Abstract
Epigenetic changes, including DNA methylation, are a common finding in cancer. In lung cancers methylation of cytosine residues may affect tumor initiation and progression in several ways, including the silencing of tumor suppressor genes through promoter methylation and by providing the targets for adduct formation of polycyclic aromatic hydrocarbons present in combustion products of cigarette smoke. Although the importance of aberrant DNA methylation is well established, the extent of DNA methylation in lung cancers has never been determined. Restriction landmark genomic scanning (RLGS) is a highly reproducible two-dimensional gel electrophoresis that allows the determination of the methylation status of up to 2000 promoter sequences in a single gel. We selected 1184 CpG islands for RLGS analysis and determined their methylation status in 16 primary non-small cell lung cancers. Some tumors did not show methylation whereas others showed up to 5.3% methylation in all CpG islands of the profile. Cloning of 21 methylated loci identified 11 genes and 6 ESTs. We demonstrate that methylation is part of the silencing process of BMP3B in primary tumors and lung cancer cell lines.
Keywords: non-small cell lung cancer, DNA methylation, RLGS, genome scanning, epigenetic
Introduction
Lung cancer is the leading cause of cancer-related death in both males and females worldwide [1]. Clinically, lung cancer can be divided into two groups: small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC) [2,3]. The latter constitutes approximately 75% of all lung cancers [2] and includes squamous cell carcinoma (SCC), adenocarcinoma (AC) and large cell carcinoma (LCC) (approximately 30%, 40% and 15%, respectively in all lung cancer cases in North America) [3].
Molecular abnormalities in lung cancer affect both growth-promoting oncogenes and growth-inhibiting tumor suppressor genes. So far mutations have been reported in the oncogene K-RAS [4–6] as well as in tumor suppressor genes p53 [7], CDKN2 [8–11] and RB [12]. In addition to genetic changes, the epigenetic change of DNA methylation, the addition of a methyl group to the cytosine ring in 5′-CpG-3′ dinucleotides, may play a significant role during lung cancer development [13–17]. DNA methylation is established and maintained by a family of DNA methyltransferases [18] and affects chromatin organization as well as gene expression [19]. A well-studied example in lung cancer is the aberrant promoter methylation of the tumor suppressor gene, CDKN2, which correlates with gene silencing [8,10,20–22]. Because most of the reports describe methylation in single cancer genes, no measurement of the overall contribution of promoter methylation in lung cancer exists. As an initial step to address this question, Zochbauer-Muller et al. showed that numerous genes, including retinoic acid receptor β-2 (RARβ), tissue inhibitor of metaloproteinnase 3 (TIMP-3), CDKN2, O6-methylguanine-DNA-methyltransferase (MGMT), death-associated protein kinase (DAPK), E-cadherin (ECAD), p14ARF and glutathione S-transferase P1 (GSTP1), were methylated at various degrees in a collection of 107 primary NSCLC [22].
Methylation changes in lung cancer appear to be early events and thus should be useful in improving early detection of potentially malignant cells [23–25]. For example, CDKN2 promoter methylation is proposed as a biomarker for early detection of lung cancer and monitoring of prevention trials [23–25]. Using sensitive PCR-based methylation analysis, methylation in CDKN2 and/or MGMT promoters was found in sputum of smokers up to 3 years before clinical diagnosis of squamous cell lung carcinoma [23–25].
There is a need for the identification of novel markers in lung cancer as well as the identification of cancer-related genes. Aberrantly methylated target sequences can guide a search for novel genes that may be useful biomarkers, as well as candidate cancer genes. In this study we use, for the first time, restriction landmark genomic scanning (RLGS) to determine frequencies of DNA methylation and to identify novel methylation targets in NSCLC samples. Transcription patterns for one methylated gene, BMP3B, were studied in greater detail in primary tumors as well as in lung cancer cell lines. We show that aberrant methylation of the CpG island of BMP3B downregulates gene transcription of this interesting gene product.
Materials and Methods
Primary Human NSCLC Samples and Cell Lines
The tumor samples were derived from patients here at The Ohio State University, James Cancer Hospital. Complete pathologic classification is available for all tumor samples studied. Tissues were collected through the Cooperative Human Tissue Network to maintain patients' confidentiality. Samples were subsequently stored in our lung tumor tissue bank. Sixteen frozen paired NSCLC tumors with normal adjacent tissue were selected for this study. We used four lung cancer cell lines, all obtained from ATCC. H23 was derived from an AC, H125 was derived from an adenosquamous carcinoma, H522 was derived from an AC, and H1115 was derived from an LCC that metastasized to the lymph node. All cell lines were cultured in RPMI-1640 medium (Gibco BRL, Rockville, MD) supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 0.1 mg/ml streptomycin (Gibco BRL).
Two-Dimensional Separation by Restriction Landmark Genome Scanning (RLGS)
RLGS was performed as described previously [26,27]. In summary, high molecular weight DNA was digested with the methylation sensitive restriction enzyme NotI (Promega, Madison, WI), end-labeled by [α-32P]dGTP and [α-32P]dCTP (Amersham, Piscataway, NJ), and then digested using the restriction enzyme EcoRV (Promega). NotI-EcoRV DNA fragments were separated in a first dimension through a 0.8% agarose tube gel, followed by an in-gel digestion with a third restriction enzyme, HinfI (New England Biolabs, Beverly MA). Finally, the DNA was separated on a second dimension 5% polyacrylamide gel; the gel was dried and exposed to X-ray film for 5 to 10 days. RLGS profiles of primary tumors and normal adjacent lung tissue were superimposed to visually detect differences in the intensities and/or presence of the radiolabeled fragments.
RLGS Analysis
The fragments in the RLGS profiles have been named on our “Master RLGS profile” (see website E-mail: http://pandora.med.ohio-state.edu/masterRLGS/). The master RLGS profile, derived from normal peripheral blood lymphocyte DNA, is divided into 63 sections. Each RLGS fragment is given a unique identifier (e.g., 3C1) that relates to the position within the RLGS profile. Therefore, data sets from different patients can be compared to identify commonly changed fragments.
Cloning of RLGS Fragments
A human NotI-EcoRV plasmid library and library mixing gels were created previously to facilitate cloning of RLGS fragments [26,28]. These mixing gels allow the determination of an address for a library clone corresponding to the RLGS fragment by identifying enhancement in the plate, row, and column mixing gels [26]. Bacterial clones were cultured in LB medium with ampicillin to isolate plasmid DNA using Qiagen miniprep kit (Qiagen, Valencia, CA). The plasmid DNA was digested with NotI and EcoRV (Promega), end-labeled with [α-32P]dGTP and [α-32P]dCTP (Amersham). Labeled DNA, 5.2 and 10.4 pg per clone, were mixed with labeled peripheral blood lymphocytes (PBL) genomic DNA from a normal healthy donor, and subsequently separated in the two-dimensional RLGS mixing gel. Enhanced intensities of the RLGS fragment of interest in these mixing gels indicated that the NotI/EcoRV clone represents the RLGS fragment of interest.
Characterization of RLGS Fragments
The confirmed plasmid clones are sequenced with M13 forward and M13 reverse primers. DNA sequences from M13 forward primer were used to perform standard nucleotide-nucleotide BLAST searches, using nonredundant (nr) and high throughput genomic sequence (htgs) databases at release time March 1, 2001 (http://www.ncbi.nlm.nih.gov/BLAST/). When possible, 2-kb genomic sequences from both sides of the NotI site were used for subsequent BLAST searches for genes or ESTs. Chromosomal location of cloned DNA fragments were obtained either directly from the information given in Genebank (http://www.ncbi.nlm.nih.gov/), or by searching the OMIM database (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM) using gene names, or by searching the BAC resource website (http://www.ncbi.nlm.nih.gov/genome/cyto/hbrc.shtml) that contains cytogenetic data of FISH-mapped and sequence-tagged BAC clones. The properties of CpG islands were determined using a web-based program http://www.itba.mi.cnr.it/webgene/, which is provided by the Institute of Advanced Biomedical Technologies (ITBA), Italy.
Southern Hybridization
Southern hybridization was performed as described previously [26]. Briefly, control samples, DNA from healthy donors, were digested by EcoRV alone and a NotI/EcoRV combination. The control sample will show the expected fragment size in the EcoRV digestion and a smaller fragment in the double digestion. All tumor samples were digested by NotI/EcoRV. The probes were prepared by restriction enzyme digestion of the clone DNA, purification of the target fragments, and subsequent labeling by random priming using the Prime IT II kit (Stratagene, La Jolla, CA). Percentage of methylation was quantified by a phosphorimager.
5-aza-2′-Deoxycytidine Treatment of Cell Lines
A total of 3x106 cells of each NSCLC cell line were seeded into T75 culture flasks and cultured with RPMI-1640 media overnight. 5-aza-2′-deoxycytidine media was freshly made each day from stock solution (10 mmol/l in 100% DMSO) (Sigma, St. Louis, MO) to a final concentration of 1 µmol/l. Cells were cultured with 5-aza-2µ-deoxycytidine medium for 24 hours and then washed with PBS twice and continued to culture with fresh medium without 5-aza-2′-deoxycytidine for 2 days (24-hour timepoint). Cells for the 72-hour timepoint were cultured in 5-aza-2′-deoxycytidine medium that was changed daily for 3 days, then washed with PBS twice and continued to culture in fresh medium without 5-aza-2′-deoxycytidine for one additional day.
Semiquantitative RT-PCR
Total RNA from cell lines with or without 5-aza-2′-deoxycytidine treatment, primary tumors and paired normal adjacent lung tissue was isolated using TRIzol (Gibco BRL) and purified by RNeasy Mini Kit (Qiagen). Three micrograms total RNA was reverse transcribed in vitro by random hexamer and oligo dT using SUPERSCRIPT first-strand synthesis kit (Gibco BRL). cDNA was amplified by PCR. Primers for RT-PCR were designed from the published cDNA sequences. Forward and reverse primers are from different exons to avoid amplification from genomic DNA.
Primer sequences were as follows: BMP3B forward: 5′-GGTGGACTTCGCAGACATCG-3′; BMP3B reverse: 5′-GATGGTGGCATGGTTGGATG-3′, product size: 130 bp. GPI forward: 5′-GACCCCCAGTTCCAGAAGCTG-3′; GPI reverse: 5′-GCATCACGTCCTCCGTCACC-3′, product size: 178 bp. In all reactions the forward primer for each pairs was end labeled by [γ-32P]ATP with T4 kinase (Gibco BRL).
The semiquantitative radioactive RT-PCR was performed with optimized conditions for both the target gene and an internal control, glucose-6-phosphate isomerase (GPI), in a single reaction tube. Amplification was stopped in the exponential range for both genes. The exponential range was determined by phosphorimager quantification of the PCR product band intensities from different amplification cycles. Each PCR reaction was carried out in 50 µl final volume containing 5 µl of 10x PCR buffer, 1.5 mM MgCl2, 10 pmol of each primer, 200 µM of each dNTP and 2.5 U Platinum Taq DNA polymerase (Gibco). The reactions were initiated with 95°C for 10 minutes to activate DNA polymerase and then followed by amplification. BMP3B was amplified at 96°C for 20 seconds, 63°C for 15 seconds, and 72°C for 15 seconds for eight cycles before addition of GPI primers and additional 22 cycles.
Statistical Methods
Tests were performed for heterogeneity in methylation across patients and for preferential methylation of certain CpG island fragments, described in detail in Ref. [30]. Briefly, the heterogeneity test is based on a comparison of the mean methylation frequency to its variance in a chi-square statistic. Preferential methylation is assessed using a standard goodness-of-fit test [29] assuming that all spots are lost at equal true frequency. Empirical null distributions for both of these statistics were obtained by performing appropriate 10,000 random permutations of the fragment/patient data. Such an approach accounts for multiple testing (i.e., multiple fragments were examined) and does not rely on asymptotic distribution assumptions.
Results
Levels of Methylation in CpG Islands of NSCLC
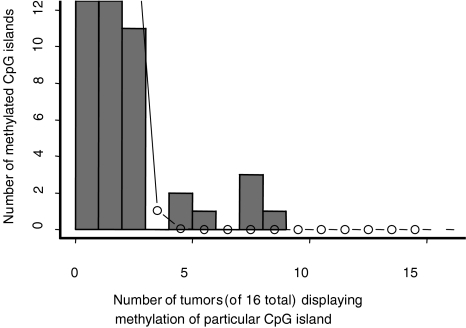
RLGS profiles from 16 matched pairs consisting of lung tumors from NSCLC patients and matched normal lung tissue were prepared using the enzyme combination NotI-EcoRV-HinfI (Figure 1A). Each tumor profile was compared against the matching normal lung RLGS profile. Previously, we had shown that the loss of an RLGS fragment in the tumor compared with the matching normal is indicative of DNA methylation in the NotI site [30,31]. The total number of methylation events out of 1184 RLGS fragments analyzed, as well as clinical data for the patients are shown in Table 1. The range of methylation in these samples is from 0% to 5.3%. Although most (12 of 16) of the tumor samples showed methylation levels below 1%, 4 of 16 patients show levels of CpG island methylation above 2%. Of these, patients 5, 11, 14, and 17 show methylation frequencies of 4.9%, 5.3%, 2.4%, and 5.0%, respectively. No obvious correlation of overall methylation frequency and clinical data can be seen. This range of variation is greater than would be expected if all patients had the same underlying methylation rate. A chi-square test (see Statistical Methods section) shows significant heterogeneity in methylation levels across the patients (P<.0001). In addition, some CpG islands are preferentially methylated, as indicated by the number of fragments showing relatively high methylation frequency (Figure 2, P<.0001). Of the total 1184 analyzed RLGS fragments, 1036 were never methylated. A total of 76 fragments were methylated in only one tumor, 53 fragments were methylated in two tumors, 9 fragments (2E61, 2F43, 3D24, 3F82, 3F85, 4D8, 4E53, 4F15, and 4F58) were methylated in three tumors, 2 fragments (2D45 and 3F16) were methylated in four tumors, 1 fragment (3F28) was methylated in five tumors, 1 fragment (3G78) was methylated in seven tumors, 5 fragments (2C35, 3C1, 3E55, 4E1, and 1 RLGS fragment not present in the RLGS master profile) were methylated in eight tumors, and 1 fragment (1F22) was methylated in nine tumor samples (Figure 2).
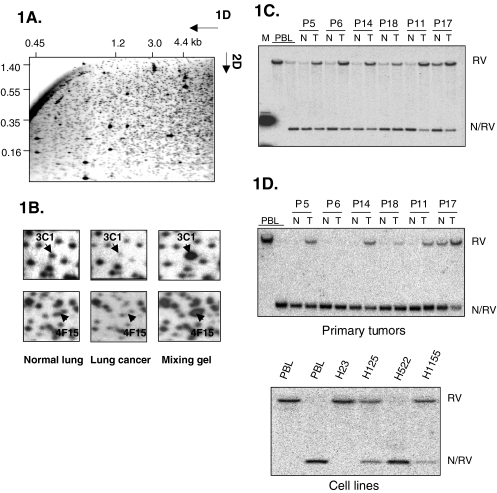
Figure 1.
RLGS and confirmation of methylation by Southern hybridization. (A) Representative RLGS profile of normal lung DNA using the enzyme combination NotI-EcoRV-HinfI. Fragment sizes for the first dimension (1D) and second dimension (2D) are given in kilobases. (B) Sections of RLGS profiles from normal lung and lung tumor from patient 17 highlighting RLGS fragment 3C1 (GNAL) and 4F15 (BMP3B) (arrows). While the lung cancer profile shows decreased intensity of 3C1 and 4F15, the mixing gel shows enhancement, indicating that the correct NotI-EcoRV clone was isolated from the library. Only differences in RLGS fragments 3C1 and 4F15 were indicated, other changes between normal and tumor profiles were not marked by arrows. (C) and (D) Southern blot analysis of primary lung cancers and matched normal adjacent tissue. Hybridization was used to confirm methylation of RLGS fragment 3C1, GNAL (C) and 4F15, BMP3B (D) in RLGS profiles of primary lung cancer is due to DNA methylation. The peripheral blood lymphocyte (PBL) DNA in the first lane of each blot was digested with EcoRV only. All other tumor (T) and normal adjacent lung (N) DNA samples were double digested by EcoRV/NotI. The majority of the DNA is digested by NotI and results in the smaller NotI-EcoRV (N/RV) fragment. Tumor samples show a much higher degree of methylation. M: indicates the marker lane. H23, H125, H522, and H1155 are lung cancer cell lines.
Table 1.
DNA Methylation in 16 Patients with Lung Cancer.
| Patient No. | Methylation events out of 1184 CpG islands | % Methylation | Age | Gender | Tumor stage | Differentiation | Tumor type |
| 1 | 5 | 0.4 | 45 | F | T2N1, IIB | Well | AC |
| 2 | 3 | 0.3 | 72 | M | T1N0, IA | Moderate | SCC |
| 3 | 7 | 0.6 | 56 | F | T2N1, IIB | Poor | LCC with features of AC |
| 5 | 58 | 4.9 | 75 | M | T2N0, IB | Moderate | SCC |
| 6 | 7 | 0.6 | 62 | F | T1N0, IA | Poor | SCC |
| 7 | 2 | 0.2 | 68 | M | T3N2, IIIA | Undifferentiated | LCC |
| 8 | 0 | 0 | 66 | M | T2N0, IB | Well | AC |
| 9 | 5 | 0.4 | 78 | F | T1N0, IA | Poor | SCC |
| 10 | 1 | 0.1 | 69 | M | T2N1, IIB | Poor | N/A |
| 11 | 63 | 5.3 | 67 | F | T2N0, IB | Poor | LCC |
| 13 | 0 | 0 | 76 | M | T1N1, IIA | Poor | SCC |
| 14 | 28 | 2.4 | 61 | M | N/A | Poor | SCC |
| 15 | 4 | 0.3 | 70 | F | T3N0, IIB | N/A | LCC with SCC and AC |
| 16 | 2 | 0.2 | 79 | F | T1N0, IA | N/A | AC (with clear cell features) |
| 17 | 59 | 5.0 | 81 | M | N/A | Well | AC |
| 18 | 8 | 0.7 | 63 | F | N/A | Poor | SCC |
SCC, squamous cell carcinoma; AC, adenocarcinoma; LCC, large cell carcinoma. N/A, no data available.
Figure 2.
Global methylation patterns in lung cancer are nonrandom. Frequency distribution of methylation events in CpG islands within 16 NSCLCs.
No correlation between global methylation frequency and any of the clinical parameters including tumor stage, differentiation, histopathological classification, age or gender could be detected. However, it is interesting to note that a subset of methylation events correlate with the histopathological features of the tumors. For example, of the 50 CpG island sequences that are methylated at least twice in tumors with clear histopathology (excluding patients No. 3, 10, and 15), 1 is found only in the two LCCs, no CpG islands was found methylated only in the four ACs, and 2 are found methylated exclusively in the seven SCCs. Other methylation events (a total of 16 CpG islands) are shared between all three subtypes. In addition seven methylation events are found in AC and LCC, nine are shared between AC and SCC and a total of 15 CpG islands are methylated in both SCC and LCC (Table 2). Validation of these methylation events in larger sample sets is required to unambiguously identify tumor-type-specific methylation events.
Table 2.
Distribution of CpG Islands Methylated at Least Twice in Various Histopathological Subgroups of Non-Small Cell Lung Cancer.
| Subgroup | Number of methylated CpG islands |
| LCC | 1 |
| AC | 0 |
| SCC | 2 |
| LCC+AC | 7 |
| AC+SCC | 9 |
| LCC+SCC | 15 |
| LCC+AC+SCC | 16 |
LCC, large cell carcinoma; AC, adenocarcinoma; SCC, squamous cell carcinoma.
Cloning of RLGS Fragments and Properties of Cloned Fragments
To further characterize some of the methylated fragments in NSCLC, the NotI/EcoRV plasmid clone library mixing gels were used to locate the corresponding clones [26]. Clones with expected insert sizes were analyzed in RLGS mixing gels to confirm that the correct fragment was cloned. Figure 1B shows two examples for RLGS fragments 3C1 and 4F15. A total of 21 fragments were cloned and sequenced. Twelve of these clones (2D14, 2D20, 2C35, 2E24, 2E61, 3B36, 3C1, 3E55, 3F16, 3F50, 3F82, and 4E53) have been identified as methylation targets in other types of malignancies [30–32]. BLAST searches identified homologies to 11 genes and six EST sequences (Table 3). Two sequences (3F16 and 3F82) show high homology to DNA-binding protein A (DBPA) and mouse early B-cell factor 3, respectively and may represent either pseudogenes or novel gene family members. The remaining two showed homology to genomic sequences. We found that 20 of 21 NotI sites are located within CpG islands. Six of the 11 CpG islands with homology to genes are located in the 5′ end of the genes. The CpG islands identified in insulin promoter factor 1 (IPF1), orthodenticle (drosophila) homolog 1 (OTX1), HOX11, T-box brain 1, monocarboxylate transporter 3 (MCT3) are located in the middle or 3′ end of the genes. Chromosomal location of all the fragments were derived from database searches of the human draft sequence of the human genome, Genebank or OMIM database. The detailed information for these 21 fragments, including fragment addresses on master profile, total methylation frequency in primary tumors, CpG island properties and location in the genes, BLAST search results, and chromosomal location of the genes, are listed in Table 3.
Table 3.
Cloned RLGS Fragments Altered in RLGS Profiles of 16 Primary Tumors.
| RLGS master address | No. of tumors methylated | Methylation found in following subgroups | CpG island | Location of CpG island in gene | Gene or EST | Accession number | Chromosomal location |
| 2C35 | 8 | AC/LCC/SCC | Yes | EST BG142595 | AL139281, NT_024073.1 | 10p12 | |
| 3C1 | 8 | AC/LCC/SCC | Yes | 5′ end | GNAL | U55180 | 18p11.21-pter |
| 3E55 | 8 | AC/LCC/SCC | Yes | 3′ end | IPF1 | NM 000209 | 13q12.1 |
| 3G78 | 7 | AC/LCC/SCC | Yes | 5′ end | TAL1 | AL135960 | 1p32 |
| 2D20 | 4 | AC/LCC/SCC | Yes | middle | OTX1 | AB037501 | 2p13 |
| 3F16 | 4 | AC/LCC/SCC | Yes | Homologous to EBF | AL354950, NT_024100.1 | 10q26 | |
| 3F50 | 4 | LCC/SCC | Yes | 3′ end | HOX11 | AJ009794 | 10q24 |
| 2E61 | 3 | SCC | Yes | EST AJ230817 | AL354000 | 17p11.2 | |
| 3B55 | 3 | LCC/SCC | Yes | 3′ end | T-box brain 1 | XM_002531 | 2q23–27 |
| 3F82 | 3 | LCC/SCC | Yes | Homologous to DBPA | M24069 | ||
| 4D8 | 3 | AC/LCC/SCC | No | EST BF928282 | AC012118, NT_010641.1 | 17q25.1 | |
| 4E15 | 3 | LCC/SCC | Yes | EST BE247619 | AC003959 | 5q | |
| 4E53 | 3 | AC/LCC/SCC | Yes | AL353195, NT_009829.1 | 13q12.2 | ||
| 4F15 | 3 | LCC/SCC | Yes | 5′ end | BMP3B | D49493, NT_008757.1 | 10q11.21–11.23 |
| 5E25 | 3 | AC/LCC/SCC | Yes | Formin 2 like | AL359918, NT_004771.1 | 1q44 | |
| 2D14 | 2 | AC/LCC | Yes | 5′ end | CD8 β1 Chain | S87068 | 2p12 |
| 3B36 | 2 | AC/SCC | Yes | 5′ end | CYP1b1 | XM_002576 | 2p21–22 |
| 3D44 | 2 | AC/SCC | Yes | AL355304, NT_019429.1 | 6q23.1–6q24.3 | ||
| 5C32 | 2 | AC/SCC | Yes | 5′ end | CD34 | M81938 | 1q32 |
| 2E24 | 1 | AC | Yes | middle | MCT3 | AF132611 | 22q12.3–13.2 |
| 3E34 | 1 | SCC | Yes | EST AI631157 | AC000068 | 22q11.2 | |
AC, adenocarcinoma; LCC, large cell carcinoma; SCC, squamous cell carcinoma.
RLGS Fragment Loss Is Due to DNA Methylation
Southern hybridization was performed as a more sensitive method to estimate the degree of methylation of the NotI site from RLGS fragments lost in tumors. Southern hybridization has the ability to detect methylation as low as 5% to 10%, whereas RLGS allows the detection of 30% methylation [31]. Southern blot hybridization was also performed to evaluate the possibility that the RLGS fragment loss was due to homozygous deletion. Corresponding NotI/EcoRV plasmid clones for four RLGS fragments were used as probes on Southern blots. These four clones included three fragments with homology to known genes 3C1 (G-α-olfactory, GNAL), 3E55 (insulin promoter factor 1, IPF1), and 4F15 (Bone morphogenetic protein 3B, BMP3B). In addition, fragment 2C35 with homology to an EST sequence was used. Representative Southern blots for RLGS fragment 3C1 (GNAL) and 4F15 (BMP3B) are shown in Figure 1C and D, respectively. Tumor samples and paired normal adjacent lung tissue DNAs were digested with NotI/EcoRV. The detection of a fragment in the NotI/EcoRV digests equal in size to the one detected with EcoRV only was scored as a methylation event in the tumor DNA. The hybridization shows that most tumor samples have a much higher degree of methylation of the NotI site than the normal counterparts. Some of the normal tissue DNAs show a small fraction of methylated sequences. These normal lung tissues were matched controls derived from the lung cancer patients and it is possible that they may contain contaminating preneoplastic cells. No homozygous deletions were detected because the hybridization signals are present in all tumor samples. Methylation of the NotI site in the 5′ end of GNAL was observed in 10 of 16 patients. Methylation of 4F15 (BMP3B), was detected in five of six tumors and thus confirmed the methylation events found by RLGS. Tumor-specific or increased methylation was observed in 11 of 16 primary NSCLC for fragment 2C35 and 11 of 16 for 3E55 (IPF1) (Figure 1C and D and data not shown).
Aberrant Transcription of BMP3B in Primary NSCLC and NSCLC Cell Lines
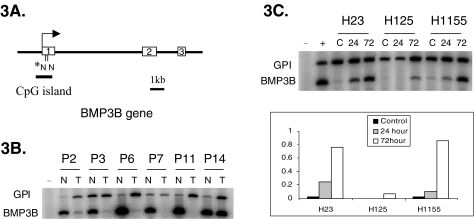
Radioactive, semiquantitative RT-PCR reactions were performed to determine the expression levels of BMP3B in six primary NSCLC samples and their paired normal lung tissue. BMP3B was found hypermethylated in a CpG island that is located in the 5′ end of the gene (Figure 3A) and was selected for further analysis. RT-PCR was performed under optimized conditions for both the target gene and the internal control gene glucose-6-phosphate isomerase (GPI). BMP3B was expressed in normal lung, whereas expression of BMP3B in all studied tumor samples was reduced (Figure 3B). Interestingly, tumors from patients Nos. 2 and 6, which did not show methylation of the NotI site exhibited very low levels of BMP3B expression, suggesting heterogeneous methylation of the CpG island. However, it is also possible, and maybe even more likely, that BMP3B is silenced by other mechanisms including mutations, deletions and/or LOH.
Figure 3.
Abnormal transcription of BMP3B NSCLC samples. (A) Schematic representation of the genomic structure of BMP3B. The location of the NotI site relative to exon 1 and the extent of the CpG island are displayed. The star indicates the NotI site identified by RLGS. (B) Semiquantitative radioactive RT-PCR was used to determine the relative expression levels of BMP3B in six patients with lung cancer. Relative expression levels of RNAs obtained from tumor (T) and normal adjacent lung (N) were determined by comparing the intensities to the internal control, GPI. Negative control (-) contains water. GPI: Glucose-6-phosphate isomerase. (C) Radioactive semiquantitative RT-PCR was used to determine the relative expression of BMP3B levels in RNAs from three cell lines (H23, H125, and H1155) after treatment with 5′-aza-2′-deoxycytidine. The upper panel shows the RT-PCR results and the lower panel shows the relative expression levels of BMP3B compared to the internal control GPI. Cell lines were treated with 1 µmol/l 5′-aza-2′-deoxycytidine for either 24 hours (24) or 72 hours (72). Untreated control cell lines (C) were harvested in exponential growth phase. Y-axis gives the relative ratio of band intensity for the target gene/GPI quantified by phosphorimager. Negative control (-) contained water, positive control (+) was normal lung tissue (NL).
To investigate the effect of CpG island methylation on the transcription of the associated genes in more detail, we used three NSCLC cell lines that are methylated in the NotI site of BMP3B promoter region. H23, H125, and H1155 show more than 50% methylation in the BMP3B promoter. The cells were treated with two different timepoints (24 and 72 hours) of 5-aza-2′-deoxycytidine (Figure 3C). Similar to the primary tumors, all three cell lines (H23, H125, H1155) did not show any detectable level of BMP3B expression at baseline. However, expression was induced after 5-aza-2′-deoxycytidine treatment in all three cell lines (Figure 3C).
Discussion
The methylation scanning properties of RLGS have previously been used for the identification of imprinted genes in the mouse genome [33,34], as well as for the identification of methylated sequences in various human malignancies [30,31,35] but not lung cancer. Genome-wide scans for methylated sequences in lung cancer have been performed by two different strategies including the use of a methylated DNA binding column [36] and arbitrarily primed PCR [37]. However, these techniques either showed a bias for methylated repetitive sequences or were limited in the number of analyzed sequences. In contrast, RLGS, a highly reproducible two-dimensional gel electrophoresis, is a genome-wide scan of DNA methylation changes in CpG islands. Established cloning protocols utilizing an arrayed plasmid clone library facilitate the rapid identification of genomic sequences corresponding to the methylated targets [26,28]. RLGS is based on the digestion of genomic DNA with the methylation sensitive restriction enzyme NotI, which can only digest unmethylated genomic DNA and does not rely on prior knowledge of the gene sequence [26,31,35].
We have used RLGS to determine the contribution of CpG island hypermethylation in NSCLC and to identify novel methylation targets. Although the importance of DNA methylation in lung tumor development had been demonstrated in several reports [23,38–40], the overall extent was previously unknown. Under the assumption that all RLGS fragments represent promoter sequences we are, for the first time, able to demonstrate that up to 5.3% of all promoter regions, or 1537 of the estimated total 29,000 CpG islands [41,42] in the tumor genome could be methylated. The variability in the range of methylation shows that NSCLC represents a heterogeneous group not only with respect to the genetic defects identified but also on the epigenetic level.
Promoter methylation in cancer-related genes is well known in lung cancer and is correlated with gene silencing in genes involved in cell cycle (CDKN2) [8–11], apoptosis (DAP) [43], metastasis H-cadherin [44] and (TIMP-3) [45], differentiation (RARβ) [46], DNA repair (MGMT) [47], and the recently identified candidate tumor suppressor gene (RASSF1A) with homology to the RAS family [38]. In this study, we have identified 21 additional genomic loci including 11 genes and six ESTs with aberrant CpG island methylation in NSCLC. None of the cloned genes has been reported to be methylated in lung cancer previously. However, it is interesting to note that CpG island 3F16 is approximately 200 kb away from MGMT (located in sequence contig NT_024100.1). MGMT is a DNA repair gene that was previously found to be methylated in 21% of NSCLCs [22]. This finding would suggest a more regional effect of methylation, similar to aberrant methylation found in chromosome 17p11.2 in the major breakpoint cluster region for medulloblastomas [30]. Another interesting target sequence is 4D8 located in chromosome 17q25.1. Chromosome 17q had previously been implicated with frequent (42%) loss of heterozygosity in NSCLC [48], suggesting that both genetic and epigenetic mechanisms could be involved in the silencing of a putative tumor suppressor gene in this region. Other fragments are derived from chromosome segments in 1p (3G78), 5q (4E15), 10q (3F50), 13q (3E55) 18p (3C1), and 22q (2E24), all within regions for which either LOH or homozygous deletions have been reported in lung cancer [49,50]. Whether any of the newly identified genes meet the expected criteria for tumor suppressor genes remains to be determined and will be the focus of future work.
We focused our studies on RLGS fragment 4F15, derived from a CpG island in the promoter region of the BMP3B gene and determined the relation of CpG island methylation to transcription. BMP3B is located in chromosome 10q11.21-11.23, a region that shows 20% to 30% LOH in NSCLCs and 51% LOH in SCLCs [51] (see online http://www.helsinki.fi/∼lgl_www/LOSS/Respiratory.html) and thus located in a candidate tumor suppressor region. BMP3B is a member of the transforming growth factor β (TGF-β) superfamily, originally identified due to their osteoinductive capacity. Members of this family are usually involved in the regulation of cell growth/differentiation during development and were shown to be dysregulated in various human malignancies. Interestingly other members of the BMP family have been shown to induce apoptosis during organ development [52–54]. In addition, two BMP family members, BMP4 and BMP2, have been shown to induce apoptosis in multiple myeloma cell lines [55] or hematopoetic cells [56], respectively. BMP2 was also shown to suppress the transformed phenotype in the human lung carcinoma cell line A549 [57].
BMP3B is highly expressed in human adult lung, brain, skeletal muscle, pancreas, and testis, an expression pattern that distinguishes it from the closest family member BMP3 [58]. BMP3B knockout mice did not show any detectable abnormalities, suggesting a redundant function with that of other members of the TGF-β family [59]. However, adult rat lung tissue does not express BMP3B suggesting the possibility of different functions of BMP3B in rodents and humans [60]. We have shown that methylation in BMP3B is correlated with transcriptional repression. The repression is reversible by treatment with the demethylating agent 5-aza-2′deoxycytidine. Thus, our data suggest a causal relationship between methylation of the BMP3B promoter and transcriptional repression. We found BMP3B downregulated in all NSCLC patient samples and cell lines, even in those without methylation in the NotI site, suggesting that methylation patterns in the CpG island are heterogeneous. This assumption was confirmed by COBRA analysis testing the methylation status in four BstUI (CGCG) restriction sites in the promoter (Dai et al. unpublished). Alternatively, other mechanisms (e.g., LOH or mutations) of gene silencing could be present. Additional work to study the complete genetic and epigenetic mutation spectrum of BMP3B in lung tumorigenesis is underway.
The identification of multiple targets for methylation opens the exciting possibility to use these methylation events as biomarkers for the early detection of lung cancer in sputum as demonstrated by others [23]. In addition, our data also indicate the possibility that certain methylation events may be specific for lung cancer or subtypes within this group and thus could serve as potential markers for the molecular classification of lung cancers and different disease stages. Validation of methylation events as possible markers for early diagnosis, as predictive markers for survival or markers that classify subtypes will require larger sets of patient samples.
Acknowledgements
We thank Julia Hall and Yue-Zhong Wu for expert technical assistance. The authors thank Ming You for helpful advice with the manuscript. Tissue samples were provided by the Comparative Human Tissue Network (CHTN), which is funded by the National Cancer Institute.
Footnotes
This work was supported in part by the National Cancer Institute, Bethesda, MD, grant P30 CA16058, by a grant from the Ohio Cancer Research Associates (G.O.), and by grant DFG15.16/1-1 (M.F.).
References
- 1.Parkin DM, Pisani P, Ferlay J. Global cancer statistics. Ca-Cancer J Clin. 1999;49:33–64. doi: 10.3322/canjclin.49.1.33. [DOI] [PubMed] [Google Scholar]
- 2.Marby M, Nelkin BD, Baylin SB. Chapter 41 Lung cancer. In: Vogelstein B, Kinzler KW, editors. The Genetic Basis of Human Cancer. New, York, NY: McGraw-Hill; 1998. pp. 671–679. [Google Scholar]
- 3.Ginsberg RJ, Vokes EE, Raben A. Section 2: Non-small cell lung cancer in chapter 30: cancer of the lung. In: DeVita VT Jr, Hellman S, Rosenberg SA, editors. Cancer: Principles and Practice of Oncology. Philadelphia, PA: Lippincott-Raven; 1997. pp. 858–865. [Google Scholar]
- 4.Sekido Y, Fong KM, Minna JD. Progress in understanding the molecular pathogenesis of human lung cancer. Biochim Biophys Acta. 1998;1378:F21–F59. doi: 10.1016/s0304-419x(98)00010-9. [DOI] [PubMed] [Google Scholar]
- 5.Slebos RJ, Rodenhuis S. The ras gene familyin human non-small-cell lung cancer. J Natl Cancer Inst Monogr. 1992;13:23–29. [PubMed] [Google Scholar]
- 6.Salgia R, Skarin AT. Molecular abnormalities in lung cancer. J Clin Oncol. 1998;16:1207–1217. doi: 10.1200/JCO.1998.16.3.1207. [DOI] [PubMed] [Google Scholar]
- 7.Chiba I, Takahashi T, Nau MM, D'Amico D, Curiel DT, Mitsudomi T, Buchhagen DL, Carbone D, Piantadosi S, Koga H, et al. Mutations in the p53 gene are frequent in primary, resected non-small cell lung cancer. Oncogene. 1990;5:1603–1610. [PubMed] [Google Scholar]
- 8.Otterson GA, Khleif SN, Chen W, Coxon AB, Kaye FJ. CDKN2 gene silencing in lung cancer by DNA hypermethylation and kinetics of p16INK4 protein induction by 5-aza 2′deoxycytidine. Oncogene. 1995;11:1211–1216. [PubMed] [Google Scholar]
- 9.Otterson GA, Kratzke RA, Coxon A, Kim YW, Kaye FJ. Absence of p16INK4 protein is restricted to the subset of lung cancer lines that retains wildtype RB. Oncogene. 1994;9:3375–3378. [PubMed] [Google Scholar]
- 10.Gazzeri S, Gouyer V, Vour'ch C, Brambilla C, Brambilla E. Mechanisms of p16INK4A inactivation in non small-cell lung cancers. Oncogene. 1998;16:497–504. doi: 10.1038/sj.onc.1201559. [DOI] [PubMed] [Google Scholar]
- 11.Kashiwabara K, Oyama T, Sano T, Fukuda T, Nakajima T. Correlation between methylation status of the p16/CDKN2 gene and the expression of p16 and Rb proteins in primary non-small cell lung cancers. Int J Cancer. 1998;79:215–220. doi: 10.1002/(sici)1097-0215(19980619)79:3<215::aid-ijc1>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 12.Xu HJ, Hu SX, Cagle PT, Moore GE, Benedict WF. Absence of retinoblastoma protein expression in primary non-small cell lung carcinomas. Cancer Res. 1991;51:2735–2739. [PubMed] [Google Scholar]
- 13.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 14.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 15.Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis. 2000;21:461–467. doi: 10.1093/carcin/21.3.461. [DOI] [PubMed] [Google Scholar]
- 16.Tycko B. Epigenetic gene silencing in cancer. J Clin Invest. 2000;105:401–407. doi: 10.1172/JCI9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warnecke PM, Bestor TH. Cytosine methylation and human cancer. Curr Opin Oncol. 2000;12:68–73. doi: 10.1097/00001622-200001000-00012. [DOI] [PubMed] [Google Scholar]
- 18.Bird A. DNA methylation de novo. Science. 1999;286:2287–2288. doi: 10.1126/science.286.5448.2287. [DOI] [PubMed] [Google Scholar]
- 19.Bird AP, Wolffe AP. Methylation-induced repression-belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 20.Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 21.Nuovo GJ, Plaia TW, Belinsky SA, Baylin SB, Herman JG. In situ detection of the hypermethylation-induced inactivation of the p16 gene as an early event in oncogenesis. Proc Natl Acad Sci USA. 1999;96:12754–12759. doi: 10.1073/pnas.96.22.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zochbauer-Muller S, Fong KM, Virmani AK, Geradts J, Gazdar AF, Minna JD. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res. 2001;61:249–255. [PubMed] [Google Scholar]
- 23.Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, Baylin SB, Herman JG. Aberrant methylation of p16 (INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci USA. 1998;95:11891–11896. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kersting M, Friedl C, Kraus A, Behn M, Pankow W, Schuermann M. Differential frequencies of p16 (INK4a) promoter hypermethylation, p53 mutation, and K-ras mutation in exfoliative material mark the development of lung cancer in symptomatic chronic smokers. J Clin Oncol. 2000;18:3221–3229. doi: 10.1200/JCO.2000.18.18.3221. [DOI] [PubMed] [Google Scholar]
- 25.Palmisano WA, Divine KK, Saccomanno G, Gilliland FD, Baylin SB, Herman JG, Belinsky SA. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000;60:5954–5958. [PubMed] [Google Scholar]
- 26.Smiraglia DJ, Frühwald MC, Costello JF, McCormick SP, Dai Z, Peltomaki P, MS Od, Cavenee WK, Plass C. A new tool for the rapid cloning of amplified and hypermethylated human DNA sequences from restriction landmark genome scanning gels. Genomics. 1999;58:254–262. doi: 10.1006/geno.1999.5840. [DOI] [PubMed] [Google Scholar]
- 27.Hatada I, Hayashizaki Y, Hirotsune S, Komatsubara H, Mukai T. A genomic scanning method for higher organisms using restriction sites as landmarks. Proc Natl Acad Sci USA. 1991;88:9523–9527. doi: 10.1073/pnas.88.21.9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plass C, Weichenhan D, Catanese J, Costello JF, Yu F, Yu L, Smiraglia D, Cavenee WK, Caligiuri MA, deJong P, Held WA. An arrayed human NotI-EcoRV boundary library as a tool for RLGS spot analysis. DNA Res. 1997;4:253–255. doi: 10.1093/dnares/4.3.253. [DOI] [PubMed] [Google Scholar]
- 29.Freund RJ, Wilson WJ. Statistical Methods. San Diego, CA: Academic Press; 1997. [Google Scholar]
- 30.Frühwald MC, O'Dorisio MS, Dai Z, Rush LJ, Krahe R, Smiraglia DJ, Pietsch T, Elsea SH, Plass C. Aberrant hypermethylation of the major breakpoint cluster region in 17p11. in medulloblastomas but not supratentorial PNETs. Genes Chromosomes Cancer. 2001;30:38–47. [PubMed] [Google Scholar]
- 31.Costello JF, Frühwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomaki P, Lang JC, Schuller DE, Yu L, Bloomfield CD, Caligiuri MA, Yates A, Nishikawa R, Su Huang H, Petrelli NJ, Zhang X, MS OD, Held WA, Cavenee WK, Plass C. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 32.Rush LJ, Dai Z, Smiraglia DJ, Gao X, Wright FA, Fruhwald MF, Costello JF, Held WA, Yu L, Krahe R, Kolitz JE, Bloomfield CD, Caligiuri MA, Plass C. Novel methylation targets in de novo acute myeloid leukemia with prevalence of chromosome 11 loci. Blood. 2001;17:3226–3233. doi: 10.1182/blood.v97.10.3226. [DOI] [PubMed] [Google Scholar]
- 33.Plass C, Shibata H, Kalcheva I, Mullins L, Kotelevtseva N, Mullins J, Kato R, Sasaki H, Hirotsune S, Okazaki Y, Held WA, Hayashizaki Y, Chapman VM. Identification of Grf1 on mouse chromosome 9 as an imprinted gene by RLGS-M. Nat Genet. 1996;14:106–109. doi: 10.1038/ng0996-106. [DOI] [PubMed] [Google Scholar]
- 34.Hayashizaki Y, Shibata H, Hirotsune S, Sugino H, Okazaki Y, Sasaki N, Hirose K, Imoto H, Okuizumi H, Muramatsu M, et al. Identification of an imprinted U2af binding protein related sequence on mouse chromosome 11 using the RLGS method. Nat Genet. 1994;6:33–40. doi: 10.1038/ng0194-33. [DOI] [PubMed] [Google Scholar]
- 35.Plass C, Yu F, Yu L, Strout MP, El-Rifai W, Elonen E, Knuutila S, Marcucci G, Young DC, Held WA, Bloomfield CD, Caligiuri MA. Restriction landmark genome scanning for aberrant methylation in primary refractory and relapsed acute myeloid leukemia; involvement of the WIT-1 gene. Oncogene. 1999;18:3159–3165. doi: 10.1038/sj.onc.1202651. [DOI] [PubMed] [Google Scholar]
- 36.Shiraishi M, Chuu YH, Sekiya T. Isolation of DNA fragments associated with methylated CpG islands in human adenocarcinomas of the lung using a methylated DNA binding column and denaturing gradient gel electrophoresis. Proc Natl Acad Sci USA. 1999;96:2913–2918. doi: 10.1073/pnas.96.6.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohno T, Kawanishi M, Inazawa J, Yokota J. Identification of CpG islands hypermethylated in human lung cancer by the arbitrarily primed-PCR method. Hum Genet. 1998;102:258–264. doi: 10.1007/s004390050689. [DOI] [PubMed] [Google Scholar]
- 38.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 39.Denissenko MF, Chen JX, Tang MS, Pfeifer GP. Cytosine methylation determines hot spots of DNA damage in the human P53 gene. Proc Natl Acad Sci USA. 1997;94:3893–3898. doi: 10.1073/pnas.94.8.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eguchi K, Kanai Y, Kobayashi K, Hirohashi S. DNA hypermethylation at the D17S5 locus in non-small cell lung cancers: its association with smoking history. Cancer Res. 1997;57:4913–4915. [PubMed] [Google Scholar]
- 41.International-Human-Genome-Sequencing-Consortium, author. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 42.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Francesco VD, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang ZY, Wang A, Wang X, Wang J, Wei MH, Wides R. The sequence of the human genome. Science. 2001;5507:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 43.Tang X, Khuri FR, Lee JJ, Kemp BL, Liu D, Hong WK, Mao L. Hypermethylation of the death-associated protein (DAP) kinase promoter and aggressiveness in stage I non-small-cell lung cancer. J Natl Cancer Inst. 2000;92:1511–1516. doi: 10.1093/jnci/92.18.1511. [DOI] [PubMed] [Google Scholar]
- 44.Sato M, Mori Y, Sakurada A, Fujimura S, Horii A. The Hcadherin (CDH13) gene is inactivated in human lung cancer. Hum Genet. 1998;103:96–101. doi: 10.1007/s004390050790. [DOI] [PubMed] [Google Scholar]
- 45.Bachman KE, Herman JG, Corn PG, Merlo A, Costello JF, Cavenee WK, Baylin SB, Graff JR. Methylation-associated silencing of the tissue inhibitor of metalloproteinase-3 gene suggest a suppressor role in kidney, brain, and other human cancers. Cancer Res. 1999;59:798–802. [PubMed] [Google Scholar]
- 46.Virmani AK, Rathi A, Zochbauer-Muller S, Sacchi N, Fukuyama Y, Bryant D, Maitra A, Heda S, Fong KM, Thunnissen F, Minna JD, Gazdar AF. Promoter methylation and silencing of the retinoic acid receptor-beta gene in lung carcinomas. J Natl Cancer Inst. 2000;92:1303–1307. doi: 10.1093/jnci/92.16.1303. [DOI] [PubMed] [Google Scholar]
- 47.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 48.Fong KM, Kida Y, Zimmerman PV, Ikenaga M, Smith PJ. Loss of heterozygosity frequently affects chromosome 17q in non-small cell lung cancer. Cancer Res. 1995;55:4268–4272. [PubMed] [Google Scholar]
- 49.Kohno T, Yokota J. How many tumor suppressor genes are involved in human lung carcinogenesis? Carcinogenesis. 1999;20:1403–1410. doi: 10.1093/carcin/20.8.1403. [DOI] [PubMed] [Google Scholar]
- 50.Nishioka M, Kohno T, Takahashi M, Niki T, Yamada T, Sone S, Yokota J. Identification of a 428-kb homozygously deleted region disrupting the SEZ6L gene at 22q12.1 in a lung cancer cell line. Oncogene. 2000;19:6251–6260. doi: 10.1038/sj.onc.1204031. [DOI] [PubMed] [Google Scholar]
- 51.Knuutila S, Aalto Y, Autio K, Bjorkqvist AM, El-Rifai W, Hemmer S, Huhta T, Kettunen E, Kiuru-Kuhlefelt S, Larramendy ML, Lushnikova T, Monni O, Pere H, Tapper J, Tarkkanen M, Varis A, Wasenius VM, Wolf M, Zhu Y. DNA copy number losses in human neoplasms. Am J Pathol. 1999;155:683–694. doi: 10.1016/S0002-9440(10)65166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trousse F, Esteve P, Bovolenta P. BMP4 Mediates apoptotic cell death in the developing chick eye. J Neurosci. 2001;21:1292–1301. doi: 10.1523/JNEUROSCI.21-04-01292.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mehler MF, Mabie PC, Zhang D, Kessler JA. Bone morphogenetic proteins in the nervous system. Trends Neurosci. 1997;20:309–317. doi: 10.1016/s0166-2236(96)01046-6. [DOI] [PubMed] [Google Scholar]
- 54.Merino R, Ganan Y, Macias D, Rodriguez-Leon J, Hurle JM. Bone morphogenetic proteins regulate interdigital cell death in the avian embryo. Ann NY Acad Sci. 1999;887:120–132. doi: 10.1111/j.1749-6632.1999.tb07927.x. [DOI] [PubMed] [Google Scholar]
- 55.Hjertner O, Hjorth-Hansen H, Borset M, Seidel C, Waage A, Sundan A. Bone morphogenetic protein-4 inhibits proliferation and induces apoptosis of multiple myeloma cells. Blood. 2001;97:516–522. doi: 10.1182/blood.v97.2.516. [DOI] [PubMed] [Google Scholar]
- 56.Kawamura C, Kizaki M, Yamato K, Uchida H, Fukuchi Y, Hattori Y, Koseki T, Nishihara T, Ikeda Y. Bone morphogenetic protein-2 induces apoptosis in human myeloma cells with modulation of STAT3. Blood. 2000;96:2005–2011. [PubMed] [Google Scholar]
- 57.Tada A, Nishihara T, Kato H. Bone morphogenetic protein 2 suppresses the transformed phenotype and restores actin microfilaments of human lung carcinoma A549 cells. Oncol Rep. 1998;5:1137–1140. doi: 10.3892/or.5.5.1137. [DOI] [PubMed] [Google Scholar]
- 58.Hino J, Takao M, Takeshita N, Konno Y, Nishizawa T, Matsuo H, Kangawa K. cDNA cloning and genomic structure of human bone morphogenetic protein-3b (Bmp-3b) Biochem Biophys Res Commun. 1996;223:304–310. doi: 10.1006/bbrc.1996.0889. [DOI] [PubMed] [Google Scholar]
- 59.Zhao R, Lawler AM, Lee SJ. Characterization of GDF-10 expression patterns and null mice. Dev Biol. 1999;212:68–79. doi: 10.1006/dbio.1999.9326. [DOI] [PubMed] [Google Scholar]
- 60.Takao M, Hino J, Takeshita N, Konno Y, Nishizawa T, Matsuo H, Kangawa K. Identification of rat bone morphogenetic protein-3b (BMP-3b), a new member of BMP-3. Biochem Biophys Res Commun. 1996;219:655–662. doi: 10.1006/bbrc.1996.0289. [DOI] [PubMed] [Google Scholar]