Abstract
Transgene-induced post-transcriptional gene silencing (PTGS) results from specific degradation of RNAs that are homologous with the transgene transcribed sequence. This phenomenon, also known as cosuppression in plants and quelling in fungi, resembles RNA interference (RNAi) in animals. Indeed, cosuppression/quelling/RNAi require related PAZ/PIWI proteins (AGO1/QDE-2/RDE-1), indicating that these mechanisms are related. Unlike Neurospora crassa qde-2 and Caenorhabditis elegans rde-1 mutants, which are morphologically normal, the 24 known Arabidopsis ago1 mutants display severe developmental abnormalities and are sterile. Here, we report the isolation of hypomorphic ago1 mutants, including fertile ones. We show that these hypomorphic ago1 mutants are defective for PTGS, like null sgs2, sgs3, and ago1 mutants, suggesting that PTGS is more sensitive than development to perturbations in AGO1. Conversely, a mutation in ZWILLE/PINHEAD, another member of the Arabidopsis AGO1 gene family, affects development but not PTGS. Similarly, mutations in ALG-1 and ALG-2, two members of the C. elegans RDE-1 gene family, affect development but not RNAi, indicating that the control of PTGS/RNAi and development by PAZ/PIWI proteins can be uncoupled. Finally, we show that hypomorphic ago1 mutants are hypersensitive to virus infection, confirming the hypothesis that in plants PTGS is a mechanism of defense against viruses.
INTRODUCTION
In plants, transgenes can be silenced through transcriptional freezing (transcriptional gene silencing) or through the specific degradation of transgene RNA (post-transcriptional gene silencing [PTGS]) (reviewed by Kooter et al., 1999; Wolffe and Matzke, 1999; Fagard and Vaucheret, 2000; Matzke et al., 2001; Vaucheret and Fagard, 2001). PTGS transgenes can trigger the degradation of homologous endogenous RNAs, a phenomenon referred to as cosuppression in plants (Napoli et al., 1990; Smith et al., 1990; van der Krol et al., 1990) and quelling in the fungus Neurospora crassa (Romano and Macino, 1992; Cogoni et al., 1996). Cosuppression and quelling strongly resemble RNA interference (RNAi), a PTGS phenomenon mediated by the introduction of double-stranded RNA (dsRNA) in animals (Fire et al., 1998; Fire, 1999). Indeed, genetic dissection of cosuppression in Arabidopsis, quelling in N. crassa, and RNAi in Caenorhabditis elegans led to the isolation of two sets of similar proteins: (1) SGS2, QDE-1, and EGO-1 (Cogoni and Macino, 1999a; Mourrain et al., 2000; Smardon et al., 2000), which are similar to the tomato RNA-dependent RNA polymerase (RdRP); and (2) AGO1, QDE-2, and RDE-1 (Tabara et al., 1999; Catalanotto et al., 2000; Fagard et al., 2000), which contain PAZ and PIWI domains (Cerutti et al., 2000). Given these findings, it was suggested that PTGS, quelling, and RNAi could be related phenomena deriving from an ancestral mechanism directed against invading nucleic acids (Catalanotto et al., 2000; Cogoni and Macino, 2000; Fagard et al., 2000). Such a role is supported by the finding that in Arabidopsis, PTGS-deficient sgs2, sgs3, and sde3 mutants are hypersusceptible to viral infection (Mourrain et al., 2000; Dalmay et al., 2001).
Models for PTGS/quelling/RNAi predict that dsRNA molecules are intermediates leading to specific RNA degradation (Bass, 2000). These dsRNAs can be supplied exogenously (RNAi) or produced by sense transgenes (cosuppression, quelling) when they are arranged as an inverted repeat (Waterhouse et al., 1998; Chuang and Meyerowitz, 2000; Smith et al., 2000) or when they produce aberrant RNA that serves as a matrix for the RdRP encoded by the SGS2/SDE1 and QDE-1 genes (Cogoni and Macino, 1999a; Dalmay et al., 2000; Mourrain et al., 2000). During RNAi in Drosophila melanogaster, the dsRNAs are degraded by an RNase III–like dsRNA endonuclease (DICER) to generate short sense, antisense, and dsRNA pieces of 21 or 22 nucleotides (Bernstein et al., 2001), which also are found in PTGS plants (Hamilton and Baulcombe, 1999). These short RNA pieces and mRNA are thought to participate in an RNA degradation complex called RISC in D. melanogaster (Bernstein et al., 2001), leading to complete RNA degradation. Interestingly, a protein similar to tomato RdRP also is required for RNAi in C. elegans (Smardon et al., 2000), indicating that, in worms as in plants and fungi, RdRP could participate not only in the synthesis of complementary RNA (and consequently in the formation of dsRNA) but also in the amplification/regeneration of a systemic signal, allowing silencing to spread throughout the organism (Palauqui et al., 1997; Fire et al., 1998; Voinnet et al., 1998).
In addition to RdRPs (Cogoni and Macino, 1999a; Mourrain et al., 2000; Smardon et al., 2000), RNases (Ketting et al., 1999; Bernstein et al., 2001), and RNA helicases (Domeier et al., 2000; Wu-Scharf et al., 2000; Dalmay et al., 2001), RNAi/quelling/cosuppression also require a set of related proteins (RDE-1 [Tabara et al., 1999], QDE-2 [Catalanotto et al., 2000], and AGO-1 [Fagard et al., 2000]) collectively designated PAZ/PIWI proteins (Cerutti et al., 2000). The role of PAZ/PIWI proteins in RNAi/quelling/cosuppression is unclear. RDE-1 was shown to be required for the formation of the interfering agent but not for interference thereafter (Grishok et al., 2000). Conversely, an additional member of this PAZ/PIWI family, DmAGO2, was identified as part of the RISC complex in D. melanogaster (Hammond et al., 2001). The role of PAZ/PIWI proteins in the control of development also is unclear. In C. elegans, the role of three related PAZ/PIWI proteins, RDE-1, ALG-1, and ALG-2, has been investigated. rde-1 mutants are defective for RNAi but are morphologically normal (Tabara et al., 1999), whereas alg-1 and alg-2 mutants are impaired in the regulation of small temporal RNAs that control developmental timing but are not defective in RNAi (Grishok et al., 2001). In N. crassa, qde-2 mutants are defective for quelling but are morphologically normal (Catalanotto et al., 2000). In Arabidopsis, the 24 known ago1 mutants show severe developmental abnormalities, including complete sterility (Bohmert et al., 1998; Camus, 1999; Fagard et al., 2000). Because all other Arabidopsis PTGS-deficient mutants (sgs1 to sgs3 and sde1 to sde4) (Elmayan et al., 1998; Dalmay et al., 2000; Mourrain et al., 2000) show no significant defect in development, the possible link between PTGS and development in plants is unclear. Whether endogenous genes that control development are regulated by part of the PTGS machinery (including AGO1 and additional steps) or AGO1 is a protein that participates independently in PTGS and development pathways is unknown.
Here, we describe new ago1 mutants that are impaired in PTGS even though they are fertile, suggesting differential sensitivity of PTGS and development to AGO1 defects. We also show that zll/pnh mutants are affected in development but not in PTGS, indicating that not all members of the AGO family participate in PTGS, as is the case for the RDE-1 family in C. elegans (Grishok et al., 2001). In addition, we show that, like the sgs2, sgs3, and sde3 mutants (Mourrain et al., 2000; Dalmay et al., 2001), ago1 mutants are hypersensitive to virus infection, indicating that all of the components of PTGS identified to date in plants are involved in virus resistance.
RESULTS
Identification of New PTGS-Deficient Mutants Showing Original Developmental Alterations
We reported previously the isolation of four classes of PTGS-deficient mutants after mutagenesis of the silenced 35S-uidA line L1 with ethyl methanesulfonate. Mutants called sgs1, sgs2, and sgs3 (for suppressor of gene silencing) showed no significant changes in development (Elmayan et al., 1998; Mourrain et al., 2000), whereas mutants called ago1 showed severe alterations of development (unexpanded cotyledons, small rosettes with narrow leaves, unique short stems with no leaves, and sterile flowers) (Bohmert et al., 1998; Fagard et al., 2000) (Figure 1). After screening for PTGS-deficient mutants, we identified three mutants (called 19-3, 33-2, and 69-4) exhibiting β-glucuronidase (GUS) activity at levels similar to that of other sgs mutants and showing developmental defects differing from those of the 24 known ago1 mutants (Figure 1). Mutant 33-2 developed a small rosette with dark green and serrated leaves, initiated flowering 10 to 15 days after the wild type, and was almost totally sterile, although occasionally it produced short siliques containing two to five seed (a maximum of 50 seed could be obtained per plant). Mutants 19-3 and 69-4 developed rosettes with dark green and serrated leaves. Rosette size was intermediate between that of mutant 33-2 and wild-type plants. These mutants initiated flowering 7 to 12 days after the wild type and were fertile: they produced 45 to 60% of the amount of seed of the wild type (depending on growth conditions).
Figure 1.
Vegetative and Reproductive Developmental Defects in PTGS-Deficient Mutants.
Newly isolated PTGS mutants (69-4/ago1-27, 19-3/ago1-25, and 33-2/ago1-26) compared with the L1 (wild type [wt]) line, from which the mutants were isolated after ethyl methanesulfonate mutagenesis, and with a representative of the original ago1 alleles (60-1/ago1-24). Photographs show plants after 12, 20, and 35 days of growth in vitro in short days and 15, 30, 34, and 40 days after their transfer to a greenhouse for further growth in long days. The newly isolated mutants had dark and highly serrated leaves. The mutants 69-4/ago1-27 and 19-3/ago1-25 were fertile, in contrast to mutants 33-2/ago1-26 and 60-1/ago1-24. The previously identified 60-1/ago1-24 mutant showed similar but stronger developmental defects and eventually died in soil (fifth and sixth photographs from the top in the right column). Only in vitro did this mutant develop a stem bearing sterile flowers (bottom photograph in the right column).
Mutants 19-3, 33-2, and 69-4 Are Alleles
Backcrosses of [GUS+] (nonsilenced) mutants 19-3, 33-2, and 69-4 to [GUS−] (silenced) line L1 yielded [GUS−] F1 plants with a wild-type phenotype. Selfing these F1 hybrids yielded 75% (70 of 96, 75 of 96, and 74 of 96, respectively) [GUS−] plants with a wild-type phenotype and 25% [GUS+] plants with the original mutant phenotype, suggesting that in each mutant a single recessive mutation was responsible for the developmental defects and for the deficiency in PTGS. Reciprocal crosses (see Methods) between these three mutants yielded [GUS+] plants with a mutant phenotype. Furthermore, no wild-type plant was found in the F2 progeny among 500 plants analyzed for each reciprocal cross, indicating that these mutants carry allelic mutations. Interestingly, F1 hybrids between 69-4 or 19-3 (which are both fertile and larger than 33-2) and 33-2 (which is sterile) were fertile and had the phenotype of 69-4 or 19-3, indicating that the weaker alleles (69-4 and 19-3) act dominantly over the stronger allele (33-2).
Mutants 19-3, 33-2, and 69-4 are Hypomorphic ago1 Mutants
To map the corresponding locus, we analyzed 800 recombinant chromosomes derived from a cross between mutant 69-4 (in ecotype Columbia) and the polymorphic ecotype Landsberg erecta. The mutation was mapped on chromosome 1 between markers GAPB and nga280 (Figure 2A). Subsequent analysis indicated that the mutation was located between markers mi291 and F11I4t7. Because this 300-kb region also carried the AGO1 gene, we used a cleaved-amplified polymorphic sequence (CAPS) marker (CER462285) located 6 kb upstream of the AGO1 gene and found that there was no recombination point between this marker and the mutation in the 800 recombinant chromosomes, strongly suggesting that the mutation was located in the AGO1 gene. Crosses between parents heterozygous for the ago1-24 mutation (the homozygous ago1-24 mutant is sterile; Figure 1) (Fagard et al., 2000) and fertile mutants 19-3 and 69-4 yielded 50% (28 of 52 and 35 of 74, respectively) [GUS−] plants with a wild-type phenotype and 50% [GUS+] plants with the 19-3 or 69-4 (fertile) phenotype, indicating that mutants 19-3 and 69-4 are ago1 alleles. Because ago1-24 mutants are sterile and show stronger developmental defects than 19-3 and 69-4, this further confirmed that the weaker allele acted dominantly. Similarly, crosses between parents heterozygous for the ago1-24 mutation and parents heterozygous for the 33-2 mutation (the homozygous 33-2 mutant is sterile) yielded 75% (27 of 37) [GUS−] plants with a wild-type phenotype and 25% [GUS+] plants with the 33-2 (sterile) phenotype, indicating that mutant 33-2 also is an ago1 allele. Selfing the fertile 19-3 × ago1-24 and 69-4 × ago1-24 F1 hybrids yielded 75% (381 of 500 and 368 of 500, respectively) plants with the 19-3 or 69-4 phenotype and 25% plants with the ago1-24 phenotype. Thus, the absence of wild-type plants in the F2 population confirmed that 19-3 and 69-4 are ago1 alleles. Similar results were obtained after crossing mutants 19-3, 33-2, and 69-4 with other known ago1 mutants (data not shown), suggesting that these new mutants are hypomorphic ago1 alleles according to the developmental defects and act as dominant alleles over null ago1 alleles.
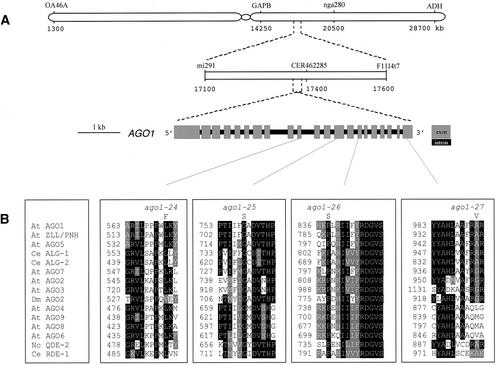
Figure 2.
Mapping and Sequences of the ago1-25, ago1-26, and ago1-27 Mutations.
(A) Physical map of the chromosomal area carrying the 69-4 (ago1-27) mutation. The ago1-27 mutation was mapped between the CAPS marker GAPB and the microsatellite nga280 on chromosome 1 and subsequently between the restriction fragment length polymorphism marker mi291 and the CAPS marker F11I4t7 (http://www.Arabidopsis.org/). No recombination point was found between the CAPS marker CER462285 (http://www.Arabidopsis.org/cereon/) and the ago1-27 mutation in the 800 recombinant chromosomes tested.
(B) Sequencing of the 69-4, 19-3, and 33-2 mutant lines indicated that they all carry mutations in the C-terminal part of the AGO1 protein. The AGO1 protein (At1g48410) is aligned with the nine other members of the Arabidopsis (At) PAZ/PIWI family: AGO2 (At1g31280), AGO3 (At1g31290), AGO4 (At2g27040), AGO5 (At2g27880), AGO6 (At2g32940), AGO7 (At1g69440), AGO8 (At5g21030), AGO9 (At5g21150), and ZLL/PNH (At5g43810); with three members of the C. elegans (Ce) PAZ/PIWI family: RDE1, ALG-1, and ALG-2; and with N. crassa (Nc) QDE-2 and D. melanogaster (Dm) AGO2 proteins. Sequence identities are indicated by closed boxes, and conservative changes are shaded. Amino acid positions are indicated at left. Dots indicate gaps introduced by the MultiAlign algorithm to maximize alignment (http://prodes.toulouse.inra.fr/multalin/multalin.html). The alignment was processed by BOXSHADE version 3.21 (http://www.ch.embnet.org/software/BOX_form.html).
We identified the mutations by sequencing the AGO1 gene in mutants 19-3, 33-2, and 69-4. Single nucleotide transitions were found in the 3′ portion of the gene, leading to the following substitutions: Gly-758 → Ser (19-3), Pro-838 → Ser (33-2), and Ala-992 → Val (69-4) (Figure 2B). These new hypomorphic ago1 alleles were named ago1-25, ago1-26, and ago1-27, respectively. Alignment of AGO1 with RDE-1 and QDE-2 (which are required for RNAi and quelling) revealed that the essential Gly-758, Pro-838, and Ala-992 also are present in RDE-1 and QDE-2 within conserved motifs (Figure 2B), suggesting that these amino acids also could participate in the silencing function of these two proteins. Interestingly, they are present as well in ALG-1 and ALG-2, which are required for development but not RNAi, but not in DmAGO2, which is part of the RISC complex (Figure 2B).
Hypomorphic Mutant 69-4 (ago1-27) Is Deficient for PTGS Like Other Null PTGS Mutants
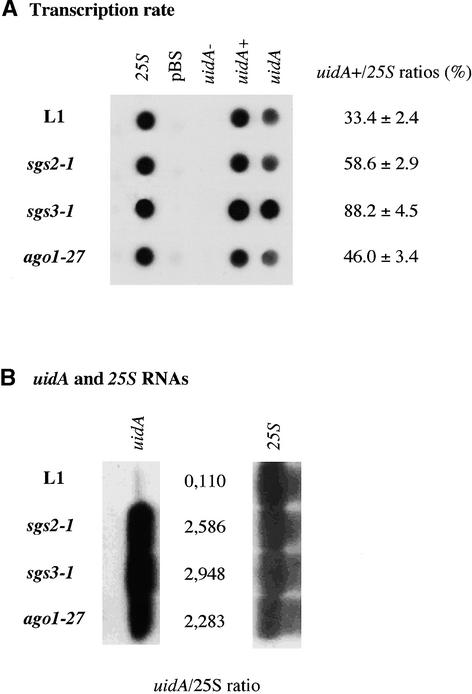
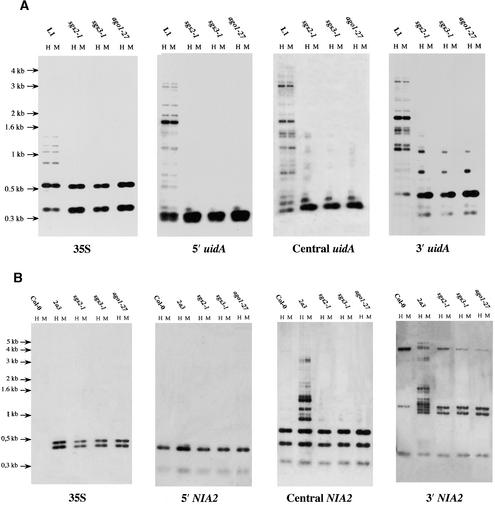
We determined that hypomorphic ago1 mutants, like sgs mutants and null ago1 mutants, were deficient for PTGS. Nuclei, RNA, and DNA extracted from mutant 69-4 (ago1-27) were analyzed by run-on RNA gel blot analysis (Figures 3A and 3B) and DNA gel blot analysis (Figure 4) and compared with line L1 and sgs2-1 and sgs3-1 mutants. The 35S-uidA transgene was transcribed at a high level in L1, sgs2-1, sgs3-1, and ago1-27 (Figure 3A), whereas uidA mRNA accumulated only in sgs2-1, sgs3-1, and ago1-27 mutants (Figure 3B). The ratio between transcription and mRNA accumulation was similar in sgs2-1, sgs3-1, and ago1-27 mutants, indicating that PTGS was affected similarly in these three mutants. In addition, methylation of the uidA coding sequence was reduced similarly in the three mutants (Figure 4A), as in mutant ago1-24 (Fagard et al., 2000). To confirm that the hypomorphic mutant ago1-27 (69-4) was generally impaired in the process of PTGS, we crossed mutant ago1-27 with line 2a3. This line carries a 35S-NIA2 transgene that triggers cosuppression of endogenous NIA genes with 100% efficiency in a wild-type background (Elmayan et al., 1998). F2 plants that are homozygous for both the ago1-27 mutation and the 2a3 locus were identified, and their progeny were analyzed. We observed that cosuppression was abolished completely (0 of 500 plants died when grown on nitrate as a sole source of nitrogen) and that methylation of the 35S-NIA2 transgene was reduced strongly in mutant ago1-27, as in mutants sgs2-1 and sgs3-1 (Figure 4B). These results indicate that the silencing properties of the ago1-27 allele could not be distinguished from that of null ago1 and sgs mutants, suggesting that this hypomorphic ago1 allele is completely deficient for PTGS.
Figure 3.
PTGS Is Abolished Completely in the ago1-27 Mutant as in the sgs2 and sgs3 Mutants.
The ago1-27 mutant was compared with the L1 line, which shows PTGS of the 35S-uidA transgene, and with representative alleles of the sgs2 and sgs3 mutants deficient for PTGS (Mourrain et al., 2000).
(A) Transcript levels were determined using run-on analysis as described by Elmayan et al. (1998). Radiolabeled RNA extracted from nuclei from adult plants were hybridized to 2 μg of double-stranded DNA (pBS, empty pBluescript KS− vector; 25S, rDNA cloned in pBS; uidA, GUS gene cloned in pBS) and 2 μg of single-stranded DNA (sense, uidA−; antisense, uidA+). The signals of three independent experiments were quantified, and the ratios of uidA+ to 25S are indicated with standard deviations.
(B) RNA accumulation was measured by RNA gel blot analysis using 10 μg of total RNA. Note the faint band visible in the L1 lane with the uidA GUS probe. The signals were quantified, and the ratios of uidA to 25S are indicated.
Figure 4.
Methylation of the uidA and NIA2 Transgenes Is Affected Similarly in the ago1-27 Mutant and in the sgs2 and sgs3 Mutants.
Methylation of the 35S-uidA transgene (A) and of the 35S-NIA2 transgene (B) was measured using the sensitive enzymes HpaII (H) and MspI (M) and probes corresponding to the transgene promoter (35S) and to the 5′, central, and 3′ regions of the transgenes, as described by Elmayan et al. (1998). In the case of the 35S-NIA2 transgene, wild-type Columbia (Col-0) DNA was included to distinguish between the endogenous and transgenic copies of the NIA2 genes. Although the silenced transgene loci were hypermethylated in a wild-type background (as shown by the high-molecular-mass bands), the sgs2-1, sgs3-1, and ago1-27 mutants exhibited reduced methylation of the transgenes.
Hypomorphic ago1-26 and ago1-27 Mutants Are Hypersusceptible to Infection by Cucumber Mosaic Virus
We reported previously that the PTGS-deficient sgs2 and sgs3 mutants are hypersusceptible to infection by cucumber mosaic virus (CMV) (Mourrain et al., 2000), a member of the cucumovirus family that infects more than 800 plant species. To determine if PTGS as a whole is a defense mechanism that helps to limit virus infection, we tested whether PTGS-deficient ago1 mutants also were hypersusceptible to CMV. The identification of hypomorphic ago1 mutants allowed us to perform such experiments. As was the case for sgs2 and sgs3 mutants, disease symptoms were much more pronounced in CMV-infected ago1-26 and ago1-27 mutants than in CMV-infected wild-type plants (Figure 5A and data not shown). When infected, both ago1-26 and ago1-27 mutants had very small stems, were completely sterile, and eventually died. In contrast, infected wild-type plants developed stems with elongated internodes and were able to form seed. We confirmed that the difference in symptom severity caused by CMV infection was attributable to an average fivefold to sixfold overaccumulation of viral RNA in ago1-26 and ago1-27, as in sgs2 and sgs3 mutants (Figure 5B).
Figure 5.
The ago1 Mutants Are Hypersusceptible to CMV.
Plants were inoculated with CMV as described by Mourrain et al. (2000). Symptoms of mock-infected and CMV-infected ago1-27 mutants (A) and virus accumulation in CMV-infected L1 plants and ago1-26 and ago1-27 mutants (B) are shown 3 weeks after infection. Five micrograms of total RNA was tested with full-length CMV and constitutive 25S probes. As in the sgs2 and sgs3 mutants (Mourrain et al., 2000), CMV accumulation was fivefold to sixfold higher in the ago1 mutants compared with the L1 line as measured by signal quantification on a phosphorimager. The accumulation correlated with increased symptoms, and the infected mutants eventually died, whereas CMV-infected L1 plants produced seed. The experiment was performed three times, and the results of one representative experiment are shown.
A Mutation in the ZWILLE/PINHEAD Gene Affects Development but Not PTGS
The AGO1 gene belongs to a multigene family of 10 members (Figure 2B). Mutants affected in a member called ZWILLE or PINHEAD (ZLL or PNH) show developmental defects resembling that of ago1 mutants, suggesting that the functions of AGO1 and ZLL/PNH partially overlap (Moussian et al., 1998; Lynn et al., 1999). To determine if ZLL/PNH also participates in PTGS, we crossed line L1 to the zll-3 mutant, which shows the strongest developmental defects (Moussian et al., 1998) and lacks a functional protein (T. Laux, personal communication). Plants homozygous for the zll-3 mutation and homozygous for the L1 locus were identified in the F2 progeny of the cross and selfed. GUS activity was measured in rosette and cauline leaves and in stems of 50 F3 plants derived from two independent F2 plants. GUS activity was similar to that found in L1 plants (Table 1), indicating that the ZLL/PNH gene is not required for PTGS at the L1 locus (at least in these tissues).
Table 1.
GUS Activity Conferred by the L1 Locus in Leaves and Stems of Wild-Type Plants and zll-3 or ago1-27 Mutants
| Genotype
|
|||
|---|---|---|---|
| Tissue | L1/L1 ZLL/ZLL AGO1/AGO1 | L1/L1 zll-3/zll-3 AGO1/AGO1 | L1/L1 ZLL/ZLL ago1-27/ago1-27 |
| Rosette leaves | 5.8 ± 4.9 | 4.7 ± 3.9 | 4850 ± 237 |
| Cauline leaves | 5.7 ± 3.9 | 6.2 ± 5.2 | 5063 ± 351 |
| Stems | 8.9 ± 4.5 | 11.1 ± 7.5 | 4756 ± 189 |
GUS activity (nmol methylumbelliferone min−1 μg−1 protein) is the average measurement (±sd) in 50 plants (see Methods for details on the obtainment of the L1/L1 zll-3/zll-3 plants).
DISCUSSION
By a combination of genetic and biochemical approaches, many components of RNAi, quelling, and cosuppression have been identified (Matzke et al., 2001). The involvement of proteins similar to DNA helicase (Cogoni and Macino, 1999b) and of proteins involved in chromatin remodeling and DNA methylation (Morel et al., 2000) indicates that quelling and PTGS have nuclear determinants. These components probably are involved in the primary production and/or in the maintenance of production of aberrant RNA that triggers the RNA degradation process. In addition, proteins similar to RdRP (Cogoni and Macino, 1999a; Mourrain et al., 2000; Smardon et al., 2000), 3′ to 5′ exonuclease RNase D (Ketting et al., 1999), dsRNA endonuclease RNase III (Bernstein et al., 2001), and RNA helicase (Domeier et al., 2000; Wu-Scharf et al., 2000; Dalmay et al., 2001) are involved in sequence-specific and systemic RNA degradation (Cogoni et al., 1996; Palauqui et al., 1997; Fire et al., 1998). Most of the RNAi/quelling/cosuppression mutants do not show developmental defects in standard conditions of growth. Indeed, Arabidopsis mutants such as sgs1, sgs2, sgs3, sde1, sde2, sde3, and sde4 are viable (Elmayan et al., 1998; Dalmay et al., 2000; Mourrain et al., 2000), as are N. crassa qde-1, qde-2, and qde-3 and C. elegans rde-1 mutants (Cogoni and Macino, 1997; Tabara et al., 1999), suggesting that PTGS, quelling, and RNAi as a whole are dispensable for development. In contrast, the 24 reported mutations in the Arabidopsis AGO1 gene have pleiotropic effects on development and fertility that strongly compromise life and reproduction in standard conditions of growth (Bohmert et al., 1998; Camus, 1999; Fagard et al., 2000). Similarly, mut-7 and ego-1 mutants impaired in RNAi also show gametogenesis defects and sterility in C. elegans (Ketting et al., 1999; Smardon et al., 2000). Thus, whether PTGS/RNAi regulate some step(s) of the expression of endogenous genes during development or simply share a common enzyme(s) or pathway(s) with development was undetermined.
Here, we report the identification and characterization of hypomorphic Arabidopsis ago1 mutants that are as impaired in PTGS as the previously identified strong ago1 mutants (Fagard et al., 2000), although they show limited developmental abnormalities (Figure 1). The identification of these hypomorphic mutants allows us to define three classes of ago1 mutants. The first class corresponds to PTGS-deficient ago1 mutants showing very strong developmental defects that strongly compromise their growth in soil and that are completely sterile in vitro or in soil (ago1-1 to ago1-24) (Bohmert et al., 1998; Camus, 1999; Fagard et al., 2000). The second class corresponds to PTGS-deficient ago1 mutants that develop vigorously in soil although they are smaller than wild-type plants and that are almost completely sterile (ago1-26) (this work). Finally, the third class corresponds to PTGS-deficient ago1 mutants that are fertile and that show limited developmental defects (ago1-25 and ago1-27) (this work). The partial uncoupling of PTGS defects and developmental defects in hypomorphic ago1 mutants indicates that PTGS is much more sensitive than development to perturbations in AGO1.
Whether AGO1 regulates development via the regulation of endogenous genes by PTGS or acts independently in PTGS and development is not known. However, it is unlikely that PTGS as a whole could regulate development because sgs and sde PTGS-deficient mutants (including null alleles) do not exhibit developmental defects comparable to those of ago1 mutants. Nevertheless, part of the PTGS machinery, including AGO1 and proteins that regulate subsequent RNA degradation steps, could participate in the regulation of some endogenous genes. The partial uncoupling between silencing and developmental defects in hypomorphic ago1 mutants could be related to the differences in transcription levels of the target transgenes and endogenes. Indeed, transgenes are expressed more strongly than endogenous genes because they are expressed under the control of the 35S promoter. Therefore, mutations in the hypomorphic ago1 mutants could trigger little perturbations in the amount and/or activity of the AGO1 protein that could have dramatic consequences in the case of transgenes because they are strongly expressed, whereas endogenous mRNA targets, which are likely to be present at lower levels, could be processed correctly with less AGO1 activity.
Alternatively, AGO1 could be a protein acting independently in PTGS and development, and the structural requirements for a functional AGO1 protein may be different in the two pathways. Mutations that result from T-DNA insertions that impede the production of a full-length protein (Bohmert et al., 1998; C. Bellini, personal communication) as well as a single change affecting Leu-571 in the ago1-24 mutant (Fagard et al., 2000) are responsible for both PTGS deficiency and very strong developmental defects. Conversely, point mutations at the 3′ end of the gene (changing Gly-758, Pro-838, and Ala-992) strongly affect PTGS but trigger weaker developmental defects (this work). Therefore, the C terminus of the protein may be much more specialized in the AGO1 silencing function. Nevertheless, the entire protein is required to produce a functional protein and allow normal development of the plant. Interestingly, mutations in AGO1 that completely impair PTGS affect amino acids that also are found in QDE-2 and RDE-1 (which are required for quelling and RNAi) in highly conserved motifs (Figure 2B). Neither N. crassa qde-2 nor C. elegans rde-1 null mutants show developmental defects (Tabara et al., 1999; Catalanotto et al., 2000). Conversely, C. elegans alg-1 and alg-2 mutants (which are defective in the production of two RDE-1 homologs) are impaired in the regulation of small temporal RNAs that control developmental timing but are not defective in RNAi (Grishok et al., 2001). Therefore, AGO1 could combine RDE-1 silencing functions and ALG-1/ALG-2 developmental functions on a single molecule, an hypothesis that is in agreement with the fact that mutations in AGO1 that are responsible for developmental abnormalities affect amino acids that also are found in ALG-1 and ALG-2 (Figure 2B). Conversely, two of the four mutations in AGO1 that impair PTGS in Arabidopsis affect amino acids that are not found in DmAGO2, which participates in the RISC complex in D. melanogaster (Figure 2B). Together, these results suggest that AGO1 functions may be more similar to those of ALG-1, ALG-2, and RDE-1 (the latter being involved in the production of the interfering agent) than those of DmAGO2 (which is involved in interference thereafter), a result that is in agreement with the fact that silencing induced by panhandle transgenes that constitutively produce dsRNA occurs as efficiently in wild-type plants as in ago1 mutants (C. Béclin, S. Boutet, and H. Vaucheret, unpublished results).
As proposed above, differences in the structural requirements for a functional AGO1 protein in PTGS and development could explain the partial uncoupling of PTGS defects and developmental defects in hypomorphic ago1 mutants. Alternatively, other members of the AGO family in Arabidopsis may compensate for (part of) the defects of AGO1 developmental functions but not AGO1 silencing functions. Indeed, AGO1 belongs to a family of 10 members in Arabidopsis (Figure 2B). Except for two members for which mutants with developmental defects have been isolated (AGO1 and ZLL/PNH), the roles of the other members of the family are unknown (Bohmert et al., 1998; Moussian et al., 1998; Lynn et al., 1999). AGO1 clearly participates in silencing because both PTGS of an exogenous 35S-uidA transgene and cosuppression of NIA transgenes and endogenous genes are abolished in ago1 mutants. Among four sequenced PTGS-deficient ago1 mutants (Fagard et al., 2000; this work), three show point mutations changing amino acids that are conserved among the 10 members (Leu-571, Gly-758, and Pro-838), a result that is not informative on the role of the other genes. However, the fourth mutation affects an amino acid (Ala-992) that is absent in seven members (AGO2, AGO3, AGO4, AGO6, AGO7, AGO8, and AGO9) and present in only three closely related members (AGO1, ZLL/PNH, and AGO5), suggesting that only these three also could participate in silencing. To answer this question, we determined whether PTGS could occur in the zll-3 mutant showing the strongest developmental defects and lacking a functional protein (T. Laux, personal communication). We found that PTGS occurs efficiently in the tissues we tested (leaves and stems). Because PTGS is inhibited in ago1 mutants but not in zll/pnh mutants, AGO1 could be the only member having a function in PTGS, and the other members could participate in (partially) redundant developmental processes, as has been shown for ZLL/PNH (Moussian et al., 1998; Lynn et al., 1999). Whether PTGS and/or development are affected in ago5 mutants needs to be determined to answer this question definitively.
Because sgs and sde PTGS-deficient mutants do not exhibit developmental defects comparable to those of ago1 mutants, the role of PTGS in regulating the expression of endogenous genes and subsequently the development of the plants is still a matter of debate. Conversely, a natural role of the PTGS machinery seems to be in limiting virus infection. Indeed, we showed that PTGS-deficient sgs2 and sgs3 mutants are hypersusceptible to infection by CMV, a single-stranded RNA cucumovirus (Mourrain et al., 2000). Subsequently, it was shown that sde3 mutants also are hypersusceptible to infection by CMV (Dalmay et al., 2001). Here, we showed that hypomorphic PTGS-deficient ago1 mutants also are hypersusceptible to infection by CMV and overaccumulate CMV RNA, like sgs2, sgs3, and sde3 mutants, indicating that AGO1 also participates in protecting plants against some viruses. Therefore, AGO1, SGS2, SGS3, and SDE3 act as components of a phenomenon that is naturally targeted against viruses and that can be activated against (trans)gene RNA under circumstances that remain to be elucidated.
METHODS
Plant Material and Growth Conditions
After seed sterilization, plants of Arabidopsis thaliana ecotype Columbia, from which were derived the silenced transgenic lines L1 (35S-uidA) and 2a3 (35S-NIA2) (Elmayan et al., 1998), the ago1-24 mutant (Fagard et al., 2000), and the zll-3 mutant (Moussian et al., 1998), were grown in sterile medium under a short day (8 hr of light/16 hr of dark) regimen at 100 μmol m−2 sec−1 Plants were transferred to soil after 6 weeks and grown under a long day regimen (16 hr of light/8 hr of dark).
Mutant Selection and Genetic Analysis
Thirty thousand plants derived from the self-progeny of 1500 ethyl methanesulfonate–mutagenized seed of the silenced 35S-uidA transgenic line L1 were grown in the greenhouse. After 4 weeks of growth, β-glucuronidase (GUS) activity (nmol 4-methylumbelliferone min−1 μg−1 total protein) was measured in the leaves as described previously (Elmayan et al., 1998), and the [GUS+] plants were retained for genetic analysis. Mutants with a wild-type phenotype (sgs1, sgs2, and sgs3) and mutants showing the phenotype of null ago1 mutants were described previously (Elmayan et al., 1998; Fagard et al., 2000; Mourrain et al., 2000). The remaining [GUS+] plants showing a different phenotype were crossed to sgs1-1, sgs2-1, and sgs3-1 mutants (Elmayan et al., 1998; Mourrain et al., 2000) and to parents heterozygous for the ago1-24 mutation (Fagard et al., 2000). Recovery of 100% [GUS−] F1 plants with a wild-type phenotype in crosses with sgs mutants and of 50% [GUS+] F1 plants with the mutant phenotype in crosses with the heterozygous parent of ago1-24 was indicative of allelism between the tested mutation and ago1. Further analyses with the newly identified hypomorphic ago1 mutants were performed after two backcrosses with the L1 line.
Introgression of the 2a3 locus into the ago1-27 background was performed as follows. The ago1-27 mutant (ago1-27/ago1-27, L1/L1, −/−) was crossed to line 2a3 (AGO1/AGO1, −/−, 2a3/2a3). F1 hybrids were selfed, and their progeny were sown on medium supplemented with hygromycin (to which resistance is conferred by the 2a3 locus). Hygromycin-resistant F2 plants exhibiting the ago1 phenotype were transferred to soil, and their progeny were harvested. Each batch of F3 seed was sown on medium supplemented with kanamycin (to which resistance is conferred by the L1 locus) or hygromycin, allowing the identification of mutants homozygous for the 2a3 locus only (ago1-27/ago1-27, −/−, 2a3/2a3) and their progeny, which are 100% kanamycin sensitive and 100% hygromycin resistant.
Introgression of the L1 locus into the zll-3 background was performed as follows. The zll-3 mutant (zll-3/zll-3, −/−) was crossed to line L1 (ZLL/ZLL, L1/L1). F1 hybrids were selfed, and their progeny were sown on medium supplemented with kanamycin (to which resistance is conferred by the L1 locus). Kanamycin-resistant F2 plants exhibiting the zll phenotype were transferred to soil, and their progeny were harvested. Each batch of F3 seed was sown on medium supplemented with kanamycin, allowing identification of mutants homozygous for the L1 locus (zll-3/zll-3, L1/L1) and their progeny, which are 100% kanamycin resistant.
Molecular Characterization
Genomic DNA was extracted by the standard cetyl-trimethyl-ammonium bromide extraction method (Mourrain et al., 2000). The cleaved- amplified polymorphic sequence marker CER462285 located 6 kb up-stream of the AGO1 gene on bacterial artificial chromosome F11A17 was derived from the Cereon database (http://www.Arabidopsis.org/cereon/). The primers used were 5′-GTGTGTGTGGTGTGTTTGCAG-3′ and 5′-CGTGAAGTCCATCCAGAAGCC-3′. The 0.8-kb amplified fragment was cut by AccI to reveal the published polymorphism between ecotypes Columbia (L1 background) and Landsberg erecta. Sequencing of the ago1 alleles was performed on several independent polymerase chain reaction products and on both strands to confirm point mutations. Methylation analysis by DNA gel blotting was performed using the methylation-sensitive HpaII and MspI restriction enzymes as described previously (Mourrain et al., 2000). RNA and run-on signals were quantified using a phosphorimager.
Acknowledgments
We thank Thomas Laux for kindly providing the zll-3 mutant. We thank Hervé Ferry, Jean-Luc Macia, Jean-Marie Pollien, and Gérard Vastra for plant culture. This work was supported partly by Rhobio and the French Ministry of Research and Technology.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010358.
References
- Bass, B. (2000). Double-stranded RNA as a template for gene silencing. Cell 101, 235–238. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., Caudy, A., Hammond, S., and Hannon, G. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 295–296. [DOI] [PubMed] [Google Scholar]
- Bohmert, K., Camus, I., Bellini, C., Bouchez, D., Caboche, M., and Benning, C. (1998). AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17, 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus, I. (1999). ARGONAUTE d'Arabiopsis thaliana Définit une Famille de Gènes Conservés chez les Eucaryotes: Impliqués dans le Contrôle du Développement. Thèse de Doctorat (Paris: de l'Université Pierre et Marie Curie).
- Cerutti, L., Mian, N., and Bateman, A. (2000). Domains in gene silencing and cell differentiation proteins: The novel PAZ domain and redefinition of the Piwi domain. Trends Biochem. Sci. 25, 481–482. [DOI] [PubMed] [Google Scholar]
- Catalanotto, C., Azzalin, G., Macino, G., and Cogoni, C. (2000). Gene silencing in worms and fungi. Nature 404, 245. [DOI] [PubMed] [Google Scholar]
- Chuang, C., and Meyerowitz, E. (2000). Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97, 4985–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni, C., and Macino, G. (1997). Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc. Natl. Acad. Sci. USA 94, 10233–10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni, C., and Macino, G. (1999. a). Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399, 166–169. [DOI] [PubMed] [Google Scholar]
- Cogoni, C., and Macino, G. (1999. b). Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science 286, 2342–2344. [DOI] [PubMed] [Google Scholar]
- Cogoni, C., and Macino, G. (2000). Post-transcriptional gene silencing across kingdoms. Curr. Opin. Genet. Dev. 10, 638–643. [DOI] [PubMed] [Google Scholar]
- Cogoni, C., Irelan, J.T., Schumacher, M., Schmidhauser, T., Selker, E.U., and Macino, G. (1996). Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. EMBO J. 15, 3153–3163. [PMC free article] [PubMed] [Google Scholar]
- Dalmay, T., Hamilton, A., Rudd, S., Angell, S., and Baulcombe, D. (2000). An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101, 543–553. [DOI] [PubMed] [Google Scholar]
- Dalmay, T., Horsefield, R., Braunstein, T., and Baulcombe, D. (2001). SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 20, 2069–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domeier, M., Morse, D., Knight, S., Portereiko, M., Bass, B., and Mango, S. (2000). A link between RNA interference and nonsense-mediated decay in Caenorhabditis elegans. Science 289, 1928–1931. [DOI] [PubMed] [Google Scholar]
- Elmayan, T., Balzergue, S., Béon, F., Bourdon, V., Daubremet, J., Guénet, Y., Mourrain, P., Palauqui, J.-C., Vernhettes, S., Vialle, T., Wostrikoff, K., and Vaucheret, H. (1998). Arabidopsis mutants impaired in cosuppression. Plant Cell 10, 1447–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard, M., and Vaucheret, H. (2000). (Trans)genes silencing in plants: How many mechanisms? Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 167–194. [DOI] [PubMed] [Google Scholar]
- Fagard, M., Boutet, S., Morel, J., Bellini, C., and Vaucheret, H. (2000). AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. USA 97, 11650–11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A. (1999). RNA-triggered gene silencing. Trends Genet. 15, 358–363. [DOI] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Grishok, A., Tabara, H., and Mello, C. (2000). Genetic requirements for inheritance of RNAi in C. elegans. Science 287, 2494–2497. [DOI] [PubMed] [Google Scholar]
- Grishok, A., Pasquinelli, A., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D., Fire, A., Ruvkun, G., and Mello, C. (2001). Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23–34. [DOI] [PubMed] [Google Scholar]
- Hamilton, A., and Baulcombe, D. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Hammond, S., Boettcher, S., Caudy, A., Kobayashi, R., and Hannon, G. (2001). Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293, 1146–1150. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Haverkamp, T.H., van Luenen, H.G., and Plasterk, R.H. (1999). Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99, 133–141. [DOI] [PubMed] [Google Scholar]
- Kooter, J., Matzke, M., and Meyer, P. (1999). Listening to the silent genes: Transgene silencing, gene regulation and pathogen control. Trends Plant Sci. 4, 340–347. [DOI] [PubMed] [Google Scholar]
- Lynn, K., Fernandez, A., Aida, M., Sedbrook, J., Tasaka, M., Masson, P., and Barton, M. (1999). The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126, 469–481. [DOI] [PubMed] [Google Scholar]
- Matzke, M., Matzke, A., Pruss, G., and Vance, V. (2001). RNA-based silencing strategies in plants. Curr. Opin. Genet. Dev. 11, 221–227. [DOI] [PubMed] [Google Scholar]
- Morel, J., Mourrain, P., Beclin, C., and Vaucheret, H. (2000). DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr. Biol. 10, 1591–1594. [DOI] [PubMed] [Google Scholar]
- Mourrain, P., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101, 533–542. [DOI] [PubMed] [Google Scholar]
- Moussian, B., Schoof, H., Haecker, A., Jurgens, G., and Laux, T. (1998). Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 17, 1799–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli, C., Lemieux, C., and Jorgensen, R. (1990). Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous gene in trans. Plant Cell 2, 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palauqui, J.-C., Elmayan, T., Pollien, J.-M., and Vaucheret, H. (1997). Systemic acquired silencing: Transgene specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16, 4738–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano, N., and Macino, G. (1992). Quelling: Transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 6, 3343–3353. [DOI] [PubMed] [Google Scholar]
- Smardon, A., Spoerke, J.M., Stacey, S.C., Klein, M.E., Mackin, N., and Maine, E.M. (2000). EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 10, 169–178. [DOI] [PubMed] [Google Scholar]
- Smith, C.J.S., Watson, C.F., Bird, C.R., Ray, J., Schuch, W., and Grierson, D. (1990). Expression of a truncated tomato polygalacturonase gene inhibits expression of the endogenous gene in transgenic plants. Mol. Gen. Genet. 224, 477–481. [DOI] [PubMed] [Google Scholar]
- Smith, N., Singh, S., Wang, M., Stoutjesdijk, P., Green, A., and Waterhouse, P.M. (2000). Total silencing by intron-spliced hairpin RNAs. Nature 407, 319–320. [DOI] [PubMed] [Google Scholar]
- Tabara, H., Sarkissian, M., Kelly, W.G., Fleenor, J., Grishok, A., Timmons, L., Fire, A., and Mello, C.C. (1999). The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99, 123–132. [DOI] [PubMed] [Google Scholar]
- van der Krol, A.R., Mur, L.A., Beld, M., Mol, J.N.M., and Stuitje, A.R. (1990). Flavonoid genes in petunia: Addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 2, 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret, H., and Fagard, M. (2001). Transcriptional gene silencing in plants: Targets, inducers and regulators. Trends Genet. 17, 29–35. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Vain, P., Angell, S., and Baulcombe, D.C. (1998). Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95, 177–187. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P.M., Graham, M.W., and Wang, M.-B. (1998). Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA 95, 13959–13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe, A.P., and Matzke, M.A. (1999). Epigenetics: Regulation through repression. Science 286, 481–486. [DOI] [PubMed] [Google Scholar]
- Wu-Scharf, D., Jeong, B., Zhang, C., and Cerutti, H. (2000). Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-box RNA helicase. Science 290, 1159–1162. [DOI] [PubMed] [Google Scholar]