Abstract
Plants respond to biotic and abiotic stresses by inducing overlapping sets of mitogen-activated protein kinases (MAPKs) and response genes. To define the mechanisms of how different signals can activate a common signaling pathway, upstream activators of SIMK, a salt stress– and pathogen-induced alfalfa MAPK, were identified. Here, we compare the properties of SIMKK, a MAPK kinase (MAPKK) that mediates the activation of SIMK by salt stress, with those of PRKK, a distantly related novel MAPKK. Although both SIMKK and PRKK show strongest interaction with SIMK, SIMKK can activate SIMK without stimulation by upstream factors. In contrast, PRKK requires activation by an upstream activated MAPKK kinase. SIMKK mediates pathogen elicitor signaling and salt stress, but PRKK transmits only elicitor-induced MAPK activation. Of four tested MAPKs, PRKK activates three of them (SIMK, MMK3, and SAMK) upon elicitor treatment of cells. However, PRKK is unable to activate any MAPK upon salt stress. In contrast, SIMKK activates SIMK and MMK3 in response to elicitor, but it activates only SIMK upon salt stress. These data show that (1) MAPKKs function as convergence points for stress signals, (2) MAPKKs activate multiple MAPKs, and (3) signaling specificity is obtained not only through the inherent affinities of MAPKK-MAPK combinations but also through stress signal–dependent intracellular mechanisms.
INTRODUCTION
Protein phosphorylation is one of the major mechanisms for controlling cellular functions in response to external signals. In eukaryotes, a specific class of Ser/Thr protein kinases, the mitogen-activated protein kinases (MAPKs), is involved in many of these processes. A general feature of MAPK cascades is their composition of three functionally linked protein kinases. A MAPK is phosphorylated and thereby activated by a MAPK kinase (MAPKK), which itself becomes activated by another Ser/Thr protein kinase, a MAPKK kinase (MAPKKK). Targets of MAPKs can be various transcription factors and protein kinases as well as upstream components of the MAPK cascade, such as MAPKKs, MAPKKKs, or the receptors themselves (Karin, 1998; Whitmarsh and Davis, 1998).
Signaling through MAPK cascades can lead to different cellular responses, including differentiation, cell division, and stress responses (Robinson and Cobb, 1997). In plants, a number of studies have demonstrated that MAPKs play roles in development, cell division, and hormone action (Ligterink and Hirt, 2001). Plant MAPKs also are involved in signaling of biotic and abiotic stresses, including cold and drought (Jonak et al., 1996) and wounding (Seo et al., 1995; Usami et al., 1995; Bögre et al., 1997; Zhang and Klessig, 1998a), and during plant–pathogen interactions (Ligterink et al., 1997; Zhang and Klessig, 1998b; Romeis et al., 1999; Cardinale et al., 2000; Droillard et al., 2000; Nühse et al., 2000).
Several MAPKKs have been isolated from different plants, including Arabidopsis AtMEK1 and AtMKK2-5 (Morris et al., 1997; Ichimura et al., 1998a, 1998b), alfalfa SIMKK (Kiegerl et al., 2000), tomato LeMEK1 (Hackett et al., 1998), tobacco NPK2, NtMEK1-2, and SIPKK (Shibata et al., 1995; Liu et al., 2000; Calderini et al., 2001; Yang et al., 2001), and maize ZmMEK1 (Hardin and Wolniak, 1998). At present, it is still unclear on which pathways most of these MAPKKs function. Recently, however, SIMKK was shown to mediate the salt-induced activation of SIMK in alfalfa (Kiegerl et al., 2000) and NtMEK2 was found to be an upstream activator of SIPK and WIPK in tobacco (Yang et al., 2001).
We reported previously the identification and characterization of SIMK as a salt stress– and elicitor-induced MAPK from alfalfa (Munnik et al., 1999; Cardinale et al., 2000) and determined SIMKK as a mediator of SIMK activation by salt stress (Kiegerl et al., 2000). In this article, we report the isolation and functional characterization of another alfalfa MAPKK termed PRKK. PRKK and SIMKK both were isolated by interaction screening with SIMK in yeast. In contrast to SIMKK, recombinant PRKK is inactive as a MAPKK and requires activation by an upstream MAPKKK. Upon pathogen elicitor but not salt stress, PRKK activated three specific MAPKs. In contrast, SIMKK activated distinct MAPKs in response to salt stress or pathogen elicitor. These results indicate that the activation of stress-responsive MAPKs underlies not only pairwise affinities between MAPKKs and MAPKs but also complex stress signal–dependent intracellular mechanisms.
RESULTS
Isolation of PRKK
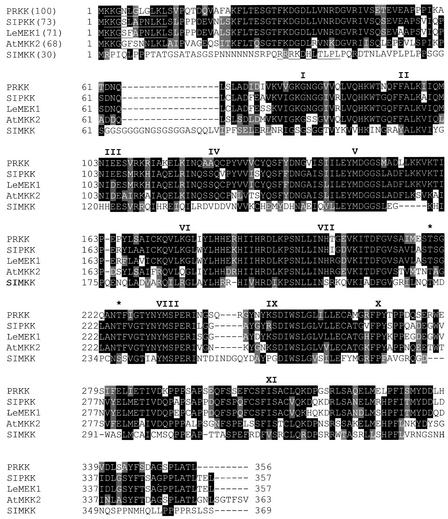
To determine the mechanisms of how common MAPK pathways can be activated by different stresses, we decided to isolate and study the upstream components of SIMK, a salt stress– and elicitor-activated MAPK from alfalfa (Munnik et al., 1999; Cardinale et al., 2000). For this purpose, SIMK was fused to the GAL4 DNA binding domain of pBD-Gal4Cam. The yeast strain PJ69-4A was cotransformed with pBD-Gal4Cam-SIMK, and a plasmid cDNA library was prepared from suspension-cultured cells of alfalfa (Hybri-ZAP), which expresses the plant genes as fusions with the GAL4 activation domain. Approximately 150,000 transformants were screened by plating on selective medium lacking adenine, Leu, and Trp. Colonies were analyzed by plasmid rescue (Robzyk and Kassir, 1992) and retransformation of PJ69-4A yeast cells, followed by testing of their ability to confer adenine and histidine prototrophy to the yeast cells when coexpressed with SIMK. Several clones were obtained that could grow on medium lacking either adenine or histidine. Sequencing of the inserts of the isolated plasmids revealed that one of the cDNAs potentially encodes a protein of the family of MAPKKs; it was named PRKK for pathogen-responsive MAPKK. PRKK has the typical features of MAPKKs, containing 11 catalytic subdomains and two putative phosphorylation sites (Figure 1, asterisks) that are targeted by the upstream activating MAPKKK.
Figure 1.
Primary Structure of PRKK, a Pathogen-Responsive Alfalfa MAPKK.
The amino acid sequence of PRKK was aligned with its closest homologs belonging to the PMKK1 subfamily of plant MAPKKs (Ligterink and Hirt, 2001). PRKK showed 73% identity to tobacco SIPKK (Liu et al., 2000), 71% identity to tomato LeMEK1 (Hackett et al., 1998), and 68% identity to Arabidopsis AtMKK2 (Ichimura et al., 1998a). For comparison, SIMKK also was included (Kiegerl et al., 2000). Identical and conserved amino acids are shaded in black and gray, respectively; dashes represent gaps. The 11 catalytic subdomains are represented by roman numerals above the respective regions, and amino acids defining the putative MAPK docking site are underlined. The putative phosphorylation sites are indicated by asterisks; note that the consensus sequence for plant MAPKKs in this region is S/T-(X)5-S/T and not S/T-(X)3-S/T, as in yeast and animal MAPKKs (Alessi et al., 1994).
Sequence comparisons of PRKK with current databases revealed highest sequence identity to SIPKK from tobacco (Liu et al., 2000), LeMEK1 from tomato (Hackett et al., 1998), and AtMKK2 from Arabidopsis (Ichimura et al., 1998a). As shown in Figure 1, the N termini of these plant MAPKKs contain a slightly modified DEJL motif (K/R-K/R-K/R-X[1-5]-L/I-X-L/I), which was shown to function as a MAPK docking site in mammals (Jacobs et al., 1999). PRKK, SIPKK, LeMEK1, and AtMKK2 belong to the PMKK1 subgroup of plant MAPKKs (Ligterink and Hirt, 2001). In contrast, SIMKK, an alfalfa MAPKK that also interacts with SIMK, belongs to the PMKK2 subfamily (Kiegerl et al., 2000) and shows only 30% identity to PRKK or any of the other MAPKKs of the PMKK1 subfamily (Figure 1).
Recombinant PRKK Is an Inactive MAPKK
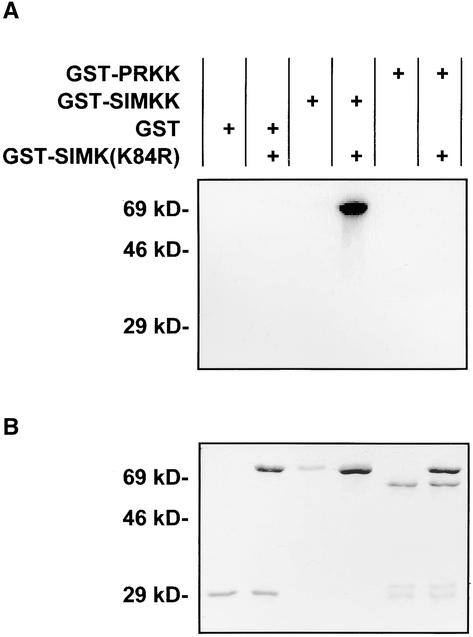
To determine whether PRKK encodes a functional MAPKK, its open reading frame was cloned into pGEX-4T-1. The bacterially expressed glutathione S-transferase (GST)-PRKK fusion protein was affinity purified and used in in vitro kinase assays together with the kinase-negative version of SIMK (GST-SIMK[K84R]) as substrate. However, GST-PRKK was unable to phosphorylate GST-SIMK(K84R) to any degree (Figure 2A). In contrast, GST-SIMKK showed strong phosphorylation of GST-SIMK(K84R) (Figure 2A). Staining of the SDS–polyacrylamide gel with Coomassie blue (Figure 2B) indicated that different amounts of the recombinant proteins were not responsible for the different activities of the MAPKKs.
Figure 2.
Recombinant PRKK Is an Inactive MAPKK.
Different abilities of GST-PRKK and GST-SIMKK to phosphorylate the kinase-negative GST fusion protein of SIMK (GST-SIMK[K84R]) in vitro. Autoradiogram (A) and Coomassie blue staining (B) of SDS-PAGE showing analyses of the products of in vitro kinase reactions between affinity-purified GST-PRKK, GST-SIMKK, or GST on GST-SIMK(K84R) as substrate. Proteins to be tested for MAPK phosphorylating activity (GST [1 μg], GST-PRKK [1 μg], or GST-SIMKK [0.5 μg]) were incubated for 30 min in kinase reaction buffer (50 mM Tris, pH 7.5, 10 mM MgCl2, 1 mM DTT, 0.1 mM ATP, and 6 μCi of γ-32P-ATP) alone or with 5 μg of MAPK (GST-SIMK[K84R]) as a substrate.
PRKK Is a MAPKK That Requires Activation through an Upstream MAPKKK
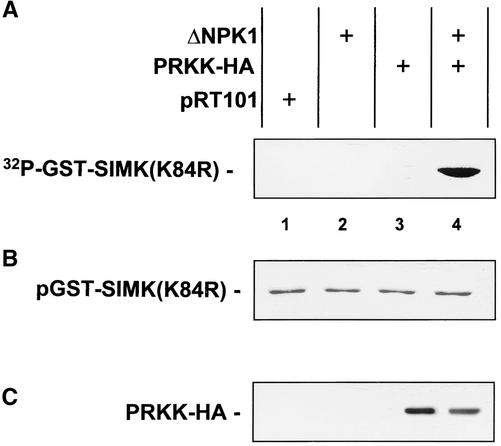
MAPKKs usually become activated by phosphorylation through MAPKKKs (Robinson and Cobb, 1997). Therefore, it was likely that recombinant GST-PRKK required activation by upstream activators for MAPKK activity. To test this possibility, PRKK was tagged with a hemagglutinin (HA) peptide and expressed in parsley protoplasts as PRKK-HA in the absence and presence of ΔNPK1, a constitutively active tobacco MAPKKK (Kovtun et al., 2000). Thereafter, PRKK-HA was immunoprecipitated from protein extracts and tested for the ability to phosphorylate SIMK. When PRKK-HA was expressed in the absence of ΔNPK1, immunoprecipitated PRKK-HA showed no MAPKK activity (Figure 3A, lane 3). In contrast, in the presence of active MAPKKK, PRKK was active and phosphorylated GST-SIMK(K84R) (Figure 3A, lane 4). As shown by Coomassie blue staining of the SDS–polyacrylamide gel used for autoradiography, equal amounts of GST-SIMK(K84R) were present in the assays (Figure 3B). Expression of the empty vector pRT101 or ΔNPK1 alone showed that the HA antibody did not immunoprecipitate endogenous MAPKK activity. Immunoblotting the protein extracts with the HA antibody further proved the specificity of the assay by showing that only PRKK-HA was recognized under these conditions (Figure 3C). These data show that PRKK is a functional enzyme that requires activation by an upstream MAPKKK for MAPKK activity.
Figure 3.
Active MAPKKK Is Required for PRKK Activation.
Approximately 106 parsley protoplasts per treatment were transformed to transiently express the constitutively active MAPKKK ΔNPK1 (Kovtun et al., 2000) (lane 2), the HA-tagged version of PRKK (lane 3), or both (lane 4). Protoplasts transformed with the empty vector pRT101 were used as controls (lane 1). After protein extraction, samples were immunoprecipitated with HA antibody. In vitro kinase assays of immunoprecipitated PRKK-HA were performed on 4 μg of kinase-negative GST-SIMK as substrate. Reaction products were analyzed by SDS-PAGE followed by autoradiography (A) and Coomassie blue staining (B). (C) shows PRKK-HA expression. The specificity of immunoprecipitation was controlled by immunoblotting extracts with HA antibody.
SIMK, MMK3, and SAMK Are Activated by PRKK in Vivo
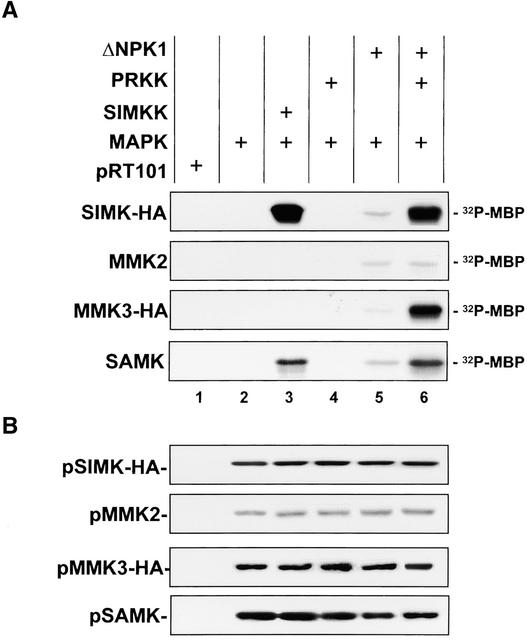
To determine whether PRKK also can activate SIMK in vivo, the two enzymes were coexpressed transiently in parsley protoplasts. Parsley cells were used for the transient expression assays because attempts to use various alfalfa cell lines were unsuccessful. SIMK was immunoprecipitated subsequently from protein extracts and analyzed for activity by in vitro kinase assays using myelin basic protein (MBP) as a substrate. The same experiment was conducted in parallel with three other alfalfa MAPKs, MMK2, MMK3, and SAMK, as well as with SIMKK as an upstream MAPK activator. No kinase activity was associated with immunoprecipitates from cells that contained the empty vector pRT101 (Figure 4A, lane 1). In cells that expressed the MAPKs alone, extremely low MBP kinase activity was associated with the immunoprecipitated MAPKs (Figure 4A, lane 2), indicating that the transiently expressed MAPKs require upstream activation. In agreement with previous data (Kiegerl et al., 2000), coexpression of the MAPKs with SIMKK resulted in the activation of SIMK and SAMK (Figure 4A, lane 3). Coexpression of the four MAPKs with PRKK did not result in significant activation of the MAPKs (Figure 4A, lane 4). When constitutively active ΔNPK1 was expressed, activation of all four tested MAPKs was observed (Figure 4A, lane 5). Coexpression of the MAPKs with ΔNPK1 and PRKK, however, resulted in a severalfold increase in the activation of SIMK, MMK3, and SAMK. No enhancement of MMK2 activation was observed under such conditions (Figure 4A, lane 6). As shown by immunoblotting, the differences in MAPK activities were not attributable to different amounts of MAPKs in the extracts (Figure 4B). These results indicate that PRKK and SIMKK are both MAPK activators but that the MAPKKs activate different and partially overlapping sets of MAPKs.
Figure 4.
PRKK Activates SIMK, MMK3, and SAMK in Vivo.
MBP kinase activities of immunoprecipitated SIMK-HA, MMK2, MMK3-HA, and SAMK when expressed transiently in protoplasts alone (lane 2), with active MAPKKK ΔNPK1 (lane 5), with SIMKK (lane 3), or with PRKK in the absence (lane 4) or presence (lane 6) of ΔNPK1. Protoplasts transformed with the empty vector pRT101 were used as a negative control (lane 1). MAPK activity was analyzed by MBP phosphorylation of immunoprecipitated SIMK-HA, MMK2, MMK3-HA, and SAMK. After SDS-PAGE, MAPK activities were determined by autoradiography (A) and MAPK expression levels were assessed by immunoblotting with HA, MMK2, or SAMK antibody (B).
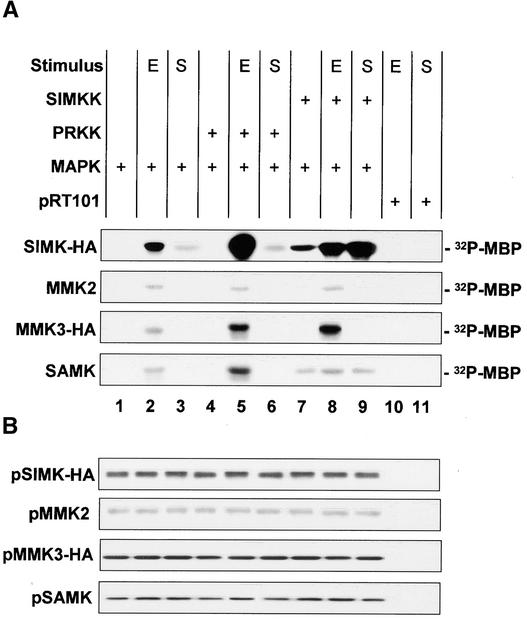
SIMKK and PRKK Both Mediate Elicitor-Induced SIMK Activation, but Only SIMKK Mediates the Salt Stress–Induced Activation of SIMK
To determine which extracellular signals are mediated by PRKK, parsley protoplasts were cotransformed with expression vectors containing either SIMKK or PRKK and any of the four MAPKs (SIMK-HA, MMK2, MMK3, or SAMK). In addition, transformed protoplasts were treated with high salt and elicitor. SIMK is activated by hyperosmotic conditions (Munnik et al., 1999) and by various fungal elicitors (Cardinale et al., 2000). Treatment of protoplasts for 10 min with 50 nM Pep13, an oligopeptide elicitor derived from the fungal pathogen Phytophthora sojae, resulted in strong activation of SIMK-HA and a weaker activation of MMK2, MMK3, and SAMK (Figure 5A, lane 2). NaCl (250 mM) induced SIMK activation exclusively at 10 min but none of the other three MAPKs (Figure 5A, lane 3). In the absence of extracellular signals, coexpression of PRKK was unable to activate any MAPK (Figure 5A, lane 4). In contrast to salt stress (Figure 5A, lane 6), coexpressed PRKK enhanced the activation of SIMK, MMK3, and SAMK upon elicitor treatment (Figure 5A, lane 5). These results indicate that PRKK is not a transmitter of the salt-induced activation of SIMK but is a mediator of the elicitor-induced activation of SIMK, MMK3, and SAMK.
Figure 5.
PRKK and SIMKK Enhance the Elicitor-Induced Activation of MAPKs.
Parsley protoplasts were transformed transiently with a combination of the following expression plasmids: pRT101-SIMKK, pRT101-PRKK, pSH9-SIMK-HA, pRT101-MMK2, pSH9-MMK3-HA, and pRT101-SAMK. MBP kinase activity of immunoprecipitated MAPK was evaluated after transient expression in protoplasts with SIMKK or PRKK in the absence or presence of 50 nM Pep13 elicitor (E; lanes 5 and 8) or 250 mM NaCl (S; lanes 6 and 9). After harvesting of cells 10 min after treatment, proteins were extracted and MAPKs were immunoprecipitated. Untreated samples were used as controls for the basal activity of overexpressed MAPKs; treated samples transformed with the empty vector pRT101 are shown to exclude nonspecific immunoprecipitation of endogenous elicitor- or salt-activated MAPKs (lanes 10 and 11).
(A) The activity of the MAPKs was analyzed by in vitro MBP kinase reactions. The phosphorylated products were analyzed by SDS-PAGE followed by autoradiography.
(B) Immunoblotting of cell extracts used in (A) with anti-HA, anti-MMK2, or anti-SAMK antibody confirmed that equal expression levels of MAPKs were present in different protoplast expression assays.
When SIMKK was coexpressed in parsley protoplasts with MAPKs in the absence of extracellular signals, activation of SIMK and SAMK was observed (Figure 4A, lane 3, and Figure 5A, lane 7). Upon elicitor treatment, SIMKK coexpression resulted in strong activation of SIMK and MMK3 and weak enhancement of SAMK (Figure 5A, lane 8). In contrast, coexpression of SIMKK in salt-stressed cells strongly enhanced the activation of SIMK but not SAMK or any of the other MAPKs (Figure 5A, lane 9). These data show that SIMKK is a mediator of elicitor-induced activation of SIMK and MMK3. Moreover, SIMKK acts as a mediator of salt stress–induced SIMK activation.
To exclude artifacts from the endogenous parsley kinases, protoplasts expressing empty vector also were analyzed after stress treatment. No MAPK activity was immunoprecipitated under any of the conditions used (Figure 5A, lanes 10 and 11), indicating that the kinase activities are not derived from endogenous MAPKs. To exclude the possibility that different amounts of MAPK protein were responsible for the differences observed in MAPK activities, the cell extracts used for immunokinase assays in Figure 5A were immunoblotted with antibodies against the MAPKs. As shown in Figure 5B, MAPK protein levels did not vary in these experiments, indicating that different levels of MAPK activities were not the result of different expression levels of the MAPKs but of activation by different upstream MAPKKs. Together, these data show that SIMKK and PRKK both mediate the elicitor-induced activation of distinct but overlapping sets of MAPKs, but SIMKK alone mediates the salt stress–induced activation of SIMK.
DISCUSSION
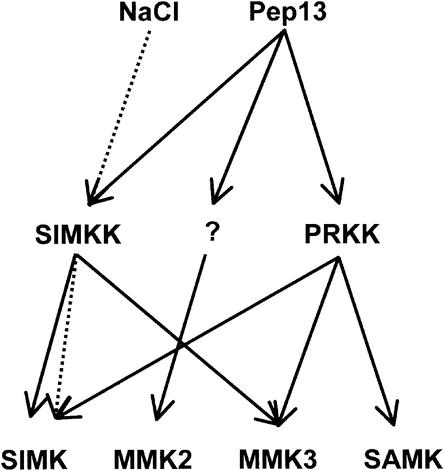
MAPKs play important roles in mediating stress responses in animals, yeast, and plants (Meskiene and Hirt, 2000). In plants, the SIMK pathway has been identified as being activated by high salt concentrations and elicitors (Munnik et al., 1999; Cardinale et al., 2000). By yeast two-hybrid interaction screening of SIMK with an alfalfa library, SIMKK and PRKK were isolated. SIMKK and PRKK are two highly different MAPKKs that are only 30% identical in primary sequences. They belong to separate MAPKK subfamilies and have distinct substrate specificities and regulatory properties. Both MAPKKs respond to elicitor, but only SIMKK mediates salt stress signaling (Figure 6). In the presence of elicitor, SIMKK and PRKK both target SIMK and MMK3, but PRKK also targets SAMK. SIMKK, but not PRKK, is a mediator of salt stress–induced SIMK activation. These studies provide insight into how different upstream signals can be integrated into common MAPK signaling pathways and how a cell distributes information to different downstream MAPK effectors.
Figure 6.
Convergence and Divergence of Salt- and Elicitor-Induced Signals at the Level of MAPKKs.
Both stimuli seem to be mediated by SIMKK, which activates SIMK in the case of salt stress and activates SIMK and MMK3 upon elicitor treatment. Although unable to mediate salt stress signaling, PRKK is capable of distributing the elicitor signal onto SIMK, MMK3, and SAMK in vivo. The MAPKK responsible for elicitor-induced MMK2 activation remains to be identified.
Analysis of SIMKK and PRKK revealed that the two MAPKKs have quite distinct properties with respect to their activation. In contrast to SIMKK, recombinant PRKK was completely inactive as a MAPKK. This result suggested that the activation of PRKK, but not of SIMKK, required phosphorylation by upstream factors. In agreement with this idea, PRKK immunoprecipitated from transformed protoplasts was completely inactive unless coexpressed with an active MAPKKK, whereas SIMKK did not require activation by an upstream MAPKKK. An explanation for the different behavior of PRKK and SIMKK might be found in the activation loops of these two enzymes. For activation, all animal and yeast MAPKKs must become phosphorylated by MAPKKKs at Ser and/or Thr residues in the SXXXS/T motif that is located between subdomains VII and VIII (Robinson and Cobb, 1997). The corresponding region of plant MAPKKs lacks the first Serine residue and has a spacing of five amino acids between the putative phosphorylation sites, with S/TXXXXXS/T as a consensus sequence (Figure 1) (Ligterink and Hirt, 2001). Interestingly, SIMKK and other MAPKKs of the PMKK2 subfamily (such as tobacco NtMEK2) share the same consensus sequence but contain an Asp residue at position 3, showing TXDXXXS (Ligterink and Hirt, 2001). Because substitution of any of the animal phosphorylation sites by acidic residues activates the respective MAPKKs, it is likely that the PMKK2 group constitutes an autoactive class of MAPKKs, but additional studies on the phosphorylation of these plant MAPKKs are required to assess the biological importance of this variation.
Previous work with tobacco identified SIPKK as an interaction partner of SIPK (Liu et al., 2000). Protein sequence analysis of all available sequences indicates that SIPKK and SIPK are the tobacco orthologs of PRKK and SIMK, respectively (data not shown). Although SIPKK interacts strongly with SIPK, the recombinant protein was unable to phosphorylate SIPK in vitro. In our analysis, recombinant GST-PRKK also was unable to phosphorylate its potential substrate SIMK. Our results suggest that PRKK requires phosphorylation and thereby activation by an upstream MAPKKK. When transiently expressed in protoplasts, PRKK was still inactive as a MAPKK. However, coexpression of PRKK with a constitutively active MAPKKK (ΔNPK1) resulted in the activation of PRKK, enabling the kinase to activate downstream MAPKs. It should be noted that in contrast to a previous study (Kovtun et al., 2000), we found that ΔNPK1 showed no specificity toward activating MAPKs (Figure 4). One possible explanation for the discrepancy in results is that the species and cell types used for the transient expression assays were different. Although Kovtun et al. (2000) derived protoplasts from differentiated Arabidopsis leaf mesophyll cells, we used actively dividing suspension-cultured parsley cells. Different cell types probably express different MAPKKs; hence, it is likely that transient expression of a constitutively active MAPKKK might activate only those heterologous MAPKs if the correct endogenous MAPKK is present in the system.
To identify the extracellular stimuli to which PRKK and SIMKK respond, parsley protoplasts were transfected transiently with expression vectors carrying different combinations of MAPKs and either PRKK or SIMKK. The protoplasts then were treated with salt stress or elicitor. Under these conditions, SIMKK was found to mediate SIMK activation by salt stress and elicitor, suggesting that SIMKK functions to integrate different upstream stimuli. This finding provides an explanation for how members of the SIMK subfamily of MAPKs can be activated by abiotic and biotic stimuli such as salt stress and elicitors (Figure 6).
Striking differences between downstream MAPK targets of SIMKK were observed when protoplasts were transformed with different combinations of MAPKs and SIMKK. In the absence of any extracellular signal, SIMKK activated SIMK and SAMK but not MMK3 (Figure 5A). However, upon elicitor treatment, SIMKK strongly activated SIMK and MMK3 but barely increased SAMK activation (Figure 5A). Moreover, in salt stress–treated cells, SIMKK enhanced the activation of SIMK but not of MMK3 or SAMK (Figure 5A). These results indicate that the outcome of signaling cascades is not influenced only by the inherent affinities between different combinations of MAPKKs and MAPKs but also by intracellular signal-dependent mechanisms. Among the best candidates for generating signaling selectivity are scaffold proteins. A number of scaffold proteins have been identified for animal and yeast MAPK cascades, showing that scaffold proteins influence the assembly, transport, and substrate specificity of MAPK modules (Whitmarsh and Davis, 1998). Preliminary evidence from our group indicates that some of these features also are found in plant MAPK scaffold proteins (J. Beyerly, H. Nakagami, and H. Hirt, unpublished results).
Combinatorial transient expression of different MAPKs with PRKK in parsley protoplasts revealed that PRKK enhances the elicitor-induced activation of SIMK, MMK3, and SAMK (Figure 5A). However, PRKK is unable to enhance the activation of any MAPK by salt stress (Figure 5A). It should be observed that elicitor-induced MMK2 activation is not mediated by either PRKK or SIMKK (Figure 5A). Therefore, another yet unidentified MAPKK must be postulated that is responsible for MMK2 activation (Figure 6). Together, our data show that at least three MAPKKs, PRKK, SIMKK, and an unknown MAPKK, are involved in pathogen signaling. This might explain our previous findings that various elicitors can activate multiple MAPK cascades (Cardinale et al., 2000). The ability of PRKK and SIMKK to activate more than one MAPK by fungal elicitor is noteworthy. In yeast and animals, single stimulus-mediated activation of multiple MAPKs occurs mainly at the level of MAPKKKs (Widmann et al., 1999). In these organisms, a number of MAPKKKs have the ability to activate multiple MAPKKs, whereas MAPKKs usually activate a single, specific MAPK (Fanger, 1999).
A possible answer to this puzzle might be found in the number of components available to assemble MAPK cascades. Database analysis of the complete Arabidopsis genome indicated that there are 37 MAPKKKs, 10 MAPKKs, and 24 MAPKs present (Wrzaczek and Hirt, 2001). Consequently, multiple MAPKKKs must converge their information at the level of relatively few MAPKKs. On the other hand, an average of 2.4 MAPKs will be activated by a given MAPKK. Although it is difficult to verify this prediction in our analysis of two MAPKKs and four MAPKs, it is clear that PRKK and SIMKK can target up to three and two MAPKs, respectively. These considerations show that we are still far from understanding the full complexity of MAPK signaling pathways. Nonetheless, the identification of PRKK and SIMKK as mediators of salt stress and elicitors proves that MAPKs are prime regulators of stress signaling in plants, making MAPKKs attractive targets for improving stress tolerance and pathogen resistance in agriculture.
METHODS
Isolation of PRKK
The amino acid sequence of PRKK was deduced from the sequence of a cDNA clone isolated by screening an alfalfa (Medicago sativa) cDNA library (Hybri-ZAP; Stratagene) using SIMK in the pBD-Gal4Cam vector (Stratagene) as a bait in a yeast two-hybrid system. Yeast colonies of strain PJ69-4A (James et al., 1996) were selected sequentially for adenine-positive and histidine-positive prototrophy on medium lacking Leu, Trp, and adenine or Leu, Trp, and histidine, respectively. Strict histidine auxotrophy was obtained by growing the cells in the presence of 10 mM 3-aminotriazole. Approximately 150,000 transformants were screened. To eliminate false-positive results, the plasmids obtained from primary positive clones were retransformed into PJ69-4A yeast and tested again for histidine or adenine auxotrophy. The PRKK clone was sequenced entirely and shown to contain a 1065-bp open reading frame, a 6-bp 5′ untranslated region, and a 401-bp 3′ untranslated region including a poly(A)+ tail.
In Vitro Kinase Assays
PRKK was cloned into pGEX-4T-1 and expressed as a glutathione S-transferase (GST) fusion protein. GST-PRKK (1 μg) or GST-SIMKK (0.5 μg) was incubated in 20 μL of kinase reaction buffer (50 mM Tris, pH 7.5, 1 mM DTT, 10 mM MgCl2, 0.1 mM ATP, and 6 μCi of γ-32P-ATP) with 5 μg of the kinase-negative SIMK GST fusion protein (GST-SIMK[K84R]). The change of the conserved Lys-84 to Arg was obtained by in vitro mutagenesis as described by Kiegerl et al. (2000). The reactions were stopped after 30 min by adding 4 × SDS loading buffer and heating for 5 min at 95°C. After SDS-PAGE, the reaction products were analyzed by autoradiography and Coomassie Brilliant Blue R250 staining.
Transient Expression Assays
The open reading frames of PRKK, SIMKK, MMK2, and SAMK were cloned into the plant expression vector pRT101 (Töpfer et al., 1987; Kiegerl et al., 2000), whereas SIMK and MMK3 were fused at their C termini to a triple hemagglutinin (HA) epitope and cloned into pSH9 (Holtorf et al., 1995). Expression vector pRT101-PRKK-HA was obtained by fusion of the triple HA epitope to the C terminus of PRKK and insertion into pRT101. The constitutively active tobacco mitogen-activated protein kinase kinase kinase ΔNPK1 was expressed under the HBT95 version of the 35S promoter (Kovtun et al., 2000). Parsley protoplasts were transformed transiently via polyethylene glycol with 5 μg each of different combinations of the following plasmids: ΔNPK1, pRT101-PRKK, pRT101-PRKK-HA, pRT101-SIMKK, pSH9-SIMK-HA, pRT101-MMK2, pSH9-MMK3-HA, and pRT101-SAMK. Then they were subjected to different treatments 12 to 16 hr after transformation (Kiegerl et al., 2000).
Immunocomplex Kinase Assays
Protein extracts were prepared as described by Bögre et al. (1999) in protein extraction buffer (25 mM Tris, pH 7.8, 75 mM NaCl, 10 mM MgCl2, 15 mM EGTA, 1 mM DTT, 1 mM NaF, 0.5 mM NaVO3, 15 mM β-glycerophosphate, 15 mM p-nitrophenylphosphate, 0.1% Tween 20, 0.5 mM phenylmethylsulfonyl fluoride, 5 μg/mL leupeptin, and 5 μg/mL aprotinin). After centrifugation at 20,000g for 45 min, equal protein amounts from the cleared supernatant were subjected to a 2-hr preincubation in the presence of mixed protein G– and protein A–Sepharose beads (20 μL, 1:1; preclearance step). The supernatant then was immunoprecipitated for 2 hr with either protein G beads and 5 μL of HA antibody (for HA-tagged SIMK and MMK3) or protein A beads and 5 μg of protein A–purified M11 antibodies for MMK2 or M24 antibody for SAMK (Jonak et al., 1995; Munnik et al., 1999; Cardinale et al., 2000). The beads were washed three times with wash buffer (50 mM Tris, pH 7.4, 250 mM NaCl, 5 mM EGTA, 5 mM EDTA, and 0.1% Tween 20) and once with kinase buffer (50 mM Tris, pH 7.5, 1 mM DTT, 10 mM MgCl2, and 0.1 mM ATP).
Kinase reactions on the immunoprecipitated mitogen-activated protein kinases were performed for 30 min at room temperature in 20 μL of kinase buffer containing 5 μg of myelin basic protein (MBP) and 2 μCi of γ-32P-ATP. The reaction was stopped by adding SDS-PAGE loading buffer, and the phosphorylation of MBP was analyzed by autoradiography after SDS-PAGE. Kinase reactions on the immunoprecipitated PRKK-HA were performed in 20 μL of kinase buffer in the presence of 6 μCi of γ-32P-ATP and 4 μg of the SIMK kinase-negative GST fusion protein GST-SIMK(K84R) (Kiegerl et al., 2000).
For protein gel blot analysis, equal amounts of total protein extracts were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and probed with either the HA antibody as recommended by the manufacturer (BABCO, Richmond, CA) or with M11 or M24 antibodies as described previously (Jonak et al., 1995; Munnik et al., 1999; Cardinale et al., 2000). Alkaline phosphatase–conjugated goat anti–mouse and anti–rabbit IgG (Sigma) were used as secondary antibodies, and the reaction was visualized by fluorography using CDP-Star (Amersham Life Sciences) as a substrate.
Treatment of Transiently Transfected Protoplasts with NaCl and Fungal Elicitor
Transiently transformed parsley (Petroselinum crispum) protoplasts were treated with 250 mM NaCl (Kiegerl et al., 2000) or 50 nM Pep13 elicitor (Ligterink et al., 1997). Samples were harvested at 10 min.
Accession Numbers
The accession numbers for PRKK are in the EMBL and MUA databases as AJ29327 and 293275, respectively.
Acknowledgments
We thank J. Sheen for providing the ΔNPK1 expression vector and I. Somssich, D. Scheel, and T. Nürnberger for providing the parsley cell culture and Pep13 elicitor. This work was supported by Grant Nos. P14631-GEN, P14114-GEN, and P13535-GEN of the Austrian Science Foundation.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010256.
References
- Alessi, D.R., Saito, Y., Campbell, D.G., Cohen, P., Sithanandam, G., Rapp, U., Ashworth, A., Marshall, C.J., and Cowley, S. (1994). Identification of the sites in MAP kinase kinase-1 phosphorylated by p74raf-1. EMBO J. 13, 1610–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bögre, L., Ligterink, W., Meskiene, I., Barker, P.J., Heberle-Bors, E., Huskisson, N.S., and Hirt, H. (1997). Wounding induces the rapid and transient activation of a specific MAP kinase pathway. Plant Cell 9, 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bögre, L., Calderini, O., Binarova, P., Mattauch, M., Till, S., Kiegerl, S., Jonak, C., Pollaschek, C., Barker, P., Huskisson, N.S., Hirt, H., and Heberle-Bors, E. (1999). A MAP kinase is activated late in plant mitosis and becomes localized to the plane of cell division. Plant Cell 11, 101–114. [PMC free article] [PubMed] [Google Scholar]
- Calderini, O., Glab, N., Bergounioux, C., Heberle-Bors, E., and Wilson, C. (2001). A novel tobacco MAP kinase kinase, NtMEK1, activates the cell cycle-regulated p43Ntf6 MAP kinase. J. Biol. Chem. 276, 18139–18145. [DOI] [PubMed] [Google Scholar]
- Cardinale, F., Jonak, C., Ligterink, W., Niehaus, K., Boller, T., and Hirt, H. (2000). Differential activation of four specific MAPK pathways by distinct elicitors. J. Biol. Chem. 275, 36734–36740. [DOI] [PubMed] [Google Scholar]
- Droillard, M.J., Thibivilliers, S., Cazale, A.C., Barbier-Brygoo, H., and Lauriere, C. (2000). Protein kinases induced by osmotic stresses and elicitor molecules in tobacco cell suspensions: Two crossroad MAP kinases and one osmoregulation-specific protein kinase. FEBS Lett. 474, 217–222. [DOI] [PubMed] [Google Scholar]
- Fanger, G.R. (1999). Regulation of the MAPK family members: Role of subcellular localization and architectural organization. Histol. Histopathol. 14, 887–894. [DOI] [PubMed] [Google Scholar]
- Hackett, R.M., Oh, S.A., Morris, P.C., and Grierson, D. (1998). A tomato MAP kinase kinase gene (accession No. AJ 000728) differentially regulated during fruit development, leaf senescence, and wounding (PGR98–151). Plant Physiol. 117, 1526. [Google Scholar]
- Hardin, S.C., and Wolniak, S.M. (1998). Molecular cloning and characterization of maize ZmMEK1, a protein kinase with a catalytic domain homologous to mitogen- and stress-activated protein kinase kinases. Planta 206, 577–584. [DOI] [PubMed] [Google Scholar]
- Holtorf, S., Apel, K., and Bohlmann, H. (1995). Comparison of different constitutive and inducible promoters for the overexpression of transgenes in Arabidopsis thaliana. Plant Mol. Biol. 29, 637–646. [DOI] [PubMed] [Google Scholar]
- Ichimura, K., Mizoguchi, T., Hayashida, N., Seki, M., and Shinozaki, K. (1998. a). Molecular cloning and characterization of three cDNAs encoding putative mitogen-activated protein kinase kinases (MAPKKs) in Arabidopsis thaliana. DNA Res. 5, 341–348. [DOI] [PubMed] [Google Scholar]
- Ichimura, K., Mizoguchi, T., Irie, K., Morris, P., Giraudat, J., Matsumoto, K., and Shinozaki, K. (1998. b). Isolation of ATMEKK1 (a MAP kinase kinase kinase)-interacting proteins and analysis of a MAP kinase cascade in Arabidopsis. Biochem. Biophys. Res. Commun. 253, 532–543. [DOI] [PubMed] [Google Scholar]
- Jacobs, D., Glossip, D., Xing, H., Muslin, A.J., and Kornfeld, K. (1999). Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 13, 163–175. [PMC free article] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak, C., Kiegerl, S., Lloyd, C., Chan, J., and Hirt, H. (1995). MMK2, a novel alfalfa MAP kinase, specifically complements the yeast MPK1 function. Mol. Gen. Genet. 248, 686–694. [DOI] [PubMed] [Google Scholar]
- Jonak, C., Kiegerl, S., Ligterink, W., Barker, P.J., Huskisson, N.S., and Hirt, H. (1996). Stress signaling in plants: A mitogen-activated protein kinase pathway is activated by cold and drought. Proc. Natl. Acad. Sci. USA 93, 11274–11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin, M. (1998). Mitogen-activated protein kinase cascades as regulators of stress responses. Ann. NY Acad. Sci. 851, 139–146. [DOI] [PubMed] [Google Scholar]
- Kiegerl, S., Cardinale, F., Siligan, C., Gross, A., Baudouin, E., Liwosz, A., Eklof, S., Till, S., Bogre, L., Hirt, H., and Meskiene, I. (2000). SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress-induced MAPK, SIMK. Plant Cell 12, 2247–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun, Y., Chiu, W.-L., Tena, G., and Sheen, J. (2000). Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 97, 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligterink, W., and Hirt, H. (2001). Mitogen-activated protein (MAP) kinase pathways in plants: Versatile signaling tools. Int. Rev. Cytol. 201, 209–215. [DOI] [PubMed] [Google Scholar]
- Ligterink, W., Kroj, T., zur Nieden, U., Hirt, H., and Scheel, D. (1997). Receptor-mediated activation of a MAP kinase in pathogen defense of plants. Science 276, 2054–2057. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Zhang, S., and Klessig, D.F. (2000). Molecular cloning and characterization of a tobacco MAP kinase kinase that interacts with SIPK. Mol. Plant-Microbe Interact. 13, 118–124. [DOI] [PubMed] [Google Scholar]
- Meskiene, I., and Hirt, H. (2000). MAP kinase pathways: Molecular plug-and-play chips for the cell. Plant Mol. Biol. 42, 791–806. [DOI] [PubMed] [Google Scholar]
- Morris, P.C., Guerrier, D., Leung, J., and Giraudat, J. (1997). Cloning and characterisation of MEK1, an Arabidopsis gene encoding a homologue of MAP kinase kinase. Plant Mol. Biol. 35, 1057–1064. [DOI] [PubMed] [Google Scholar]
- Munnik, T., Ligterink, W., Meskiene, I., Calderini, O., Beyerly, J., Musgrave, A., and Hirt, H. (1999). Distinct osmo-sensing protein kinase pathways are involved in signalling moderate and severe hyper-osmotic stress. Plant J. 20, 381–388. [DOI] [PubMed] [Google Scholar]
- Nühse, T., Peck, S.C., Hirt, H., and Boller, T. (2000). Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK 6. J. Biol. Chem. 275, 7521–7526. [DOI] [PubMed] [Google Scholar]
- Robinson, M.J., and Cobb, M.H. (1997). Mitogen-activated protein kinase pathways. Curr. Opin. Cell Biol. 9, 180–186. [DOI] [PubMed] [Google Scholar]
- Robzyk, K., and Kassir, Y. (1992). A simple and highly efficient procedure for rescuing autonomous plasmids from yeast. Nucleic Acids Res. 20, 3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis, T., Piedras, P., Zhang, S., Klessig, D.F., Hirt, H., and Jones, J.D. (1999). Rapid Avr9- and Cf-9–dependent activation of MAP kinases in tobacco cell cultures and leaves: Convergence of resistance gene, elicitor, wound, and salicylate responses. Plant Cell 11, 273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, S., Okamoto, M., Seto, H., Ishizuka, K., Sano, H., and Ohashi, Y. (1995). Tobacco MAP kinase: A possible mediator in wound signal transduction pathways. Science 270, 1988–1992. [DOI] [PubMed] [Google Scholar]
- Shibata, W., Banno, H., Ito, Y., Hirano, K., Irie, K., Usami, S., Machida, C., and Machida, Y. (1995). A tobacco protein kinase, NPK2, has a domain homologous to a domain found in activators of mitogen-activated protein kinases (MAPKKs). Mol. Gen. Genet. 246, 401–410. [DOI] [PubMed] [Google Scholar]
- Töpfer, R., Matzeit, V., Gronenborn, B., Schell, J., and Steinbiss, H.H. (1987). A set of plant expression vectors for transcriptional and translational fusions. Nucleic Acids Res. 15, 5890–5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami, S., Banno, H., Ito, Y., Nishihama, R., and Machida, Y. (1995). Cutting activates a 46-kilodalton protein kinase in plants. Proc. Natl. Acad. Sci. USA 92, 8660–8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmarsh, A.J., and Davis, R.J. (1998). Structural organization of MAP-kinase signaling modules by scaffold proteins in yeast and mammals. Trends Biochem. Sci. 23, 481–485. [DOI] [PubMed] [Google Scholar]
- Widmann, C., Gibson, S., Jarpe, M.B., and Johnson, G.L. (1999). Mitogen-activated protein kinase: Conservation of a three-kinase module from yeast to human. Physiol. Rev. 79, 143–180. [DOI] [PubMed] [Google Scholar]
- Wrzaczek, M., and Hirt, H. (2001). Plant MAP kinase pathways: How many and what for? Biol. Cell 93, 1–7. [DOI] [PubMed] [Google Scholar]
- Yang, K.Y., Liu, Y., and Zhang, S. (2001). Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc. Natl. Acad. Sci. USA 98, 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., and Klessig, D.F. (1998. a). The tobacco wounding-activated mitogen-activated protein kinase is encoded by SIPK. Proc. Natl. Acad. Sci. USA 95, 7225–7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., and Klessig, D.F. (1998. b). Resistance gene N-mediated de novo synthesis and activation of a tobacco mitogen-activated protein kinase by tobacco mosaic virus infection. Proc. Natl. Acad. Sci. USA 95, 7433–7438. [DOI] [PMC free article] [PubMed] [Google Scholar]