Abstract
We describe the identification of a mutant in the Arabidopsis accession Columbia (Col-0) that exhibits enhanced downy mildew (edm1) susceptibility to several Peronospora parasitica isolates, including the RPP7-diagnostic isolate Hiks1. The mutation was mapped to chromosome IV and characterized physically as a 35-kb deletion spanning seven genes. One of these genes complemented the mutant to full wild-type resistance against all of the Peronospora isolates tested. This gene (AtSGT1b) encodes a predicted protein of 39.8 kD and is an Arabidopsis ortholog of yeast SGT1, which was described originally as a key regulatory protein in centromere function and ubiquitin-mediated proteolysis. AtSGT1b contains three tetratricopeptide repeats at the N terminus followed by a bipartite chord-containing SGT domain and an SGT-specific domain at the C terminus. We discuss the role of AtSGT1b in disease resistance and its possible involvement in ubiquitin-mediated proteolysis in plants.
INTRODUCTION
Resistance of plants to biotrophic pathogens is controlled by a complex regulatory system with many features that suggest an ancient origin (for reviews, see Dangl and Jones, 2001; Holub, 2001). Specific molecular recognition of pathogen avirulence (Avr) determinants by receptor-like plant proteins, encoded by resistance (R) genes, triggers signal transduction processes that activate a variety of defense reactions in infected plants. These inducible defense responses include an oxidative burst resulting from the generation of highly reactive oxygen intermediates (Lamb and Dixon, 1997; Torres et al., 2001), irreversible membrane damage (Woods et al., 1988), hypersensitive death of host cells (Lam et al., 1999), increased expression of defense-associated genes (Maleck et al., 2000; Schenk et al., 2000), and synthesis of antimicrobial metabolites such as phytoalexins (Glazebrook et al., 1997).
Two key defense regulators that are required for the function of multiple R genes have been identified in Arabidopsis: NDR1 (Century et al., 1995) encodes a potentially membrane-associated protein of unknown function (Century et al., 1997), and EDS1 (Parker et al., 1996) encodes a soluble protein that has homology with eukaryotic lipases (Falk et al., 1999). Signaling through both EDS1 and NDR1 activates a common set of defense responses, including the synthesis of salicylic acid, an important component in local and systemic disease resistance (Feys and Parker, 2000).
Many R genes have been shown to preferentially use either NDR1 or EDS1 to confer resistance against either bacterial or eukaryotic pathogens in Arabidopsis (Aarts et al., 1998). Most of the R genes known at the time from Arabidopsis were used in this analysis, all of which encode receptor-like protein products that contain a carboxyl Leu-rich repeat domain (LRR) and a central nucleotide binding site (NB) (referred to hence as NB-LRR genes). Aarts et al. (1998) observed that R gene functional subsets appeared to correlate with different structural subclasses of NB-LRR genes: the NDR-dependent subclass encodes proteins with an N-terminal coiled-coil domain (CC), whereas the EDS1-dependent subclass encodes proteins with an N terminus that resembles cytoplasmic domains of the Drosophila Toll and mammalian interleukin 1 transmembrane receptors (TIR domain). Therefore, it was proposed that EDS1 and NDR1 relay alternative defense signals that originate from different subclasses of R gene products and converge upstream of salicylic acid accumulation.
Several exceptions to this model have been defined subsequently using isolates of Peronospora parasitica (downy mildew) and Erysiphe spp (powdery mildew). For example, EDS1 was selected and characterized as a gene required for downy mildew resistance conferred by RPP5 in cotyledons and adult leaves of the Arabidopsis accession Landsberg erecta (Ler-0) (Parker et al., 1997), whereas an allele of RPP5 in Columbia (Col-0), designated RPP4 (Tör et al., 1994; Van der Biezen et al., 2002), requires NDR1 in cotyledons (Century et al., 1995) as well as EDS1. RPP8 (McDowell et al., 1998) encodes a CC-NB-LRR protein, and its function is not affected by a single mutation in NDR1. RPP8-mediated resistance is suppressed weakly in an eds1 ndr1 double mutant (McDowell et al., 2000), demonstrating that full wild-type resistance is achieved by the accumulation of distinct defense responses, each contributing partially to resistance and regulated by NDR1, EDS1, or an as yet unknown component. Finally, the powdery mildew resistance gene RPW8 encodes a coiled-coil protein that requires EDS1 but not NDR1 (Xiao et al., 2001).
Similar to RPP8, resistance conferred by RPP7 (in Col-0 and recognizing isolate Hiks1) was largely unaffected by single mutations in defense regulators, including the two described above, as well as others that affect phytoalexin production or salicylic acid, jasmonic acid, and ethylene signaling (Glazebrook et al., 1997; Warren et al., 1999; McDowell et al., 2000). McDowell and colleagues (2000) described double mutant analysis that suggested that RPP7-mediated resistance also was achieved by cumulative defense responses. RPP7 has yet to be isolated; however, it has been mapped to a 100-kb interval that spans one of the largest groups of NB-LRR genes in Arabidopsis. None of these is in the TIR subclass, but several share high sequence homology with RPP8 and RPP13 (Holub, 2001; A. Cuzick and E.B. Holub, unpublished data).
Here, we describe the identification of a 35-kb deletion mutant in Col-0 designated edm1 (enhanced downy mildew) that largely suppresses RPP7 resistance to Hiks1 as well as resistance to several additional Col-0–incompatible Peronospora isolates. The deletion spans seven genes, including one that encodes a protein similar to yeast SGT1, which was described by Kitagawa et al. (1999) as a key regulatory protein in centromere function and ubiquitin-mediated proteolysis.
RESULTS
Phenotypic Characterization of Col-edm1
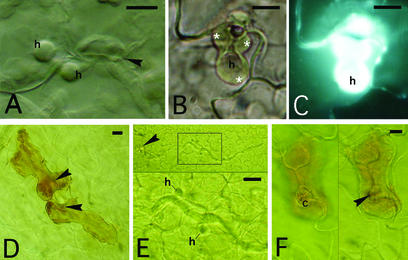
The wild-type phenotype of RPP7-mediated resistance in Arabidopsis to infection by the Peronospora isolate Hiks1 has been described macroscopically as a barely visible host response (minute, necrotic flecks) that completely restricts parasite reproduction (Holub et al., 1994). Trypan blue staining of whole cotyledons 7 days after inoculation revealed membrane disruption of host cells bordering the penetrating hyphae (McDowell et al., 2000). The resistance is visible microscopically as complete cessation of parasite growth within the first 24 to 48 hr of infection, with the penetrating hyphae terminating soon after producing one or two haustoria in adjacent mesophyll cells (Figure 1A). A rapid host response becomes visible 48 hr after inoculation, as indicated by callose and autofluorescent deposits (probably phenolic compounds) in host cells surrounding the penetrating hyphae. Callose ensheathment of haustoria and host cell collapse occur in later stages of the wild-type defense response (Figures 1B and 1C).
Figure 1.
Cotyledons of Wild-Type Col-0, Col-edm1, and Col-edm1::SGT1b Infected with the Peronospora Isolate Hiks1.
(A) Twinned haustoria (h) produced by the parasite in adjacent mesophyll cells before a visible host response in wild-type Col-0 photographed 1 day after inoculation.
(B) Callose ensheathment (white asterisks) of a haustorium in the wild type photographed 7 days after inoculation.
(C) Same as (B) but fluorescing under blue light excitation from aniline blue staining of callose.
(D) H2O2 accumulation detected with DAB staining 1 day after inoculation in mesophyll cells of the wild type.
(E) No detection of H2O2 with DAB staining in Col-edm1 1 day after inoculation.
(F) DAB staining 1 day after inoculation of mesophyll cells of Col-edm1::SGT1b.
Arrowheads indicate penetration sites between anticlinal walls of the epidermal cells (higher focal plane). Bars = 10 μm.
The edm1 mutant disrupts RPP7-mediated resistance significantly and is an example of a single mutant in Col-0 that is susceptible to Hiks1. The mutant was selected from screening fast neutron–treated Col-5 (glabrous1 mutant of Col-0) with the Peronospora isolate Cala2, which is recognized in Col-0 by RPP2 (Holub et al., 1994; Tör et al., 1994). Our extensive attempts to select further fast neutron or ethyl methanesulfonate (EMS) mutant alleles of edm1 using Hiks1 were unsuccessful (see Methods).
Col-edm1 supported enhanced sporulation after infection with seven Col-0–incompatible Peronospora isolates that are each recognized by different RPP genes (Table 1). The level of susceptibility (measured as the amount of sporulation) varied for each isolate, ranging from heavy (>20 sporangiophores per cotyledon; full susceptibility) produced by Cala2 to low (<10 sporangiophores per cotyledon) produced by Wand1 and Wela3. The other four isolates, including Hiks1, produced moderate sporulation (∼15 sporangiophores per cotyledon) in the mutant. The morphology of Col-edm1 is similar to that of the wild type; however, flowering time appears to be delayed by 1 week or more in the mutant relative to the wild type when plants are grown in either short or long days.
Table 1.
Reproduction in Arabidopsis (Wild-Type Col-0 and Col-edm1) of Seven Col-0–Incompatible Isolates of Peronospora (Downy Mildew), Each Recognized by a Different Host Resistance (RPP) Determinant
| Col-0
|
Col-edm1
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate | RPP Locusa | R-Gene Classb | Meanc | SEM | No. | IPd | Mean | SEM | No. | IP | Col-rar1e IP | Col-ndr1e IP |
| Cala2 | RPP2-IV | TIR-NB-LRR | 0.74 | 0.23 | 72 | R | 19.70 | 0.17 | 58 | H | wt | wt |
| Emoy2 | RPP4-IV | TIR-NB-LRR | 1.86 | 0.21 | 74 | L2 | 15.78 | 0.75 | 50 | M | M | M |
| Hiks1 | RPP7-I | Non-TIR | 0.00 | 0.00 | 116 | N | 14.10 | 0.77 | 102 | M | R | R |
| Wela3 | RPP6-I | Unknown | 0.00 | 0.00 | 68 | N | 6.39 | 0.59 | 52 | L6 | L4 | L2 |
| Cand5 | nd-polygenic | Unknown | 0.93 | 0.29 | 52 | R | 15.00 | 0.70 | 94 | M | H | H |
| Hind4 | nd-II | Unknown | 2.18 | 0.29 | 66 | L2 | 11.70 | 0.92 | 35 | M | L10 | L4 |
| Wand1 | nd-II | Unknown | 0.01 | 0.01 | 57 | R | 5.73 | 0.55 | 52 | L6 | H | L2 |
Recognition of Peronospora. Chromosomes are indicated by roman numerals. nd, not designated.
For review, see Holub (2001).
Mean number of sporangiophores per cotyledon, counted up to a maximum of 20.
IP, interaction phenotype interpreted from quantitative data: N, no sporulation; R, rare sporangiophore (<1 per cotyledon); L, low (1 to 10 sporangiophores, with nearest incremental number indicated); M, medium (11 to 16 sporangiophores); H, heavy (>16 sporangiophores); wt, same as the wild type.
Phenotypes for Hiks1 were taken from rar1 and ndr1 controls in Table 2. All other phenotypes were taken from the previous report by Warren et al. (1999). (Note that the rar1 allele used in this study was referred to as pbs2.)
Seedlings of wild-type Col-0, Col-edm1, and Col-rpp7.1 (a fully susceptible EMS mutant of RPP7) (Table 2) were compared for differences in their capacity to generate H2O2 24 hr after inoculation with Hiks1 using 3,3′-diaminobenzidine (DAB) to stain the infected tissue. This compound captures H2O2 and forms a reddish-brown polymer at sites of peroxidase activity (Thordal-Christensen et al., 1997), providing a means to detect an oxidative burst in host cells surrounding penetrating hyphae. More than 100 cotyledons from mutant and wild-type seedlings were inspected. DAB staining was observed in two to four cells per infection site in every wild-type cotyledon, with an average of 10 infection sites per cotyledon. No DAB staining of infection sites was observed in cotyledons of either mutant or in noninoculated control cotyledons. Examples are shown in Figures 1D and 1E. Thus, edm1 disrupts RPP7 function upstream of the oxidative burst.
Table 2.
Genetic Characterization of the edm1 Mutant of Arabidopsis (Col-0) That Exhibits Enhanced Downy Mildew after Inoculation with Peronospora Isolate Hiks1
| Selfed or F1a
|
F2
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Arabidopsis Genotype | Mean | SEM | No. | IPc | Nd | L | M + H | Observed | Predicted | χ2 | P |
| Col-0 (RPP7)b | 0.0 | 0.0 | 380 | N | |||||||
| Col-rpp7.1 | 20.0 | 0.0 | 207 | H | |||||||
| Col-edm1 | 13.7 | 0.8 | 274 | M | |||||||
| Col-ndr1 | 0.1 | 0.1 | 154 | R | |||||||
| Col-rar1.21 | 0.1 | 0.1 | 254 | R | |||||||
| Col-rpp7.1 × Col-edm1 | 2.5 | 0.5 | 48 | L2 | 157 | 6 | 144 | 163:144 | 173:134 (9:7) | 1.3 | 0.25 |
| Col-edm1 × Col-0 | 0.0 | 0.0 | 92 | N | 40 | 2 | 12 | 42:12 | 41:14 (3:1) | 0.2 | 0.64 |
| Col-rpp7.1 × Col-0 | 0.8 | 0.4 | 10 | L1 | 28 | 19 | 12 | 38:12 | 38:12 (3:1) | 0.0 | 1.00 |
a Selfed seed from parents used in crosses (bottom half) or control accessions; F1 seed correspond to segregating F2 populations. Values shown are mean numbers of sporangiophores per cotyledon.
b RPP7 is the R gene that confers resistance to Hiks1 in Col-0.
c IP, interaction phenotype interpreted from quantitative data: N, no sporulation; R, rare sporangiophore (<1 per cotyledon); L, low (1 to 10 sporangiophores, with nearest incremental number indicated); M, medium (11 to 16 sporangiophores); H, heavy (>16 sporangiophores).
d Number of progeny in N, L, M, or H sporulation classes, which was used to calculate the observed resistant (N + L) to susceptible (M + H) ratio.
We also tested Col-edm1 for altered resistance to isolates of the bacterial pathogen Pseudomonas syringae strain DC3000 expressing different Avr determinants: avrRpm1 and avrPphB eliciting NDR1-dependent resistance conferred by RPM1 and RPS5, respectively; and avrRps4 eliciting EDS1-dependent resistance conferred by RPS4 (Aarts et al., 1998). In all cases, bacterial growth was found to be of similar magnitude in wild-type and edm1 plants. Growth of avrRpm1 or avrPphB expressing bacteria in ndr1.1 seedlings and growth of avrRps4 expressing bacteria in pad4.1 seedlings were significantly higher (data not shown). Thus, RPM1, RPS4, and RPS5 are not influenced by edm1.
Genetic Characterization of the edm1 Mutation
An rpp7 EMS mutant (Col-rpp7.1) was crossed to the edm1 mutant. The resulting F1 seedlings exhibited wild-type resistance after inoculation with Hiks1 (Table 2), indicating that both mutations were recessive and that the edm1 mutations affected a gene(s) other than RPP7. This was confirmed using the isolate Cala2 to test F2 seedlings from an outcross between Col-edm1 and the Cumbrian accession Keswick (Ksk-1); Col-0 and Ksk-1 share RPP2 alleles that recognize this isolate (Holub et al., 1994). The segregation in this experiment was 107 resistant to 25 susceptible (3:1; χ2 = 0.65, P = 0.45), suggesting that edm1 was the only susceptibility determinant segregating in this cross.
For fine-scale mapping of edm1, we took advantage of extensive polymorphic loci already available from the Arabidopsis research community between Col-0 and another standard accession, Ler-0 (see Methods). Ler-0 was predicted previously to carry a functional RPP7 allele because we were unable to select a Hiks1-susceptible recombinant individual from more than 5000 F2 Col-0 × Ler-0 seedlings (McDowell et al., 2000). The F2 Col-edm1 × Ler-0 segregated in a ratio of 800 resistant to 70 susceptible seedlings after inoculation with Hiks1. We expected ∼217 susceptible seedlings as a result of the recessive edm1 mutation; thus, our result suggested that Ler-0 carries a second resistance gene (designated RPP27) that is EDM1 independent. We tentatively mapped both loci using 20 Hiks1-susceptible segregants (i.e., homozygous for Col-0 mutant alleles of edm1 and rpp27) from this outcross population (see Methods). The EDM1 locus then was identified by cross-referencing the closely linked markers (F17A8 and SC5) from one of the loci with the single edm1 locus that could be mapped in the Col-edm1 × Ksk-1 outcross population using Cala2.
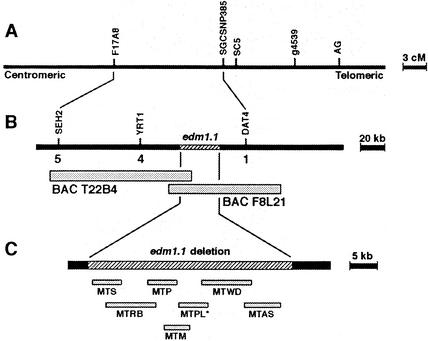
A total of 310 Hiks1-susceptible F2 Col-edm1 × Ler-0 seedlings was used to fine map EDM1 within an interval spanned by F17A8 and SC5 (Figure 2A). New markers were generated from the sequence information of two bacterial artificial chromosomes (BACs) that span the EDM1 region (T22B4 and F8L21), including markers YRT1 and DAT4, which were used to identify five key recombinant individuals (Figure 2B). Further attempts to generate markers within this interval revealed a 35-kb deletion, determined by the lack of polymerase chain reaction (PCR) amplification and DNA gel blot hybridization (Figure 3).
Figure 2.
Map-Based Cloning of Arabidopsis SGT1b.
(A) Genetic map of the edm1 locus indicating molecular markers SC5 and F17A8 that were used initially to define the mapping interval. cM, centimorgan.
(B) The BAC contig spanning the EDM1 locus, with markers used to define the interval shown above the bar and the number of recombinant individuals with each marker shown below the bar.
(C) Constructs used to complement the Col-edm1 mutant. The clone that restored wild-type resistance in the mutant is indicated by the asterisk. The constructs each contain a single gene from the deleted region and are as follows: MTS, At4 g11220, which has similarity to a seed maturation protein from soybean; MTRB, At4 g11230, which has strong similarity to the respiratory burst oxidase homolog F; MTP, At4 g11240, a phosphatase type 1 protein; MTM, At4 g11250, a putative protein that may contain an SRF-type MADS box domain; MTPL, At4 g11260, which is described in the text as AtSGT1b; MTWD, At4 g11270, which contains two regions with similarity to prokaryotic membrane lipoprotein lipid attachment sites and six WD-40 repeats; and MTAS, At4 g11280, with similarity to 1-aminocyclopropane-1-carboxylic acid synthase.
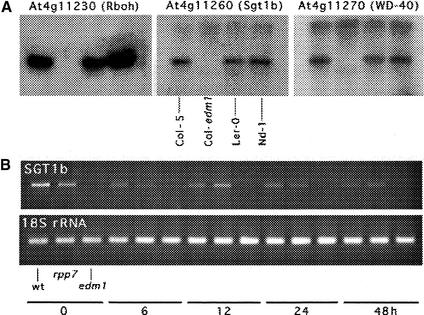
Figure 3.
DNA Gel Blot Hybridization of Col-edm1 Mutant, and Expression Analysis of SGT1b.
(A) Genomic DNA from wild-type Col-5, Col-edm1, Ler-0, and Nd-1 was digested with BglII, separated by agarose gel electrophoresis, and transferred to a nylon membrane. The membrane was probed with a 32P-labeled PCR product generated from the indicated genes in the locus. Three deleted genes are shown as examples. Gene numbers are given above the panels, and the characteristic feature of each gene is shown in parentheses. Rboh, respiratory burst oxidase homolog F; WD-40, repeat motif of the gene.
(B) Total RNA was isolated from wild-type Col-0 (wt), Col-rrp7.1, and Col-edm1 at 0, 6, 12, 24, and 48 hr after inoculation with Hiks1. RT-PCR analyses were performed with gene-specific primers using 0.5 μg of total RNA as a template. The PCR products were separated on a 1.5% agarose gel and stained with ethidium bromide. The RT-PCR products were of the expected sizes, and their sequences were identical to that of AtSGT1b.
An SGT1-like Gene Restores Wild-Type Resistance in Col-edm1
Transgenic complementation of the edm1 deletion was performed with seven constructs that contained a single gene from the deleted region (Figure 2C). All of these constructs were used to transform Col-edm1 plants. T2 seedlings from a minimum of 10 independent transformants were assessed for restoration of resistance after inoculation with Hiks1, Cala2, and Cand5. The construct MTPL, which contains a 4950-bp DNA fragment encompassing an SGT1-like gene (At4 g11260), restored wild-type resistance in Col-edm1 to all three isolates and restored H2O2 production (Figure 1F). T2 seedlings from the complementing lines all segregated for the EDM1 and edm1 phenotypes (mostly 3:1), correlating completely with Basta resistance and sensitivity, respectively (data not shown).
Sequence and Expression Analysis of SGT1b
Database searches with the SGT1-like gene identified several expressed sequence tags (ESTs) that were obtained and resequenced. One of these was confirmed as a full-length cDNA, and comparison of the genomic and cDNA sequences allowed us to define 10 exons and 9 introns (data available from http://mips.gsf.de/). Further database searches identified another SGT1-like homolog in Arabidopsis Col-0 located ∼30 centimorgan telomeric to At4 g11260 on the BAC clone F9D16. An Arabidopsis EST corresponding to this gene was referred to previously by Shirasu et al. (1999) as Arabidopsis SGT1. Hence, we refer to this homolog as AtSGT1a, and the gene that complements the edm1 deletion is designated AtSGT1b.
Blast searches demonstrated that AtSGT1b has high homology with AtSGT1a, a gene from Rubus idaeus, a rice SGT1 gene, and the yeast SGT1 gene (Figure 4). Overall, AtSGT1b shares 72, 64, 60, and 32% amino acid identity, respectively, with the predicted protein products of the other genes. AtSGT1b encodes a predicted protein of 358 amino acids (molecular mass of 39.8 kD) that is rich in Glu (12.85%), Ala (12.29%), Lys (11.17%), and Gln (10.61). Hydropathy plot analyses indicated that AtSGT1b is a soluble protein with no obvious transmembrane domains. AtSGT1a, AtSGT1b, and rice SGT1 have three tetratricopeptide repeats (TPRs) (Goebl and Yanagida, 1991) at the N-terminal region. The R. idaeus cDNA encodes two copies of TPR repeats, whereas the TPR motif is absent in yeast SGT1. The central region of AtSGT1b shares a bipartite CS domain described by Shirasu et al. (1999) with AtSGT1a, rice SGT1, and R. idaeus SGT1 but not with yeast SGT1. An SGT-specific (SGTS) domain described by Kitagawa et al. (1999) includes ∼100 amino acids at the C-terminal end in SGT1 proteins from species including those represented in Figure 4 as well as human, Drosophila melanogaster, and Caenorhabditis elegans.
Figure 4.
Comparison of SGT1 Amino Acid Sequences from Plants and Yeast.
SGT1-like protein sequences from budding yeast (Saccharomyces cerevisiae), Arabidopsis (AtSGT1a and AtSGT1b), rice (Oryza sativa), and Rubus idaeus were aligned using Pileup (Wisconsin Package Version 10.0; Genetics Computer Group, Madison, WI). Black or dark gray boxes with white letters indicate identity or similarity to yeast SGT1, respectively. Identity or similarity among the four plant SGT1 sequences exclusively is highlighted in light gray with black letters. The three TPRs (which occur only in the plant sequences), the bipartite CS motifs, and the SGTS are indicated above the sequences.
The presence of several ESTs in the database indicated that AtSGT1b is expressed constitutively in noninfected tissue of Arabidopsis. This was confirmed and compared with infected tissue by isolating total RNA from wild-type Col-5, Col-rpp7.1, and Col-edm1 at 0, 6, 12, 24, and 48 hr after inoculation with Hiks1. Reverse transcriptase–mediated (RT) PCR analysis (Figure 3B) was performed using gene-specific primers. We detected AtSGT1b mRNA in Col-5 at 0 hr after inoculation, showing that it is expressed constitutively. Amplification in Col-rpp7.1 indicated that AtSGT1b expression is not RPP7 dependent. The lack of amplification in the edm1 deletion mutant demonstrates that the primers used did not detect AtSGT1a mRNA and therefore were AtSGT1b specific. A time course of AtSGT1b expression in Col-5 and Col-rpp7.1 was very similar at 0, 6, 12, 24, and 48 hr.
DISCUSSION
The striking similarity of three structural domains (TPR, CS, and SGTS) in AtSGT1b to other SGT1 proteins (Figure 4) suggests a highly conserved function of this protein across kingdoms. AtSGT1b has three TPRs at the N-terminal region. The TPR is a degenerate 34–amino acid repeated motif widely reported in a growing number of eukaryotic and prokaryotic proteins (Lamb et al., 1995). TPR-containing proteins are known to form amphipathic α-helices that mediate protein–protein interaction and have been shown to interact with other TPR or non-TPR proteins (Goebl and Yanagida, 1991; Das et al., 1998). TPR proteins can be involved in a wide array of cellular functions, including cell cycle regulation, transcription control, stress response, protein kinase inhibition, protein transport, Rac-mediated activation of NADPH oxidase, and protein folding (Lamb et al., 1995). In Arabidopsis, a TPR-containing protein, SPY, has been shown to be involved in gibberellic acid signal transduction (Jacobsen et al., 1996).
The central bipartite CS domain and C-terminal SGTS domain provide a plausible link between a defense signal relayed from a R–Avr interaction and the SCF complex, which may lead to proteolysis. The CS domain of AtSGT1b is important for interacting with regulatory proteins such as RAR1, which was described originally by Shirasu et al. (1999) in barley. Arabidopsis RAR1 was cloned recently as a defense regulator of several downy mildew and bacterial resistance genes (Muskett et al., 2002; Tornero et al., 2002). The SGTS domain is the most conserved sequence of AtSGT1b, showing high homology with domains in yeast SGT1 and other plant SGT1 proteins (Figure 4). Azevedo et al. (2002) have reported that both AtSGT1a and AtSGT1b interact directly with Arabidopsis RAR1. In addition, they also have shown that barley SGT1 coimmunoprecipitated with the subunits of the SCF complex, including SKP1, CUL1, and two COP9 subunits (CSN4 and CSN5). Recently, Kitagawa et al. (1999) demonstrated that SGT1 in yeast is required for centromere function and associates with the SCF complex via the SGTS domain. Similarly, the human SGT1 homolog SIP has been shown to interact with the SCF complex for the degradation of β-catenin (Matsuzawa and Reed, 2001).
Null mutations in the single-copy yeast SGT1 gene are lethal, presumably as a result of its involvement in centromere function (Kitagawa et al., 1999), whereas the deletion of Atsgt1b in the Col-edm1 mutant did not affect viability, presumably because of redundant function provided by AtSGT1a. Therefore, the double mutation of sgt1a and sgt1b in Arabidopsis may be lethal. If this is the case, examples of disease resistance that rely on both SGT1 genes may be revealed only by investigating combined mutations of upstream components that are shown to relay independently via SGT1a or SGT1b. As described below, the rar1 mutation of Arabidopsis will be useful for such investigations.
A biphasic oxidative burst, resulting from a rapid release of reactive oxygen intermediates (ROI), has been implicated as an essential component of plant defense signaling during the early stages of pathogen infection (Lamb and Dixon, 1997). The first oxidative burst has been observed as a response to infection in both susceptible and resistant hosts, whereas the second burst appears to correlate only with disease resistance that is typically associated with host–pathogen interactions involving a matching combination of R and avr gene products (Grant et al., 2000). Localization of ROI in intact plant tissues has been shown with DAB staining, because DAB polymerizes instantly as it comes into contact with H2O2 in the presence of peroxidase (Thordal-Christensen et al., 1997). Using this approach, we have shown that H2O2 accumulation does not occur in Peronospora-infected tissue of the edm1 mutant, even though this is a common feature of downy mildew resistance in wild-type Arabidopsis, and that AtSGT1b restored the wild-type phenotype when expressed as a transgene in the mutant. This finding suggests that AtSGT1b is essential for the production of ROI in defense signaling, acting upstream of ROI production.
Seven isolates of Peronospora that are avirulent in Col-0 can infect and sporulate in the Col-edm1 mutant. Restoration of wild-type resistance when AtSGT1b was expressed as a transgene in the mutant demonstrates that this gene plays a pivotal role in downy mildew resistance of Arabidopsis. Quantifying parasite reproduction enabled a measurement of residual resistance in the mutant relative to the wild type after inoculation with each isolate. Full susceptibility was observed only in edm1 with the isolate Cala2 (Table 1), indicating that some degree of alternative (non-SGT1b) defense signaling exists to explain the residual resistance observed with the other isolates. This included RPP7-mediated resistance because edm1 exhibited less sporulation than the control rpp7 EMS mutant, which was fully susceptible (Table 2). Alternative defense signaling also was indicated by the discovery of RPP27 in Ler-0 and from P. syringae inoculations in which the different examples of bacterial resistance were affected by the edm1 mutation. AtSGT1a may provide an optional means for some R genes to engage with the SCF complex. Otherwise, the alternative resistance probably is achieved via a non-SCF–mediated response.
The full-resistance (no sporulation) phenotype conferred by RPP7 is similar to examples described in other accessions, including the first example that was described in Rld-0 (Koch and Slusarenko, 1990) and eventually associated with a CC-NB-LRR allele of RPP13 (Bittner-Eddy et al., 2000). Other similar examples, RPP5 and RPP8, which are TIR-NB-LRR and CC-NB-LRR genes, respectively, were characterized in the accession Ler-0 (Parker et al., 1997; McDowell et al., 1998). Defense signaling conferred by RPP5 appears to be regulated simply by EDS1 (Aarts et al., 1998), whereas single mutations (apart from R gene loss-of-function alleles) that cause moderate to full suppression of RPP7, RPP8, or RPP13 resistance have not been described previously (McDowell et al., 2000; Bittner-Eddy and Beynon, 2001).
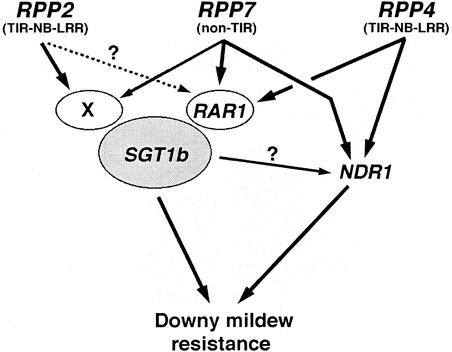
Therefore, the major role of AtSGT1b in RPP7-mediated resistance provides an important advance in the mutational analysis of defense signaling in plants. This is summarized in Figure 5 by a working model that revises a model that we described previously (McDowell et al., 2000). RPP2, RPP4, and RPP7 provided contrasting examples of defense regulation in the previous model, and an important question remained regarding whether a common regulator existed among the examples. In the revised model, SGT1b is a major defense regulator in all three examples; RAR1 and a postulated Gene X explain an intermediate link between signaling from the R gene proteins and SGT1b; and NDR1 provides an alternative defense signal that could explain the residual resistance observed in the single mutants sgt1b and rar1 on RPP4- and RPP7-mediated resistance (Table 1). Gene X is postulated between RPP2 and SGT1b, interacting directly with the latter protein, because RAR1 is not required for this resistance. This unknown protein could be highly dissimilar from RAR1 if, for example, it interacts with SGT1b at the TPR domain instead of the CS domain of SGT1b. Similarly, Gene X could explain why the rar1.21 null allele exhibits only a minor impact on RPP7-mediated resistance, as indicated by a rare sporulation phenotype (Table 2) and confirmed by microscopy (Tornero et al., 2002). Further mutant screening in a rar1 background for enhanced susceptibility to Hiks1 will be useful in attempts at selection of Gene X mutants.
Figure 5.
Model Summarizing a Central Role of AtSGT1b in RPP-Mediated Disease Resistance.
RPP2, RPP4, and RPP7 are three contrasting examples of downy mildew (Peronospora) resistance genes. NDR1, RAR1, and an unknown defense component (Gene X) may regulate distinct, accumulative defense responses to achieve wild-type resistance. This would explain the residual resistance observed in rar1 after inoculation with the isolate Cala2 (recognized by RPP2) or Hiks1 (recognized by RPP7). The model has been modified from the summary of mutant analyses described previously (McDowell et al., 2000).
Double and triple mutant analysis is required to verify the current model. For example, trypan blue staining of cotyledons indicated that rar1 ndr1 permits enhanced colonization of tissue compared with either single mutation (McDowell et al., 2000; Tornero et al., 2002). Quantifying sporulation in this double mutant will provide a measure of the relative contribution that Gene X provides to RPP7-mediated resistance. Other critical examples for testing the model include sgt1b ndr1, which should be fully susceptible to Hiks1 (RPP7), and sgt1b rar1, which should not differ significantly from sgt1b in response to either Hiks1 or Emoy2 (RPP4). An alternative method of quantifying the amount of hyphal growth or biomass of Peronospora in host tissue has been developed (Molina et al., 1998) and could be used to compare the quantitative effects of the various mutant combinations in adult leaves.
METHODS
Pathogen Isolates and Pathology Methods
Methods for subculturing Peronospora parasitica and preparing inoculum for experiments were modified from Holub et al. (1994). All isolates were maintained in Wassilewskija-eds1 (Parker et al., 1996), and 10- to 14-day-old seedlings were inoculated (instead of 7-day-old seedlings) to produce large quantities of inoculum. Plants of this age had sufficient height above the soil that enabled clean preparation of spore suspensions with minimal debris. For experiments, 7-day-old seedlings were inoculated as recommended previously. Asexual sporulation was assessed visually 7 days after inoculation by counting low numbers of sporangiophores on both sides of each cotyledon up to a maximum of 10 and recording higher levels as either medium (∼11 to 16 sporangiophores per cotyledon) or high (>16 sporangiophores per cotyledon). Mean sporulation of different host/parasite combinations was calculated using the actual number for low sporulation and 15 and 20 for medium and heavy sporulation, respectively. This method is based on minimal sampling recommended from a rigorous (double-blind) statistical experiment performed by Mert (2001), in which a minimum of 50 cotyledons sampled for a given host interaction ensured significant distinction between no sporulation and rare sporulation, among different levels of low sporulation (e.g., a mean of two versus a mean of six), and between medium and heavy levels of sporulation. Pseudomonas syringae experiments were performed as described by Tornero and Dangl (2001).
Arabidopsis Germplasm
Arabidopsis thaliana Columbia-5 (Col-5), Landsberg erecta (Ler-0), and Keswick (Ksk-1) used in this study are available from the Nottingham Arabidopsis Stock Centre (http://nasc.nott.ac.uk/). Available mutants were obtained from corresponding authors of previous publications: Col-ndr1 (Century et al., 1997), Col-Atrar1.21 (Tornero et al., 2002), and Wassilewskija-eds1.1 (Parker et al., 1996). Ethyl methanesulfonate (EMS)– and fast neutron–treated M2 seed was purchased from Lehle Seed (Round Rock, TX).
Mutant Screening
Col-edm1 was selected from a screen of M2 seedlings (derived from ∼6000 M1 plants) using the isolate Cala2. The seedlings were sown in 24- × 36-cm trays at a density of ∼2000 seedlings per tray and inoculated at 7 days old as described above. We attempted to find additional edm1 alleles by screening fast neutron– and EMS-treated Col-5 (M2 seed pools from ∼6000 and 20,000 M1 plants for each treatment, respectively) with Hiks1 instead of Cala2. No mutant was found that exhibited the same broad-spectrum susceptibility as edm1, whereas at least 11 rpp7 mutants were identified (seven EMS, including rpp7.1 [described above], and four fast neutron), as well as fast neutron mutations in two additional loci (EDM2 and EDM3; data not shown) that were all fully susceptible to Hiks1 but wild-type resistant to the other isolates.
Light Microscopy
Seedlings infected with Hiks1 and noninoculated controls were removed (severed at the hypocotyl) from soil and placed in Eppendorf tubes containing 100% methanol for 3 to 12 hr. The methanol was replaced with a saturated solution of chloral hydrate to further clear the tissue for 2 to 4 hr. The chloral hydrate was replaced with lactoglycerol (lactic acid:glycerol:water, 1:1:1) for indefinite storage, and seedlings were mounted on glass slides in lactoglycerol for microscopy. For staining, seedlings were placed in different solutions before methanol treatment: aniline blue solution (0.1% [w/v] in 1 M Gly/NaOH buffer, pH 9.5) for 1 to 2 hr to stain callose, and 3,3′-diaminobenzidine stain according to the method recommended by Thordal-Christensen et al. (1997) for the detection of H2O2. An Olympus microscope (Tokyo, Japan) equipped with differential interference contrast optics and UV illumination (UV2A and B-3A excitation filters) was used.
Map-Based Cloning of AtSGT1b
We tentatively mapped two loci (EDM1 and RPP27) using genomic DNA from 20 Hiks1-susceptible F2 segregants that were selected from an outcross between Col-edm1 and Ler-0 (i.e., homozygous for Col-0 alleles of edm1 and rpp27). Cleaved amplified polymorphic sequence markers, which are distributed throughout the genome and polymorphic between Col-0 and Ler-0, were obtained from the Arabidopsis research community (http://arabidopsis.org/aboutcaps.html). One locus was linked closely to g4026 and RPP7 on chromosome I, and the other was linked to g4539 on chromosome IV. Only Col-5 DNA was detected at both markers in all 20 of these F2 samples (no recombination), whereas markers from other chromosome arms exhibited at least 40% recombination with the Hiks1 susceptibility phenotype. Genomic DNA from 20 Cala2-susceptible F2 Col-edm1 × Ksk-1 seedlings was used to establish EDM1 as the locus on chromosome IV. All of these seedlings had inherited Col-0 DNA at marker g4539 (cosegregating with Cala2 susceptibility), whereas g4026 showed 40% recombination. RPP27, therefore, was identified as the chromosome I locus; molecular characterization of this resistance gene will be presented elsewhere.
A total of 310 Hiks1-susceptible F2 Col-edm1 × Ler-0 seedlings were used to fine map EDM1 within an interval spanned by markers F17A8 and SC5. Sequences of bacterial artificial chromosome (BAC) clones were retrieved, and primers were designed using the software Vector NTI (InforMax, Bethesda, MD) and information from the Cereon Genomics SNP/INDEL database (Cambridge, MA) to generate markers, including SEH2, YRT1, and DAT4. SEH2 was amplified using primers 5′-TGAAACAAGAGAGAAGAATTGAACA-3′ and 5′-TGACAACTAAATCCCTCTTCCTC-3′, and the polymorphism between Col-0 and Ler-0 was detected by the presence of a deletion of ∼20 bp. YRT1 was amplified using primers 5′-CATTTTCCCACGGTCTTT-3′ and 5′-TCCTCCTTCACCAAACTCTTG-3′, and the product was cleaved with RsaI to reveal the polymorphism. DAT4 was amplified using primers 5′-TGTAATATCTTTGAAATAAAATGGAGTT-3′ and 5′-ACTTTTTCTCAATTTCATCGTTTGT-3′, and the polymorphism was detected by NdeI restriction digestion. Two overlapping BAC clones (T22B4 and F8L21) that spanned the EDM1 interval were obtained from the ABRC (Columbus, OH). BAC DNA was prepared using the large construction kit according to the manufacturer's instructions (Qiagen Ltd., UK).
Agrobacterium-Mediated Transformation
Small constructs containing single genes were generated by digesting F8L21 DNA as follows: with NcoI and SalI to release a 4958-bp fragment (At4 g11220); with BspMI to release an 8996-bp fragment (At4 g11230); with SapI to give a 4201-bp fragment (At4 g11240); with PstI to give a 4007-bp fragment (At4 g11250); with NcoI to yield a 4946-bp fragment (At4 g11260); with Eco47III to give an 8638-bp fragment (At4 g11270); and with both Eco47III and BspEI to give a 6260-bp fragment (At4 g11280). The fragments were gel purified, blunt ended using T4 DNA polymerase, and ligated into the SmaI site of the binary vector pCAMBIA3300 (http://www.cambia.org/), which carries the BAR gene. Ligated products were electroporated into Escherichia coli strain DH10B, and positive clones were identified by polymerase chain reaction (PCR) and restriction analyses. The constructs were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation, and the edm1 plants then were transformed with each of the constructs. Transformants were selected by spraying 7-day-old plants grown in soil with 0.04% Basta (AgrEvo, Norfolk, UK ). These selected plants were self-pollinated to produce T2 seed, which were then tested with Hiks1, Cala2, and Cand5 isolates of Peronospora using the methods described above.
Reverse Transcriptase–Mediated PCR and DNA Sequencing
Total RNA was isolated from infected and noninfected cotyledons using the RNAeasy (Qiagen) kit according to the manufacturer's instructions. Single-tube reverse transcriptase–mediated (RT) PCR was performed on 0.5 μg of total RNA using RT-PCR beads (Amersham Pharmacia Biotech). Reactions were performed in a volume of 50 μL using the SGT1b-specific primers 5′-ATGGCCAAGGAATTA-GCAGA-3′ and 5′-CTCAATACTCCCACTTCTTGA-3′ by incubating at 42°C for 20 min and denaturing at 95°C for 5 min followed by a standard touchdown PCR regime of 94°C for 30 sec, 65°C for 30 sec (with a 1°C decrement for every cycle), and 72°C for 1 min (10 cycles); and 94°C for 30 sec, 56°C for 30 sec, and 72°C for 1 min (22 cycles). The 18S primers were used as a control to assess the integrity of RNA. Similar reactions were performed with the 18S primers, but a standard cycling regime of 94°C for 30 sec, 56°C for 30 sec, and 72°C for 1 min (16 cycles) was used. A 5-μL aliquot of each reaction was analyzed by agarose gel electrophoresis.
All DNA sequencing reactions were performed using BigDye Termination Kits (Applied Biosystems, Foster City, CA) and separated on an ABI 377 sequencer. Expressed sequence tag clones and RT-PCR products were sequenced using the universal M13 and the gene-specific primers 5′-ATGGCCAAGGAATTAGCAGA-3′, 5′-CTC-AATACTCCCACTTCTTGA-3′, and 5′-CTGAACATCGGTTTGGCA-GG-3′.
Bioinformatics
Gene sequences from the deleted region were studied using Arabidopsis sequence databases that provide a initial prediction of annotations (TIGR [http://www.tigr.org] and MIPS [http://mips.gsf.de/]). Bioinformatic software used to refine predictions included PIX (http://hgmp.mrc.ac.uk), Interpro (http://www.ebi.ac.uk/), and SMART (http://smart.embl-heidelberg.de/). The sequence of the full-length cDNA corresponding to AtSGT1b was compared with the sequence of the genomic DNA using the alignment program of Vector NTI (InforMax, Inc.), and the coordinates of the exons and introns were confirmed as provided in MIPS.
Accession Numbers
The accession number for the full-length cDNA of the SGT1-like gene described in this article is AA712212. The accession numbers for the sequences shown in Figure 4 are NP_014700.1 (budding yeast SGT1), CAA23023.1 (Arabidopsis AtSGT1a), CAB51410.1 (Arabidopsis AtSGT1b), AF192467.1 (rice SGT1), and AJ251317.1 (Rubus idaeus SGT1).
Acknowledgments
We thank Jim Beynon, John McDowell, Jonathan Jones, and Mike Wilson for helpful discussions and encouragement. This work was supported by grants from the Biotechnology and Biological Science Research Council (207/ICR07554 and core funding to Horticulture Research International) to M.T., P.G., and E.B.H.; from the National Research Initiative Competitive Grants Program of the U.S. Department of Agriculture to J.L.D.; from a Deutsche Forschungsgemeinschaft fellowship to T.E.; and by training grants from the Biotechnology and Biological Science Research Council, European Union, and the Turkish government to A.C. and E.S., and F.M.-T. and C.C., respectively.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.001123.
References
- Aarts, N., Metz, M., Holub, E.B., Staskawicz, B.J., Daniels, M.J., and Parker, J.E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene mediated pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 10306–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., and Shirasu, K., and Schulze-Lefert, P. (2002). The RAR1 interactor SGT1, an essential component of R gene triggered resistance. Science 295, 2073–2076. [DOI] [PubMed] [Google Scholar]
- Bittner-Eddy, P.D., and Beynon, J.L. (2001). The Arabidopsis downy mildew resistance gene, RPP13-Nd, functions independently of NDR1 and EDS1 and does not require the accumulation of salicylic acid. Mol. Plant-Microbe Interact. 14, 416–421. [DOI] [PubMed] [Google Scholar]
- Bittner-Eddy, P.D., Crute, I.R., Holub, E.B., and Beynon, J.L. (2000). RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J. 21, 177–188. [DOI] [PubMed] [Google Scholar]
- Century, K.S., Holub, E.B., and Staskawicz, B.J. (1995). NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc. Natl. Acad. Sci. USA 92, 6597–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century, K.S., Holub, E.B., and Staskawicz, B.J. (1997). NDR1, a pathogen induced component required for Arabidopsis disease resistance. Science 278, 1963–1965. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Das, A.K., Cohen, P.W., and Barford, D. (1998). The structure of the tetratricopeptide repeats of protein phosphatase 5: Implications for TPR-mediated protein-protein interactions. EMBO J. 17, 1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk, A., Feys, B.J., Frost, L.N., Jones, J.D.G., Daniels, M.J., and Parker, J.E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis, has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96, 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J., and Parker, J.E. (2000). Interplay of signaling pathways in plant disease resistance. Trends Genet. 16, 449–455. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J., Zook, M., Mert, F., Kagan, I., Rogers, E.E., Crute, I.R., Holub, E.B., and Ausubel, F.M. (1997). Phytoalexin deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics 146, 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebl, M., and Yanagida, M. (1991). The TPR snap-helix: A novel protein repeat motif from mitosis to transcription. Trends Biochem. Sci. 16, 173–177. [DOI] [PubMed] [Google Scholar]
- Grant, M., Brown, I., Adams, S., Knight, M., Ainslie, A., and Mansfield, J. (2000). The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J. 23, 441–450. [DOI] [PubMed] [Google Scholar]
- Holub, E.B. (2001). The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2, 516–527. [DOI] [PubMed] [Google Scholar]
- Holub, E.B., Beynon, J.L., and Crute, I.R. (1994). Phenotypic and genotypic characterization of interactions between isolates of Peronospora parasitica and accessions of Arabidopsis thaliana. Mol. Plant-Microbe Interact. 7, 223–239. [Google Scholar]
- Jacobsen, S.E., Binkowski, K.A., and Olszewski, N.E. (1996). SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Natl. Acad. Sci. USA 93, 9292–9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa, K., Skowyra, D., Elledge, S.J., Harper, J.W., and Hieter, P. (1999). SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol. Cell 4, 21–23. [DOI] [PubMed] [Google Scholar]
- Koch, E., and Slusarenko, A. (1990). Arabidopsis is susceptible to infection by a downy mildew. Plant Cell 2, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, E., Pontier, D., and Pozo, O. (1999). Die and let live: Programmed cell death in plants. Curr. Opin. Plant Biol. 2, 502–507. [DOI] [PubMed] [Google Scholar]
- Lamb, C., and Dixon, R.A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 251–275. [DOI] [PubMed] [Google Scholar]
- Lamb, J.R., Tuguenreich, S., and Hieter, P. (1995). Tetratrico peptide repeat interactions: To TPR or not to TPR. Trends Biochem. Sci. 20, 257–259. [DOI] [PubMed] [Google Scholar]
- Maleck, K., Levine, A., Eulgem, T., Morgan, A., Schmid, J., Lawton, K.A., Dangl, J.L., and Dietrich, R.A. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26, 403–410. [DOI] [PubMed] [Google Scholar]
- Matsuzawa, S., and Reed, J. (2001). Siah1, SIP and EBI collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Mol. Cell 7, 915–926. [DOI] [PubMed] [Google Scholar]
- McDowell, J.M., Dhandaydham, M., Long, T.A., Aarts, M.G.M., Goff, S., Holub, E.B., and Dangl, J.L. (1998). Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 10, 1861–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell, J.M., Cuzick, A., Can, C., Beynon, J., Dangl, J.L., and Holub, E.B. (2000). Downy mildew (Peronospora parasitica) resistance genes in Arabidopsis vary in functional requirements for NDR1, EDS1, NPR1 and salicylic acid accumulation. Plant J 22, 523–529. [DOI] [PubMed] [Google Scholar]
- Mert, F. (2001). Quantification of Enhanced Downy Mildew Susceptibility and Camalexin Accumulation in Arabidopsis thaliana. PhD Dissertation (London, UK: Imperial College of Biotechnology, Science and Medicine at Wye, University of London).
- Molina, A., Hunt, M.D., and Ryals, J.A. (1998). Impaired fungicide activity in plants blocked in disease resistance signal transduction. Plant Cell 10, 1903–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskett, P.R., Kahn, K., Austin, M.J., Moisan, L.J., Sadanandom, A., Shirasu, K., Jones, J.D.G., and Parker, J.E. (2002). Arabidopsis RAR1 exerts rate-limiting control of R gene–mediated defenses against multiple pathogens. Plant Cell 14, 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.E., Holub, E.B., Frost, L.N., Falk, A., Gunn, N.D., and Daniels, M.J. (1996). Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8, 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.E., Coleman, M.J., Szabo, V., Frost, L.N., Schmidt, R., Van der Biezen, E.A., Moores, T., Dean, C., Daniels, M.J., and Jones, J.D.G. (1997). The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the Toll and Interleukin-1 receptors with N and L6. Plant Cell 9, 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk, P.M., Kazan, K., Wilson, I., Anderson, J.P., Richmond, T., Somerville, S.C., and Manners, J.M. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 97, 11655–11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu, K., Lahaye, T., Tan, M.-W., Zhou, F., Azevado, C., and Schulze-Lefert, P. (1999). A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 99, 355–366. [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen, H., Zhang, Z., Wei, Y., and Collinge, D.B. (1997). Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during barley-powdery mildew interaction. Plant J. 11, 1187–1194. [Google Scholar]
- Tör, M., Holub, E.B., Brose, B., Musker, R., Gunn, N., Can, C., Crute, I.R., and Beynon, J.L. (1994). Map positions of three loci in Arabidopsis thaliana associated with isolate specific recognition of Peronospora parasitica. Mol. Plant-Microbe Interact. 7, 214–222. [Google Scholar]
- Tornero, P., and Dangl, J.L. (2001). A high throughput method for quantifying growth of phytopathogenic bacteria in Arabidopsis thaliana. Plant J. 28, 475–481. [DOI] [PubMed] [Google Scholar]
- Tornero, P., Merit, P., Sadanandom, A., Shirasu, K., Innes, R.W., and Dangl, J.L. (2002). RAR1 and NDR1 contribute quantitatively to the function of Arabidopsis disease resistance genes in both simple and nonlinear pathways. Plant Cell 14, 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, M.A., Dangl, J.L., and Jones, J.D.G. (2001). Arabidopsis gp91-phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99, 517-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Biezen, E.A., Freddie, C.T., Kahn, K., Parker, J.E., and Jones, J.D.G. (2002). Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 29, 439–451. [DOI] [PubMed] [Google Scholar]
- Warren, R.F., Merritt, P.M., Holub, E., and Innes, R.W. (1999). Identification of three putative signal transduction genes involved in R gene-specified disease resistance in Arabidopsis. Genetics 152, 401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods, A.M., Fagg, J., and Mansfield, J.W. (1988). Fungal development and irreversible membrane damage in cells of Lactuca sativa undergoing the hypersensitive reaction to the downy mildew fungus Bremia lactucae. Physiol. Mol. Plant Pathol. 32, 483–497. [Google Scholar]
- Xiao, S., Ellwood, S., Calis, O., Patrick, E., Li, T., Coleman, M., and Turner, J.G. (2001). Broad spectrum resistance in Arabidopsis thaliana mediated by RPW8. Science 291, 118–120. [DOI] [PubMed] [Google Scholar]