Abstract
Plant disease resistance (R) genes mediate specific pathogen recognition, leading to a successful immune response. Downstream responses include ion fluxes, an oxidative burst, transcriptional reprogramming, and, in many cases, hypersensitive cell death at the infection site. We used a transgenic Arabidopsis line carrying the bacterial avirulence gene avrRpm1 under the control of a steroid-inducible promoter to select for mutations in genes required for RPM1-mediated recognition and signal transduction. We identified an allelic series of eight mutants that also were allelic to the previously identified pbs2 mutation. Positional cloning revealed this gene to be AtRAR1, the Arabidopsis ortholog of barley RAR1, a known mediator of R function. AtRAR1 is required for both full hypersensitive cell death and complete disease resistance mediated by many, but not all, tested R genes. Double mutant analysis of Atrar1 in combination with the R signal intermediate ndr1 suggests that AtRAR1 and NDR1 can operate in both linear and parallel signaling events, depending on the R gene function triggered. In Atrar1 null plants, the levels of RPM1-myc are reduced severely, suggesting that AtRAR1 may regulate R protein stability or accumulation.
INTRODUCTION
Plant recognition of pathogens is mediated by large families of highly polymorphic disease resistance (R) genes (Dangl and Jones, 2001; Jones, 2001). The products of these genes function to recognize, directly or indirectly, the products of pathogen-encoded avirulence (Avr) genes (Nimchuk et al., 2001). Recognition stimulates a signal transduction cascade leading to the activation of multiple defense responses, including, in many cases, hypersensitive plant cell death (HR) at the site of infection (reviewed by Heath, 2000). Most R products contain a central nucleotide binding site and C-terminal Leucine-rich repeat domains (NB-LRR). There are ∼150 NB-LRR proteins encoded in the complete Arabidopsis genome (Arabidopsis Genome Initiative, 2000). The N termini of these proteins contain either potential coiled-coil (CC) or Toll–Interleukin 1 receptor homology (TIR) domains.
Genetic screens in Arabidopsis have defined several loci required for R function. There is evidence from these studies that the NB-LRR class of R proteins trigger multiple signaling pathways. Many, but not all, CC-NB-LRR proteins require NDR1, a protein of undefined biochemical function (Century et al., 1995, 1997). In contrast, all tested members of the TIR class require the EDS1 protein (Parker et al., 1996). EDS1 encodes a protein of unknown function, although it has homology with lipases (Falk et al., 1999). Whether or not these pathways converge into a simple linear signal transduction cascade is unknown, but it is unlikely given the fact that no locus defined by mutant phenotype is required for the function of all NB-LRR proteins.
RPM1 conditions resistance to Pseudomonas syringae strains expressing either avrRpm1 or the sequence-unrelated avrB (Bisgrove et al., 1994; Grant et al., 1995). Thus, RPM1 recognizes one of two different Avr proteins. This recognition occurs inside the plant cell, because P. syringae uses the evolutionarily conserved type III secretion system to deliver disease effector proteins, including AvrRpm1 and AvrB, into the host cell (He, 1998). RPM1 is a peripheral plasma membrane protein (Boyes et al., 1998), and both AvrRpm1 and AvrB are among a class of P. syringae disease effector proteins that are myristoylated when expressed in the plant cell and are targeted to the plant plasma membrane (Nimchuk et al., 2000).
We recently described a conditional screen for mutants affecting the function of the Arabidopsis RPM1 gene (Tornero et al., 2002). Expression of transgenic avrRpm1 or avrB (Gopalan et al., 1996; Tornero et al., 2002) in an RPM1 background leads to a whole seedling cell death response similar to the HR, enabling facile isolation of mutants unable to activate the RPM1 pathway. We isolated mutations in five loci required for RPM1-mediated resistance after the conditional expression of avrRpm1 in mutagenized seedlings, in addition to a large number of rpm1 alleles (Tornero et al., 2002). One of these signaling loci, originally termed lra1 (for loss of recognition of avrRpm1), was defined by eight loss-of-function alleles. Here, we demonstrate that these lra1 mutations are allelic to the previously described pbs2-1 mutation (Warren et al., 1999) and define the Arabidopsis ortholog of barley RAR1. The barley RAR1 gene was identified originally by means of genetic screens targeting the barley powdery mildew disease resistance gene Mla12 (Freialdenhoven et al., 1994; Shirasu et al., 1999).
Barley RAR1 is required for full HR and complete resistance mediated by many, but not all, highly related Mla R alleles. Interestingly, the amino acid differences between RAR1-dependent and RAR1-independent Mla alleles can be as little as 5% (Halterman et al., 2001; Zhou et al., 2001). We detail the effects of null Atrar1 alleles in signaling mediated by the CC-NB-LRR proteins RPM1, RPS2, and RPS5 and the TIR-NB-LRR protein RPS4. Each recognizes P. syringae expressing the appropriate avr gene (RPM1, Grant et al., 1995; RPS2, Bent et al., 1994; Mindrinos et al., 1994; RPS5, Warren et al., 1998; RPS4, Gassmann et al., 1999).
We also address the effects of Atrar1 mutations on the presumed CC-NB-LRR gene, RPP7, that conditions recognition of the Peronospora parasitica (Pp) isolate Hiks1 (Holub et al., 1994) and present double mutant analysis using Atrar1 in combination with ndr1. Atrar1 and ndr1 null mutations have additive effects on resistance mediated by some R genes, indicating that AtRAR1 and NDR1 can function in separate signal transduction pathways. Our data additionally support the notion that RAR1 and NDR1 can act in a single pathway. Finally, we provide evidence that the levels of an epitope-tagged RPM1 protein are reduced considerably in plants with an Atrar1 mutation. We conclude that the relative importance of Atrar1 and ndr1 in CC-NB-LRR R gene function in the Arabidopsis accession Columbia (Col-0) is dependent on the R gene in question.
RESULTS
Using a recently described conditional expression system (Tornero et al., 2002), we isolated eight mutants that exhibited severely attenuated avrRpm1-induced cell death (Figure 1). These were all alleles of one gene, which we originally named LRA1. All eight mutants were susceptible to Pseudomonas syringae pv tomato (Pst) DC3000(avrRpm1) (see below), confirming that the loss of avrRpm1-transgene responsiveness reflected an inability to respond to AvrRpm1, regardless of the delivery system.
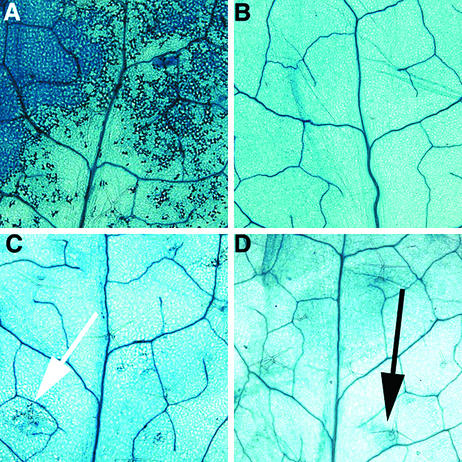
Figure 1.
lra1 Mutants Do Not Respond to Inducible Expression of avrRpm1.
Three-week-old plants containing an inducible avrRpm1 expression system were sprayed with estradiol and stained with trypan blue 24 hr later. Trypan blue stains leaf veins and dead cells, revealing the region undergoing HR.
(A) Line a11 (RPM1, LRA1). Note the extent of cell death.
(B) Line a11r (rpm1-1, LRA1) contains the transgene from a11 crossed into the isogenic rpm1-1 allele. Note the lack of cell death.
(C) and (D) lra1-1 (RPM1, lra1-1) (C) and lra1-2 (RPM1, lra1-2) (D). The arrows point to areas of faint staining. Note that the lra1-1 mutant is Atrar1-21 and the lra1-2 mutant is Atrar1-22.
We next asked whether the spectrum of R functions altered by lra1 was related to any of the known loci required for R-dependent responses. Therefore, we tested all lra1 alleles for resistance to isogenic Pst DC3000 strains expressing avrRpt2, avrRps4, or avrPphB. Recognition of these Avr genes is mediated RPS2, RPS4, and RPS5, respectively (Bent et al., 1994; Mindrinos et al., 1994; Grant et al., 1995; Warren et al., 1998). Recognition of Pst DC3000(avrRpt2) and Pst DC3000(avrPphB) was compromised, but recognition of Pst DC3000(avrRps4) was affected only negligibly (see below). Similarly, we inoculated all of the lra1 alleles with Pp isolates Cala2, Hiks1, and Emwa1 to assess the function of the R genes RPP2, RPP7, and RPP4, respectively. Only RPP4 function was compromised (data not shown). Genetic analysis indicated that lra1 is allelic to pbs2-1 (Warren et al., 1999) and maps to chromosome V. The previous linkage of pbs2-1 to markers on chromosome I (Warren et al., 1999) may be attributable to a γ-ray–induced translocation involved in the induction of this mutant (P. Merritt and R.W. Innes, unpublished data).
Using 120 F2 plants, we were able to localize lra1-1 to several overlapping bacterial artificial chromosome and P1 clones on the bottom arm of chromosome V (see Methods). An ortholog of the barley RAR1 gene was contained in this genetic interval (Shirasu et al., 1999). Attempts at polymerase chain reaction amplification of the AtRAR1 open reading frame from the pbs2-1 mutant failed, suggesting that AtRAR1 was disrupted in this mutant. Therefore, we amplified and sequenced AtRAR1 from all eight lra1 mutants. This analysis revealed a single nucleotide substitution in each mutant (see below). In agreement with the Shirasu and Parker groups, we renamed the Col-0 alleles (starting with pbs2-1) Atrar1-20 to Atrar1-28.
As shown in Figure 2, the predicted AtRAR1 protein includes the zinc-coordinating CHORD I and CHORD II domains and the central CCCH domain described previously for barley RAR1 (Shirasu et al., 1999). The deduced barley and Arabidopsis proteins are 60% identical. The point mutations in the ethyl methanesulfonate–derived Atrar1-21 to Atrar1-28 alleles produce either premature stop codons or Cys-to-Tyr exchanges (Figure 2B) that are expected to disrupt the demonstrated zinc binding coordinated by these residues (Shirasu et al., 1999). Because both early stops and disruption of zinc binding could destabilize the AtRAR1 protein, we analyzed the levels of AtRAR1 protein in all eight lra1 mutants and in the original pbs2-1 mutant.
Figure 2.
LRA1/PBS2 Is AtRAR1.
(A) Structure of the AtRAR1 gene. The predicted protein is portrayed in single-letter amino acid abbreviations. The mutations found in Atrar1-21 to Atrar1-28 are underlined. Nucleotides not present in the mRNA are shown in italic type.
(B) Relationship of Atrar1 mutations to the RAR1 protein domain structure. “C to Y” indicates a missense mutation resulting in a Cys-to-Tyr substitution. “STOP” indicates mutations that produce a stop codon. The allele number follows each mutation.
(C) AtRAR1 protein expression in the allelic series. Total proteins were extracted from each of the mutants and analyzed by protein gel blot analysis using anti-RAR1 serum. Equal loading was ensured by Ponceau staining. The top arrow indicates a molecular mass marker of 33 kD, and the bottom arrow indicates a molecular mass marker of 25 kD. Note that there is no detectable protein in the Atrar1-20 mutant or in the mutants produced by stop codons (Atrar1-21, -22, -24, -25, and -28), and there is less detectable protein where a mutation from Cys to Tyr has occurred (Atrar1-23, -26, and -27).
Protein gel blot analysis using a polyclonal antibody raised against the full AtRAR1 protein revealed no detectable RAR1 protein in Atrar1-20. Neither did we identify this protein in the mutants produced by a stop codon (Atrar1-21, -22, -24, -25, and -28). We detected AtRAR1 protein in the mutants produced by a missense mutation (Cys to Tyr) in all cases. Note that the levels of protein are lower where mutations in the CHORD I domain have occurred (Atrar1-23 and -26), compared with a mutation in the CHORD II domain (Atrar1-27). In the case of the CHORD I domain, it is possible that the Cys residues are required for proper folding and stability, possibly also for function. The levels in Atrar1-27 strongly suggest that the Cys residues in CHORD II are required for proper folding and function.
We investigated in detail the effects of Atrar1 alleles on signaling by various R genes. In preliminary tests, all alleles behaved essentially the same (data not shown); thus, we focused on the null alleles Atrar1-20 and Atrar1-21. The Atrar1-20 allele severely attenuated, but did not eliminate, the induction of HR mediated by RPM1, RPS2, and RPS5 (Table 1). The RPM1-mediated HR, typically visible 5 to 8 hr after inoculation, was delayed by several hours. Moreover, only 20% of inoculated leaves had collapsed by 18 to 22 hr after inoculation (tissue collapse caused by the susceptible response becomes visible beginning ∼24 hr after inoculation at the inoculum dose used).
Table 1.
HR in rar1 Mutant Plants
| No. of Leaves Exhibiting Visible HRa
|
||
|---|---|---|
| Bacterial Genotypeb | Col-0 | rar1-20 |
| avrB::Ωc | 1 of 53 | 2 of 45 |
| avrRpt2 | 55 of 61 | 24 of 55 |
| avrRpm1 | 59 of 59 | 12 of 60 |
| avrPphB | 65 of 68 | 17 of 60 |
Number of leaves showing visible tissue collapse out of the total number of leaves injected at 18 to 24 hr after injection.
Pst strains expressing the indicated avirulence genes were injected at an OD600 of 0.075 (∼3.75 × 107 colony-forming units/mL).
This strain expresses a nonfunctional avr gene and is unable to induce a visible HR in Col-0 plants.
The HR mediated by RPS2 and RPS5 typically occurs 16 to 20 hr after inoculation, but these too were attenuated severely in the Atrar1-20 mutant (Table 1). We observed identical results for Atrar1-21 (data not shown). The inherent weakness of the HR induced by RPS4 in the Col-0 genetic background (Gassmann et al., 1999; our unpublished data) made it impossible to test in this assay. In summary, for all three R genes tested, the HR was reduced dramatically, but not eliminated entirely, indicating that RAR1 contributes significantly to the HR.
Next, we quantified the effect of Atrar1-21 (and of several other alleles; data not shown) on R function by measuring bacterial growth after dip inoculation of seedlings (Tornero and Dangl, 2001). Col-0 expresses all of the relevant R genes, and we used null or strong loss-of-function rpm1, rps2, rps4, and rps5 alleles as fully susceptible controls. As shown in Figure 3, disease resistance controlled by RPM1 and RPS5 was eliminated in Atrar1-21. RPS2 function was nearly, but not completely, eliminated. We reproducibly observed ∼10 to 20% residual RPS2 function. Atrar1-21 had only a very minor effect on RPS4 function in this assay. These data extend those reported previously for the Atrar1-20 allele (Warren et al., 1999).
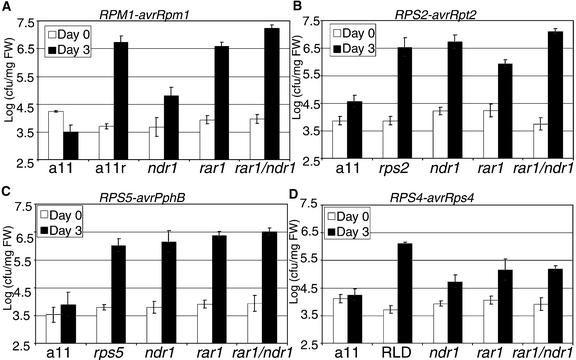
Figure 3.
AtRAR1 and NDR1 Act Differently to Control R Function.
Wild-type and mutant Arabidopsis lines were inoculated with Pst DC3000 strains containing the indicated avr genes. On day 0 (white columns) and day 3 (black columns), bacteria were extracted from the plants and enumerated. Bacterial numbers are expressed as the logarithm of colony-forming units per milligram of fresh weight (cfu/mg FW). The average and se of four independent replicates are shown. These experiments were performed three times with similar results. ndr1-1 was described previously (Century et al., 1995).
(A) The susceptible control is a11r, an isogenic a11 derivative but in a rpm1-1 background (Grant et al., 1995).
(B) The susceptible control is the isogenic rps2-101C allele (Mindrinos et al., 1994).
(C) The susceptible control is the isogenic rps5-2 allele (Warren et al., 1998).
(D) The susceptible control is accession RLD (Hinsch and Staskawicz, 1996).
We also compared the effect of the ndr1-1 mutation on the function of these R genes, because our original genetic analysis (see above) suggested that the Col-0 Atrar1 alleles we isolated affected the same spectrum of R functions as ndr1-1 (Century et al., 1995). RPM1 function was diminished only partially in an ndr1-1 background, whereas RPS2 and RPS5 functions were eliminated (Figure 3). The level of RPS4 function in Col-0 retained in the Atrar1-21 allele was statistically the same as that retained in the ndr1-1 mutant and the parental line a11 (Student's t test; α = 0.05) (Figure 3D). Interestingly, this is not the case for RPS4 function measured in Landsberg erecta–derived Atrar1 alleles (Muskett et al., 2002). These changes in disease resistance were mirrored by the appearance of visible disease symptoms in all susceptible interactions (data not shown). In addition, there was no effect of the Atrar1 mutation on the growth of virulent Pst DC3000 or virulent Pp (Noco2 isolate), so the Atrar1 mutants do not express enhanced disease susceptibility phenotypes.
We examined cell death and hydrogen peroxide production at the cellular level in ndr1 and Atrar1 mutants inoculated with Pst DC3000(avrRpm1). As reported previously by Century et al. (1995) and as illustrated in Figure 4, ndr1-1 retained the ability to induce an RPM1-dependent HR (cf. Figures 4A and 4E). Interestingly, the ndr1-1 mutant does not support an obvious RPM1-dependent oxidative burst, as indicated by greatly reduced staining of the inoculated zone with 3,3′-diaminobenzidine (DAB; Figure 4F) (Shapiro and Zhang, 2001). Consistent with previous analyses of barley rar1 mutants, we found no DAB staining in Atrar1-21 (Figure 4H).
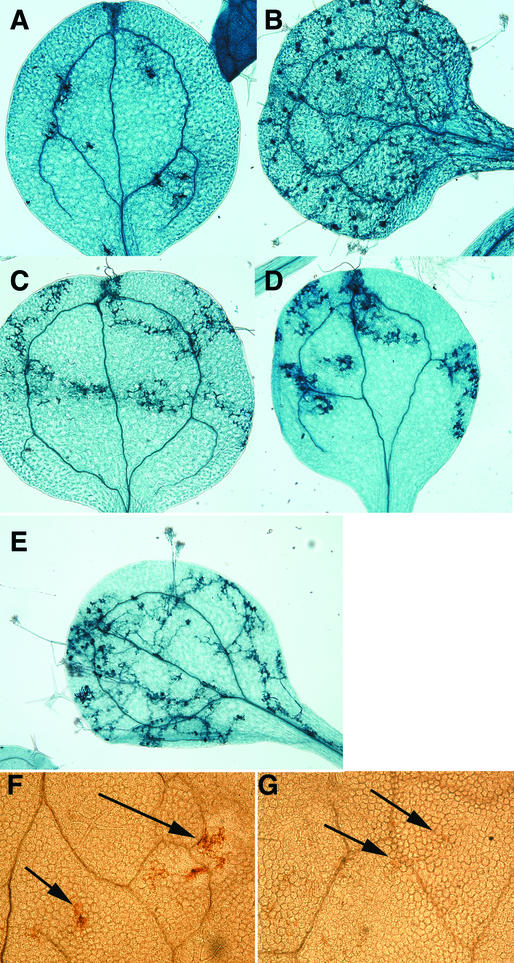
Figure 4.
Staining of HR and Reactive Oxygen Intermediates in Atrar1 and Related Mutants upon Pst DC3000(avrRpm1) Inoculation.
Plants were inoculated as described in Table 1 on the right side of the leaf and stained with trypan blue 20 hr after inoculation ([A], [C], [E], [G], and [I]) or with DAB 1.5 hr after inoculation ([B], [D], [F], [H], and [J]). Trypan blue stains veins and dead cells dark blue, and DAB stains total peroxides as a brown precipitate, as indicated by the arrow in panel (B).
(A) and (B) a11.
(C) and (D) a11r.
(E) and (F) ndr1-1.
(G) and (H) Atrar1-21.
(I) and (J) Atrar1-21 ndr1-1.
To address further the relative contributions of RAR1 and NDR1 to HR and disease resistance, we constructed an Atrar1-21 ndr1-1 double null mutant. The double mutant expressed the Atrar1 phenotype for severe attenuation of RPM1-dependent HR, as shown in Figure 4I. Additionally, the double mutant also resembled the fully susceptible Atrar1 single mutant in bacterial growth assays (Figure 3).
Thus, AtRAR1 appears to act in the same pathway as NDR1 during RPM1-dependent responses, and its activity is required for both the residual RPM1-dependent HR and the residual disease resistance observed in ndr1-1 mutants. RPS5 function is eliminated fully in either single mutant, and the double mutant phenotype resembles either single mutant phenotype (Figure 3). This is consistent with AtRAR1 and NDR1 acting in the same pathway, although no relative order can be implied. In contrast, RPS2 function in the Atrar1 ndr1-1 double mutant resembles that of the fully susceptible ndr1-1 single mutant. Thus, we conclude that AtRAR1 is more important than NDR1 in the transduction of RPM1 function, that the reverse is true for RPS2 function, and that both are essential for RPS5 function. These data suggest that the relative contributions of AtRAR1 and NDR1 to resistance mediated by a given R gene can vary.
The apparently complex functional relationship between RAR1 and NDR1 prompted us to analyze another R function measurable in the Col-0 background. We chose the RPP7 gene, which conditions resistance to the Hiks1 isolate of the obligate biotrophic oomycete parasite Pp (Holub et al., 1994). RPP7 function normally is associated with a small HR at the attempted infection site, as shown in Figure 5A using trypan blue staining. In contrast, a lack of host cell response and full sporulation were observed in an rpp7 mutant (Figure 5B). As noted previously (McDowell et al., 2000), RPP7 function was altered only slightly by ndr1-1, in that fungal hyphae elongated and the host responded with HR in cells surrounding those hyphae (Figure 5C). This phenotype is termed trailing necrosis (Morel and Dangl, 1998). We also observed trailing necrosis in Atrar1-21 mutants after Pp Hiks1 infection (Figure 5D) and no sporulation, as described previously (Warren et al., 1999).
Figure 5.
Staining of HR Sites and Production of Reactive Oxygen Intermediates Is Altered during RPP7-Mediated Responses.
Plants were inoculated with Pp strain Hiks1 by spraying with an aqueous suspension containing 5 × 104 oospores/mL and then stained with trypan blue 5 days after inoculation ([A] to [E]) or with DAB 2 days after inoculation ([F] and [G]). The arrows in panels (F) and (G) point to sites of hyphae penetration.
(A) and (F) a11.
(B) rpp7.
(C) ndr1-1.
(D) and (G) Atrar1-21.
(E) Atrar1-21 ndr1-1.
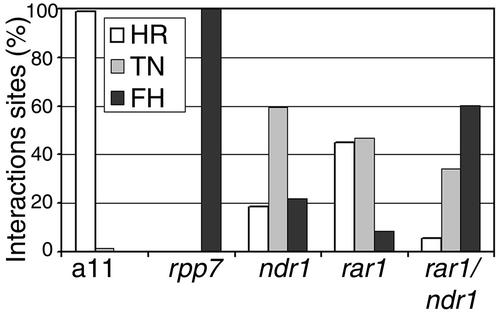
The Atrar1-21 ndr1-1 double mutant exhibited markedly reduced RPP7 function, resulting in low but reproducible production of sporangiophores and a reduction in trailing necrosis compared with either single mutant (Figure 5E). To quantitate these interactions, we compiled infection sites into three phenotypic classes (Figure 6) (Morel and Dangl, 1998): HR, trailing necrosis, and free hyphae without associated host cell response. The Atrar1-21 ndr1-1 double mutant compromised RPP7 function much more than either single mutant. We conclude from these data that RAR1 and NDR1 act in separable pathways to partially mediate RPP7 function. Yet, even the Atrar1-21 ndr1-1 double mutant retained significant RPP7 function. Thus, although both RAR1 and NDR1 are necessary for complete RPP7 function, they are not sufficient for it.
Figure 6.
Atrar1 and ndr1-1 Affect RPP7 Function Additively.
Plants were inoculated with Pp strain Hiks1 as described in Figure 5. Seven days after inoculation, the plants were stained with trypan blue, and the interactions sites were classified as HR, trailing necrosis (TN), and free hyphae (FH). A minimum of 591 interactions per genotype from three independent experiments are represented.
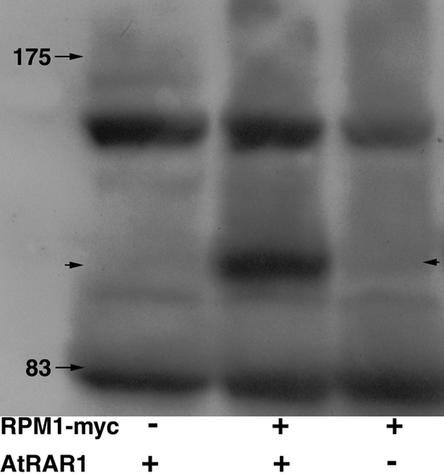
Shirasu and colleagues (1999) suggested that RAR1 functions as part of a protein complex that regulates protein degradation. We reported previously that an RPM1-myc protein expressed by the native RPM1 promoter disappears just before the onset of HR triggered by each of the bacterial avr-R combinations assayed here (Boyes et al., 1998). Therefore, we crossed the RPM1-myc transgene used in that study to Atrar1-21 and selected F2 progeny that carried the null mutant allele (Atrar1-21/Atrar1-21, RPM1/RPM1, and RPM1-myc/RPM1-myc). We also created the corresponding controls (AtRAR1/AtRAR1, RPM1/RPM1, and RPM1-myc/RPM1-myc) by introgression. Each line carries the conditional avrRpm1 expression system and the same RPM1-myc transgene. Surprisingly, the levels of RPM1-myc that we detected in the Atrar1-21 mutant were reproducibly much lower than the levels in AtRAR1 plants (Figure 7). This striking result strongly suggests that RAR1 function is required for the accumulation and/or stability of RPM1-myc. The very low levels of RPM1-myc that we observed in Atrar1-21 precluded the determination of its disappearance after pathogen inoculation.
Figure 7.
RPM1-myc in Atrar1 Mutants.
Plants were harvested at 5 weeks of age, and total extracts were prepared as described by Boyes et al. (1998). Arrows with numbers indicate the positions of molecular mass markers (kD). The arrows at both sides of the figure indicate the position of RPM1-myc. Three independent experiments were performed, and each gave similar results.
DISCUSSION
The signaling pathways that lead to disease resistance are complex. Previous work had established that some Arabidopsis R genes require the function of NDR1 and others require EDS1. Additionally, some but not all R genes require salicylic acid accumulation and NPR1/NIM1 function (reviewed by Glazebrook, 2001). At least one R gene, RPP7, appears to use NDR1 and EDS1 in combination to mediate some or all of its function (McDowell et al., 2000; but see below). Finally, ethylene- and jasmonic acid–dependent signals also can influence R function (Clarke et al., 2000). Here, we provide compelling evidence that the Arabidopsis ortholog of barley RAR1 also is required for the action of several, but not all, tested R genes. Thus, a key step in R signaling is conserved evolutionarily. We further demonstrate that AtRAR1 and NDR1 contribute differently to the overall efficiency of the defense response, depending on the R function being assayed. This relative contribution can be simple and linear or quantitative and separable.
AtRAR1 is a single-copy gene. Perhaps surprisingly, all of the mutations were either stop codons or changes in the very conserved Cys residues (Shirasu et al., 1999). This may reflect a screening bias or may imply that weak loss-of-function Atrar1 alleles have only weak effects on R function and would be missed in screens relying on strong loss-of-function phenotypes for mutant detection. Alternatively, there may be some internal functional redundancy in the RAR1 protein. For example, the CHORD I and CHORD II domains could act independently in R signaling. If this is the case, it follows that only strong loss-of-function or null phenotypes affecting both CHORD I and CHORD II function would be isolated. These results are in contrast to the original definition of RAR1 in barley, for which two partial loss-of-function alleles were described and about which it was speculated that RAR1 might be essential (Shirasu et al., 1999).
We further establish that the functions of some Col-0 R genes are not altered grossly by the Atrar1 mutation. RPS4, RPP7, and RPP2 are not totally compromised in the Col-0 Atrar1-20 and Atrar1-21 backgrounds (Warren et al., 1999; but see Muskett et al., 2002, for analysis of RPS4 function in Landsberg erecta). The deduced RPS4 protein is of the TIR-NB-LRR subclass, as is RPP2 (Holub, 2001). RPP7 has not been isolated, but it is defined by several allelic loss-of-function ethyl methanesulfonate alleles (A. Cuzick and E. Holub, unpublished data). It has been mapped to a 100-kb region that is rich in CC-NB-LRR genes (Holub, 2001; A. Cuzick and E. Holub, unpublished data). There are no TIR-NB-LRR genes in this region. If one of the CC-NB-LRR genes proves to encode RPP7, then examples of both R protein structural subclasses will exist that do not require RAR1 for signaling. Conversely, RPP4, which requires AtRAR1, was shown recently to belong to the TIR-NB-LRR class (van der Biezen et al., 2002). Thus, the requirement for RAR1 function in Col-0 does not correlate with the structural subclass of the R protein in question.
Several aspects of our data strongly support the conclusion that AtRAR1 quantitatively contributes to R signaling in conjunction with EDS1 and NDR1:
(1) At least three R gene functions in Col-0 (RPM1, RPS2, and RPS5) are altered significantly in Atrar1 mutants. Each of these requires NDR1 to function, as measured by the restriction of bacterial growth, but this effect is quantitative. Furthermore, we can differentiate Atrar1 from ndr1-1 based on their effects on HR induction: the ndr1-1 null mutation still produces an essentially wild-type RPM1-dependent HR, whereas Atrar1 mutations greatly attenuate, but do not abolish, this response. We also observed stochastic reduction in the HR mediated by RPM1 and RPS2 in Atrar1 null mutants. Some leaves in each experiment consistently expressed a seemingly wild-type response. Therefore, AtRAR1 is required for the restriction of pathogen growth, but it contributes differentially to the HR triggered by any given R gene.
(2) At least three R gene functions in Col-0 (RPS4, RPP2, and RPP7) are not compromised obviously in Atrar1 mutants (Warren et al., 1999; see above). RPS4 requires EDS1 for signaling (Gassmann et al., 1999). RPP2, a TIR-NB-LRR protein active against Pp isolate Cala2, also is largely EDS1 dependent (Parker et al., 1996).
(3) AtRAR1 can act in combination with NDR1. We demonstrated that Atrar1-21 ndr1-1 double mutants expressed the Atrar1-21 phenotype with respect to RPM1 function, the ndr1-1 phenotype with respect to RPS2 function, the phenotype of either single mutant with respect to RPS5 function, and an additive phenotype with respect to RPP7 function. Figure 8 presents an interpretation of these results in which the size of the font represents the relative contribution of each gene to the particular R function tested. Thus, the relative contributions of AtRAR1 and NDR1 to resistance are dependent on which R function is assayed. Both RPP2 and RPP4 are suppressed very modestly in Atrar1-20, Atrar1-21, and ndr1-1 single mutants (Warren et al., 1999; our unpublished data). Moreover, the Atrar1-21 ndr1-1 double mutant does not enhance the suppression of these R functions significantly (data not shown), as it did for RPP7. Thus, there is a level of specificity achieved in the quantitative level of resistance that is determined by a combination of particular R proteins with NDR1, AtRAR1, and EDS1. This conclusion is puzzling at first because the specificity of R genes is assumed to lie exclusively in the R protein itself. Nevertheless, the genetically defined requirements for R gene function produce a “fingerprint” that informs the eventual phenotypic output. This information might allow the plant to use the same R genes in different contexts, multiplying the effectiveness of the NB-LRR system as a whole.
Figure 8.
Genetic Requirements for the R Gene Functions Tested.
The font size reflects the relative contribution of each locus to the function of each R protein listed at top. A larger font implies that the null mutant compromises R function severely, and a smaller font implies a moderate effect. A locus placed in the same vertical orientation implies a single pathway. Note that the order of action can be inferred clearly only in the case of RPM1. Loci side by side imply that no relationship was determined, and split arrows represent parallel pathways.
Recent work by Shirasu and colleagues (Azevedo et al., 2002) has shown that the AtRAR1 protein interacts physically with a plant ortholog of the yeast SGT1 protein (Kitigawa et al., 1999). SGT1 is a regulatory component of the SCF complex (Skp1, Cullin, F-box) (reviewed by Deshaies, 1999; Bachmair et al., 2001) that acts as an E3 ligase in ubiquitination of target proteins. Modification of target proteins by ubiquitin, or ubiquitin-like molecules, can lead to their degradation via the proteasome or can serve to regulate function directly (Hicke, 2001). The simplest model for the role of RAR1 in R function is that it directs either the removal of a negative regulator (Gray et al., 1999) or the activation of a positive regulator (Wang et al., 2001) by recruitment of that factor to the SCF via SGT1 and subsequent ubiquitination.
Our finding that RPM1-myc levels are reduced severely in the Atrar1-21 background supports this general model but does not address whether AtRAR1 acts positively and directly on RPM1-myc stability or whether it acts by removing a negative regulator of RPM1-myc stability. It is possible that the main function of AtRAR1 is to regulate the steady state levels of some, but not all, R proteins present in the plant cell. However, this model is difficult to reconcile with the finding that R proteins that differ by as little as 5% differ in their requirements for barley RAR1 (Zhou et al., 2001).
The evidence presented by our colleagues (Azevedo et al., 2002; Muskett et al., 2002; Tör et al., 2002) and in this report suggests that, in addition to the response specificity encoded in the R protein, additional specificity operates at the level of post-translational regulation. The fact that RAR1-dependent and RAR1-independent barley R proteins differ by <5% (Zhou et al., 2001) focuses attention on how those amino acid differences mediate the subsequent signaling or regulation of R protein stability and/or accumulation.
METHODS
Plant Lines
The transgenic Arabidopsis thaliana Columbia lines a11 and a11r have been described (Tornero et al., 2002). Briefly, these lines allow the conditional expression of avrRpm1 upon the application of estradiol. The background of a11 is Columbia (RPM1), and that of a11r is rpm1-1 (Grant et al., 1995). The Atrar1-21 line used for all described experiments was backcrossed twice to its parental line, a11. Plants were grown in a short day regimen, as described by Ritter and Dangl (1996). Mutant lines used were ndr1-1 (a null allele; Century et al., 1997), pbs2-1 (Warren et al., 1999), rps2-101C (Mindrinos et al., 1994), rps5-2 (Warren et al., 1998), and a strong loss-of-function allele of rpp7 (McDowell et al., 2000). We constructed double mutant lines by crossing Atrar1-21 with ndr1-1, selecting F2 plants that were rar1 based on phenotype against Pseudomonas syringae pv tomato (Pst) DC3000(avrRpm1) and confirmed by sequencing of the mutation and F2 plants that were ndr1 based on polymerase chain reaction (PCR)–based molecular markers as described by Rusterucci et al. (2001).
Pathogen Strains and Quantitation of Bacterial Growth in Leaves
Pst DC3000 derivatives containing pVSP61 (empty vector), avrRpm1, avrB, avrB::Ω (a disrupted nonfunctional version of avrB), avrRpt2, avrPphB, or avrRps4 were maintained as described (Ritter and Dangl, 1996). Plant inoculations and counting of the bacteria were performed as described (Tornero and Dangl, 2001). Where indicated, high concentrations of bacteria (OD600 = 0.075, 3.75 × 107 colony-forming units/mL) were infiltrated into the bottom part of the leaf with a blunt syringe to test for the induction of hypersensitive plant cell death.
Peronospora parasitica isolate Hiks1 was maintained and inoculated on 10-day-old plants as described (Holub et al., 1994). Inoculated plants were kept under a sealed propagator lid to achieve high RH in a growth chamber at 19°C under an 8-hr light period (100 to 160 μE·m−2·sec−1). To evaluate the infection, plants were stained with trypan blue (Koch and Slusarenko, 1990) 5 days after inoculation and observed with a microscope. Infection sites were scored as “free hyphae,” “trailing necrosis,” or “hypersensitive response” (Morel and Dangl, 1998). Free hyphae was defined as hyphal growth without detectable plant cell death. Trailing necrosis was defined as hyphal growth with surrounding plant cell death. Hypersensitive response was defined as plant cell death at the infection site with no hyphal growth beyond the infection site.
Genetic Analysis
Allelism between lra1 mutants and pbs2-1 (Warren et al., 1999) was determined by standard genetic crosses followed by analysis of F1 and F2 progeny for resistance to estradiol treatment (induction of the avrRpm1 transgene) as described (Tornero et al., 2002) or by hand inoculations of Pst DC3000(avrRpm1). Mapping populations were established by crossing Landsberg erecta with lra1-1 (Atrar1-21) and lra1-8 (Atrar1-28) and analyzing the response of F2 individuals to infection by Pst DC3000(avrRpm1). Plants that were susceptible to these bacteria were allowed to self and were retested in the F3 generation, and a sample of their genomic DNA was extracted by conventional methods (Ausubel et al., 1987). DNA from 35 of these plants was used in PCR amplification of known PCR-based molecular markers (www.arabidopsis.org) to obtain approximate mapping positions.
Subsequently, we refined this interval using newly developed molecular markers (available upon request) and a total of 120 F2 individuals, localizing lra1 to several overlapping bacterial artificial chromosome (BAC) and P1 clones. The genetic interval containing lra1 was defined by a single recombinant event between a marker derived from nucleotides 50,446 to 50,174 on P1 clone MIO24 and lra1 and by three recombinant events between a marker derived from nucleotides 16,930 to 17,292 on BAC K2OJ1 and lra1. Note that the first marker is 43,400 nucleotides 3′ of the AtRAR1 open reading frame, because the nucleotide numbering of this BAC is inverted relative to that of BAC K2OJ1. Attempts to amplify the AtRAR1 open reading frame from the pbs2-1 allele failed, consistent with a deletion or rearrangement of AtRAR1 in the pbs2-1 mutant. Therefore, we sequenced the AtRAR1 gene (nucleotides 5276 to 7046 of BAC MIO24; primers available upon request) from all eight lra1 mutants and found the mutations described.
Analysis of Protein Levels
Generation of the anti-RAR1 antiserum is described by Azevedo et al. (2002). For protein gel blot analysis, tissue from 3-week-old plants was extracted with a buffer containing 50 mM Tris-HCl, pH 7.5, 10% glycerol, 50 mM NaCl, 10 mM MgCl2, 5 mM EDTA, 1 mM DTT, 5 mM 2-mercaptoethanol, 1 × proteinase inhibitor (Sigma), and 0.3% insoluble polyvinylpyrrolidone. Protein concentrations were determined by Bradford (1976) assay (Bio-Rad). For immunodetection, 40-μg protein samples were electrophoresed on 15% polyacrylamide gels and run in the presence of 0.38 M Tris and 0.1% SDS. Proteins were transferred from the gels to nitrocellulose filters by electroblotting, incubated with primary anti-RAR1 antibody and horseradish peroxidase–conjugated secondary antibody, and detected with enhanced chemiluminescence (ECL+; Amersham).
For detection of RPM1-myc, we used plants that carry a transgenic RPM1-myc expressed from the native RPM1 promoter (Boyes et al., 1998). This line was introgressed into the a11 line. The resulting line has two functional RPM1 genes. Similarly, we introgressed Atrar1-21 into this line. Protein extraction and protein gel blot analysis were performed as described (Boyes et al., 1998).
Microscopy
Trypan blue staining was performed according to Koch and Slusarenko (1990). Stained leaves were observed with an Eclipse E800 microscope (Nikon, Tokyo, Japan) equipped with a Spot charge-coupled device camera (Diagnostic Instruments, Sterling Heights, MI). The images shown were taken with a ×4 objective using transmitted light. Images were processed using Spot software (version 2.1; Diagnostic Instruments) and Photoshop (version 5.5; Adobe Systems, Mountain View, CA). Images from whole leaves (Figure 4) were taken by directly scanning the mounted tissue with a Microtek 8700 color scanner (Redondo Beach, CA). Hydrogen peroxide was detected by staining with 3,3′-diaminobenzidine using a modification of the protocol described by Thordal-Christensen et al. (1997). In brief, leaves were excised 90 min after inoculation and vacuum infiltrated with a solution (1 mg/mL) of 3,3′-diaminobenzidine. Subsequently, the leaves were kept under high humidity and darkness for 5 hr, cleared with ethanol, and mounted on slides.
Acknowledgments
We thank Swapna Putcha for her help with genetic mapping and Dr. Jane Parker for sharing map information before publication. This work was funded by National Science Foundation Grant IBN-9724075, renewed under the Arabidopsis 2010 project as IBN-0114795, and by U.S. Department of Agriculture National Research Initiative Competitive Grants Program Grant 99-35301-7848 to J.L.D. and National Institutes of Health Grant R01GM46451 to R.W.I. We gratefully acknowledge a Syngenta Collaborative Research Grant to J.L.D., a postdoctoral fellowship from the Spanish Ministry of Science and Education to P.T., and a U.S. Department of Agriculture National Needs predoctoral fellowship to P.M. A.S. and K.S. are supported by grants from The Gatsby Charitable Foundation.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.001032.
References
- Arabidopsis Genome Initiative (2000). Analysis of the genome of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1987). Current Protocols in Molecular Biology. (New York: John Wiley & Sons).
- Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., and Schulze-Lefert, P. (2002). The RAR1 interactor SGT1 is an essential component of R-gene triggered disease resistance. Science, in press. [DOI] [PubMed]
- Bachmair, A., Novatchkova, M., Potuschak, T., and Eisenhaber, F. (2001). Ubiquitination in plants: A post-genomic look at a post-translational modification. Trends Plant Sci. 6, 463–470. [DOI] [PubMed] [Google Scholar]
- Bent, A.F., Kunkel, B.N., Dahlbeck, D., Brown, K.L., Schmidt, R., Giraudat, J., Leung, J., and Staskawicz, B.J. (1994). RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science 265, 1856–1860. [DOI] [PubMed] [Google Scholar]
- Bisgrove, S.R., Simonich, M.T., Smith, N.M., Sattler, N.M., and Innes, R.W. (1994). A disease resistance gene in Arabidopsis with specificity for two different pathogen avirulence genes. Plant Cell 6, 927–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes, D.C., Nam, J., and Dangl, J.L. (1998). The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc. Natl. Acad. Sci. USA 95, 15849–15854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Century, K.S., Holub, E.B., and Staskawicz, B.J. (1995). NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc. Natl. Acad. Sci. USA 92, 6597–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century, K.S., Shapiro, A.D., Repetti, P.P., Dahlbeck, D., Holub, E., and Staskawicz, B.J. (1997). NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278, 1963–1965. [DOI] [PubMed] [Google Scholar]
- Clarke, J.D., Volko, S.M., Ledford, H., Ausubel, F.M., and Dong, X. (2000). Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12, 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Deshaies, R.J. (1999). SCF and cullin/RING H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Falk, A., Feys, B., Frost, L.N., Jones, J.D.G., Daniels, M.J., and Parker, J.E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96, 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freialdenhoven, A., Scherag, B., Hollricher, K., Collinge, D.B., Thordal-Christensen, H., and Schulze-Lefert, P. (1994). Nar-1 and Nar-2, two loci required for Mla12-specified race-specific resistance to powdery mildew in barley. Plant Cell 6, 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann, W., Hinsch, M., and Staskawicz, B. (1999). The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 20, 265–277. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2001). Genes controlling expression of defense responses in Arabidopsis: 2001 status. Curr. Opin. Plant Biol. 4, 301–308. [DOI] [PubMed] [Google Scholar]
- Gopalan, S., Bauer, D.W., Alfano, J.R., Lonlello, A.O., He, S.Y., and Collmer, A. (1996). Expression of the Pseudomonas syringae avirulence protein AvrB in plant cells alleviates its dependence on the hypersensitive response and pathogenicity (Hrp) secretion system in eliciting genotype-specific hypersensitive cell death. Plant Cell 8, 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, M.R., Godiard, L., Straube, E., Ashfield, T., Lewald, J., Sattler, A., Innes, R.W., and Dangl, J.L. (1995). Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269, 843–846. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., Ma, H., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halterman, D., Zhou, F., Wei, F., Wise, R.P., and Schulze-Lefert, P. (2001). The MLA6 coiled-coil, NBS-LRR protein confers AvrMla6-dependent resistance specificity to Blumeria graminis f. sp. hordei in barley and wheat. Plant J. 25, 335–348. [DOI] [PubMed] [Google Scholar]
- He, S.-Y. (1998). Type III secretion systems in plant and animal pathogenic bacteria. Annu. Rev. Phytopathol. 36, 363–392. [DOI] [PubMed] [Google Scholar]
- Heath, M.C. (2000). Hypersensitive response-related death. Plant Mol. Biol. 44, 321–334. [DOI] [PubMed] [Google Scholar]
- Hicke, L. (2001). Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell. Biol. 2, 195–201. [DOI] [PubMed] [Google Scholar]
- Hinsch, M., and Staskawicz, B.J. (1996). Identification of a new Arabidopsis disease resistance locus RPS4, and cloning of the corresponding avirulence gene, avrRps4, from Pseudomonas syringae pv. pisi. Mol. Plant-Microbe Interact. 9, 55–61. [DOI] [PubMed] [Google Scholar]
- Holub, E. (2001). The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2, 516–527. [DOI] [PubMed] [Google Scholar]
- Holub, E.B., Beynon, J.L., and Crute, I.R. (1994). Phenotypic and genotypic characterization of interactions between isolates of Peronospora parasitica and accessions of Arabidopsis thaliana. Mol. Plant-Microbe Interact. 7, 223–239. [Google Scholar]
- Jones, J.D.G. (2001). Putting knowledge of plant disease resistance genes to work. Curr. Opin. Plant Biol. 4, 281–287. [DOI] [PubMed] [Google Scholar]
- Kitigawa, K., Skowyra, D., Elledge, S.J., Harper, J.W., and Hieter, P. (1999). SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin complex. Mol. Cell 4, 21–33. [DOI] [PubMed] [Google Scholar]
- Koch, E., and Slusarenko, A.J. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell, J.M., Cuzick, A., Can, C., Benyon, J., Dangl, J.L., and Holub, E.B. (2000). Downy mildew (Peronospora parasitica) resistance genes in Arabidopsis vary in functional requirements for NDR1, EDS1, NPR1, and salicylic acid accumulation. Plant J. 22, 523–530. [DOI] [PubMed] [Google Scholar]
- Mindrinos, M., Katagiri, F., Yu, G.-L., and Ausubel, F.M. (1994). The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78, 1089–1099. [DOI] [PubMed] [Google Scholar]
- Morel, J.-B., and Dangl, J.L. (1998). Suppressors of the Arabidopsis lsd5 cell death mutation identify genes involved in regulating disease resistance responses. Genetics 151, 305–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskett, P.J., Kahn, K., Austin, M.J., Moisan, L.J., Sadanandom, A., Shirasu, K., Jones, J.D.G., and Parker, J.E. (2002). Arabidopsis RAR1 exerts rate-limiting control of R gene–mediated defense against multiple pathogens. Plant Cell 14, 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk, Z., Marois, E., Kjemtrup, S., Leister, R.T., Katagiri, F., and Dangl, J.L. (2000). Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 101, 353–363. [DOI] [PubMed] [Google Scholar]
- Nimchuk, Z., Rohmer, L., Chang, J.H., and Dangl, J.L. (2001). Knowing the dancer from the dance: R gene products and their interactions with other proteins from host and pathogen. Curr. Opin. Plant Biol. 4, 288.–294. [DOI] [PubMed] [Google Scholar]
- Parker, J.E., Holub, E.B., Frost, L.N., Falk, A., Gunn, N.D., and Daniels, M.J. (1996). Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8, 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter, C., and Dangl, J.L. (1996). Interference between two specific pathogen recognition events mediated by distinct plant disease resistance genes. Plant Cell 8, 251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusterucci, C., Aviv, D.H., Holt, B.F., Dangl, J.L., and Parker, J.E. (2001). The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13, 2211–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, A.D., and Zhang, C. (2001). The role of NDR1 in avirulence gene-directed signaling and control of programmed cell death in Arabidopsis. Plant Physiol. 127, 1089–1101. [PMC free article] [PubMed] [Google Scholar]
- Shirasu, K., Lahaye, T., Tan, M.-W., Zhou, F., Azevedo, C., and Schulze-Lefert, P. (1999). A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 99, 355–366. [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen, H., Zhang, Z., Wei, Y., and Collinge, D.B. (1997). Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during barley-powdery mildew interaction. Plant J. 11, 1187–1194. [Google Scholar]
- Tör, M., et al. (2002). An SGT1-like gene in Arabidopsis is essential for defense signalling conferred by several downy mildew (Peronospora parasitica) resistance genes including RPP7. Plant Cell 14, 993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero, P., and Dangl, J.L. (2001). A high throughput method for quantifying growth of phytopathogenic bacteria in Arabidopsis thaliana. Plant J. 28, 475–481. [DOI] [PubMed] [Google Scholar]
- Tornero, P., Chao, R.A., Luthin, W.N., Goff, S.A., and Dangl, J.L. (2002). Large-scale structure-function analysis of the Arabidopsis RPM1 disease resistance protein. Plant Cell 14, 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen, E.A., Freddie, C.T., Kahn, K., Parker, J.E., and Jones, J.D.G. (2002). Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signaling components. Plant J. 29, 439–451. [DOI] [PubMed] [Google Scholar]
- Wang, C., Deng, L., Hong, M., Akkaraju, G.R., Inoue, J., and Chen, Z.J. (2001). TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412, 346–351. [DOI] [PubMed] [Google Scholar]
- Warren, R.F., Henk, A., Mowery, P., Holub, E., and Innes, R.W. (1998). A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell 10, 1439–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, R.F., Merritt, P.M., Holub, E.B., and Innes, R.W. (1999). Identification of three putative signal transduction genes involved in R gene-specified disease resistance in Arabidopsis. Genetics 152, 401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, F., Kurth, J., Wei, F., Elliot, C., Vale, G., Yahiaoui, N., Keller, B., Somerville, S., Wise, R.P., and Schulze-Lefert, P. (2001). Cell-autonomous expression of barley Mla1 confers race-specific resistance to the powdery mildew fungus via a Rar1-independent signaling pathway. Plant Cell 13, 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]