Abstract
Salt stress is a major environmental factor influencing plant growth and development. To identify salt tolerance determinants, a genetic screen for salt overly sensitive (sos) mutants was performed in Arabidopsis. We present here the characterization of sos4 mutants and the positional cloning of the SOS4 gene. sos4 mutant plants are hypersensitive to Na+, K+, and Li+ ions. Under NaCl stress, sos4 plants accumulate more Na+ and retain less K+ compared with wild-type plants. SOS4 encodes a pyridoxal kinase that is involved in the biosynthesis of pyridoxal-5-phosphate, an active form of vitamin B6. The expression of SOS4 cDNAs complements an Escherichia coli mutant defective in pyridoxal kinase. Supplementation of pyridoxine but not pyridoxal in the growth medium can partially rescue the sos4 defect in salt tolerance. SOS4 is expressed ubiquitously in all plant tissues. As a result of alternative splicing, two transcripts are derived from the SOS4 gene, the relative abundance of which is modulated by development and environmental stresses. Besides being essential cofactors for numerous enzymes, as shown by pharmacological studies in animal cells, pyridoxal-5-phosphate and its derivatives are also ligands for P2X receptor ion channels. Our results demonstrate that pyridoxal kinase is a novel salt tolerance determinant important for the regulation of Na+ and K+ homeostasis in plants. We propose that pyridoxal-5-phosphate regulates Na+ and K+ homeostasis by modulating the activities of ion transporters.
INTRODUCTION
Salt stress affects plant growth and development in many different ways. Excess salt causes ion toxicity inside the cell. High concentrations of salt in the root medium also create hyperosmotic stress that impedes water absorption and transport. Secondary stresses such as nutritional imbalance and oxidation often occur as a consequence of ion toxicity and hyperosmotic stress (Zhu, 2001). Plants respond to salt stress by changing gene expression pattern, metabolic activity, and ion and water transport to minimize stress damage and to reestablish ion and water homeostasis (Serrano and Gaxiola, 1994; Hasegawa et al., 2000).
Reestablishing ion homeostasis is of critical importance for plant adaptation to salt stress (Niu et al., 1995; Tyerman and Skerrett, 1999). Various ion transporters are the terminal determinants of ion homeostasis and are regulated tightly at the transcriptional and post-transcriptional levels. Under normal physiological conditions, plants maintain a relatively high K+ concentration and low Na+ concentration in their cytosol (Binzel et al., 1988). This cytosolic high K+/Na+ ratio results from selective K+ uptake over Na+, preferential exclusion of Na+, and compartmentation of Na+ into the vacuole.
Plant cells take up K+ from the extracellular medium using K+ channels and cotransporters (Rodríguez-Navarro, 2000). The K+ channel AKT1 is essential for high-affinity K+ uptake in the presence of NH4+ (Hirsch et al., 1998) and displays a high K+/Na+ selectivity at physiological external concentrations of K+ and Na+ (Gaymard et al., 1996). The K+ transporter HKT1 from wheat is energized by Na+ when expressed in yeast and Xenopus oocytes (Schachtman and Schroeder, 1994; Rubio et al., 1995; Schachtman et al., 1997), implying that it might mediate Na+ influx into plant cells. In addition, nonselective cation channels are thought to constitute a major Na+ influx system (Amtmann and Sanders, 1999; Davenport and Tester, 2000).
Limiting Na+ entry into the cell probably is one of the most important mechanisms to maintain a low Na+ concentration in the cytosol. On the other hand, once Na+ gets inside the cell, it can be exported to the extracellular space by plasma membrane Na+/H+ antiporters and to the vacuole by tonoplast Na+/H+ antiporters (Barkla and Pantoja, 1996; Blumwald et al., 2000). Na+ extrusion from the cell against its electrochemical gradient by plasma membrane Na+/H+ antiporters is an active process driven by the downhill movement of H+ into the cell. Plasma membrane Na+/H+ antiporter activities have been detected in different plant species (Blumwald et al., 2000). Recently, a putative plasma membrane Na+/H+ antiporter, SOS1, was cloned from Arabidopsis (Shi et al., 2000). Mutations in SOS1 render Arabidopsis plants extremely sensitive to Na+ stress (Wu et al., 1996).
Na+ compartmentation into the vacuole is an economical means of avoiding the deleterious effects of Na+ in the cytosol because the vacuolar Na+ serves as an osmoticum to maintain osmotic homeostasis. The transport of Na+ into vacuoles is mediated by tonoplast Na+/H+ antiporters. Although the presence of vacuolar Na+/H+ antiporter activity in plants was demonstrated some time ago (Blumwald and Poole, 1985), the molecular nature of the antiporters was not revealed until recently (Apse et al., 1999; Gaxiola et al., 1999; Quintero et al., 2000). The important role of the vacuolar Na+/H+ antiporters for plant salt tolerance was supported by the finding that the overexpression of one of them, AtNHX1, improved plant salt tolerance (Apse et al., 1999; Frommer et al., 1999).
Ion transporter activities are modulated rapidly in response to several environmental stimuli (Zimmermann et al., 1999). Evidence suggests that plant ion channel activities are modulated by a variety of cellular factors, including second messengers such as cytosolic calcium, pH, and nucleotides. Ion channels are known to interact with signaling proteins (i.e., protein kinases and phosphatases, cytoskeletal components, GTP binding proteins, and 14-3-3 proteins) (Schroeder and Hedrich, 1989; Blatt and Thiel, 1993; Nürnberger et al., 1997; Zimmermann et al., 1999). Direct ligand binding to the channel proteins is an important mechanism for the regulation of transport activities. In mammalian cells, the P2X receptors are ligand-gated ion channels that open in milliseconds in response to the binding of extracellular ATP and conduct the flow of Na+, K+, and Ca2+ across cell membranes (Brake and Julius, 1996; Burnstock, 1996). Pharmacological studies indicate that pyridoxal-5-phosphate (PLP) and its derivatives antagonize the activation of P2X receptors (Ralevic and Burnstock, 1998). Although PLP is known to be an antagonist of the ATP-gated ion channel in animal cells and a well-known cofactor for numerous enzymes, nothing is known about its function in plants, especially for the regulation of ion transporter activities.
We have been using a mutational approach to the study of salt tolerance by screening for Arabidopsis salt overly sensitive (sos) mutant. This approach has yielded sos mutants that fall into three complementation groups: sos1, sos2, and sos3 (Zhu, 2000). SOS1 encodes a putative plasma membrane Na+/H+ antiporter, the transcript level of which is upregulated specifically by salt stress (Shi et al., 2000). SOS2 encodes a Ser/Thr protein kinase (Liu et al., 2000). SOS3 encodes a myristoylated calcium binding protein that presumably senses the cytosolic calcium signal elicited by salt stress (Liu and Zhu, 1998; Ishitani et al., 2000). SOS3 interacts physically with and activates the kinase activity of SOS2 (Halfter et al., 2000). Salt stress upregulation of SOS1 expression is partly under the control of the SOS3-SOS2 regulatory pathway (Shi et al., 2000).
To identify additional salt tolerance determinants in Arabidopsis, we performed a genetic screen for new sos mutants using higher levels of salt stress than were used previously for the sos1, sos2, and sos3 screen. This screen yielded two new groups of sos mutants, designated sos4 and sos5. We report here the characterization of sos4 mutants and the positional cloning and characterization of the SOS4 gene. sos4 mutant plants are hypersensitive to NaCl, KCl, and LiCl but not to CsCl stress. In response to NaCl stress, sos4 mutant plants accumulate more Na+ and less K+ than do wild-type plants. SOS4 encodes a pyridoxine/pyridoxal/pyridoxamine (PN/PL/PM) kinase homolog that functions in the biosynthesis of PLP. We propose that PLP or its derivatives may regulate ion channels and transporters that are important for salt tolerance.
RESULTS
Isolation of sos4 Mutants
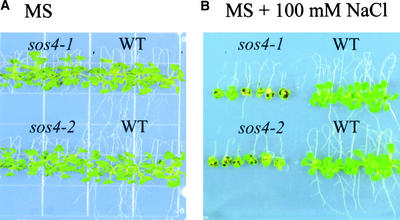
We previously screened for sos mutants using a root-bending assay at 50 and 75 mM NaCl (Zhu et al., 1998). To identify novel salt tolerance determinants, a root-bending assay at 100 mM NaCl was used to screen for sos mutants that may not be allelic to sos1, sos2, or sos3 (Wu et al., 1996; Liu and Zhu, 1997, 1998). Approximately 60,000 seedlings from ethyl methanesulfonate– or fast neutron–mutagenized M2 seed were screened on Murashige and Skoog (1962) (MS) nutrient medium supplemented with 100 mM NaCl. Two allelic mutants, designated sos4-1 and sos4-2, were identified and chosen for detailed characterization. sos4-1 was recovered from an ethyl methanesulfonate–mutagenized population, whereas sos4-2 originated from fast neutron–mutagenized plants. Figure 1 shows the phenotypes of sos4-1 and sos4-2 mutant seedlings under NaCl stress. On MS nutrient medium, the aerial parts of both mutants were indistinguishable from those of the wild type, but the roots of the mutants grew more slowly than did wild-type roots (Figure 1A). Upon transfer to medium supplemented with 100 mM NaCl, the growth of both the shoot and the root of mutant plants was inhibited to a greater extent than the growth of those of wild-type plants (Figure 1B). The greater sensitivity of sos4 mutants also was indicated by the dark color of the leaves, which reflects anthocyanin accumulation caused by stress damage.
Figure 1.
Phenotypes of Wild-Type and sos4 Mutant Seedlings Grown on Vertically Placed MS Agar Plates with or without NaCl Supplement.
Seedlings were grown first on vertical MS agar plates for 5 days before being transferred to vertical agar plates without (A) or with (B) 100 mM NaCl. The plates were placed upside down for root bending. The photographs were taken 2 weeks after seedling transfer. WT, wild type.
Both mutants were backcrossed with wild-type plants, and the resulting F1 seedlings exhibited the wild-type level of salt tolerance in root-bending assays. The F2 progeny from self-fertilized F1 plants showed an ∼3:1 segregation ratio of wild type to mutant, indicating that both sos4-1 and sos4-2 are caused by recessive mutations in a single nuclear gene (data not shown). sos4-1 was crossed to sos4-2, and the resulting F1 seedlings all behaved like the mutant in the root-bending assay. Therefore, sos4-1 and sos4-2 are allelic. Pairwise crosses with sos1, sos2, or sos3 mutants were performed, and the results show that the sos4 mutants are not allelic to any of them (data not shown).
sos4 Plants Are Hypersensitive to Na+, Li+, and K+, but Not to Cs+, and Resistant to High Concentrations of Mannitol
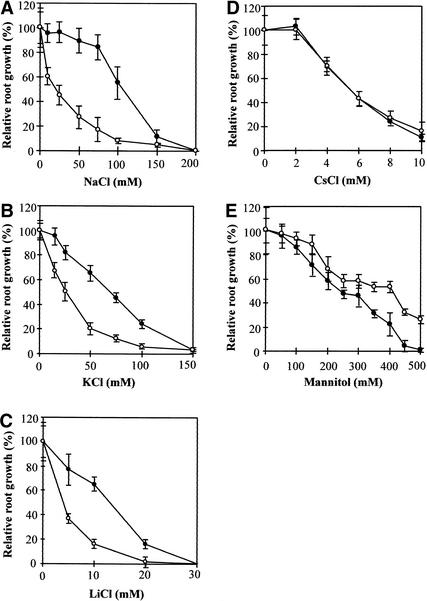
Because the two sos4 alleles showed virtually identical phenotypes either with or without salt stress, the sos4-1 mutant was characterized in detail. All subsequent characterization was performed using a sos4-1 mutant that had been backcrossed with the wild type at least three times. As a convenient and accurate indicator of Arabidopsis seedling growth, root elongation was used to determine the response of sos4-1 to various salt and mannitol treatments. Figure 2A shows that sos4-1 is hypersensitive to NaCl stress. The concentration of NaCl that decreased the root elongation rate by 50% relative to the control without salt was estimated. These concentrations for sos4-1 and wild-type seedlings were ∼23 and 100 mM, respectively. Interestingly, sos4-1 also was hypersensitive to KCl stress (Figure 2B), distinct from sos1, sos2, or sos3 mutants. Growth analysis of sos4-1 on LiCl medium indicated that sos4-1 was hypersensitive to Li+, a more toxic analog of Na+ (Figure 2C). Although Cs+ also is a toxic cation related to Na+, sos4-1 was not hypersensitive to CsCl (Figure 2D). The hypersensitivity of sos4-1 to NaCl, KCl, and LiCl but not to CsCl suggests that the altered salt sensitivity is not attributable to Cl−. Unlike sos1, sos2, sos3, and sos4-1 mutant plants can grow normally on low-potassium culture medium, as do wild-type plants (data not shown).
Figure 2.
Sensitivity of sos4 Seedlings to Various Salt and Osmotic Stresses as Measured by Relative Root Growth.
Four-day-old seedlings were transferred to vertical agar plates containing MS medium or MS medium supplemented with NaCl (A), KCl (B), LiCl (C), CsCl (D), or mannitol (E) at the concentrations indicated. Root elongation at day 7 after transfer is presented as a percentage relative to elongation on the MS medium. Closed circles, wild type; open circles, sos4. Values are averages ±sd (n = 15).
To determine whether sos4-1 plants are hypersensitive to general osmotic stress, root elongation in response to different concentrations of mannitol was measured. Figure 2E shows that sos4-1 seedlings were not hypersensitive to osmotic stress caused by mannitol. Instead, sos4-1 plants displayed a more tolerant phenotype to high concentrations of mannitol (Figure 2E).
SOS4 Is Involved in Na+ and K+ Homeostasis in Plants
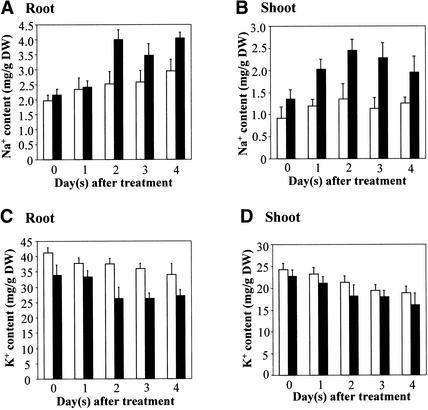
Na+ and K+ homeostasis is critical for salt tolerance. The hypersensitivity of sos4 mutants to NaCl and KCl salts but not to general osmotic stress suggests that SOS4 may be involved in the regulation of Na+ and K+ homeostasis. To investigate the role of SOS4 in controlling Na+ and K+ homeostasis in Arabidopsis, we compared Na+ and K+ accumulation in the root and shoot of sos4 mutant and wild-type plants in response to salt stress. Even without NaCl treatment, both the mutant and the wild-type plants had rather high Na contents (Figure 3). This was likely because the plants were grown in Turface soil and were watered with full-strength Hoagland solution. As shown in Figure 3A, there was no significant difference in root Na+ contents between sos4-1 and wild-type plants either without NaCl treatment or 1 day after treatment. However, after NaCl treatment for 2 days or longer, sos4-1 roots accumulated more Na+ than did wild-type roots (Figure 3A). The shoots of sos4-1 mutant plants had a slightly higher Na+ content even without salt treatment and much more increased Na+ accumulation after salt stress treatment compared with the levels in wild-type shoots (Figure 3B). sos4-1 roots had a slightly lower K+ content before or 1 day after NaCl treatment. After longer periods of NaCl treatment, the lower K+ content in sos4-1 roots became more pronounced (Figure 3C). K+ contents in sos4-1 shoots were not significantly different from wild-type levels, although the mutant levels appeared to be slightly lower (Figure 3D). The lower K+ content and higher Na+ content in the roots of sos4 mutant plants indicate a role of SOS4 in regulating K+ and Na+ transport. The increased Na+ accumulation in sos4 shoots also suggests that SOS4 may affect Na+ translocation from the root to the shoot.
Figure 3.
Na+ and K+ Contents in Wild-Type and sos4-1 Mutant Plants Treated with 100 mM NaCl.
(A) Na+ content in root.
(B) Na+ content in shoot.
(C) K+ content in root.
(D) K+ content in shoot.
DW, dry weight. Open bars, wild type; closed bars, sos4-1. Error bars represent sd (n = 6).
Molecular Cloning of the SOS4 Gene
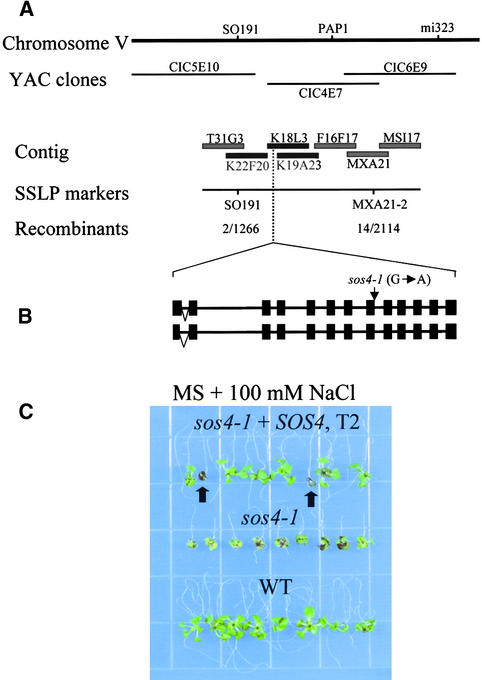
To map the sos4 mutation, a segregating F2 population was obtained from a cross between sos4-2 mutants in the Columbia background and wild-type plants in the Landsberg background. A total of 1057 sos4 mutant plants were selected from the F2 population, and DNA was extracted from each plant for genetic mapping. sos4 was mapped to chromosome V between the simple sequence length polymorphism (SSLP) markers PHYC and DFR. For fine mapping, a new SSLP marker, MXA21-2, was developed. On the basis of the analysis of 2114 recombinant chromosomes, sos4 was delimited to a region between the markers SO191 and MXA21-2 (Figure 4A). To further localize the SOS4 gene, three transformation-competent artificial chromosome clones in this region, K22F20, K18L3, and K19A23, were introduced into sos4-2 mutant plants by Agrobacterium tumefaciens– mediated transformation. Root-bending assays performed on T2 transgenic plants revealed that clone K18L3 complemented the NaCl-hypersensitive phenotype of sos4-2. In contrast, neither K22F20 nor K19A23 complemented the sos4 mutant (data not shown). These results show that SOS4 resides in the ∼8-kb region in K18L3 that is not overlapped by K19A23. Examination of the genomic DNA sequence in this region identified a predicted gene encoding a PL kinase–like protein. Genomic DNA corresponding to this predicted gene was amplified from sos4-1 mutant plants and sequenced. A single base pair mutation of G to A was found in the sos4-1 mutant allele (Figure 4B). We failed to amplify this gene from the sos4-2 mutant by polymerase chain reaction (PCR), suggesting that the gene was deleted in this mutant allele as a result of fast neutron bombardment.
Figure 4.
Positional Cloning of the SOS4 Gene.
(A) Map location of SOS4. SOS4 is located at the middle of chromosome V. Adjacent SSLP and cleaved-amplified polymorphic sequence markers are indicated. The contigs were assembled based on information in the Arabidopsis database (http://www.arabidopsis.org). The numbers of recombinant and total chromosomes tested are given for the respective loci. The transformation-competent artificial chromosome clones used for complementation tests in the region of the SOS4 locus are indicated as solid black bars.
(B) Structure of the SOS4 gene. Two types of SOS4 gene organization were found. The intron and exon organization of the SOS4 gene shown was determined by comparison of the cDNAs obtained by reverse transcriptase–mediated PCR and genomic sequences from the Arabidopsis database. The alternative splicing in the first intron is indicated by inverted triangles. The arrow indicates the location of the sos4-1 mutation. Closed boxes indicate exons, and lines between boxes indicate introns.
(C) Complementation of sos4-1 by the wild-type SOS4 gene. Five-day-old seedlings of the wild type, sos4-1, and sos4-1 transformed with the wild-type SOS4 gene (T2) grown on MS agar medium were transferred to a vertical MS agar plate containing 100 mM NaCl placed upside-down. The photograph was taken 10 days after transfer. Arrows indicate sos4 mutants that were segregated from the T2 population.
WT, wild type; YAC, yeast artificial chromosome.
To determine whether the mutated gene is responsible for the sos4 mutant phenotype, an ∼7.0-kb HindIII genomic fragment spanning the entire SOS4 gene was cloned from wild-type plants and introduced into sos4-1. Ten independent kanamycin-resistant T1 transformants were selected. Root-bending assays on T2 transgenic plants produced from these transformants show that the T2 progeny segregated for wild-type and sos4 mutant phenotypes on MS medium containing 100 mM NaCl (Figure 4C). Analysis of the transgene by PCR confirmed cosegregation of the transgene with the wild-type phenotype. These results clearly demonstrate that sos4-1 was complemented and that the salt hypersensitivity of sos4 mutants was caused by mutations in the PL kinase–like gene.
SOS4 Encodes a PL Kinase Homolog
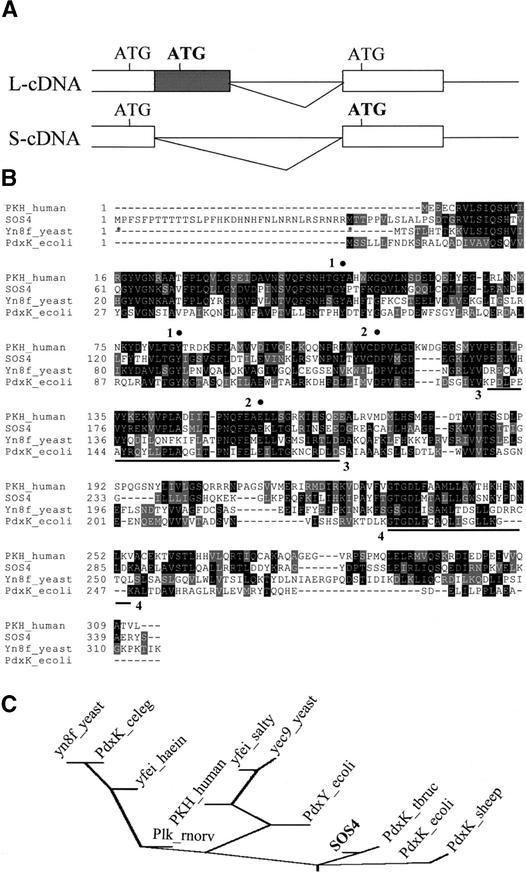
SOS4 cDNAs were cloned by reverse transcriptase–mediated PCR. Alignment of the cDNA and genomic DNA sequences revealed that the SOS4 gene consists of 13 exons and 12 introns (Figure 4B), which is different from the computer-based annotation by the Arabidopsis Genome Initiative (2000). The G-to-A substitution in the sos4-1 mutant allele disrupts the splicing acceptor site of the eighth intron (Figure 4B). This would cause a splicing defect, resulting in the inclusion of the eighth intron in the SOS4 transcript and consequently a premature stop codon that exists within the intron sequence. Interestingly, two types of SOS4 cDNAs were obtained, which reveals alternative splicing within the first intron (Figure 4B). The two cDNAs thus differ in the first exon; the long cDNA (L-cDNA) includes ∼100 bp that is spliced out in the short cDNA (S-cDNA) (Figure 5A).
Figure 5.
SOS4 Encodes a PL Kinase Homolog.
(A) Schemes of alternative splicing in the first intron. The closed box indicates the alternatively spliced region. The triangles represent the intron to be spliced out. The locations of the ATG codons are presented, and the possible ATG start codons are shown in boldface.
(B) Alignment of SOS4 with E. coli PN/PL/PM kinase PdxK, human homolog of PL kinase PKH, and yeast putative homolog Yn8fp. Amino acids identical in at least two proteins are highlighted in black, and conservative substitutions are highlighted in gray. The sequences were aligned by the program CLUSTAL W (http://dot.imgen.bcm.tmc.edu:9331/multi-align/Options/clustalw.html). Asterisks below the SOS4 sequence indicate two possible start Met residues resulting from the alternative splicing. Conserved residues and domains that might be involved in substrate binding or catalysis are marked with filled circles and underlined, respectively, and the numbering schemes for the conserved residues were described in the text.
(C) Phylogenetic analysis of SOS4 and other PN/PL/PM kinase homologs. Kinase activities have been demonstrated directly only for E. coli PdxK and PdxY, human PKH, and T. brucei PdxK. The other sequences are putative homologs identified by BLAST searches. Multiple sequence alignment was performed with CLUSTAL W. Evolutionary distances were calculated by the neighbor-joining method, and the phylogenetic tree was drawn by the program DRAWGRAM (http://bioweb.pasteur.fr/seqanal/phylogeny/phylip-uk.html). celeg, Caenorhabditis elegans; haein, Haemophilus influenzae; rnorv, Rattus norvegicus; salty, Salmonella typhimurium.
The two cDNAs correspond to the two transcripts that are detected in RNA gel blot analysis (see below). The L-cDNA contains three ATGs, whereas the S-cDNA contains two ATGs (Figure 5A). The first ATG in both cDNAs is not predicted to be a start codon because there would be an in-frame stop codon. The second and third ATGs are in frame in L-cDNA, and both of them could serve as translation initiation codons. Because the second ATG present in L-cDNA is spliced out in S-cDNA, the second ATG in S-cDNA (i.e., the third ATG in L-cDNA) is predicted to be the start codon. Therefore, it is possible that the two transcripts could be translated into two proteins; the larger protein would include an extra 34 amino acid residues at the N terminus compared with the smaller protein (Figure 5B). The larger protein is predicted to contain 343 amino acid residues with a deduced molecular mass of 38.2 kD and a theoretical pI of 6.56. The smaller protein is predicted to be a 34.0-kD protein with 309 amino acid residues and a theoretical pI of 5.55. Hydropathy analysis and secondary structure predictions show that both proteins are hydrophilic and predicted to reside in the cytoplasm.
The deduced amino acid sequence of SOS4 shows substantial similarity to those of human, yeast, and bacteria PL kinases (Figure 5B). Over a stretch of 304 amino acid residues, SOS4 has 46% identity and 66% similarity to human PL kinase (Hanna et al., 1997) and 45% identity and 65% similarity to sheep PL kinase (Maras et al., 1999). Sequence alignment among the human PL kinase, the Escherichia coli PN/PL/PM kinase PdxK, and a putative PL/PN kinase from Saccharomyces cerevisiae reveals conserved motifs that may be involved in substrate binding or catalysis (Figure 5B). With respect to ATP binding, PL kinase has been shown to contain domains common to many kinases. A motif search revealed that a region in SOS4 (region 4) is similar to the signature motif found in the PfkB superfamily of carbohydrate kinases (Wu et al., 1991), which may be involved in ATP binding. The ATP binding domain was suggested to be composed of the consensus sequence GXGXXAX13-26K, which is found in many kinases with the Lys at the 24th residue downstream from the consensus Ala (Tsay and Robinson, 1991). In PL kinases, a similar motif exists, and it was identified experimentally as the ATP binding domain in the sheep brain PL kinase using the bisubstrate analog adenosine tetraphosphate as an affinity label (Dominici et al., 1988).
The sequence from residue 265 to 287 of SOS4 (region 4), GXGXXXXAX14K, shows a very similar arrangement. Candidate Asp and Glu residues (Figure 5B, region 2), which may act as general bases in phosphate transfer (Scott and Phillips, 1997), are conserved in the sequences analyzed. Two conserved Tyr residues (Figure 5B, region 1) were reported to be important for PL binding because one of them cannot be modified after PL binding (Scholz and Kwok, 1989). A motif search (at http://www.motif.genome.ad.jp) also revealed a putative PLP binding domain in SOS4 (residues 170 to 207) (Figure 5B, region 3), which probably is important for the formation of the reaction product. Phylogenetic analysis indicates that SOS4 is related most closely to PdxK kinases from Trypanosoma brucei and E. coli (Figure 5C).
SOS4 Complements an E. coli Mutant Defective in PdxK
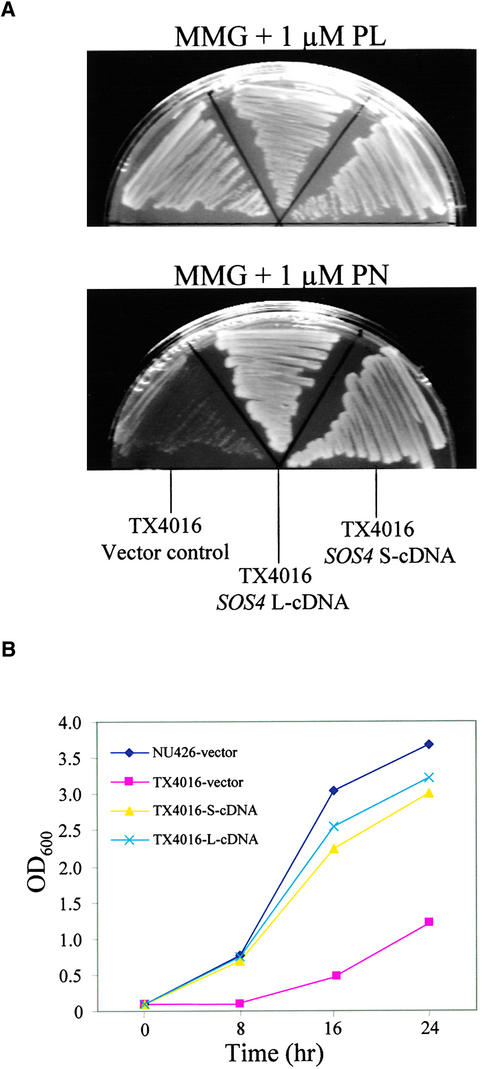
In E. coli, PdxK encodes a PN/PL/PM kinase that functions in the salvage pathway for the biosynthesis of the coenzyme PLP by using PL, PN, and PM taken up from the growth medium (Yang et al., 1996, 1998). The de novo pathway for PLP synthesis in E. coli is thought to form 4-phosphohydroxy-l-threonine from erythrose 4-phosphate by a series of reactions, one of which is catalyzed by the PdxB dehydrogenase (Schoenlein et al., 1989; Lam and Winkler, 1990). The E. coli mutant strain TX4016 (pdxB pdxK), which is defective in PLP biosynthesis through both the de novo and salvage pathways, grows poorly in MMG minimal medium (see Methods) without supplements or supplemented with 1 μM PN, but it grows well in MMG medium supplemented with 1 μM PL because of the function of PdxY, a PL-specific PL kinase (Yang et al., 1996, 1998).
Because SOS4 is phylogenetically more closely related to PdxK than to other PL kinases, we determined whether SOS4 could rescue the mutant phenotype of TX4016. Both L-cDNA and S-cDNA were cloned into an E. coli expression vector and introduced into the mutant strain. Figure 6A shows that both L-cDNA and S-cDNA of SOS4 restored the growth of TX4016 on MMG solid medium containing 1 μM PN to a level similar to that on MMG medium supplemented with 1 μM PL. In MMG liquid medium containing 1 μM PN, the growth of TX4016 was much poorer than that of wild-type E. coli strain NU426 (Figure 6B). In contrast, the growth of TX4016 mutant cells expressing SOS4 L-cDNA or S-cDNA was restored to a level similar to that of wild-type E. coli cells (Figure 6B). These results suggest that SOS4 is functionally homologous with E. coli PdxK.
Figure 6.
SOS4 Complements the PdxK-Defective E. coli Mutant.
(A) Complementation of TX4016 (pdxB pdxK) cells by L-cDNA and S-cDNA of SOS4. E. coli TX4016 was transformed with plasmids containing L-cDNA or S-cDNA or with the empty vector. Transformants were grown on medium supplemented with 1 μM PL or PN.
(B) Growth curves of TX4016 transformants containing SOS4 L-cDNA (blue crosses), SOS4 S-cDNA (yellow triangles), or vector only (red squares) and wild-type strain NU426 transformed with vector only (black diamonds).
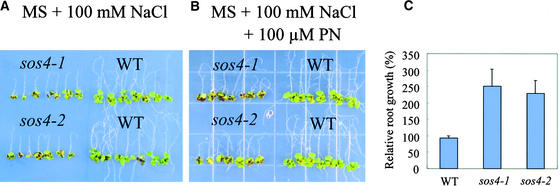
PN Improves the Salt Tolerance of sos4
To determine whether different forms of vitamin B6 could improve the salt tolerance of sos4 mutant plants, a root-bending assay was performed on 100 mM NaCl medium supplemented with 100 μM PN, PL, or PLP. Seeds were germinated first on MS agar medium, and the seedlings then were transferred to different media for root-bending tests. Seven days after being transferred to NaCl medium, sos4 seedlings had very little new root growth without the vitamin supplements (Figure 7A). However, with 100 μM PN supplement, sos4 seedlings exhibited significant new root growth on the NaCl medium, although the supplement did not restore mutant growth to the wild-type level (Figure 7B). Quantitative measurements confirmed that the supplemented PN had very little effect on the root growth of wild-type seedlings but significantly improved the root growth of both sos4-1 and sos4-2 grown on MS medium with 100 mM NaCl (Figure 7C). This finding suggests that PN specifically rescued the sos4 phenotype rather than played a general role in the promotion of root growth under salt stress. Not surprisingly, PLP failed to significantly rescue the root growth of sos4 under salt stress, because PLP is known to be incapable of passing through the cell membrane (Lam et al., 1992). Importantly, PL did not significantly improve the root growth of sos4 under NaCl stress, suggesting that SOS4 is a PL kinase.
Figure 7.
PN Improves the Tolerance of sos4 to NaCl.
(A) Root-bending assay of sos4-1, sos4-2, and wild-type plants on MS agar medium supplemented with 100 mM NaCl.
(B) Root-bending assay of sos4-1, sos4-2, and wild-type plants on MS agar medium supplemented with 100 mM NaCl and 100 μM PN.
(C) Quantitative measurement of relative root growth. Four-day-old seedlings were transferred to MS agar medium supplemented with 100 mM NaCl or MS agar medium supplemented with 100 mM NaCl and 100 μM PN and cultured for 7 days. Relative root growth is presented as a percentage of root elongation on MS medium supplemented with 100 mM NaCl and 100 μM PN relative to that on MS medium supplemented with 100 mM NaCl only. Error bars represent standard deviation (n = 16).
In (A) and (B), photographs were taken on day 7 after the seedlings were transferred to the test media. WT, wild type.
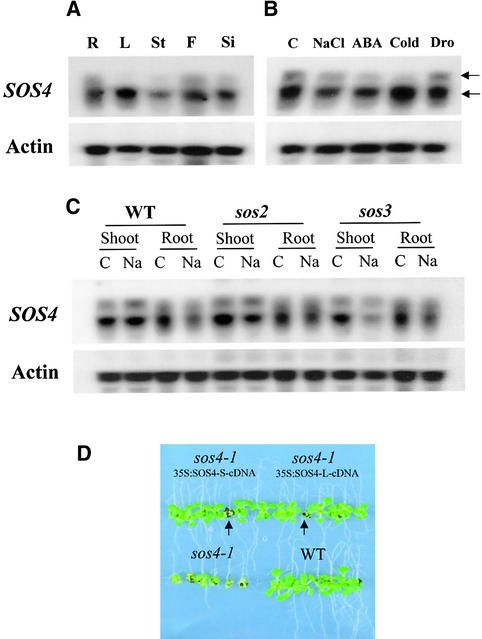
SOS4 Expression Is Regulated by Stress
RNA gel blot analysis was performed to investigate the regulation and organ-specific expression of SOS4. Two transcripts were detected, and the short transcript (∼1.3 kb) was found to be more abundant than the long transcript (∼1.4 kb) under all treatment conditions and in all organs tested (Figure 8). The short transcript was detected in all organs, including root, leaf, stem, flower, and silique, with the highest abundance in leaf (Figure 8A). The long transcript was detected in root, flower, and silique but very weakly in stem; it was not detectable in leaf (Figure 8A). As shown in Figure 8B, SOS4 expression was modulated by NaCl, abscisic acid, and cold treatment but not by drought. Both the long and short SOS4 transcripts were downregulated by NaCl and abscisic acid treatment (Figure 8B). This downregulation by NaCl treatment occurred in root but not in shoot (Figure 8C). Under cold stress, the short transcript was increased significantly but the long transcript became virtually undetectable (Figure 8B).
Figure 8.
Expression of the SOS4 Gene.
Two transcripts (indicated by arrows) were detected by RNA gel blot analysis. Actin is shown as a loading control.
(A) Expression of SOS4 in different plant parts. R, root; L, leaf; St, stem; F, flower; Si, silique.
(B) SOS4 expression under different stress conditions. C, control (MS salt only); NaCl, 300 mM NaCl for 5 hr; ABA, 100 μM abscisic acid for 3 hr; Cold, 0°C for 24 hr; Dro, dehydration for 30 min.
(C) Comparison of SOS4 expression in roots or shoots of the wild type (WT), sos2-1 mutant, and sos3-1 mutant. C, control (MS salt only); Na, 300 mM NaCl for 5 hr.
(D) Overexpression of either SOS4 L-cDNA or SOS4 S-cDNA complements the salt-hypersensitive phenotype of sos4-1. Five-day-old seedlings of wild-type (WT), sos4-1, and sos4-1 transgenic plants (T2) harboring SOS4 L-cDNA or SOS4 S-cDNA grown on MS agar medium were transferred to a vertical MS agar plate containing 100 mM NaCl. The photograph was taken 10 days after transfer. Arrows indicate sos4-1 mutants that were segregated from the T2 transgenic population.
To determine whether SOS4 downregulation by NaCl was affected by the SOS2 or the SOS3 gene, SOS4 expression was determined in sos2 and sos3 mutants. Although in general there was no significant difference between SOS4 expression in the wild type and the sos2 or sos3 mutant, there appeared to be downregulation by NaCl of the short transcript in the shoot of mutants (Figure 8C). The roots and leaves used in Figure 8A were collected from 3-week-old plants, but the shoots and roots of the wild type and mutants used in Figure 8C were collected from 10-day-old seedlings. The leaves from young seedlings (Figures 8B and 8C) had significant levels of the long transcript, but the leaves from adult plants (Figure 8A) did not express the long transcript. Together, these results show that there are two SOS4 transcripts as a result of alternative splicing and that they are regulated differentially by stress and development.
To determine whether both of these transcripts are functional in Arabidopsis, sos4 mutant complementation by the two types of cDNAs was tested. As shown in Figure 8D, each one of the cDNAs, when overexpressed, rescued the salt-hypersensitive phenotype of sos4-1. This result indicates that both transcripts can be translated into a functional protein in Arabidopsis. Nevertheless, the two transcripts could have subtle functional differences, such as different translation efficiencies, that might be important in controlling PLP homeostasis during plant development and in response to stress.
Tissue Expression Pattern of SOS4
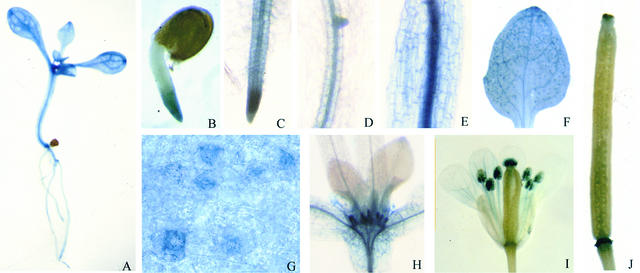
A promoter–β-glucuronidase (GUS) fusion analysis was performed to investigate the potential developmental and tissue-specific expression of SOS4. GUS staining was detected throughout the transgenic plants harboring the SOS4 promoter–GUS fusion construct (Figure 9), suggesting that SOS4 is expressed ubiquitously. Right after seed germination, GUS expression was found in the emerging radicle, and it was particularly strong at the tip (Figure 9B). In young seedlings, GUS staining was detected in root, hypocotyl, and leaf, with stronger staining in vascular tissues (Figures 9C to 9F). Interestingly, high expression of GUS was detected in guard cells when visualized under high magnification (Figure 9G), which appeared as dark blue spots of GUS staining in the leaf (Figure 9F). Strong expression also was observed in stipules (Figure 9H). In flowers, strong expression was found in pollen grains within the anthers and in the stigma (Figure 9I). GUS expression also was detected in siliques, with strong staining at the tip and base (Figure 9J). These expression patterns indicate a ubiquitous function of SOS4 in Arabidopsis, consistent with the role of PLP as an essential cofactor for numerous enzymes in cellular metabolism and as a regulator of ion transport.
Figure 9.
Localization of SOS4 Promoter–GUS Activity in Arabidopsis Transgenic Plants.
(A) Ten-day-old seedling.
(B) Radicle at 2 days after germination.
(C) Root tip.
(D) Primary root.
(E) Hypocotyl.
(F) Cotyledon.
(G) High expression of GUS in guard cells.
(H) Strong GUS staining in stipules.
(I) Flower.
(J) Silique.
Samples in (A) to (H) were from young seedlings.
DISCUSSION
Plant salt tolerance is a complex trait involving many genes. Even though the entire genome sequence of Arabidopsis is known, the functional identification of salt tolerance determinants in this model plant remains a formidable challenge. Forward genetic screens based on plant phenotypes are very powerful because the approach does not depend on previous knowledge and therefore has the potential to reveal unexpected salt tolerance determinants and unsuspected connections.
The SOS4 gene was discovered through a genetic screen for mutants with reduced salt tolerance. sos4 mutant plants are hypersensitive to Na+, Li+, and K+ ions. Upon salt stress, sos4 mutant plants accumulate more Na+ and retain less K+ than do wild-type plants. These results suggest that SOS4 is important for Na+ and K+ homeostasis in plants. Map-based cloning revealed that SOS4 encodes a putative PL kinase. Functional complementation of the E. coli mutant pdxB pdxK demonstrated that SOS4 is in fact a PdxK homolog. Although a connection between PL kinase and salt tolerance has not been suspected previously, our results show that this enzyme is an important salt tolerance determinant in plants.
In E. coli and human cells, PL kinase has been shown to catalyze the biosynthesis of PLP (Yang et al., 1996; Hanna et al., 1997). PLP and its derivatives are known to be antagonists of ATP-gated P2X receptor ion channels in animals (Ralevic and Burnstock, 1998). The antagonistic role indicates that PLP may bind to the P2X receptor and block the ATP binding site in the ion channel. In plants, a number of studies have shown the importance of the nucleotide ATP in the function of K+ channels (Spalding and Goldsmith, 1993; Wu and Assmann, 1995). The presence of a putative cyclic nucleotide binding site in K+ channels also has been reported (Sentenac et al., 1992; Daram et al., 1997). Although the function of the putative cyclic nucleotide binding site in the regulation of channel activity has not been determined, presumably it is important for KAT1 activity because deletion of this region abolished the function of KAT1 (Marten and Hoshi, 1997). However, neither PLP binding nor the effect of PLP on plant ion channels has been investigated. It is possible that the KCl and NaCl hypersensitivity of sos4 mutant plants is caused by the PLP regulation of K+ and Na+ channels or transporters. Related to the potential function of SOS4 in regulating ion channel activities is the observation that the expression of SOS4 promoter–GUS was higher in guard cells than in mesophyll cells.
PLP is an essential cofactor for numerous cellular enzymes (Schneider et al., 2000). PLP-dependent enzymes play a major role in the metabolism of amino acids and are found in various pathways, ranging from the interconversion of amino acids to the biosynthesis of antibiotic compounds (Schneider et al., 2000). One probably universal response of plants to osmotic stress is the accumulation of osmolytes such as Pro, Gly betaine, and ectoine (McCue and Hanson, 1990). Pathways leading to osmolyte synthesis are connected to those in basic metabolism in which numerous enzymatic reactions are involved (Bohnert and Jensen, 1996). There is no doubt that some of the enzymes are PLP dependent. However, no obvious changes in Pro accumulation in control or NaCl-treated seedlings were observed in sos4 compared with the wild type (data not shown), possibly as a result of pathway redundancy for both the synthesis of PLP (see below) and the synthesis of these amino acids. Interestingly, although sos4 plants are more sensitive to NaCl stress, they exhibited obviously enhanced tolerance to high concentrations of mannitol, as measured by root growth. Mannitol as an impermeable osmolyte was used to impose osmotic stress. Among several possibilities, it seems likely that this enhanced tolerance to high concentrations of mannitol in sos4 may relate to a potentially reduced production of ethylene in sos4 roots under osmotic stress conditions. It is known that PLP is a cofactor required by 1-aminocyclopropane-1-carboxylic acid synthase, a key enzyme in the ethylene biosynthesis pathway (Capitani et al., 1999). A previous study has indicated that ethylene production under drought stress inhibits the elongation of primary roots (Spollen et al., 2000).
Because PLP is essential for basic cellular metabolism, the complete disruption of PLP biosynthesis in plants is expected to be lethal. However, null mutations in the SOS4 gene are not lethal, suggesting that SOS4 is not the only gene responsible for PLP biosynthesis in Arabidopsis. Our feeding experiments showed that PL could not rescue the salt tolerance defect of sos4, which is consistent with SOS4 being a PL kinase. In contrast, PN significantly rescued the sos4 mutant phenotype, suggesting that there is a PN kinase in Arabidopsis that converts PN to PNP, because PNP can be converted further to PLP through the action of PN oxidase. It was reported that the hydroxymethylpyrimidine kinase of E. coli involved in thiamine biosynthesis has kinase activities for PL, PM, and PN (Mizote and Nakayama, 1989). Although one gene in Arabidopsis encoding a putative hydroxymethylpyrimidine kinase shows only 29% identity and 49% similarity to SOS4 over a stretch of 107 amino acids, it is possible that this protein or even other proteins in Arabidopsis function as PN kinase in the salvage pathway of PLP biosynthesis. Therefore, PLP biosynthesis in sos4 probably is disrupted incompletely. It is possible that PLP production in sos4 mutants is unbalanced, resulting in the inappropriate regulation of PLP-dependent enzymes or ion transporters. PLP amounts in plant cells presumably are controlled tightly by the regulation of the expression of PLP biosynthetic enzymes. In the case of SOS4, its expression is regulated by alternative splicing. Although both cDNAs from the alternative splicing could complement the E. coli mutant and the sos4 mutant when expressed ectopically, they may have different levels of translation efficiency. The alternative splicing is modulated by development and various environmental stresses, suggesting that it is potentially important for PLP homeostasis.
METHODS
Isolation of Mutants and Genetic Analysis
Arabidopsis thaliana ecotype Columbia carrying the homozygous recessive glabrous (gl1) mutation (Koornneef et al., 1982) was used as the parental strain for mutant isolation. Ethyl methanesulfonate– or fast neutron–mutagenized M2 seed were surface-sterilized in a solution of Clorox plus 0.01% Triton X-100 for 10 min, washed with sterilized water three times, and suspended in sterile 0.3% low-melting-point agarose. The seed were planted in rows onto agar medium containing Murashige and Skoog (1962) (MS) salts with 3% Suc and 1.2% agar, pH 5.7. The plates were stored at 4°C for 48 hr to synchronize germination and then incubated at 22°C under continuous illumination. Plates were placed in a vertical position to allow roots to grow along the agar surface toward gravity. Four-day-old seedlings were transferred onto a second medium that was supplemented with 100 mM NaCl, and putative mutants were identified using the root-bending assay of Wu et al. (1996). Putative mutant seedlings were picked up 1 week later and transferred onto a 0.6% agar medium without NaCl. When appropriate, seedlings were transplanted to pots and grown to maturity. Growth conditions were as described (Wu et al., 1996).
Mutants were backcrossed to the wild type for at least three generations to reduce other mutations from the background. The mutants also were crossed to each other and to sos1-1, sos2-1, and sos3-1 for allelic tests. The salt sensitivity of F1 and F2 seedlings arising from the crosses was determined by the root-bending assay (Wu et al., 1996).
Growth Measurement and Ion Content Determination
Four-day-old wild-type and mutant seedlings grown on vertical MS agar plates were transferred to various agar media for stress treatment and growth measurements as described (Wu et al., 1996). For feeding tests, the seedlings were transferred to vertical MS agar plates containing 100 mM NaCl or 100 mM NaCl plus 100 μM pyridoxine, 100 μM pyridoxal, or 100 μM pyridoxal-5-phosphate for the root-bending assay (Wu et al., 1996). For ion content measurement, plants were grown for 4 weeks in Turface soil (Profile Products LLC, Buffalo Grove, IL) and fertilized with 1 × Hoagland solution (Hoagland and Arnon, 1938). Salt treatments were performed by immersing the pots in one-tenth-strength MS salts plus 100 mM NaCl solution for the number of days indicated in Figure 3. Shoots and roots were harvested separately for ion content measurement. Materials were collected and dried at 80°C for at least 2 days and weighed. The samples were digested with HNO3, and the Na+ and K+ concentrations were assayed by atomic emission spectrophotometry (model 3100; Perkin-Elmer, Norwalk, CT).
Genetic Mapping
The sos4-2 mutant in the Columbia background was crossed to wild-type Landsberg erecta. The F2 population from selfed F1 individuals was screened for sos4 mutants by the root-bending assay (Wu et al., 1996). A total of 1076 homozygous sos4 seedlings were selected and used for mapping with simple sequence length polymorphism (SSLP) markers (Bell and Ecker, 1994). The primer sequences of the SSLP markers nga76, PHYC, SO191, and DFR on chromosome V are available in TAIR (http://www.arabidopsis.org). For fine mapping, the SSLP marker MXA21-2 was developed based on the genomic sequence of the P1 clone MXA21 on chromosome V. The primer pairs for MXA21-2 are as follows: forward primer, 5′-GAAGAAAAA-ATAATATTAGAGTC-3′; reverse primer, 5′-TCCCGTCCCGAGTTATGA-CC-3′.
Nucleotide Analysis
To identify the SOS4 locus, oligonucleotide primers were designed to allow polymerase chain reaction (PCR) fragments to cover the predicated genes with sufficient overlaps. The fragments were amplified from both the wild type and sos4 mutants using genomic DNA prepared from respective strains. The resulting PCR products were sequenced from both strands. DNA sequences of the wild type and mutants were compared with identity mutations and putative mutations verified by independent PCR amplifications.
Complementation Tests
The transformation-competent artificial chromosome (TAC) clones K22F20, K18L3, and K19A23 were obtained from ABRC (Columbus, OH). TAC plasmids were introduced into Agrobacterium tumefaciens strain GV 3101 and transferred into sos4-2 mutant plants using the vacuum infiltration method (Bechtold et al., 1993). An ∼7.0-kb genomic DNA fragment containing the SOS4 promoter, coding region, and 3′ untranslated region obtained from HindIII partial digestion of TAC clone K18L3 was subcloned into the HindIII site of binary vector pBIN19. The construct was used to transform sos4-1 mutant plants as described above. T1 transgenic plants were selected on MS medium containing 40 mg/L kanamycin and transferred to soil to grow to maturity. The transgenic plants were confirmed further by PCR amplification using the specific primer pairs against the SOS4 gene and the vector. For complementation tests, 10 T2 transgenic lines were subjected to the root-bending assay (Wu et al., 1996).
cDNA Isolation and Overexpression
cDNA containing the complete SOS4 open reading frame was amplified by reverse transcriptase–mediated PCR using RNA isolated from Columbia wild-type plants as a template. The SOS4-specific primer pair containing XbaI and SacI sites at the termini are 5′-CTC-ATGGGTCAAACAGAAGC-3′ and 5′-TCACCTGCTTCAGCTGTATC-3′. The PCR product was cloned into pCR-Blunt II-TOPO vector (Invitrogen, Carlsbad, CA) and sequenced. Full-length cDNA was subcloned into the XbaI and SacI sites of pIG 121-Hm to obtain SOS4 over-expression constructs under the control of the 35S promoter of Cauliflower mosaic virus. The constructs were introduced into A. tumefaciens, which was used to transform Columbia wild-type and sos4-1 mutant plants as described above. The characterization of T2 transgenic plants harboring overexpression constructs was performed using the root-bending assay (Wu et al., 1996).
Escherichia coli Complementation
SOS4 cDNAs were ligated into the multicloning sites in pJF118 (Fürste et al., 1986). The resulting plasmids were introduced into E. coli mutant strain Tx4016 (pdxB pdxK, derived from wild-type strain NU426) (Yang et al., 1998). For growth tests, the plasmid-containing E. coli TX4016 grown on Luria-Bertani agar medium was streaked onto freshly prepared MMG solid medium (0.81 mM MgSO4, 9.51 mM citric acid, 57.41 mM K2HPO4, 16.74 mM NaNH5PO4, 0.4% [w/v] glucose) supplemented with 1 mM isopropylthio-β-galactoside and 1 μM pyridoxal or 1 μM pyridoxine (Yang et al., 1998). The plate was incubated at 37°C for 36 hr before being photographed. The E. coli was inoculated into the same liquid medium to obtain growth curves by measuring OD600 at the times indicated in Figure 6B.
RNA Gel Blot Analysis
Arabidopsis seedlings were grown on MS agar medium under continuous light (Wu et al., 1996), and 10-day-old seedlings were treated with NaCl, abscisic acid, and low temperature as described previously (Shi et al., 2000). For the drought treatment, 10-day-old seedlings were transferred from MS agar medium to filter paper and kept in a flow hood for 30 min. Determination of gene expression in roots and shoots was performed as described previously (Shi et al., 2000). For the collection of plant parts, wild-type plants were grown in Turface soil to facilitate root harvesting. Roots and leaves were collected from 3-week-old-seedlings, and stems, flowers, and siliques were collected after plants flowered. RNA isolation and RNA gel blot analysis were performed according to Zhu et al. (1998).
Promoter–β-Glucuronidase Analysis
An ∼1.9-kb promoter region of the SOS4 gene was amplified by PCR from genomic DNA with the following primer pair introducing a HindIII site at the 5′ end and a BamHI site at the 3′ end to facilitate cloning: 5′-GTGTGAAGCTTTGATATTCTCTGAG-3′ and 5′-GAGACT-TTTTAACTAAAGCTCACTG-3′. The fragment was cloned into HindIII-BamHI sites of pCAMBIA1391Z to obtain a transcriptional fusion of the SOS4 promoter and the β-glucuronidase coding sequence. Transgenic plants harboring this construct were generated as described above. For β-glucuronidase assay, materials were stained at 37°C overnight in 1 mg/mL 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 0.03% Triton X-100, and 0.1 M sodium phosphate buffer, pH 7.0.
Accession Numbers
The sequence accession numbers for SOS4 and the homologs shown in Figure 5C are as follows: Arabidopsis SOS4, GenBank AF400125; Sheep PdxK (PdxK_sheep), SP P82197; E. coli PdxK (PdxK_ecoli), GenBank U53700; Trypanosoma brucei PdxK (PdxK_tbruc), GenBank U96712; E. coli PdxY (PdxY_ecoli), DDBJ D90807; human PKH (PKH_human), GenBank U89606; Saccharomyces cerevisiae Yec9p (yec9_yeast), SW P39988; Salmonella typhimurium Yfei (yfei_salty), SW P40192; Rattus norvegicus Plk (Plk_rnorv), GenBank AF020346; Haemophilus influenzae Yfei (yfei_haein), SW P44690; Caenorhabditis elegans PdxK (PdxK_celeg), GenBank AF003142; and S. cerevisiae Yn8fp (yn8f_yeast), SW P53727.
Acknowledgments
We thank Dr. Malcolm E. Winkler for kindly providing the E. coli strains NU426 and TX4016. This work was supported by National Institutes of Health Grant No. R01GM59138 to J.-K.Z.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010417.
References
- Amtmann, A., and Sanders, D. (1999). Mechanisms of Na+ uptake by plant cell. Adv. Bot. Res. 29, 75–112. [Google Scholar]
- Apse, M.P., Aharon, G.S., Snedden, W.A., and Blumwald, E. (1999). Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiporter in Arabidopsis. Science 285, 1256–1258. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Barkla, B.J., and Pantoja, O. (1996). Physiology of ion transport across the tonoplast of higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 127–157. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316, 1194–1199. [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Binzel, M.L., Hess, F.D., Bressan, R.A., and Hasegawa, P.M. (1988). Intracellular compartmentation of ions in salt adapted tobacco cells. Plant Physiol. 86, 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatt, M.R., and Thiel, G. (1993). Hormonal control of ion channel gating. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44, 543–567. [Google Scholar]
- Blumwald, E., and Poole, R.J. (1985). Na+/H+ antiport in isolated tonoplast vesicles from storage tissue of Beta vulgaris. Plant Physiol. 78, 163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumwald, E., Aharon, G.S., and Apse, M.P. (2000). Sodium transport in plant cells. Biochim. Biophys. Acta 1465, 140–151. [DOI] [PubMed] [Google Scholar]
- Bohnert, H.J., and Jensen, R.G. (1996). Strategies for engineering water-stress tolerance in plants. Trends Biotechnol. 14, 89–97. [Google Scholar]
- Brake, A.J., and Julius, D. (1996). Signaling by extracellular nucleotides. Annu. Rev. Cell Dev. Biol. 12, 519–541. [DOI] [PubMed] [Google Scholar]
- Burnstock, G. (1996). Development and perspectives of the purinceptor concept. J. Auton. Pharmacol. 16, 295–302. [DOI] [PubMed] [Google Scholar]
- Capitani, G., Hohenester, E., Feng, L., Storici, P., Kirsch, J.F., and Jansonius, J.N. (1999). Structure of 1-aminocyclopropane-1-carboxylate synthase, a key enzyme in the biosynthesis of the plant hormone ethylene. J. Mol. Biol. 294, 745–756. [DOI] [PubMed] [Google Scholar]
- Daram, P., Urbach, S., Gaymard, F., Sentenac, H., and Cherel, I. (1997). Tetramerization of the AKT1 plant potassium channel involves its C-terminal cytoplasmic domain. EMBO J. 16, 3455–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport, R.J., and Tester, M. (2000). A weakly voltage-dependent, nonselective cation channel mediates toxic sodium influx in wheat. Plant Physiol. 122, 823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominici, P., Scholz, G., Kwok, F., and Churchich, J.E. (1988). Affinity labeling of pyridoxal kinase with adenosine polyphosphopyridoxal. J. Biol. Chem. 263, 14712–14716. [PubMed] [Google Scholar]
- Frommer, W.B., Ludewig, U., and Rentsch, D. (1999). Taking transgenic plants with a pinch of salt. Science 285, 1222–1223. [DOI] [PubMed] [Google Scholar]
- Fürste, J.P., Pansegrau, W., Frank, R., Blocker, H., Scholz, P., Bagdasarian, M., and Lanka, E. (1986). Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48, 119–131. [DOI] [PubMed] [Google Scholar]
- Gaxiola, R.A., Rao, R., Sherman, A., Grisafi, P., Alper, S.L., and Fink, G.R. (1999). The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc. Natl. Acad. Sci. USA 96, 1480–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaymard, F., Cerutti, M., Horeau, C., Lemaillet, G., Urbach, S., Ravallec, M., Devauchelle, G., Sentenac, H., and Thibaud, J.B. (1996). The baculovirus/insect cell system as an alternative to Xenopus oocytes: First characterization of the AKT1 K+ channel from Arabidopsis thaliana. J. Biol. Chem. 271, 22863–22870. [DOI] [PubMed] [Google Scholar]
- Halfter, U., Ishitani, M., and Zhu, J.-K. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 97, 3735–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna, M.C., Turnert, A.J., and Kirkness, E.F. (1997). Human pyridoxal kinase: cDNA cloning, expression, and modulation by ligands of the benzodiazepine receptor. J. Biol. Chem. 272, 10756–10760. [DOI] [PubMed] [Google Scholar]
- Hasegawa, P.M., Bressan, R.A., and Pardo, J.M. (2000). The dawn of plant salt tolerance genetics. Trends Plant Sci. 5, 317–319. [DOI] [PubMed] [Google Scholar]
- Hirsch, R.E., Lewis, B.D., Spalding, E.P., and Sussman, M.R. (1998). A role for the AKT1 potassium channel in plant nutrition. Science 280, 918–921. [DOI] [PubMed] [Google Scholar]
- Hoagland, D.R., and Arnon, D.I. (1938). The water culture method for growing plants without soil. Calif. Agr. Expt. Sta. Circular 347, 39. [Google Scholar]
- Ishitani, M., Liu, J., Halfter, U., Kim, C.S., Shi, W., and Zhu, J.K. (2000). SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12, 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Dellaert, L.W.M., and van der Veen, J.H. (1982). EMS- and radiation-induced mutation frequencies at individual loci in Arabidopsis thaliana (L.) Heynh. Mutat. Res. 93, 109–123. [DOI] [PubMed] [Google Scholar]
- Lam, H.M., and Winkler, M.E. (1990). Metabolic relationships between pyridoxine (vitamin B6) and serine biosynthesis in Escherichia coli K-12. J. Bacteriol. 172, 6518–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, H.M., Tancula, E., Dempsey, W.B., and Winkler, M.E. (1992). Suppression of insertions in the complex pdxJ operon of Escherichia coli K-12 by lon and other mutations. J. Bacteriol. 174, 1554–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., and Zhu, J.-K. (1997). An Arabidopsis mutant that requires increased calcium for potassium nutrition and salt tolerance. Proc. Natl. Acad. Sci. USA 94, 14960–14964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., and Zhu, J.-K. (1998). A calcium sensor homolog required for plant salt tolerance. Science 280, 1943–1945. [DOI] [PubMed] [Google Scholar]
- Liu, J., Ishitani, M., Halfter, U., Kim, C.-S., and Zhu, J.-K. (2000). The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA 97, 3730–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maras, B., Valiante, S., Orru, S., Simmaco, M., Barra, D., and Churchich, J.E. (1999). Structure of pyridoxal kinase from sheep brain and role of the tryptophanyl residues. J. Protein Chem. 18, 259–268. [DOI] [PubMed] [Google Scholar]
- Marten, I., and Hoshi, T. (1997). Voltage-dependent gating characteristics of the K+ channel KAT1 depend on the N and C termini. Proc. Natl. Acad. Sci. USA 94, 3448–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCue, K.F., and Hanson, A.D. (1990). Drought and salt tolerance: Towards understanding and application. Biotechnology 8, 358–362. [Google Scholar]
- Mizote, T., and Nakayama, H. (1989). Purification and properties of hydroxymethylpyrimidine kinase from Escherichia coli. Biochim. Biophys. Acta 991, 109–113. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Niu, X., Bressan, R.A., Hasegawa, P.M., and Pardo, J.M. (1995). Ion homeostasis in NaCl stress environments. Plant Physiol. 109, 735–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberger, T., Wirtz, W., Nennstiel, D., Hahlbrock, K., Jabs, T., Zimmermann, S., and Scheel, D. (1997). Signal perception and intracellular signal transduction in plant pathogen defense. J. Recept. Signal Transduct. Res. 17, 127–136. [DOI] [PubMed] [Google Scholar]
- Quintero, F.J., Blatt, M.R., and Pardo, J.M. (2000). Functional conservation between yeast and plant endosomal Na+/H+ antiporters. FEBS Lett. 471, 224–228. [DOI] [PubMed] [Google Scholar]
- Ralevic, V., and Burnstock, G. (1998). Receptors for purines and pyrimidines. Pharmacol. Rev. 50, 413–492. [PubMed] [Google Scholar]
- Rodríguez-Navarro, A. (2000). Potassium transport in fungi and plants. Biochim. Biophys. Acta 1469, 1–30. [DOI] [PubMed] [Google Scholar]
- Rubio, F., Gassmann, W., and Schroeder, J.I. (1995). Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270, 1660–1663. [DOI] [PubMed] [Google Scholar]
- Schachtman, D.P., and Schroeder, J.I. (1994). Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature 370, 655–658. [DOI] [PubMed] [Google Scholar]
- Schachtman, D.P., Kumar, R., Schroeder, J.I., and Marsh, E.L. (1997). Molecular and functional characterization of a novel low-affinity cation transporter (LCT1) in higher plants. Proc. Natl. Acad. Sci. USA 94, 11079–11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, G., Käck, H., and Lindqvist, Y. (2000). The manifold of vitamin B6 dependent enzymes. Structure 8, R1–R6. [DOI] [PubMed] [Google Scholar]
- Schoenlein, P.V., Roa, B.B., and Winkler, M.E. (1989). Divergent transcription of pdxB and homology between the pdxB and sera gene products in Escherichia coli K-12. J. Bacteriol. 171, 6084–6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz, G., and Kwok, F. (1989). Brain pyridoxal kinase: Photoaffinity labeling of the substrate-binding site. J. Biol. Chem. 264, 4318–4321. [PubMed] [Google Scholar]
- Schroeder, J.I., and Hedrich, R. (1989). Involvement of ion channels and active transport in osmoregulation and signaling of higher plant cells. Trends Biochem. Sci. 14, 187–192. [DOI] [PubMed] [Google Scholar]
- Scott, T.C., and Phillips, M.A. (1997). Characterization of Trypanosoma brucei pyridoxal kinase: Purification, gene isolation and expression in Escherichia coli. Mol. Biochem. Parasitol. 88, 1–11. [DOI] [PubMed] [Google Scholar]
- Sentenac, H., Bonneaud, N., Minet, M., Lacroute, F., Salmon, J.M., Gaymard, F., and Grignon, C. (1992). Cloning and expression in yeast of a plant potassium ion transport system. Science 256, 663–665. [DOI] [PubMed] [Google Scholar]
- Serrano, R., and Gaxiola, R. (1994). Microbial models and salt stress tolerance in plants. Crit. Rev. Plant Sci. 13, 121–138. [Google Scholar]
- Shi, H., Ishitani, M., Kim, C., and Zhu, J.-K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 97, 6896–6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding, E.P., and Goldsmith, M.H.M. (1993). Activation of K+ channels in the plasma membrane of Arabidopsis by ATP produced photosynthetically. Plant Cell 5, 477–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spollen, W.G., LeNoble, M.E., Samuel, T.D., Bernstein, N., and Sharp, R.E. (2000). Abscisic acid accumulation maintains maize primary root elongation at low water potentials by restricting ethylene production. Plant Physiol. 122, 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay, Y.H., and Robinson, G.W. (1991). Cloning and characterization of ERG8, an essential gene of Saccharomyces cerevisiae that encodes phosphomevalonate kinase. Mol. Cell. Biol. 11, 620–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyerman, S.D., and Skerrett, M. (1999). Root ion channels and salinity. Sci. Hortic. 78, 175–235. [Google Scholar]
- Wu, L.F., Reizer, A., Reizer, J., Cai, B., Tomich, J.M., and Saier, M.H., Jr. (1991). Nucleotide sequence of the Rhodobacter capsulatus fruK gene, which encodes fructose-1-phosphate kinase: Evidence for a kinase superfamily including both phosphofructokinases of Escherichia coli. J. Bacteriol. 173, 3117–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S., Ding, L., and Zhu, J.-K. (1996). SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8, 617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, W.H., and Assmann, S.M. (1995). Is ATP required for K+ channel activation in guard cells? Plant Physiol. 107, 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Zhao, G., and Winkler, M.E. (1996). Identification of the pdxK gene that encodes pyridoxine (vitamin B6) kinase in Escherichia coli K-12. FEMS Microbiol. Lett. 141, 89–95. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Tsui, H.C., Man, T.K., and Winkler, M.E. (1998). Identification and function of the pdxY gene, which encodes a novel pyridoxal kinase involved in the salvage pathway of pyridoxal 5′-phosphate biosynthesis in Escherichia coli K-12. J. Bacteriol. 180, 1814–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.-K. (2000). Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol. 124, 941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.-K. (2001). Plant salt tolerance. Trends Plant Sci. 6, 66–71. [DOI] [PubMed] [Google Scholar]
- Zhu, J.-K., Liu, J., and Xiong, L. (1998). Genetic analysis of salt tolerance in Arabidopsis: Evidence for a critical role of potassium nutrition. Plant Cell 10, 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, S., Ehrhardt, T., Plesch, G., and Müller-Röber, B. (1999). Ion channels in plant signaling. Cell. Mol. Life Sci. 55, 183–203. [DOI] [PMC free article] [PubMed] [Google Scholar]