Abstract
Plant growth and development depends on the activity of a continuously replenished pool of stem cells within the shoot apical meristem to supply cells for organogenesis. In Arabidopsis, the stem cell–specific protein CLAVATA3 (CLV3) acts cell nonautonomously to restrict the size of the stem cell population, but the hypothesis that CLV3 acts as an extracellular signaling molecule has not been tested. We used genetic and immunological assays to show that CLV3 localizes to the apoplast and that export to the extracellular space is required for its function in activating the CLV1/CLV2 receptor complex. Apoplastic localization allows CLV3 to signal from the stem cell population to the organizing center in the underlying cells.
INTRODUCTION
Plants, unlike animals, continuously produce organs from their growing tips, which are called apical meristems. In the shoot apical meristem (SAM), which generates all of the aboveground structures of the plant, a few cells at the apex are maintained as a pluripotent stem cell population. As the stem cells divide, their progeny are displaced toward the flanks of the meristem, where they become incorporated into organ primordia. The maintenance of a functional SAM requires precise coordination between the loss of stem cells from the meristem through organogenesis and their replacement through cell division.
The Arabidopsis CLAVATA1 (CLV1), CLV2, and CLV3 genes are required to restrict the amount of stem cell accumulation in both shoot and floral meristems. Plants with loss-of-function mutations in any of the CLV genes form greatly enlarged shoot and floral meristems, causing stem overgrowth and the production of extra flowers and floral organs (Clark et al., 1993, 1995; Kayes and Clark, 1998). The CLV1 gene encodes a Leu-rich repeat receptor Ser/Thr kinase (Clark et al., 1997), a member of a large class of proteins found in both plants and animals, many of which are involved in cell signaling. CLV2 encodes a Leu-rich repeat receptor–like transmembrane protein with a short cytoplasmic tail (Jeong et al., 1999). Together, CLV1 and CLV2 form components of a membrane-bound receptor signal transduction complex (Trotochaud et al., 2000). CLV3 encodes a 96–amino acid predicted extracellular protein (Fletcher et al., 1999) that can act in a cell nonautonomous manner (Fletcher et al., 1999; Brand et al., 2000). CLV3 binds to CLV1 and CLV2 and is required for the formation of the active receptor complex, indicating that CLV3 acts as a ligand that signals through CLV1 and CLV2 (Trotochaud et al., 2000).
CLV3 and CLV1 expression is restricted to subsets of shoot and floral meristem cells. CLV3 mRNA is expressed in stem cells, which are found primarily in the epidermal and subepidermal cell layers of shoot and floral meristems (Fletcher et al., 1999). CLV1 is expressed in the underlying cells, in a domain partially overlapping that of CLV3 (Clark et al., 1997). It has been hypothesized that CLV3 is made in the overlying cell layers and moves to the underlying cells to activate the CLV pathway. However, the subcellular localization of the components of the CLV signaling pathway has not been reported. Moreover, the expression data do not exclude the possibility that activation occurs intracellularly within cells that express both the ligand and the receptor complex. We have now devised genetic and immunological assays to reveal the localization and site of activation of the CLV pathway. We show that CLV3 is transported through the secretory pathway, is localized to the apoplast, and activates the CLV pathway in the extracellular space.
RESULTS
CLV3 Is Localized to the Extracellular Space
CLV3 contains an 18–amino acid N-terminal hydrophobic region that presumably acts as a signal peptide to direct the protein into the secretory pathway (Fletcher et al., 1999). No other hydrophobic regions are present, indicating that CLV3 is a soluble protein. Retention or sorting signals are not apparent in its amino acid sequence; thus, CLV3 is predicted to be exported through the secretory pathway to the extracellular space by the default pathway for soluble plant proteins (Denecke et al., 1990).
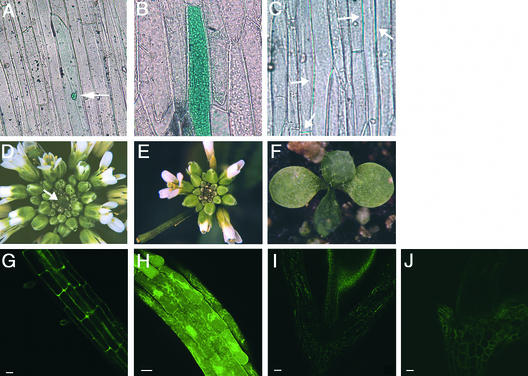
To test this prediction, we generated a translational fusion of full-length CLV3 to the N terminus of green fluorescent protein/β-glucuronidase (GFP/GUS) (CLV3-G) and a translational fusion of a truncated CLV3 protein, lacking the putative signal peptide, fused to the N terminus of GFP/GUS (CLV3Δsp-G). These fusion constructs were transformed transiently into leek epidermal cells by particle bombardment (Sanford et al., 1993). A control construct containing a nuclear localization signal upstream of GFP/GUS was confined to the nucleus (Figure 1A), whereas the CLV3Δsp-G fusion protein was detected in the cytoplasm (Figure 1B). In contrast, signal from the CLV3-G fusion protein was observed only between cells (Figure 1C). This result indicates that CLV3 is localized in the plasma membrane and/or the apoplastic space and that the presence of the signal peptide is required for its extracellular localization.
Figure 1.
CLV3 Protein Is Localized to the Apoplast.
(A) to (C) Phase-contrast optics used to detect GUS.
(A) Cell transiently expressing the GFP/GUS protein alone, which is confined to the nucleus (arrow).
(B) Cells transiently expressing the CLV3Δsp-GFP/GUS fusion protein, which is present throughout the cytoplasm.
(C) Cells transiently expressing the CLV3-GFP/GUS fusion protein, which is detected only between cells (arrows).
(D) Inflorescence apex of a clv3-2 mutant plant showing massive meristem enlargement (arrow) and production of supernumerary flowers and floral organs.
(E) Inflorescence apex of a clv3-2 mutant plant stably expressing the 35S::CLV3-G fusion protein. This transgenic T2 plant resembles the wild type, indicating that CLV3 function has been restored by the transgene.
(F) Seedling with a prematurely terminated meristem from a 35S::CLV3-G clv3-2 line. The shoot apical meristem has formed a terminal leaf.
(G) GFP fluorescence in a root from a 35S::CLV3-G wild-type T2 transgenic plant.
(H) GFP fluorescence in a root from a 35S::CLV3Δsp-G wild-type T2 transgenic plant.
(I) Autofluorescence in the cell wall of an untransformed wild-type hypocotyl.
(J) GFP fluorescence in the hypocotyl of a 35S::CLV3-G wild-type T2 transgenic plant.
Bars = 20 μm in (G) and (H) and 50 μm in (I) and (J).
Next, we determined the subcellular location of these fusion proteins in stably transformed plants. Transgenic plants expressing CLV3-G showed GFP fluorescence only in the extracellular space (Figures 1G and 1J). In contrast, no autofluorescence was detected in roots of untransformed wild-type plants (data not shown), whereas autofluorescence in hypocotyls was detected as punctate spots (Figure 1I). Introduction of CLV3-G into clv3-2 null mutant plants partially or fully rescued the mutant phenotype (Figure 1E; cf. with the untransformed clv3-2 plant in Figure 1D). A subset of the transgenic plants prematurely terminated SAM activity after the production of a few lateral organs (Figure 1F), a typical CLV3 gain-of-function phenotype. Among the CLV3-G clv3-2 T2 lines analyzed, the extent of phenotypic rescue correlated with the level of GUS staining observed (data not shown). Similar effects were observed among 35S::CLV3 transgenic lines, indicating that the amount of CLV3 ligand is the rate-limiting factor in determining the size of the stem cell population (Brand et al., 2000).
In contrast, transgenic plants carrying the truncated CLV3Δsp-G construct displayed high levels of GFP fluorescence within each cell, indicating that the truncated form of CLV3 is not secreted from Arabidopsis cells (Figure 1H). Moreover, expression of CLV3Δsp-G caused no detectable phenotype. These results show that deletion of the first 18 amino acids of CLV3 inactivate the protein, most likely by blocking its delivery into the secretory pathway. However, we could not exclude the possibility that the truncated protein is inactive because of the deletion of amino acids required for interaction with the receptor complex, nor could we determine in what compartment of the secretory pathway CLV3 activates the CLV pathway. Indeed, the Drosophila Fringe protein, another regulatory molecule with a predicted signal peptide, was demonstrated recently to function in the Golgi apparatus instead of the extracellular space (Munro and Freeman, 2000).
Apoplastic Localization of CLV3 Is Required for Its Function
To unequivocally determine whether CLV3 activates the CLV pathway in the extracellular space, we needed to block the delivery to the apoplast but maintain transit through the same compartments of the secretory pathway. This can be achieved by redirecting the protein to the vacuole. In the secretory pathway of plants, proteins destined for secretion and many vacuolar proteins are transported through the same compartments of the endomembrane system until they reach the Golgi apparatus. In the trans-Golgi network, they are finally separated in different vesicle populations destined to the vacuole or to the plasma membrane for secretion. Therefore, if we fused vacuolar sorting signals to CLV3, the chimeric protein still would traffic through the same compartments as endogenous CLV3, but it would not be delivered to the apoplast. Then, we could test whether this had any effect on the activity of CLV3. The vacuolar sorting signals would have an effect only if the activation of the CLV pathway by CLV3 took place in the extracellular space.
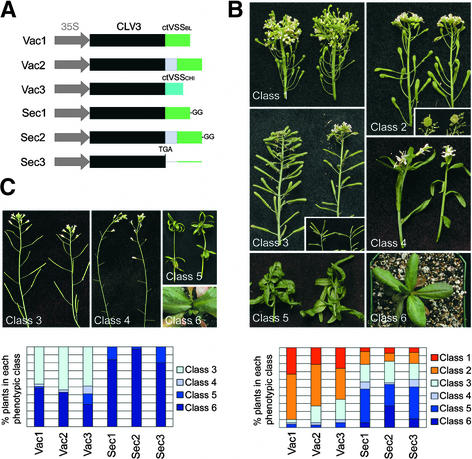
We made translational fusions of CLV3 to the C-terminal vacuolar sorting signals (ctVSS) from barley lectin and tobacco chitinase (Vac1 and Vac3; Figure 2A). These sorting signals are small peptides shown to be sufficient to redirect secreted proteins to the vacuole (Bednarek and Raikhel, 1991; Neuhaus et al., 1991). A third vacuole-targeted fusion protein (Vac2) contains a T7 peptide tag (CLV3-T7) that provides a linker region between the CLV3 sequence and the barley lectin ctVSS. The addition of the peptide tag enabled us to determine the subcellular localization of CLV3 with anti-T7 antibodies (see below). In control experiments, we assayed the activity of CLV3 and CLV3-T7 fused to a mutated version of the barley lectin ctVSS, consisting of the entire ctVSS with two additional Gly residues at the C terminus (Sec1 and Sec2). This mutated version of the ctVSS is not functional as a vacuolar sorting signal (Dombrowski et al., 1993); consequently, it does not block the secretion of CLV3. We compared the effect of overexpressing these five constructs with that of a construct overexpressing CLV3 with no additional tags (Sec3).
Figure 2.
Targeting to the Vacuole Blocks the Activity of CLV3.
(A) Scheme of the fusion constructs. The C-terminal vacuolar sorting signal from barley lectin (ctVSSbl) was fused at the C terminus of CLV3 (Vac1) or CLV3 fused to a T7 peptide tag (Vac2). The ctVSS from tobacco chitinase A (ctVSSchi) was fused at the C terminus of CLV3 (Vac3). The ctVSSbl with two additional Gly residues (GG), which no longer functions as a vacuole-sorting signal, was attached either at the C terminus of CLV3 (Sec1) or CLV3-T7 (Sec2). Sec3 is obtained from Vac2 by deleting a single nucleotide that inserts a stop codon after the final amino acid of CLV3. All constructs are driven by the 35S promoter of Cauliflower mosaic virus.
(B) clv3-2 plants transformed with the constructs described in (A). Primary transformants were grouped into six phenotypic classes according to the severity of meristem termination. Class 1 plants showed no transgene activity and resembled clv3-2 mutants. Class 2 plants showed weak transgene activity and displayed the phenotype of weak clv3 alleles. (The inset shows the mass of cells accumulating in the SAM of these plants 10 weeks after transplanting.) Class 3 plants showed intermediate transgene activity that complemented the clv3 phenotype. These plants looked essentially like untransformed wild-type plants, although the SAMs terminated prematurely in some cases (inset). Most class 4 plants showed SAM termination before the production of floral organs, although some flowers formed and set seed. Class 5 and class 6 plants showed strong transgene activity, and all of the SAMs were terminated before producing flower organs (class 5) or inflorescence meristems (class 6). The graph at bottom shows for each construct the percentage of transformed plants that belong to each phenotypic class.
(C) Wild-type Columbia plants transformed with the constructs described in (A). Primary transformants were grouped into classes 3, 4, 5, and 6 as described in (B). The graph at bottom shows for each construct the percentage of transformed plants that belong to each phenotypic class.
We measured the activity of the six constructs by transforming them either into clv3-2 plants or into wild-type plants of the Columbia ecotype and scoring their effects on shoot and floral meristem activity. A range of phenotypes resulting from various degrees of SAM inhibition was detected (Figures 2B and 2C). We classified the T1 transgenic plants into six phenotypic classes based on the extent of SAM termination: from plants that showed the enlarged meristems of clv3-2 mutants (class 1) to plants that showed termination of the SAM before the formation of an inflorescence stem (class 6). The remaining classes had phenotypes intermediate between these two extremes (Figures 2B and 2C). Because the constructs tested complemented the clv3-2 mutation and their effect on shoot and floral meristem termination in the wild-type background was additive with that of endogenous CLV3 (see below), we conclude that the CLV3 fusion proteins have the same activity as CLV3.
In both the clv3-2 and wild-type backgrounds, the predicted vacuolar CLV3 fusion proteins containing the functional ctVSS (Vac1, Vac2, and Vac3) showed highly reduced activity compared with the predicted secreted ctVSS (Sec1, Sec2, and Sec3). In the clv3-2 background (Figure 2B), 65 to 90% of the plants transformed with the Vac constructs showed either no phenotype (class 1 plants, indistinguishable from clv3-2 mutants) or very weak complementation (class 2, similar to weak clv3 alleles), whereas only 17 to 21% of the plants transformed with the Sec constructs had similar phenotypes. Conversely, 0 to 6% of the plants transformed with the Vac constructs and 48 to 54% of the plants transformed with the Sec constructs showed strong phenotypes (classes 5 and 6). Because of the additive effect of the endogenous CLV3, the difference between the secreted and the vacuole-targeted CLV3 constructs was even more apparent in the wild-type background (Figure 2C): 80 to 97% of the plants carrying Sec constructs terminated meristem activity during vegetative development (class 6). Of the remaining plants that developed an inflorescence meristem (class 5), only a few developed any flowers, and all of these lacked carpels. Thus, we did not obtain progeny from these transformants. In contrast, 51 to 59% of plants carrying the Vac constructs produced progeny, and 42 to 46% were indistinguishable from the wild type (class 3).
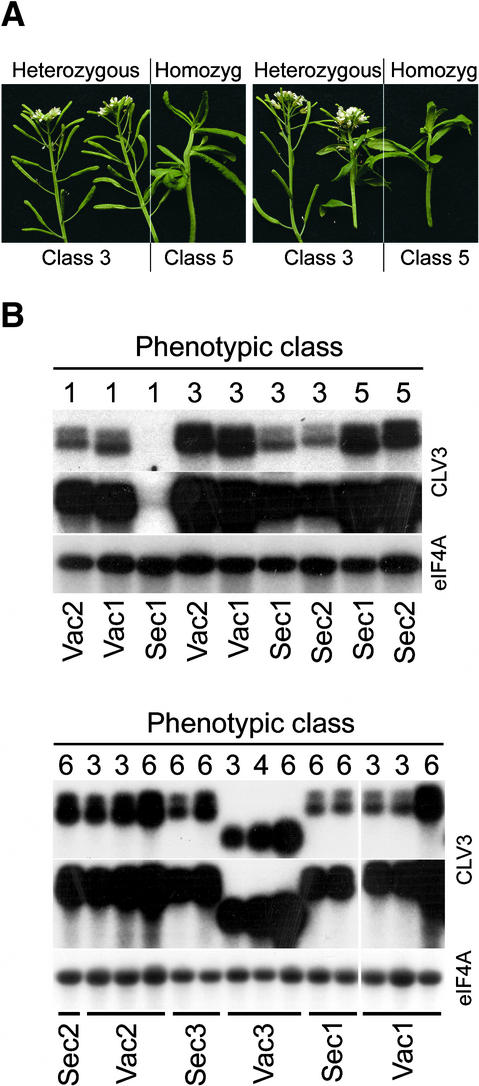
In many cases, we observed a stronger phenotype when the transgene copy number was increased to homozygosity in the subsequent generations (Figure 3A), demonstrating that the amount of expressed CLV3 fusion protein correlates directly with the severity of the phenotype. To determine whether the differences observed in transgene activity between Vac and Sec constructs were attributable to differences in expression, we analyzed the transgene mRNA levels in the transformed plants. The Sec and Vac constructs are almost identical in sequence (Sec3 and Vac2 differ by a single nucleotide), suggesting that transgene transcription and stability should be similar. As shown in Figure 3B, Sec constructs caused stronger meristem termination phenotypes than did Vac constructs when plants with similar levels of transgene expression were compared.
Figure 3.
The Extent of Meristem Termination Is Related Directly to Transgene Expression Level.
(A) Progeny from two independent primary transformants are shown. Eight weeks after kanamycin-resistant T2 plants were transplanted to soil, segregation of class 3 and class 5 phenotypes was observed. The T2 plants were genotyped by scoring the segregation of kanamycin resistance in their progeny (class 5 plants eventually set some seed). Plants with class 3 phenotypes were heterozygous for the transgene, whereas plants with class 5 phenotypes were homozygous.
(B) RNA gel blot analysis of transgene expression. Five micrograms of total RNA from rosette leaves of plants transformed with the CLV3 fusion constructs was hybridized with probes corresponding to CLV3 and eIF4A. Two different exposures of the hybridization with the CLV3 probe are shown (20 min and 2 hr). The mRNA abundance of the eIF4A transcript was used as a loading control. The top panel shows primary transformants in a clv3-2 background; the bottom panel shows primary transformants in the wild-type Columbia background. The phenotypic class of the plant is indicated at top, and the genotype is indicated at bottom. All constructs except for Vac3 contain the 3′ untranslated region of barley lectin and two transcription termination sites, which explains the larger transcript sizes and doublet bands in these lanes compared with the Vac3 lanes.
Most Sec-transformed plants showed detectable transgene activity (100% in the wild-type background and 93 to 95% in the clv3-2 background; Figure 2), and the few plants that showed no activity had very low or undetectable levels of transgene expression (Figure 3B). In contrast, 42 to 44% of the Vac-transformed plants in the wild-type background and 21 to 33% of those in the clv3-2 background showed no detectable activity of the transgene, even though there was considerable expression (see overexposure of blots in Figure 3B). Vac-transformed plants showed activity only at very high levels of expression of the transgene that probably cause saturation of the vacuolar sorting pathway and secretion of the protein (see below).
These results demonstrate that the reduced activity of Vac constructs is not caused by a decrease in transgene expression. Instead, our data indicate that sorting to the vacuole mediated by functional ctVSS blocks CLV3 activity, providing genetic evidence that CLV3 is a secreted protein that activates the CLV signaling pathway in the apoplast and not in an intermediate compartment of the secretory pathway.
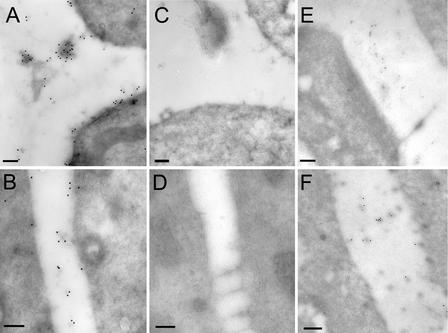
To confirm these genetic results, we analyzed the localization of the T7-tagged CLV3 fusion proteins Sec2 and Vac2 by immunoelectron microscopy. Our prediction was that the fusion proteins would be localized in the apoplast in those plants that showed transgene activity. We analyzed the localization of Sec2 in clv3-2–transformed plants that showed reversion to the wild-type phenotype (class 3; Figure 2B) and therefore should express amounts of the tagged CLV3 similar to that of the endogenous CLV3. In these plants, the tagged CLV3 was localized to the apoplast (Figures 4A and 4B), consistent with the genetic data described above. The Sec2 fusion protein has the same biological activity as CLV3 and contains no additional targeting signals. Thus, its localization in the apoplast likely reflects that of the endogenous CLV3 protein. Control sections showed no labeling (Figures 4C and 4D).
Figure 4.
CLV3 Is Localized in the Extracellular Space.
(A) and (B) Localization of a Sec2 fusion protein was analyzed in roots of a transgenic line that showed complementation of the clv3-2 mutation.
(C) Section from an untransformed clv3-2 mutant plant showing no labeling.
(D) Section from the same plant as in (A) and (B) incubated with no primary antibody showing no labeling.
(E) Vac2 transformant with no transgene activity showing no immunolabeling in the cell wall.
(F) Vac2 transformants with complementation of the clv3-2 mutation showing immunolabeling in the cell wall.
(A) to (D) show streptavidin conjugated to 15-nm gold particles; (E) and (F) show streptavidin conjugated to 10-nm gold particles. Bars = 200 nm.
Plants that had very high expression of vacuole-targeted CLV3 also showed activity of the transgene. We hypothesized that in these plants, extreme overexpression of the Vac constructs saturated the vacuolar sorting pathway, leading to partial secretion of the fusion protein, as has been shown for other proteins (Frigerio et al., 1998). Indeed, immunolabeling of the cell wall was observed in Vac2-transformed plants with high levels of transgene expression and an obvious effect of the transgene on meristem termination (class 3; Figure 4F). In contrast, no immunolabeling was detected in the apoplast of Vac2-transformed plants that showed no transgene activity (class 1; Figure 4E). Only occasionally did we observe weak labeling in the vacuole of Vac2-transformed plants (data not shown), suggesting that CLV3 is degraded in the lytic environment of this organelle. Alternatively, the processing of the ctVSS in the vacuole may affect the antigenicity of the T7 tag.
DISCUSSION
Several developmental regulatory proteins have been shown to move between plant cells. One is the maize homeodomain protein KNOTTED1 (KN1) (Vollbrecht et al., 1991), which is required to maintain proper SAM activity (Kerstetter et al., 1997; Vollbrecht et al., 2000). KN1 is found throughout the SAM, but kn1 mRNA is not present in the epidermal layer, indicating that KN1 must move outward from the underlying cells (Jackson et al., 1994). Several proteins involved in flower development also have cell nonautonomous activities associated with protein movement. The snapdragon MADS domain transcription factors DEFICIENS and GLOBOSA have been shown to traffic between cells (Perbal et al., 1996), as has the Arabidopsis LEAFY protein (Sessions et al., 2000).
There are two ways in which endogenous proteins can move between plant cells. One is for the protein to enter the secretory pathway of the cell and be exported into the apoplastic space between the cell walls. The other is for the protein to move symplastically through the plasmodesmata, elongated channels that span the cell wall to connect the cytoplasm of adjacent cells. The SAM appears to be composed of a network of symplastic fields (van der Schoot and Rinne, 1999). The angiosperm SAM is organized into clonally distinct tunica and corpus cell layers (Steeves and Sussex, 1989), and symplastic continuity mediated by plasmodesmatal connections occurs within but not between these layers (Gisel et al., 1999).
Experiments using fluorescent tracer dyes to track symplasm permeability in the SAM of birch seedlings revealed that the tunica is subdivided further into a central and a peripheral symplastic field, and these fields coordinate morphogenetic events during development (Rinne and van der Schoot, 1998). It has even been proposed that CLV1, which is expressed in the center of the SAM in the corpus layers (Clark et al., 1997), characterizes a central symplastic field of the corpus (van der Schoot and Rinne, 1999). Each of the above-mentioned proteins is proposed to move via the symplasm, and symplastic movement via plasmodesmata has been demonstrated for KN1 (Lucas et al., 1995). Thus, symplastic trafficking appears to be an important means of delivering developmental regulatory proteins to their destinations.
Our results indicate that apoplastic protein movement also is important for proper growth and development, in particular for proper SAM function. We have demonstrated both genetically and immunologically that CLV3 is a secreted protein that activates the CLAVATA pathway in the extracellular space. Secretion enables the apoplastic movement of CLV3 and can account for its nonautonomous regulation of stem cell accumulation as a secreted ligand for the CLV1/CLV2 receptor signaling complex.
CLV3 is therefore an example of a key developmental regulator in plants for which apoplastic localization is required for its function, which is to communicate cell fate information between different regions of Arabidopsis shoot and floral meristems. CLV3 signals from the stem cell population in the overlying SAM cells to the “organizing center” in the underlying cells, cutting across the tunica and corpus layers, which are symplastically isolated from one another. Thus, apoplastic signaling via small secreted polypeptides may be one method plants use to coordinate the behavior of cells in different symplastic domains.
Our results are consistent with a model for CLV signal transduction in which the CLV3 protein is secreted from stem cells at the apex of the meristem, travels through the extracellular space, and binds to the CLV1/CLV2 receptor complex at the plasma membrane of the underlying cells (Figure 5). Activation of the CLV signal transduction pathway downregulates the homeodomain-containing transcription factor gene WUSCHEL (WUS) that promotes stem cell activity (Laux et al., 1996; Mayer et al., 1998). Signaling via CLV3 limits the WUS-mediated stem cell–promoting signal back to the stem cell population, maintaining the appropriate amount of stem cell activity throughout development. Further experiments to detect CLV3 protein in shoot and floral meristem tissue by immunolocalization will enable us to determine the range of CLV signaling required to restrict WUS expression in Arabidopsis apices.
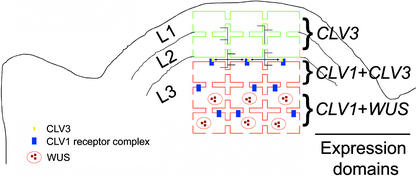
Figure 5.
Model of CLV3 Localization and Action.
The Arabidopsis SAM consists of three clonally distinct cell layers. The L1 epidermal layer and the L2 subepidermal layer are each single cell layers (outlined in green), whereas the interior L3 cells make up the bulk of the meristem (cells outlined in red). CLV3 is expressed at the apex of the SAM, predominantly in the superficial L1 and L2 cell layers and in a few cells of the L3. CLV1, which encodes a receptor for CLV3, is expressed in the underlying L3 cell layers. CLV3 is localized to the extracellular space and is predicted to travel through the apoplast from the L1 and L2 to the CLV1-expressing cells in the L3, which are isolated symplastically from the tunica (L1 and L2). Signaling by CLV3 through the CLV1 receptor complex causes the downregulation of WUS, restricting the WUS expression domain to the interior cells of the meristem.
METHODS
Constructs
For the transient assays, CLV3 cDNA with or without signal sequence was cloned as N-terminal fusions to green fluorescent protein (GFP) via a unique NcoI site in pAVA120 (von Armin et al., 1998). For stable transformation of Arabidopsis, the CLV3 cDNA was cloned into pRJG23 (Grebenok et al., 1997), which contains a double 35S promotor followed by GFP and β-glucuronidase (GUS). The nuclear localization signal from pRJG23 was removed by digestion with HindIII and SalI and replaced with full-length CLV3 (CLV3-G) or CLV3 without the signal sequence (CLV3Δsp-G). The pRJG23 constructs carrying the CLV3 cDNA then were digested with SacI and SalI, and this fragment carrying the 35S promoter of Cauliflower mosaic virus and CLV3 was cloned into the SalI-SacI site of the binary vector pBGF-0 (Chytilova et al., 1999).
For the vacuolar sorting assays, the 15–amino acid C-terminal propeptide of barley lectin (CTPPBL: 5′-VFAEAIAANSTLVAE-3′) was fused at the C terminus of either CLV3 (Vac1) or CLV3 fused to a T7 peptide tag (Vac2). The seven–amino acid C-terminal propeptide from tobacco chitinase A (CTPPCHI: 5′-GLLVDTM-3′) was fused at the C terminus of CLV3 (Vac3). A mutated CTPPBL (5′-VFAEAIAANSTLVAEGG-3′), which is not functional as a vacuolar sorting signal, was attached at the C terminus of either CLV3 (Sec1) or CLV3-T7 (Sec2). The Sec3 construct was made by deleting a single nucleotide in Vac2, which introduces a stop codon after the final amino acid of CLV3. The Sec1, Sec2, Sec3, Vac1, and Vac2 constructs all contain the 3′ untranslated region from barley lectin.
Transformations
Arabidopsis thaliana plants were transformed by floral dip (Clough and Bent, 1998). Thirty-three primary transformants were obtained for CLV3-pBGF in clv3-2, 18 for CLV3-pBGF in Landsberg erecta, and 5 for CLV3Δsp-pBGF in Landsberg erecta. In the Sec and Vac clv3-2 transformation experiments, >40 primary transformants per construct were obtained and phenotyped 8 weeks after transplanting to soil. The clv3-2 transformation was repeated three times with similar results. In the Sec and Vac Columbia (wild-type) transformation experiments, 40 primary transformants per construct were phenotyped 8 weeks after transplanting to soil. The Columbia transformation was repeated twice with similar results.
GUS and GFP Expression Analysis
Transient expression assays in leek epidermal cells using particle bombardment (Bio-Rad, Richmond, CA) were performed as described previously (Restrepo et al., 1990). Cells were incubated for 48 hr at 25°C, and the location of GUS activity was determined using 5-bromo-4-chloro-3-indolyl-β-d-glucuronide. For GFP and GUS visualization, Arabidopsis tissue was examined using a Zeiss Axiophot microscope (Jena, Germany) with a 12-bit MicroMax cooled charge-coupled device camera operated by IPLab software (Scanalytics, Fairfax, VA). For GFP localization in tissues, transgenic plants were germinated on Murashige and Skoog (1962) medium with kanamycin for 7 days, and tissues were put on depression slides for observation using a Zeiss 510 UV/visible light confocal microscope. Images were processed using Adobe Photoshop (Mountain View, CA).
RNA Analysis
Rosette leaves from plants transformed with the CLV3 fusion constructs were collected. Total RNA was extracted from the leaves (Logemann et al., 1987), and 5 μg was analyzed by RNA gel blotting for transgene expression using a probe to detect CLV3 mRNA.
Immunoelectron Microscopy
Cryosections of Arabidopsis root tips were prepared (Sanderfoot et al., 1999) and used for immunolabeling experiments. Immunolabeling was performed as described previously (Sanderfoot et al., 1999; Zheng et al., 1999). An anti-T7 monoclonal antibody followed by rabbit anti-mouse IgG and biotinylated goat anti-rabbit secondary antibody were visualized with streptavidin conjugated to gold particles. Control experiments were performed on cryosections of clv3 mutants with anti-T7 monoclonal antibody and in plants transformed with CLV3-T7 with no primary antibody. The results of all control experiments were negative.
Acknowledgments
We thank Jim Tepperman for advice on GUS staining and Denise Schichnes for help with confocal microscopy. We thank members of our laboratories for helpful discussions during the course of this work. J.C.F. was supported by the U.S. Department of Agriculture (Grant 5335-21000-013-D). N.V.R. was supported by the Department of Energy (Grant DE-FG02-91ER-20021) and the National Science Foundation (Grant MCB-0296080).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.002196.
References
- Bednarek, S.Y., and Raikhel, N.V. (1991). The barley lectin carboxyl-terminal propeptide is a vacuolar protein sorting determinant in plants. Plant Cell 3, 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, U., Fletcher, J.C., Hobe, M., Meyerowitz, E.M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. [DOI] [PubMed] [Google Scholar]
- Chytilova, E., Macas, J., and Galbraith, D.W. (1999). Green fluorescent protein targeted to the nucleus, a transgenic phenotype useful for studies in plant biology. Ann. Bot. 83, 645–654. [Google Scholar]
- Clark, S.E., Running, M.P., and Meyerowitz, E.M. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119, 397–418. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Running, M.P., and Meyerowitz, E.M. (1995). CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121, 2057–2067. [Google Scholar]
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Denecke, J., Botterman, J., and Deblaere, R. (1990). Protein secretion in plant cells can occur via a default pathway. Plant Cell 2, 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski, J.E., Schroeder, M.R., Bednarek, S.Y., and Raikhel, N.V. (1993). Determination of the functional elements within the vacuolar targeting signal of barley lectin. Plant Cell 5, 587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher, J.C., Brand, U., Running, M.P., Simon, R., and Meyerowitz, E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914. [DOI] [PubMed] [Google Scholar]
- Frigerio, L., de Virgilio, M., Prada, A., Faoro, F., and Vitale, A. (1998). Sorting of phaseolin to the vacuole is saturable and requires a short C-terminal peptide. Plant Cell 10, 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisel, A., Barella, S., Hempel, F.D., and Zambryski, P.C. (1999). Temporal and spatial regulation of symplastic trafficking during development in Arabidopsis thaliana apices. Development 126, 1879–1889. [DOI] [PubMed] [Google Scholar]
- Grebenok, R.J., Peirson, E., Lambert, G.M., Gong, F.-C., Afonso, C.L., Haldeman-Cahill, R., Carrington, J.C., and Galbraith, D.W. (1997). Green florescent protein fusions for efficient characterization of nuclear targeting. Plant J. 11, 573–586. [DOI] [PubMed] [Google Scholar]
- Jackson, D., Veit, B., and Hake, S. (1994). Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120, 405–413. [Google Scholar]
- Jeong, S., Trotochaud, A.E., and Clark, S.E. (1999). The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11, 1925–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayes, J.M., and Clark, S.E. (1998). CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125, 3843–3851. [DOI] [PubMed] [Google Scholar]
- Kerstetter, R.A., Laudencia-Chingcuanco, D., Smith, L.G., and Hake, S. (1997). Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124, 3045–3054. [DOI] [PubMed] [Google Scholar]
- Laux, T., Mayer, K.F.X., Berger, J., and Jurgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96. [DOI] [PubMed] [Google Scholar]
- Logemann, J., Schell, J., and Willmitzer, L. (1987). Improved method for the isolation of RNA from plant tissues. Anal. Biochem. 163, 16–20. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J., Bouche-Pillon, S., Jackson, D.P., Nguyen, L., Baker, L., Ding, B., and Hake, S. (1995). Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270, 1980–1983. [DOI] [PubMed] [Google Scholar]
- Mayer, K.F.X., Schoof, H., Haecker, A., Lenhard, M., Jurgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. [DOI] [PubMed] [Google Scholar]
- Munro, S., and Freeman, M. (2000). The Notch signalling pathway regulator Fringe acts in the Golgi apparatus and requires the glycosyltransferase motif DxD. Curr. Biol. 10, 813–820. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Neuhaus, J.M., Sticher, L., Meins, J.F., and Boller, T. (1991). A short C-terminal sequence is necessary and sufficient for the targeting of chitinases to the plant vacuole. Proc. Natl. Acad. Sci. USA 88, 10362–10366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal, M.-C., Haughn, G., Saedler, H., and Schwarz-Sommer, Z. (1996). Non-cell-autonomous function of the Antirrhinum floral homeotic proteins DEFICIENS and GLOBOSA is exerted by their polar cell-to-cell trafficking. Development 122, 3433–3441. [DOI] [PubMed] [Google Scholar]
- Restrepo, M.A., Freed, D.D., and Carrington, J.C. (1990). Nuclear transport of plant polyviral protein. Gene 111, 229–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinne, P.L.H., and van der Schoot, C. (1998). Symplastic fields in the tunica of the shoot apical meristem coordinate morphogenetic events. Development 125, 1477–1485. [DOI] [PubMed] [Google Scholar]
- Sanderfoot, A.A., Kovaleva, V., Zheng, H.G., and Raikhel, N.V. (1999). The t-SNARE AtVAM3p resides on the prevacuolar compartment in Arabidopsis in root cells. Plant Physiol. 121, 929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford, J.C., Smith, F.D., and Russell, J.A. (1993). Optimizing the biolistic process for different biological applications. Methods Enzymol. 217, 483–509. [DOI] [PubMed] [Google Scholar]
- Sessions, A., Yanofsky, M.F., and Weigel, D. (2000). Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science 289, 779–781. [DOI] [PubMed] [Google Scholar]
- Steeves, T.A., and Sussex, I.M. (1989). Patterns in Plant Development. (New York: Cambridge University Press).
- Trotochaud, A., Jeong, S., and Clark, S.E. (2000). CLAVATA3, a multimeric ligand for the CLAVATA1 receptor-kinase. Science 289, 613–617. [DOI] [PubMed] [Google Scholar]
- van der Schoot, C., and Rinne, P. (1999). Networks for shoot design. Trends Plant Sci. 4, 31–37. [DOI] [PubMed] [Google Scholar]
- Vollbrecht, E., Veit, B., Sinha, N., and Hake, S. (1991). The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 350, 241–243. [DOI] [PubMed] [Google Scholar]
- Vollbrecht, E., Reiser, L., and Hake, S. (2000). Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127, 3161–3172. [DOI] [PubMed] [Google Scholar]
- von Armin, A.G., Deng, X.W., and Stacey, M.G. (1998). Cloning vectors for the expression of green fluorescent proteins in transgenic plants. Gene 221, 35–43. [DOI] [PubMed] [Google Scholar]
- Zheng, H.G., Fischer von Mallard, G., Kovaleva, V., Stevens, T.H., and Raikhel, N.V. (1999). The plant vesicle-associated SNARE AtVTI1a likely mediates vesicle transport from the trans-Golgi network to the prevacuolar compartment. Mol. Biol. Cell 10, 2251–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]