were not considered because such proteins should have been predicted as signal anchors by SignalP-HMM. Some of the proteins that were discarded at this step are membrane proteins with an lTP, such as PsbW, PsbX, and PsbY1-Y2.
Among the remaining sequences, potential lumenal proteins with the TAT motif R-R-xh(h/u) were collected (Figure 9B). The twin arginines in the set of 201 known lumenal proteins are located 18 to 32 amino acid residues upstream of the lTP cleavage site. Consequently, we used the presence of two adjacent arginines, present within −18 to −32 relative to the lTP cleavage site, as a criterion. They also were required to be at least 35 residues downstream of the N terminus of the precursor protein. This resulted in sets of 27, 52, 62, and 93 TAT proteins, depending on the −3,−1 motif requirement. For the p motif, only 6% of the 211 known stromal proteins were falsely predicted as lumenal with a TAT motif, and the sensitivity was 83% (Table 5). In the set of 93 proteins (p motif), only 30% were annotated functionally in MIPS. Therefore, all proteins were passed manually through all major functional domain predictors, increasing (often very limited) annotation to 56% of the proteins. Then, 22 proteins (24%) with known or predicted functions obviously incompatible with lumenal localization were excluded; the resulting list of 71 proteins is shown in Table 6. These 22 false positives included mostly proteins involved in transcription and translation and some potential nonchloroplast proteins. Forty-four percent of the remaining 71 proteins lack functional annotation (Figure 10B), and determining their function might help to fully understand the functions of the lumenal proteome.
Table 6.
Seventy-One Predicted Lumenal Proteins with a TAT Motif, According to the Scheme Shown in Figure 10A, Using a Permissive Cleavage Site of the lTPa
| Name | Length (eTP lTP) | Position of RR from N Terminus | lTP Cleavage Site Motif (s/p) | Functional Annotation (According to MIPS) | Additional Functional Annotation (This Article) and Match to 2-D Gels |
|---|---|---|---|---|---|
| At1g03600 | 68 | 41 | a | Unknown protein | Spot 207, putative new photosystem II protein |
| At1g05420 | 86 | 55 | a | Hypothetical protein | |
| At1g06430 | 73 | 48 | r | Cell division protease FtsH, putative | |
| At1g06680 | 77 | 55 | r | 23-kD polypeptide of OEC | |
| At1g14150 | 65 | 43 | r | Unknown protein | Homology with OEC16 |
| At1g15510 | 64 | 36 | p | Hypothetical protein | Pentatricopeptide repeat and prokaryotic membrane lipoprotein attachment |
| At1g17210 | 64 | 36 | a | Hypothetical protein | |
| At1g18170 | 94 | 74 | p | Hypothetical protein | Putative FKBP isomerase |
| At1g21500 | 59 | 41 | r | Unknown protein | |
| At1g26360 | 62 | 34 | p | Hypothetical protein | Epoxide hydrolase signature |
| At1g35210 | 90 | 62 | p | Hypothetical protein | |
| At1g49630 | 90 | 61 | p | Hydrogenase protein, putative | |
| At1g55580 | 60 | 42 | s | Hypothetical protein | |
| At1g70350 | 81 | 54 | r | Hypothetical protein | |
| At1g71200 | 93 | 72 | p | Hypothetical protein | Related to myc protein |
| At1g73530 | 73 | 46 | p | Hypothetical protein | |
| At1g74070 | 74 | 45 | a | Hypothetical protein | Putative cyclophilin |
| At1g76450 | 80 | 57 | r | Unknown protein | Spot 70 |
| At1g77090 | 63 | 47 | s | Unknown | Spot 108, OEC23 related |
| At2g01400 | 85 | 63 | Hypothetical protein | Related to Lon protease Brevibacillus 402504 | |
| At2g15570 | 68 | 45 | a | Putative thioredoxin m | |
| At2g20270 | 62 | 39 | a | Putative glutaredoxin | |
| At2g23670 | 71 | 49 | a | Hypothetical protein | |
| At2g26340 | 80 | 59 | r | Unknown protein | |
| At2g30790 | 76 | 56 | a | Putative photosystem II OEC23 protein | See Results and Discussion |
| At2g34860 | 93 | 64 | r | Unknown protein | Related to chaperone HSP40/DnaJ |
| At2g37240 | 68 | 45 | s | Unknown protein | Related to protein 13834657 Mus |
| At2g37660 | 86 | 62 | p | Unknown protein | Related to 3β HSD isomerase |
| At2g39080 | 72 | 40 | p | Unknown protein | |
| At2g39470 | 73 | 51 | s | Unknown protein | Spot 212, OEC23 related |
| At2g40400 | 93 | 67 | r | Unknown protein | Related to protein 7572912 |
| At2g47390 | 98 | 51 | s | Unknown protein | |
| At3g01440 | 74 | 50 | a | Hypothetical protein | Related to OEC16 |
| At3g03760 | 77 | 59 | p | Unknown protein | Related to 13569546 |
| At3g05020 | 66 | 42 | a | Acyl carrier protein 1 precursor | |
| At3g10060 | 82 | 60 | r | Unknown protein | Spot 210, putative isomerase |
| At3g10130 | 83 | 49 | s | Unknown protein | Related to 13877685 and 13424114 from Caulobacter |
| At3g11630 | 69 | 51 | a | Putative 2-Cys peroxiredoxin | Spot 74 (Table 2) |
| At3g16000 | 95 | 73 | r | Myosin heavy chain–like protein | |
| At3g52960 | 71 | 52 | a | Peroxiredoxin-like protein | Spot 68 (Table 2) |
| At3g55330 | 74 | 50 | a | Putative protein | Spot 213, OEC23 related |
| At3g56140 | 90 | 65 | a | Putative protein | Related to 4586056 |
| At3g57680 | 99 | 74 | r | C-terminal protease-like protein | |
| At4g05180 | 82 | 60 | a | Oxygen-evolving enhancer protein 3 precursor-like protein (OEC16) |
Spot 208 |
| At4g09010 | 82 | 53 | r | Putative protein | Spot 205 to 206, putative ascorbate peroxidase |
| At4g15120 | 95 | 58 | p | Hypothetical protein | Related to 11994735 |
| At4g15510 | 104 | 79 | r | Hypothetical protein | Spot 19, OEC23 related |
| At4g19830 | 78 | 52 | r | Putative protein | Putative FKBP isomerase |
| At4g21280 | 75 | 53 | a | Photosystem II OEC protein 3-like (OEC16) | Spot 203 |
| At4g25130 | 71 | 44 | r | Protein-methionine-S-oxide reductase | |
| At4g26500 | 75 | 48 | p | Putative protein | protein Duf and BolA family |
| At4g29590 | 85 | 54 | r | Putative protein | Related to methyltransferases |
| At4g31390 | 61 | 30 | a | Predicted protein | Related to ABC transporter |
| At4g31560 | 75 | 45 | s | Putative protein | |
| At4g32020 | 71 | 42 | a | Putative protein | Related to 13272401 |
| At4g34020 | 80 | 52 | p | Putative protein | ThiJ family |
| At4g34120 | 72 | 42 | a | Putative protein | CBS domain (protein interaction) |
| At4g36530 | 63 | 41 | a | Putative protein | Epoxide hydrolase signature |
| At5g04900 | 63 | 36 | a | Putative protein | |
| At5g11450 | 95 | 70 | s | Putative protein | Spot 22, pea map OEC23 related |
| At5g11550 | 60 | 42 | p | Putative protein | |
| At5g13410 | 86 | 61 | s | Putative protein | Spot 80, putative FKBP isomerase |
| At5g17710 | 67 | 43 | p | Chloroplast GrpE protein | Spot 89 |
| At5g23120 | 78 | 55 | r | Photosystem II stability/assembly factor Hcf136 | Spot 123 |
| At5g27860 | 71 | 52 | p | Putative protein | |
| At5g39830 | 92 | 67 | p | DegP protease-like protein | DegP8 |
| At5g45680 | 79 | 58 | r | Putative protein | Putative FKBP isomerase |
| At5g50110 | 66 | 40 | p | Putative protein | |
| At5g55570 | 68 | 37 | s | Unknown protein | |
| At5g62840 | 63 | 37 | r | Putative protein | |
| At5g64040 | 86 | 63 | s | Photosystem I reaction center subunit psaN precursor (PSI-N) (sp P49107) |
Spot 201 |
aFrom the 93 proteins originally predicted, 22 (24%) were removed manually because they were likely to be false positives. Matches to experimentally identified protein spots are noted.
The prediction for the set of potential lumenal proteins without a TAT motif was not as good as that for the TAT substrates, and the false positive rate was between 0.14 (p) and 0.09 (r), with sensitivities of 0.60 (p) and 0.40 (r). Therefore, these results are only summarized in Table 5.
DISCUSSION
The thylakoid lumen in chloroplasts is still a poorly characterized compartment, and only a few dozen proteins have been identified. A number of functions of the lumenal proteome are well known, such as water splitting by the OEC (involving OEC16, OEC23, and OEC33) of photosystem II, electron transport (e.g., involving PC), and regulation of the xanthophyll cycle (violaxanthin deepoxidase). A number of additional lumenal proteins have been identified with known functional domains, such as an isomerase (TLP40), a carbonic anhydrase, a photosystem II “assembly factor” (Hcf136), a protease (DegP1), and the thylakoid-processing peptidase and the D1-processing peptidase (CtpA). In addition, recent systematic searches identified a number of lumenal proteins of unknown function (Kieselbach et al., 1998; Peltier et al., 2000).
In this study, we sought to characterize the thylakoid lumen proteome further by a unique combination of experimentation and localization prediction. We also determined the relative expression levels, both to estimate the dynamic resolution of our experimental setup and to obtain insight into stoichiometries between different proteins and their paralogs.
Copurification of Peripheral Proteins
In addition to the lumenal proteins, we identified many proteins that are attached to the stromal side of the membrane as part of their function. It was natural that these peripheral proteins copurify with the lumenal proteins because they are released easily by either Yeda press or the sonication used to open the lumenal compartment. It is important to emphasize that few proteins were identified on the gels that can be considered true contaminants, such as the abundant proteins from the stroma or from outside of the chloroplasts. This is the result of multiple wash steps with buffers of low ionic strength and purification of the membranes on Percoll gradients. We did attempt to reduce the copurification of proteins located at the stromal side of the membrane by using different sonication times and energies. Overall, Yeda press treatment of the lumenal proteome gave the “purest” lumenal proteome (as judged by the low amounts of released CF1α,β [∼5 to 10% of the total]) while still obtaining good efficiency of extraction, as judged by the protein yield per milligram of chlorophyll and the amount of extracted membrane-bound OEC33. It is important to remember that the lumenal proteins can be soluble or bound peripherally (e.g., OEC33) to the lumenal side of the thylakoid membrane or both; thus, a minimal amount of energy is needed to release these proteins. We have considered protease treatments of thylakoids before extraction of lumenal proteins (as is often used for localization of individual proteins in combination with protein gel blotting), but this is likely to produce many breakdown products that will copurify with the lumen, thereby making the maps irreproducible and the identification process very difficult.
Thus, the maps in the study represent the soluble lumenal proteins and weakly, peripherally attached proteins at either side of the thylakoid membrane. Proteins that interact more tightly with the membrane, either through TM domains or lipid anchors, are not present on the 2-D gels.
Expression of Known Lumenal Proteins
From the 2-D gels of Arabidopsis, we identified 30 lumenal proteins and an additional 12 potential lumenal proteins. The expression levels of these identified proteins varied greatly, with a molar range of 1 to 10,000 (Figure 2). However, using narrow-gradient gel multidimensional chromatography combined with on-line nano-ESI/MS/MS measurements, it is technically possible to identify these proteins of lower abundance and reach a dynamic resolution of 1:100,000 molar ratio or better.
We identified all of the very abundant known lumenal proteins involved in photosynthesis, such as OEC16, OEC23, OEC33, PC, and PsaN, with relative expression levels of 50 to 100 compared with Hcf136. We should note that the expression levels of these abundant proteins were underestimated by a factor 3 to 5 because of saturation of the stains. We also identified the abundant nonphotosynthetic lumenal proteins described in earlier studies, such as Hcf136, TLP40, and TL16.5, with relative expression levels of ∼1 to 10.
Expression of Paralogs of Photosynthetic Proteins and a New Putative Photosystem II Protein
A closely related paralog was identified for OEC33, PC, and an OEC16 at a relatively high expression level (5 to 50). The pairs of PC and the OEC16 paralogs were well conserved at the primary sequence level (with 83 and 73% identity of the mature protein, respectively) (Figures 3A and 3B); thus, it is possible that they do function in photosynthesis. However, it is unclear why both members of each pair are expressed.
In addition, we identified seven paralogs of OEC23 on the Arabidopsis maps, and an eighth was identified earlier on our pea maps (spot 22). A ninth member was identified in the database and was 88% identical as OEC23, but it is unlikely to be expressed. The group of seven expressed OEC paralogs is relatively distant from OEC23 (and its nonexpressed homolog), with 18 to 31% identity; therefore, it is quite likely that they do not function in the OEC, which leaves their function completely unknown. Expression levels were 20 to 300 times lower than that of OEC23.
The presence of the paralogs of OEC16, OEC23, OEC33, PC, and others indicates gene duplication. Attempts to date these gene duplication events suggested that some of these duplications occurred around the time of the divergence of Brassica and the Rosidae (data not shown).
A very basic (pI 9.3) lumenal protein without predicted TM domains and with homology with a cyanobacterial 11-kD protein in photosystem II was identified in both pea and Arabidopsis. We tentatively assigned it as a new photosystem II protein, but its precise localization in the lumen and its function remain to be determined.
Expression of Multiple Isomerases and Proteases
Isomerases belong to two large families of proteins: the protein disulfide isomerases (PDI) and the peptidyl-prolyl cis-trans isomerases or rotamases. In addition to thiol-disulfide exchange, PDIs catalyze the GSH-dependent reduction of dehydroascorbate, bind ions and hormones, serve as a subunit of prolyl hydroxylase, and show triglyceride transferase activity (Gilbert, 1998). The PDI-like activity is mediated either by the thioredoxin-like CXXC motif with two vicinal cysteines or by a single, highly reactive cysteine; in the latter case, GSH can interact with the mixed disulfide instead of the second cysteine and release the substrate protein (Walker and Gilbert, 1997). The peptidyl-prolyl cis-trans isomerases or rotamases, which catalyze the cis-trans isomerization of the amide bond between a proline residue and the preceding amino acid residue, are composed of at least three different groups: the cyclophilins inhibited specifically by cyclosporins, the FKBPs inhibited by FK506 and rapamycin, and the parvulins inhibited by juglone (Schiene and Fischer, 2000).
Five isomerases were identified in the lumen (Table 1), and one is listed in Table 2. They can function in folding of the thylakoid proteins or in signaling (such as TLP40) or possibly are part of the antioxidant defense system via a connection to 2-Cys peroxiredoxins (see next paragraph). Only TLP40 has been described in more detail (Fulgosi et al., 1998; Vener et al., 1999): it is characterized by a cyclophilin-like C-terminal segment of 20 kD, a predicted N-terminal leucine zipper, and a potential phosphatase binding domain (Fulgosi et al., 1998). The isolated protein possesses peptidyl-prolyl cis-trans isomerase protein folding activity, and TLP40 also exerts an effect on the dephosphorylation of several key proteins of photosystem II. The genome-wide prediction of lumenal proteins identified four additional chloroplast isomerases with potential lTP with a TAT motif. One additional isomerase with a TAT motif and an isomerase targeted via the Sec (or other) pathway were found with Swiss-Prot, increasing the set of potential lumenal isomerases to 12. The relationships in this isomerase family are shown in Figure 4 in the form of a rooted tree with three main clades. None of these isomerases were identical to other known chloroplast isomerases, such FKBP13 (Luan et al., 1994a), ROC4 in the stroma (Lippuner et al., 1994), cyclophilin pCyP B (Luan et al., 1994b), RB60, a protein disulfide isomerase involved in translation regulation (Kim and Mayfield, 1997; Trebitsh et al., 2000), or any of the other known cytosolic isomerases (ROC1, -2, -3, -5, or -6) (Chou and Gasser, 1997). On the basis of the presence of a closely spaced cysteine pair, it is possible that spots 110 and 211 and At5g45680 (indicated in Figure 4) have a specific function as an oxidative folding catalyst. These three isomerases also are clustered. Five of the isomerases have no cysteines at all and likely function as prolyl cis-trans isomerases only, whereas the four remaining isomerases have one or two cysteines and possibly play a role in oxidation/reduction reactions.
Antioxidant Metabolism in the Thylakoid: A Network of m-Type Thioredoxins, Peroxiredoxins, and Ascorbate Peroxidase
Light-driven charge separations in the photosystems and side reactions in photosynthetic electron transport such as the Mehler reaction, as well as photorespiration in the peroxisomes, produce reactive oxygen species that need to be quenched to prevent extensive damage to proteins as well as membrane lipids. Different carotenoids present in the photosystems and their antennae, as well as a network of superoxide dismutases and pools of vitamin E, ascorbate, and GSH, form a strong antioxidant network in and around the thylakoid membrane (Noctor et al., 2000). However, it is not clear how much this network extends into the thylakoid lumen.
The lumenal protein violaxanthin deepoxidase is involved in the xanthophyll cycle and uses ascorbate as a cofactor, implying directly that ascorbate must be present in the lumen. However, no evidence for ascorbate in the lumen has been shown, which might relate to experimental difficulties of its identification rather than to its absence from the lumen. It is not clear how ascorbate is transported into the lumen, and neither is it clear if oxidized ascorbate can be rereduced in the lumen (either spontaneously or by enzymatic reaction) or if it is exported in its oxidized form (as monodehydroascorbate or dehydroascorbate) back into the stroma, where it is reduced by monodehydroascorbate reductase or dehydroascorbate reductase.
On the 2-D map, we identified a putative ascorbate peroxidase with all of the characteristics of a lumenal protein and distinct from the known ascorbate peroxidases soluble in the stroma and one bound to the thylakoid membrane (Jespersen et al., 1997; Yoshimura et al., 1999) and identified earlier on our 2-D gels from pea. Its precursor has a typical lTP with a TAT motif, and the experimentally determined N terminus corresponded with the prediction by SignalP, suggesting that this ascorbate peroxidase is in fact located in the thylakoid lumen.
A number of additional members of the antioxidative defense system were identified on the 2-D maps. They are a known thylakoid-located 2 Cys peroxiredoxin (2-CP) (Baier et al., 2000), an uncharacterized peroxiredoxin-like protein, and three m-type thioredoxins (m1, m2, and m4) functionally distinct from the f-type thioredoxins involved in the activation of Calvin cycle enzymes (Issakidis-Bourguet et al., 2001). 2-CPs are peroxidases that detoxify alkyl hydroperoxides (resulting from lipid oxidation) by reduction to their corresponding alcohols. Antisense plants with reduced 2-CP levels showed severe damage to the photosynthetic apparatus and upregulation specifically of the known ascorbate peroxidases in the stroma and thylakoid, without much effect on GSH metabolism, superoxide dismutases, or calatase (Baier and Dietz, 1999; Baier et al., 2000). Interestingly, the peroxiredoxins link both to the ascorbate pathway and to the cyclophilins, because it has been shown in mammalian tissue that cyclophilin A binds to peroxiredoxins, suggesting that cyclophilin A acts as an immediate donor of peroxiredoxins (Lee et al., 2001). If this is relevant for the chloroplast homologs, it would link several cyclophilins to different peroxiredoxins.
The N termini of m2 and m4 thioredoxins on our map were sequenced and confirmed chloroplast localization. Recently, four m-type thioredoxins in Arabidopsis have been implicated directly in antioxidant defense (Issakidis-Bourguet et al., 2001). By complementation assays with a mutant yeast strain, it was found that m1, m2, and m4 thioredoxin and one x-type thioredoxin, but not the f-type thioredoxins, induced hydrogen peroxide tolerance. Unexpectedly, the fourth m-type thioredoxin, m3, had a hypersensitizing effect on oxidative stress. Overall, the authors concluded that these m-type and x-type thioredoxins can serve as electron donors for the reduction of hydroperoxides (Issakidis-Bourguet et al., 2001). Possibly, these thioredoxins form direct redox couples with the peroxiredoxins, as was shown for Arabidopsis thioredoxin m3 with cytosolic thioredoxins (Verdoucq et al., 1999). It is interesting that we identified m1, m2, and m4 on our map but not m3, considering that only these three confer hydrogen peroxide tolerance in yeast. As is evident from the list of thioredoxins and peroxiredoxins in Table 2, we are not certain if these proteins are lumenal proteins. Indeed, it seems that 2-CP is located at the stromal side of the membrane (K.J. Dietz, personal communication); the precise thylakoid localization of the other proteins remains to be determined.
Peripheral Proteins Involved in Translation, Stress Response, and Proteolysis
All but one of the set of 32 peripheral proteins (Table 3) were predicted by TargetP to be in the chloroplast (but not in the lumen) by the criteria of the prediction scheme in Figure 10A. A set of six (putative) fibrillins were identified, and some have been shown to be central in storing carotenoids of different plastids and accumulating plastoglobules (in chromoplasts, etioplasts, and chloroplasts) (Deruere et al., 1994; Rey et al., 2000). Overexpression and antisense suppression of different fibrillins affected development and stress response (Monte et al., 1999; Rey et al., 2000). Clearly, the fibrillins are sufficiently abundant to be identified on the 2-D gels, opening the way to study differential expression and the fibrillins' relationship to chloroplast development and stress in more detail. Because many proteins are translated at the thylakoid surface, it is not surprising to find a number of mRNA binding proteins (Table 3). Five mRNA binding proteins were identified, three of which have been described recently; however, their precise role is unclear (Nakamura et al., 2001).
Finally, we should focus attention on ClpS1 (Table 3). Recently, we identified ClpS1 as a member of a thylakoid-associated Clp protease complex of 350 kD with two (putative) heptameric rings. ClpS1 shares weak homology with the ClpP proteins, and we suggested that ClpS1 is not part of the ring structure but is positioned at the axial opening of the ClpP/R core (Peltier et al., 2001). The identification of ClpS1 on the 2-D gel confirms the thylakoid association and suggests that it could function to link the large protease complex to the thylakoid surface, in agreement with a postulated role in the degradation of cytochrome b6f subunits (Majeran et al., 2000).
Gene Annotation Errors and Generic Methods for Reannotation
For ∼30% of the proteins listed in Table 1, significant errors in gene annotation were identified, varying from no annotation of the gene to mistakes in intron/exon boundaries, missed exons, and truncated N termini. These genes were reassigned using overlapping ESTs (if available) and verified by sequences obtained by MS/MS. To match the genomic sequences with overlapping ESTs, at least two World Wide Web sites are now available (http://bioinformatics.iastate.edu/cgi-bin/gs.cgi and http://www.tigr.org/docs/tigr-scripts/nhgi_scripts/), and it is expected that many of these misassignments will be corrected rapidly with overlapping ESTs if they are available. However, if no or limited matching ESTs are available for Arabidopsis, it will be very beneficial to reconstruct first one (or more) orthologous gene in another species and use that reconstructed gene to map it to the genomic Arabidopsis sequence, as was demonstrated in this study. To date, this interspecies matching can be done only manually (to our best of knowledge). Finally, we noticed discrepancies in gene annotation (intron/exon boundaries, etc.) between the MIPS and National Center for Biotechnology Information (NCBI) databases (e.g., spots 103, 106, and 211), but this will be resolved in coming years by The Arabidopsis Information Resource (TAIR) (S. Rhee, curator TAIR, personal communication).
TAT Proteins with a KR Motif?
BLAST searching with the known lumenal proteins indicated that targeting pathways generally are conserved between orthologous proteins. In the case of the TAT-depen-dent protein in spot 104, we observed that the twin arginines in five orthologs were altered to KR while preserving the other features of a TAT motif. This finding suggested that in some cases TAT substrates also can contain KR. We observed several additional cases showing this phenomenon, such as spot 71. Interestingly, very recently, a similar observation was made with an E. coli protein (Hinsley et al., 2001). Further experimentation is needed to confirm that the orthologs with KR in fact translocate via the TAT pathway.
Use of Cross-Correlation Plots as a Systematic Method for the Identification of Post-Translational Modifications, Large Annotation Errors, and Alternative Splicing
Cross-correlation between the theoretical and experimental pI and molecular mass values generally showed very good agreement between theory and experimentation, after removal of the presequences, for both pea and Arabidopsis. However, discrepancies existed for a few spots and suggested dimerization and post-translational modifications. One very clear case of alternative splicing was discovered, and comparison with EST data showed that one of the forms was expressed preferentially in both pea and Arabidopsis. The MALDI-TOF MS data covered the protein sequence nearly completely, thus providing strong evidence that the annotation of the sequence was correct. Interesting alternative splicing changed only the C terminus of the protein, strongly affecting the pI value of this lumenal protein. It is unclear if the expression of the acidic (pI 5.3) and basic (pI 9.3) forms relates to the strong fluctuation of pH in the lumen (between 3.5 and 7.0) in conjunction with the day and night cycles. This demonstrates that the cross-correlation plots can help to identify annotation errors that affect pI or molecular mass as well as unexpected splicing events. Moreover, it indicates the presence of positive cleavable presequences as well as oligomerization and certain post-translational modifications.
Evaluation of the Genome-Wide Localization Prediction of the Lumenal Proteome
Both TargetP and SignalP have become very popular localization predictors in the plant biology field and are used widely for protein localization and presequence prediction in many published articles (Arabidopsis Genome Initiative, 2000), reviews (Adam et al., 2001), and database entries. A relatively new subcellular localization program is Predotar, which is focused completely on distinguishing between mitochondrial and chloroplast-targeted proteins but makes no cleavage site predictions (http://www.inra.fr/Internet/Produits/Predotar/). We observed that the performance of Predotar was significantly less accurate than that of TargetP (Tables 1 to 3), and the program seemed biased toward well-known proteins involved in photosynthesis. SignalP has been used regularly to determine potential thylakoid lumenal proteins, although it was developed specifically to identify bacterial and ER signal peptides. In our earlier proteomics study of pea thylakoids, we evaluated the performance of these predictors and speculated that it would be possible to predict a large fraction of the lumenal proteome if additional constraints based on experimentally identified lumenal proteins and their N termini were included (Peltier et al., 2000; van Wijk, 2000).
In this study, therefore, we extensively explored the use of these localization predictors (TargetP and SignalP-HMM) in combination with additional constraints derived from (1) a set of 211 experimentally identified known nucleus-encoded chloroplast proteins collected from SWISS-PROT release 38 that are not located in the lumen (this is the test set used to train TargetP; for a complete list, see http://www.cbs.dtu.dk/services/TargetP/datasets/datasets.html); (2) 41 experimentally identified nonredundant lumenal proteins (from this article and other studies) and 160 orthologs in higher plants and green algae, providing a set of 201 lumenal proteins (109 proteins have a typical TAT motif in the lTP); and (3) the use of TMHMM to remove TM proteins.
TargetP predicted 3646 proteins localized in plastids (using all five reliability classes), and 38% have no annotated function in MIPS. On the basis of TargetP prediction of an earlier test set (Emanuelsson et al., 2000), this prediction is expected to be ∼70% accurate, with a sensitivity of 85%. Analysis with TM prediction programs indicates that ∼500 proteins (45% have no annotation) of these 3646 proteins have at least one TM domain and are located in the thylakoid membrane or in the inner chloroplast envelope. The other 3146 proteins (38% have no annotation) are soluble proteins and are located in the plastid stroma or thylakoid lumen (O. Emanuelsson and G. von Heijne, unpublished data). TargetP correctly predicted chloroplast localization for 89% of the 174 complete lumenal orthologs when including the algae and 94% without the green algae. Predotar correctly predicted 67%.
Because there was no reason to select either the Gram-negative or the Gram-positive version of SignalP (based on either performance [Table 1] or theory), we used the union of the predictions for Gram-negative and Gram-positive SignalP-HMM predictions. We progressed stepwise through our prediction strategy while monitoring the accuracy by measuring false positives in the set of 211 known stromal proteins and monitoring the sensitivity by measuring the success rate in the set of 41 known lumenal proteins and the set of orthologs (Table 5). The end result was two sets of potential lumenal proteins, one targeted through the TAT pathway and the other targeted through the Sec and possibly other pathways. Within each set, proteins were sorted into four groups (p, s, a, and r) based on the amino acids in the −3 and −1 positions directly upstream of the lumen cleavage site. The thylakoid-processing peptidase that is responsible for processing of the lTPs is sensitive to the amino acids in the −3 and −1 positions, as was demonstrated by mutagenesis studies in both higher plants and C. reinhardtii (Bassham et al., 1994; Kuras et al., 1995). The prediction process was quite successful, although not perfect, for the proteins with a TAT motif, with a false positive rate of 0.06 and a sensitivity of 0.83 using the permissive filter for the −3 and −1 positions, and 93 lumenal proteins with a TAT motif were predicted. Application of more stringent filters reduced the sensitivity and improved the accuracy only slightly. The prediction for the non-TAT proteins was not as strong, with a false positive rate of 0.14 and a sensitivity of 0.60 using the p motif (Table 5). The better prediction of the TAT proteins is attributable to the presence of the TAT motif, which is a relatively unique signature. When analyzing both sets of predicted proteins manually, the accuracy was lower because of the presence of nonchloroplast proteins in the TargetP prediction. These false positives were not taken into account in the accuracy and sensitivity measurements because these were measured on chloroplast proteins directly.
Given the better performance, we present only the set of predicted TAT proteins. Only 30% of the 93 predicted proteins were annotated with some function in MIPS, and a manual annotation using all major domain predictors increased annotation to 56%. Twenty-two of the 93 predicted proteins were removed manually because functional domains or experimental data (from the literature and the databases) seemed incompatible with their predicted lumenal localization. A summary of the functions of the predicted proteins is shown in Figure 10B. Forty-four percent of the proteins have no assigned function, 18% have a role in photosynthesis (some well known and those for the paralogs unknown), 20% function in protein processing, proteolysis, and (un)folding, and 7% function in antioxidative defense and nonphotosynthetic redox reactions. One ABC transporter subunit was predicted, and 10% of the predicted proteins are involved in different metabolic processes.
To improve on the genome-wide prediction for lumenal proteins, a specific lTP predictor needs to be trained using the set of 174 lumenal proteins. Finally, as discussed elsewhere in this paper, the Arabidopsis gene annotation is erroneous in a significant number of cases, which can affect localization prediction. Thus, careful annotation will help to make a better prediction.
METHODS
Plant Growth
Arabidopsis thaliana, ecotype Columbia, was grown on soil in a temperature-controlled growth chamber under a 10-hr-light/14-hr-dark cycle at 21/16°C light/dark temperatures at a light intensity of ∼100 μmol·m−2·sec−1.
Isolation of the Thylakoid Lumen
Leaves from Arabidopsis were homogenized in ice-cold medium A (50 mM Hepes-KOH, pH 7.5, and 100 mM sorbitol supplemented with 50 μg·mL−1 Pefablok, 2 μg·mL−1 antipain, and 2 μg·mL−1 leupeptin), filtered through four layers of Miracloth (Calbiochem; 22 μm), and centrifuged for 3 min at 9000g. Thylakoid pellets were resuspended in medium A with a cocktail of nine protease inhibitors (antipain [40 μg·mL−1], leupeptin [5 μg·mL−1], pepstatin [0.7 μg·mL−1], Pefablok [5 mg·mL−1], E64 [10 μg·mL−1], chymostatin [20 μg·mL−1], bestatin [40 μg·mL−1], phosphoramidon [5 μg·mL−1], and apoprotin [2 μg·mL−1]) and centrifuged on a Percoll gradient (0 to 90%) in medium A for 4 min at 3000g. The thylakoid band was collected and washed three times in the same medium A. Thylakoids were passed twice through a Yeda press (at 100 bar), and thylakoid membranes were removed subsequently by ultracentrifugation (60 min at 150,000g). The membrane-free supernatant was collected and concentrated to 20 mg protein·mL−1. Alternatively the membrane-free supernatant was concentrated 10-fold and precipitated in 90% aceton and the lumenal pellet was resuspended at 20 mg protein·mL−1.
Construction and Analysis of Denaturing Two-Dimensional Gels
Lumenal proteins (250 μg for silver stain and 1 to 1.5 mg for Coomassie Brilliant Blue R 250 stain) were solubilized in 7 M urea, 2 M thiourea, 2 mM Tributyl phosphine, 4% 3-([3-cholamidopropyl]dimethylammonio)-1-propanesulfonic acid, 0.5% Triton X-100, and 2% pharmalyte, pH 3 to 10 (map pH 4 to 7), or 2% immobilized pH gradient buffer, pH 6 to11 (map pH 7 to 11) (Rabilloud, 1998). Immobiline DryStrips (Amersham Pharmacia, Uppsala, Sweden) (pH 4 to 7 and pH 6 to 11) were rehydrated overnight in sample suspension at room temperature. Isoelectric focusing in the first dimension, gradient Tricine-SDS-PAGE in the second dimension, and staining and image analysis of the gels were performed as described by Peltier et al. (2000). Expression levels were calculated using the two-dimensional analysis software Melanie (Bio-Rad).
Mass Spectrometry, Database Searching, Sequence Analysis, Localization Prediction, Sequence Alignments, and Matching with Expressed Sequence Tags
Stained protein spots were excised from the gel, washed, reduced, and digested with modified trypsin (Promega) according to Shevchenko et al. (1996). The peptides then were extracted and dissolved in 20 μL of 5% formic acid, and 0.5 μL was applied to the matrix-assisted laser desorption ionization time-of-flight target plate by the dried droplet method using α-cyano-4-hydrocynnamic acid as a matrix. When necessary, the samples were concentrated using microcolumns (Gobom et al., 1999) and eluted directly onto the matrix-assisted laser desorption ionization target. The mass spectra were obtained manually or in automated mode using a matrix-assisted laser desorption ionization time-of-flight mass spectrometer (REFLEX II from Bruker Daltonics (Billerica, MA) or Voyager-DE-STR from Applied Biosystems [Foster City, CA]). The spectra were calibrated internally using tryptic peptides from autodigestion and were annotated with the program m/z from Proteometrics (www.proteometrics.com) or automatically using PSI software (Applied Biosystems). The latest versions of the National Center for Biotechnology Information nonredundant database were searched with the resulting peptide mass lists, manually using the search engine ProFound (www.proteometrics.com) or automatically using MS-Fit (Protein Prospector) and PSI solutions (Applied Biosystems). The search strategy was in principle as described previously (Peltier et al., 2000).
To further analyze the samples, the remainder of the extracted peptides were desalted, concentrated on microcolumns (Poros R2 or R3; Applied Biosystems), and eluted directly into nanoelectrospray needles (Protana A/S, Odense, Denmark) with 1.2 μL of 50% methanol and 1% formic acid (Wilm et al., 1996). The spectra were acquired on an electrospray tandem mass spectrometer (Q-TOF; Micromass, Manchester, UK). The instrument was calibrated with 1 μg·μL−1 NaI in 50% isopropanol. Alternatively, the spectra were used to search the public databases with the program Mascot (http://www.matrix-science.com/). The mass spectrometry spectra usually were interpreted using MassLynx and PepSeq (Micromass), and the resulting sequences were used to search different public databases using FASTA3 (www2.ebi.ac.uk/fasta3).
Manual predictions for chloroplast localization and chloroplast and lumenal transit peptides were made using the search engines TargetP (www.cbs.dtu.dk/services/TargetP), SignalP (www.cbs.dtu.dk/services/SignalP/), and Predotar (www.inra.fr/Internet/Produits/Predotar).
Data Acquisition and Localization Prediction
Arabidopsis protein sequences for theoretical analyses were obtained on November 30, 2000, via ftp (ftp://ftpmips.gsf.de/cress/arabiprot/). The set used in this study is similar but not identical to the arabi_all_proteins_v211200.tfa set at this ftp site. This set corresponds to the set presented by the Arabidopsis Genome Initiative (2000). Entries At4g341{31, 35, 38} in the set of November 30 have been replaced by At4g34130, and At5g33{390, 400, 410, 420, 430, 440} have changed their names to {W120, C190, C205, C235, W250, W305}ECH54M, respectively.
All 25,460 sequences were processed through subcellular localization predictor TargetP (version 1.01, using plant version; http://www.cbs.dtu.dk/services/TargetP/) (Emanuelsson et al., 2000), and the 3646 proteins that were predicted to contain a chloroplast transit peptide were processed further through signal peptide predictor SignalP (version 2.0, using HMM version; http://www.cbs.dtu.dk/services/SignalP-2.0/) (Nielsen et al., 1997, 1999) and transmembrane predictor TMHMM (version 2.0; http://www.cbs.dtu.dk/services/) (Sonnhammer et al., 1998; Krogh et al., 2001). Various constraints were applied (see Results) to obtain the final predicted set of lumenal proteins. The scheme of the prediction approach is shown in Figure 10A.
Multialignments and Construction of Phylogenetic Trees
Multialignment and construction of phylogenetic trees were performed using ClustalW (www2.ebi.ac.uk/clustalw/) and MultAlin (http://www.toulouse.inra.fr/multalin.html). Phylogenetic trees were calculated using parsimony and neighbor-joining distance methods (Phylip; Department of Genetics, University of Washington, Seattle) with bootstrap values to test the statistical robustness of the trees, essentially as described by Peltier et al. (2001).
Miscellaneous
Protein determinations were performed according to Bradford (1976). Chlorophyll concentrations were determined spectroscopically in 80% acetone (Porra et al., 1989).
Accession Numbers
The accession number for the sequence of the moss Physcomitrella mentioned in Figure 4 is 7046852; the accession numbers for the soybean ESTs mentioned in Figure 5 are AW508389, BE823905, and BE607927. The SWISS-PROT accession number for the Arabidopsis sequence (spot 71) mentioned in Figure 5 is P83050 and is assigned OEC23 related protein.
NOTE ADDED IN PROOF
During proofreading of this paper, a description of an isomerase with a TAT motif, an isomerase targeted via the Sec (or other) pathway, and other lumenal proteins appeared online (Schubert, M., Petersson, U.A., Haas, B.J., Funk, C., Schröder, W.P., and Kieselbach, T. [2002]. Proteome map of the chloroplast lumen of Arabidopsis thaliana. J. Biol. Chem., in press).
Acknowledgments
Dr. Per-Ingvar Ohlsson at Umea University is greatly acknowledged for his excellent Edman sequencing analysis. This study was supported by a postdoctoral fellowship to J.-B.P. from the Wenner-Grenska Samfundet, the Nordisk Kontaktorgan för Jordsbrukforskning, the Cell Factory for Functional Genomics of the Swedish Foundation for Strategic Research for general support and support of J.Y., and the Swedish National Research Council to K.J.v.W. We thank the Hasselblad Foundation for generous financial support for mass spectrometers to K.J.v.W. P.R. and D.E.K. are members of the Center for Experimental Bioinformatics sponsored by the Danish National Research Foundation. G.v.H. was supported by grants from the Swedish Foundation for Strategic Research and the Swedish Research Council.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010304.
References
- Adam, Z., Adamska, I., Nakabayashi, K., Ostersetzer, O., Haussuhl, K., Manuell, A., Zheng, B., Vallon, O., Rodermel, S.R., Shinozaki, K., and Clarke, A.K. (2001). Chloroplast and mitochondrial proteases in Arabidopsis: A proposed nomenclature. Plant Physiol. 125, 1912–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Baier, M., and Dietz, K.J. (1999). Protective function of chloroplast 2-cysteine peroxiredoxin in photosynthesis: Evidence from transgenic Arabidopsis. Plant Physiol. 119, 1407–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier, M., Noctor, G., Foyer, C.H., and Dietz, K.J. (2000). Antisense suppression of 2-cysteine peroxiredoxin in Arabidopsis specifically enhances the activities and expression of enzymes associated with ascorbate metabolism but not glutathione metabolism. Plant Physiol. 124, 823–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham, D.C., Creighton, A.M., Karnauchov, I., Herrmann, R.G., Klosgen, R.B., and Robinson, C. (1994). Mutations at the stromal processing peptidase cleavage site of a thylakoid lumen protein precursor affect the rate of processing but not the fidelity. J. Biol. Chem. 269, 16062–16066. [PubMed] [Google Scholar]
- Berks, B.C., Sargent, F., De Leeuw, E., Hinsley, A.P., Stanley, N.R., Jack, R.L., Buchanan, G., and Palmer, T. (2000). A novel protein transport system involved in the biogenesis of bacterial electron transfer chains. Biochim. Biophys. Acta 1459, 325–330. [DOI] [PubMed] [Google Scholar]
- Blackstock, W., and Mann, M., eds (2000). Proteomics: A Trends Guide. (Amsterdam, The Netherlands, Elsevier Science).
- Bradford, M.M. (1976). A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Chou, I.T., and Gasser, C.S. (1997). Characterization of the cyclophilin gene family of Arabidopsis thaliana and phylogenetic analysis of known cyclophilin proteins. Plant Mol. Biol. 35, 873–892. [DOI] [PubMed] [Google Scholar]
- Dalbey, R.E., and Robinson, C. (1999). Protein translocation into and across the bacterial plasma membrane and the plant thylakoid membrane. Trends Biochem. Sci. 24, 17–22. [DOI] [PubMed] [Google Scholar]
- Deruere, J., Romer, S., d'Harlingue, A., Backhaus, R.A., Kuntz, M., and Camara, B. (1994). Fibril assembly and carotenoid overaccumulation in chromoplasts: A model for supramolecular lipoprotein structures. Plant Cell 6, 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., and von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Froderberg, L., Rohl, T., van Wijk, K., and de Gier, J.L. (2001). Complementation of bacterial SecE by a chloroplastic homologue. FEBS Lett. 498, 52–56. [DOI] [PubMed] [Google Scholar]
- Fulgosi, H., Vener, A.V., Altschmied, L., Herrmann, R.G., and Andersson, B. (1998). A novel multi-functional chloroplast protein: Identification of a 40 kDa immunophilin-like protein located in the thylakoid lumen. EMBO J. 17, 1577–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, H.F. (1998). Protein disulfide isomerase. Methods Enzymol. 290, 26–50. [DOI] [PubMed] [Google Scholar]
- Gobom, J., Nordhoff, E., Mirgorodskaya, E., Ekman, R., and Roepstorff, P. (1999). Sample purification and preparation technique based on nano-scale reversed-phase columns for the sensitive analysis of complex peptide mixtures by matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom. 34, 105–116. [DOI] [PubMed] [Google Scholar]
- Hinsley, A.P., Stanley, N.R., Palmer, T., and Berks, B.C. (2001). A naturally occurring bacterial Tat signal peptide lacking one of the ‘invariant’ arginine residues of the consensus targeting motif. FEBS Lett. 497, 45–49. [DOI] [PubMed] [Google Scholar]
- Hynds, P.J., Plucken, H., Westhoff, P., and Robinson, C. (2000). Different lumen-targeting pathways for nuclear-encoded versus cyanobacterial/plastid-encoded Hcf136 proteins. FEBS Lett. 467, 97–100. [DOI] [PubMed] [Google Scholar]
- Issakidis-Bourguet, E., Mouaheb, N., Meyer, Y., and Miginiac-Maslow, M. (2001). Heterologous complementation of yeast reveals a new putative function for chloroplast m-type thioredoxin. Plant J. 25, 127–135. [DOI] [PubMed] [Google Scholar]
- Itzhaki, H., Naveh, L., Lindahl, M., Cook, M., and Adam, Z. (1998). Identification and characterization of DegP, a serine protease associated with the luminal side of the thylakoid membrane. J. Biol. Chem. 273, 7094–7098. [DOI] [PubMed] [Google Scholar]
- Jensen, O.N., Larsen, M.R., and Roepstorff, P. (1998). Mass spectrometric identification and microcharacterization of proteins from electrophoretic gels: Strategies and applications. Proteins (suppl.), 74–89. [DOI] [PubMed]
- Jespersen, H.M., Kjaersgard, I.V., Ostergaard, L., and Welinder, K.G. (1997). From sequence analysis of three novel ascorbate peroxidases from Arabidopsis thaliana to structure, function and evolution of seven types of ascorbate peroxidase. Biochem. J. 326, 305–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra, K., and Cline, K. (1999). Protein import and routing systems of chloroplasts. Plant Cell 11, 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieselbach, T., Hagman, Å., Andersson, B., and Schroder, W.P. (1998). The thylakoid lumen of chloroplasts: Isolation and characterization. J. Biol. Chem. 273, 6710–6716. [DOI] [PubMed] [Google Scholar]
- Kim, J., and Mayfield, S.P. (1997). Protein disulfide isomerase as a regulator of chloroplast translational activation. Science 278, 1954–1957. [DOI] [PubMed] [Google Scholar]
- Knott, T.G., and Robinson, C. (1994). The secA inhibitor, azide, reversibly blocks the translocation of a subset of proteins across the chloroplast thylakoid membrane. J. Biol. Chem. 269, 7843–7846. [PubMed] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E.L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Kuras, R., Buschlen, S., and Wollman, F.A. (1995). Maturation of pre-apocytochrome f in vivo: A site-directed mutagenesis study in Chlamydomonas reinhardtii. J. Biol. Chem. 270, 27797–27803. [DOI] [PubMed] [Google Scholar]
- Lee, S.P., Hwang, Y.S., Kim, Y.J., Kwon, K.-S., Kim, H.J., Kim, K., and Chae, H.Z. (2001). Cyclophilin a binds to peroxiredoxins and activates their peroxidase activity. J. Biol. Chem., 276, 29826–29832. [DOI] [PubMed] [Google Scholar]
- Lippuner, V., Chou, I.T., Scott, S.V., Ettinger, W.F., Theg, S.M., and Gasser, C.S. (1994). Cloning and characterization of chloroplast and cytosolic forms of cyclophilin from Arabidopsis thaliana. J. Biol. Chem. 269, 7863–7868. [PubMed] [Google Scholar]
- Luan, S., Albers, M.W., and Schreiber, S.L. (1994. a). Light-regulated, tissue-specific immunophilins in a higher plant. Proc. Natl. Acad. Sci. USA 91, 984–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan, S., Lane, W.S., and Schreiber, S.L. (1994. b). pCyP B: A chloroplast-localized, heat shock-responsive cyclophilin from fava bean. Plant Cell 6, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran, W., Wollman, F.A., and Vallon, O. (2000). Evidence for a role of ClpP in the degradation of the chloroplast cytochrome b(6)f complex. Plant Cell 12, 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann, M., and Pandey, A. (2001). Use of mass spectrometry–derived data to annotate nucleotide and protein sequence databases. Trends Biochem. Sci. 26, 54–61. [DOI] [PubMed] [Google Scholar]
- Mant, A., Kieselbach, T., Schroder, W.P., and Robinson, C. (1999). Characterisation of an Arabidopsis thaliana cDNA encoding a novel thylakoid lumen protein imported by the delta pH-dependent pathway. Planta 207, 624–627. [DOI] [PubMed] [Google Scholar]
- Meurer, J., Plucken, H., Kowallik, K.V., and Westhoff, P. (1998). A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 17, 5286–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte, E., Ludevid, D., and Prat, S. (1999). Leaf C40.4: A carotenoid-associated protein involved in the modulation of photosynthetic efficiency? Plant J. 19, 399–410. [DOI] [PubMed] [Google Scholar]
- Mori, H., Summer, E.J., Ma, X., and Cline, K. (1999). Component specificity for the thylakoidal Sec and ΔpH-dependent protein transport pathways. J. Cell Biol. 146, 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, T., Ohta, M., Sugiura, M., and Sugita, M. (2001). Chloroplast ribonucleoproteins function as a stabilizing factor of ribosome-free mRNAs in the stroma. J. Biol. Chem. 276, 147–152. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. (1997). A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst. 8, 581–599. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Brunak, S., and von Heijne, G. (1999). Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng. 12, 3–9. [DOI] [PubMed] [Google Scholar]
- Noctor, G., Veljovic-Jovanovic, S., and Foyer, C.H. (2000). Peroxide processing in photosynthesis: Antioxidant coupling and redox signalling. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 1465–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, A., and Mann, M. (2000). Proteomics to study genes and genomes. Nature 405, 837–846. [DOI] [PubMed] [Google Scholar]
- Peltier, J.B., Friso, G., Kalume, D.E., Roepstorff, P., Nilsson, F., Adamska, I., and van Wijk, K.J. (2000). Proteomics of the chloroplast: Systematic identification and targeting analysis of lumenal and peripheral thylakoid proteins. Plant Cell 12, 319–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier, J.B., Ytterberg, J., Liberles, D.A., Roepstorff, P., and van Wijk, K.J. (2001). Identification of a 350-kDa ClpP protease complex with 10 different Clp isoforms in chloroplasts of Arabidopsis thaliana. J. Biol. Chem. 276, 16318–16327. [DOI] [PubMed] [Google Scholar]
- Porra, R.J., Thompson, W.A., and Kriedemann, P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394. [Google Scholar]
- Rabilloud, T. (1998). Use of thiourea to increase the solubility of membrane proteins in two-dimensional electrophoresis. Electrophoresis 19, 758–760. [DOI] [PubMed] [Google Scholar]
- Rey, P., Gillet, B., Romer, S., Eymery, F., Massimino, J., Peltier, G., and Kuntz, M. (2000). Over-expression of a pepper plastid lipid-associated protein in tobacco leads to changes in plastid ultrastructure and plant development upon stress. Plant J. 21, 483–494. [DOI] [PubMed] [Google Scholar]
- Robinson, C., and Bolhuis, A. (2001). Protein targeting by the twin-arginine translocation pathway. Natl. Rev. Mol. Cell. Biol. 2, 350–356. [DOI] [PubMed] [Google Scholar]
- Schiene, C., and Fischer, G. (2000). Enzymes that catalyse the restructuring of proteins. Curr. Opin. Struct. Biol. 10, 40–45. [DOI] [PubMed] [Google Scholar]
- Schneider, T., and Stephens, R.M. (1990). Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuenemann, D., Amin, P., Hartmann, E., and Hoffman, N.E. (1999). Chloroplast SecY is complexed to SecE and involved in the translocation of the 33-kDa but not the 23-kDa subunit of the oxygen-evolving complex. J. Biol. Chem. 274, 12177–12182. [DOI] [PubMed] [Google Scholar]
- Settles, A.M., Yonetani, A., Baron, A., Bush, D.R., Cline, K., and Martienssen, R. (1997). Sec-independent protein translocation by the maize Hcf106 protein. Science 278, 1467–1470. [DOI] [PubMed] [Google Scholar]
- Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Sonnhammer, E.L.L., von Heijne, G., and Krogh, A. (1998). A hidden Markov model for predicting transmembrane helices in proteins sequences. In Sixth International Conference on Intelligent Systems for Molecular Biology, J. Glasgow, T. LittleJohn, F. Major, R. Lathrop, D. Sankoff, and C. Sensen, eds (Menlo Park, CA: The American Association for Artificial Intelligence Press), pp. 175–182. [PubMed]
- Summer, E.J., Mori, H., Settles, A.M., and Cline, K. (2000). The thylakoid ΔpH-dependent pathway machinery facilitates RR-independent N-tail protein integration. J. Biol. Chem. 275, 23483–23490. [DOI] [PubMed] [Google Scholar]
- Thompson, S.J., Kim, S.J., and Robinson, C. (1998). Sec-independent insertion of thylakoid membrane proteins: Analysis of insertion forces and identification of a loop intermediate involving the signal peptide. J. Biol. Chem. 273, 18979–18983. [DOI] [PubMed] [Google Scholar]
- Thompson, S.J., Robinson, C., and Mant, A. (1999). Dual signal peptides mediate the signal recognition particle/Sec-independent insertion of a thylakoid membrane polyprotein, PsbY. J. Biol. Chem. 274, 4059–4066. [DOI] [PubMed] [Google Scholar]
- Trebitsh, T., Meiri, E., Ostersetzer, O., Adam, Z., and Danon, A. (2000). The protein disulfide isomerase-like RB60 is partitioned between stroma and thylakoids in Chlamydomonas reinhardtii chloroplasts. J. Biol. Chem. 276, 4564-4569. [DOI] [PubMed] [Google Scholar]
- van Wijk, K.J. (2000). Proteomics of the chloroplast: Experimentation and prediction. Trends Plant Sci. 5, 420–425. [DOI] [PubMed] [Google Scholar]
- van Wijk, K.J. (2001). Challenges and prospects of plant proteomics. Plant Physiol. 126, 501–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vener, A.V., Rokka, A., Fulgosi, H., Andersson, B., and Herrmann, R.G. (1999). A cyclophilin-regulated PP2A-like protein phosphatase in thylakoid membranes of plant chloroplasts. Biochemistry 38, 14955–14965. [DOI] [PubMed] [Google Scholar]
- Verdoucq, L., Vignols, F., Jacquot, J.P., Chartier, Y., and Meyer, Y. (1999). In vivo characterization of a thioredoxin h target protein defines a new peroxiredoxin family. J. Biol. Chem. 274, 19714–19722. [DOI] [PubMed] [Google Scholar]
- von Heijne, G., Steppuhn, J., and Hermann, S.G. (1989). Domain structure of mitochondrial and chloroplast targeting peptides. Eur. J. Biochem. 80, 535–545. [DOI] [PubMed] [Google Scholar]
- Walker, K.W., and Gilbert, H.F. (1997). Scanning and escape during protein-disulfide isomerase-assisted protein folding. J. Biol. Chem. 272, 8845–8848. [DOI] [PubMed] [Google Scholar]
- Walker, M.B., Roy, L.M., Coleman, E., Voelker, R., and Barkan, A. (1999). The maize tha4 gene functions in sec-independent protein transport in chloroplasts and is related to hcf106, tatA, and tatB. J. Cell Biol. 147, 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm, M., Shevchenko, A., Houthaeve, T., Breit, S., Schweigerer, L., Fotsis, T., and Mann, M. (1996). Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379, 466–469. [DOI] [PubMed] [Google Scholar]
- Yates III, J.R. (2000). Mass spectrometry: From genomics to proteomics. Trends Genet. 16, 5–8. [DOI] [PubMed] [Google Scholar]
- Yoshimura, K., Yabuta, Y., Tamoi, M., Ishikawa, T., and Shigeoka, S. (1999). Alternatively spliced mRNA variants of chloroplast ascorbate peroxidase isoenzymes in spinach leaves. Biochem. J. 338, 41–48. [PMC free article] [PubMed] [Google Scholar]