Abstract
The hydra mutants of Arabidopsis are characterized by a pleiotropic phenotype that shows defective embryonic and seedling cell patterning, morphogenesis, and root growth. We demonstrate that the HYDRA1 gene encodes a Δ8-Δ7 sterol isomerase, whereas HYDRA2 encodes a sterol C14 reductase, previously identified as the FACKEL gene product. Seedlings mutant for each gene are similarly defective in the concentrations of the three major Arabidopsis sterols. Promoter::reporter gene analysis showed misexpression of the auxin-regulated DR5 and ACS1 promoters and of the epidermal cell file–specific GL2 promoter in the mutants. The mutants exhibit enhanced responses to auxin. The phenotypes can be rescued partially by inhibition of auxin and ethylene signaling but not by exogenous sterols or brassinosteroids. We propose a model in which correct sterol profiles are required for regulated auxin and ethylene signaling through effects on membrane function.
INTRODUCTION
Sterols are essential components of fungal, plant, and animal membranes. They regulate fluidity and interact with lipids and proteins within the membrane, and they are the precursors for the brassinosteroid (BR) hormones in plants (Hartmann, 1998). The sterol biosynthetic pathway in plants, therefore, can be viewed as comprising two parts: one branch produces the bulk membrane sterols (the principal sterols in Arabidopsis being stigmasterol, campesterol, and sitosterol), and the second part represents the BR synthesis branch. Sterol biosynthesis has been well characterized in yeast, supported by a powerful system of genetic analysis. In animals, and more recently in plants, sterol biosynthetic enzyme function has been confirmed via the functional complementation of yeast mutants (Gachotte et al., 1996). Functional analysis of sterol function in plants has involved a range of approaches, but recently, genetic studies have provided useful information on the requirement for particular enzymes in sterol and BR biosynthesis and, for BRs, perception and signal transduction (Clouse, 2000; Diener et al., 2000; Schaeffer et al., 2001).
In animals, sterols appear to be important to maintain correct cell-signaling activities. For example, drugs such as the σ ligand SR31747A, which inhibits the activity of the σ receptor (emopamil binding protein [EBP], which has Δ8-Δ7 sterol isomerase activity), cause defects in a diversity of cellular processes, including the inhibition of mammalian lymphocyte proliferation in response to mitogens (Derocq et al., 1995) and the inhibition of graft rejection in mouse via the modulation of gene expression (Carayon et al., 1995), and they may influence lipoprotein functions leading to immunosuppressive effects (Dussossoy et al., 1999). In plants, a lack of detailed pharmacological studies has precluded analogous investigations of the role of sterols in plant cell biology.
However, mutational and transgenic studies have given new insight into the roles of sterols in plant development. sterol methyltransferase1 (smt1) mutants accumulate cholesterol at the expense of sitosterol and cannot be rescued by BRs (Diener et al., 2000). Transgenic tobacco plants expressing a 35S::SMT2 gene accumulate sitosterol at the expense of campesterol, causing reduced stature and growth, and can be rescued by BRs (Schaeffer et al., 2001). Cosuppression of SMT2 causes low sitosterol and high campesterol, accompanied by reduced growth and low fertility, which is not modified by BRs. The reduced growth is linked with reduced cell division rather than with the reduced cell elongation that many BR mutants exhibit (Schaeffer et al., 2001). The identification of sterol binding (Steroidogenic Acute Regulatory Transfer Protein [StART]) domains (Kallen et al., 1998; Ponting and Aravind, 1999) in a number of patterning proteins, and in particular some homeodomain proteins such as PHABULOSA (PHB/ATHB14), PHAVOLUTA (PHV/ATHB9), GLABRA2 (GL2/ATHB10), REVOLUTA (REV), and ARABIDOPSIS THALIANA MERISTEM LAYER1 (ATML1), suggests that sterols may modulate regulatory protein function (McConnell et al., 2001). At present, however, a direct role for this domain in these genes remains to be proven.
Two groups have published data on a mutant in the early part of the sterol biosynthetic pathway, which is not part of the BR pathway and which is not rescued by the exogenous application of BRs (Jang et al., 2000; Schrick et al., 2000). This is the fackel (fk) mutant of Arabidopsis, which encodes a defective sterol C14 reductase, resulting in a pleiotropic phenotype characterized by defective cell shape and division control in embryogenesis. Schrick et al. (2000) have postulated that there is a novel, as yet unknown, sterol signaling molecule that is not produced in these mutants that can explain the phenotype.
In performing a mutational screen to identify genes required for embryo and seedling development in Arabidopsis, we previously identified a class of seedling-lethal dwarfs that we termed the hydra (hyd) mutants (Topping et al., 1997). Recessive mutations at two loci, designated hyd1 and hyd2, resulted in pleiotropic phenotypes, including defective cell shape, multiple cotyledons, and short roots and hypocotyls. Because of the phenotypic similarities of the two mutants, we predicted that they are defective in the same or similar pathways required for normal embryo and seedling development.
To gain more insight into the molecular basis of HYD gene function, we aimed to clone the hyd1 and hyd2 genes, and in this article, we report the identity of the genes. We show that the hyd1 mutant is defective in the plant ortholog of yeast erg2 and mammalian EBP, Δ8-Δ7 sterol isomerase. We also show that hyd2 is allelic to fk. Using these mutants, we provide evidence for a role for Δ8-Δ7 sterol isomerase activity in eukaryotic morphogenesis and show that sterols are required for correct auxin and ethylene signaling in Arabidopsis, defects that may account for many of the developmental abnormalities seen in the mutants.
RESULTS
hyd1 and hyd2 Are Sterol Biosynthetic Mutants
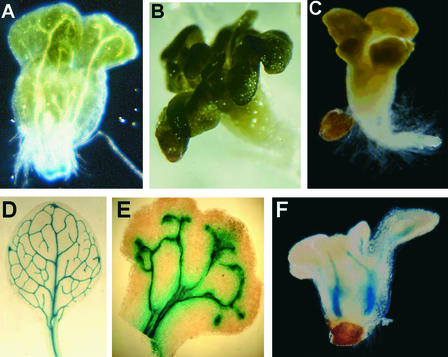
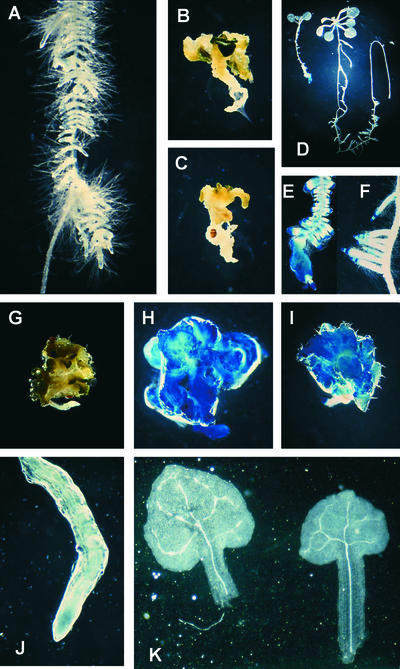
In a screen of T-DNA and ethyl methanesulfonate mutants of Arabidopsis, we identified a class of recessive seedling-lethal mutants defined by two loci, hyd1 and hyd2 (Topping et al., 1997). We showed that hyd1 mutants have a pleiotropic phenotype. They are unable to regulate cell size and shape, and they fail to undergo correct morphogenesis during embryonic and postembryonic development (Figure 1). Although apical-basal pattern elements are present (shoot, hypocotyl, and root), the radial pattern is defective, with supernumerary cell layers and aberrant vascular patterning. Multiple leaf-like cotyledons are produced (Figures 1A to 1C).
Figure 1.
Morphology and Vascular Patterning of hyd Seedlings.
(A) hyd1 seedling 3 days after germination. Magnification ×20.
(B) hyd1 seedling 7 days after germination. Magnification ×12.
(C) hyd2 seedling 5 days after germination. Magnification ×15.
(D) Cleared wild-type leaf showing PIN1::GUS expression in vascular tissues. Magnification ×8.
(E) hyd2 leaf showing PIN1::GUS expression in vascular tissues. Magnification ×30.
(F) hyd2 seedling 3 days after germination showing VT-1 promoter trap expression in vascular tissues. Magnification ×15.
The hyd2 mutant seedling phenotype is very similar to that of hyd1. In allelism tests, we found that hyd2 is allelic to the fk mutant, which is defective in the gene encoding the sterol biosynthetic enzyme C-14 sterol reductase (Jang et al., 2000; Schrick et al., 2000). These authors describe the embryonic phenotype, which is essentially identical to that of hyd1 (Topping et al., 1997). Using reverse transcriptase–mediated polymerase chain reaction, it was found that the hyd2 allele of fk contained no detectable FK transcript and probably is a null allele caused by a T-DNA insertion that cosegregates with the mutation. Like hyd1, hyd2/fk is dwarfed, with multiple cotyledons, and is seedling lethal. It exhibits severely abnormal vascular patterning, as revealed microscopically and with the aid of the vascular tissue–specific β-glucuronidase (GUS) markers VT-1 (a promoter trap marker) (Topping et al., 1997) and PIN1::GUS (which is a fusion of the PIN1 gene promoter and the GUS reporter gene) (Figures 1E and 1F) (L. Gälweiler and K. Palme, unpublished data). Commonly, there are a duplicated vascular system and discontinuities in vascular strands (Figure 1F).
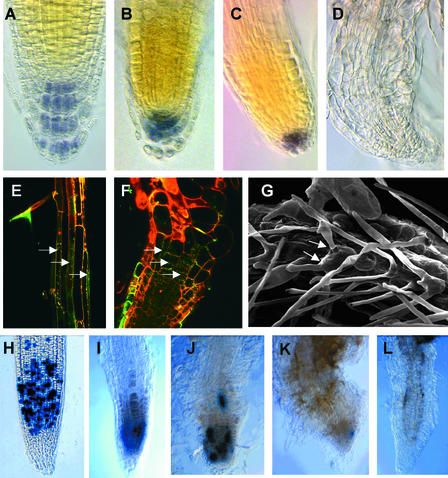
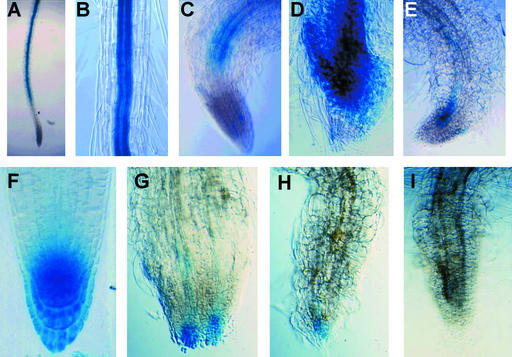
A distinction between the hyd1 and hyd2/fk mutants is the more severe root-defective phenotype of hyd2/fk. Although the hyd1 root is short and may cease cell division within 2 weeks after germination, roots may continue to grow very slowly until the seedling dies, at ∼30 to 40 days after germination (Topping et al., 1997). In contrast, the hyd2/fk root invariably stops growth completely at ∼10 to 14 days after germination. Microscopic examination showed a loss of correct cell patterning of the meristem and root cap, with a loss of Lugol staining of the columella, indicating a loss of that tissue (Figures 2A to 2D).
Figure 2.
Root Growth and Development in hyd Seedlings.
(A) to (D) Lugol-stained roots of wild-type ([A]; 14 days after germination), hyd1 ([B]; 14 days after germination), hyd2 ([C]; 10 days after germination), and hyd2 ([D]; 14 days after germination) to reveal columella. Magnification ×200.
(E) and (F) Confocal images of wild-type (E) and hyd2 (F) roots expressing GL2::GFP fusion in epidermal cell files (arrows). Note in hyd2 the expression in all cell files and the abnormal cell shape. Magnification ×200.
(G) Scanning electron micrograph of a hyd2 root showing ectopic root hairs (arrows) and abnormal root hair shape. Bar = 100 μM.
(H) to (L) CyclinB::GUS expression in mitotic cells of roots of wild type (H) and hyd2 at 3 days after germination (I), 12 days after germination (J), 15 days after germination (K), and 18 days after germination (L). Magnification ×100.
Root cell file patterning also was abnormal. The alternating expression of the cell file marker GL2::green fluorescent protein (GFP), expressed in atrichoblast (non-hair-forming) files in the wild type (Masucci et al., 1996) (Figure 2E), was seen in all root epidermal cell files in a hyd2/fk background (Figure 2F). Interestingly, all cell files produced root hairs, and up to five root hairs can form from individual trichoblasts (Figure 2G). This finding indicates a defect in the mechanisms controlling GL2 gene expression, and because the GL2 gene is proposed to act as a negative regulator of hair cell formation (Masucci et al., 1996), it suggests that GL2 protein activity is either suppressed or bypassed in the mutants.
To investigate further the meristematic activity of the hyd1 and hyd2/fk seedling roots, heterozygotes were crossed with Arabidopsis transgenic for a cyclinB::GUS gene fusion containing a cyclin destruction box, which marks individual mitotic cells (Hauser and Bauer, 2000). Although the hyd1 mutant root showed no obvious change in the frequency of GUS-positive cells during the 18 days after germination, hyd2/fk mutants showed a reduced frequency of GUS-positive cells between days 10 and 14 after germination, corresponding to the observed disorganization at the root tip at this time (Figures 2H to 2L). This finding indicates a loss of mitotic activity of the hyd2 root meristem during development.
Two independent hyd1 alleles, hyd1-1 and hyd1-2, were identified previously (Topping et al., 1997). We cloned the hyd1-2 allele, which was T-DNA tagged (Errampalli et al., 1991). A genomic fragment of 2.6 kb flanking the T-DNA in hyd1-2 was cloned by plasmid rescue (Behringer and Medford, 1992) and found to be 100% homologous with the 5′ region of an expressed sequence tag that was identified as a cDNA encoded by the Arabidopsis Δ8-Δ7 sterol isomerase gene (Grebenok et al., 1998). The gene is on chromosome 1. The enzyme Δ8-Δ7 sterol isomerase is located one step downstream of the C-14 sterol reductase in the sterol biosynthetic pathway. The predicted protein has high homology with the mammalian EBP, which is localized to the endoplasmic reticulum and the nuclear membrane (Dussossoy et al., 1999) and has been demonstrated to have Δ8-Δ7 sterol isomerase activity (Silve et al., 1996a).
A full-length expressed sequence tag clone was obtained, and a genomic clone encompassing the gene was isolated from an Arabidopsis Landsberg genomic library (Voytas et al., 1990). The T-DNA was found to have inserted into the first intron of the gene, which most likely resulted in a null mutation. This finding is supported by the observed lack of detectable HYD1 transcript in the hyd1-2 mutant, as determined by reverse transcriptase–mediated polymerase chain reaction (data not shown). Sequencing of a second mutant allele (hyd1-1) revealed the presence of a point mutation that causes a Gly-to-Asp substitution at amino acid 66, which falls within a predicted membrane-spanning domain.
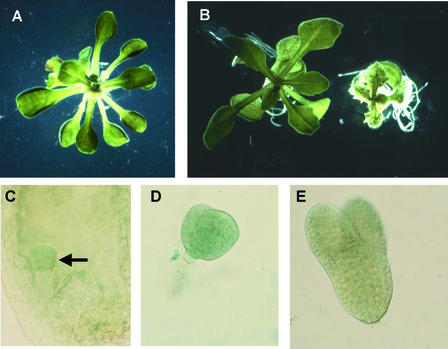
To determine whether a wild-type allele of the sterol isomerase gene complements the hyd1 mutation, hyd1-2 seedlings, which are heterozygous for the T-DNA insertional mutation carrying the NPT-II selectable marker, were transformed with a wild-type allele of the gene under the transcriptional control of 2 kb of its own promoter and carrying a selectable HPT gene, which confers hygromycin resistance. One hundred percent of 40 transgenic F2 progeny, which contained both the mutation and the wild-type allele (identifiable as being resistant to both kanamycin and hygromycin), were rescued phenotypically (Figure 3A). Transformation of hyd1-2 homozygous mutants with the wild-type allele linked to a phosphomannose isomerase selectable marker also led to phenotypic rescue.
Figure 3.
HYD1 Gene Function.
(A) Phenotype of a transgenic hyd1 mutant seedling expressing a wild-type allele of the HYD1 sterol isomerase gene under the transcriptional control of its own promoter. Magnification ×4.
(B) Wild-type seedling (left) and wild-type seedling transformed with an antisense HYD1 gene under the transcriptional control of the CaMV35S promoter (right). Magnification ×4.
(C) to (E) Embryonic expression of the HYD1::GUS promoter fusion in globular-stage (arrow) ([C]; magnification ×100), heart-stage ([D]; magnification ×200) and torpedo-stage ([E]; magnification ×200) embryos.
This finding demonstrates that the sterol isomerase was able to complement the mutation. The wild-type gene under the transcriptional control of the 35S gene promoter of Cauliflower mosaic virus (CaMV35S) (compared with the HYD1 promoter) was able to rescue the hyd1 mutant only partially, presumably because that promoter is not expressed early in embryogenesis when the hyd1 defects are established. Antisense expression of the sterol isomerase gene under the transcriptional control of the CaMV35S promoter caused a dwarfed phenotype (Figure 3B). Together, these results confirm that mutation of the Δ8-Δ7 sterol isomerase gene is responsible for the hyd1 phenotype.
A 2-kb fragment upstream of the sterol isomerase coding region was fused to the gusA reporter gene and introduced into wild-type Arabidopsis for functional analysis. GUS activity in transgenic plants was detectable in a developmentally regulated pattern during embryogenesis. At the globular and heart stages, GUS activity was constitutive in the embryo proper but was not detectable in the suspensor (Figures 3C and 3D). Torpedo-stage embryos showed lower levels of GUS activity (Figure 3E), and cotyledon- and mature-stage embryos had virtually no detectable GUS expression. These data are consistent with the mutant analysis that indicated the requirement for the sterol isomerase gene early in embryogenesis, and in the embryo proper, but not in the suspensor (Topping et al., 1997).
It has been reported previously that fk mutants have abnormal sterol profiles (Jang et al., 2000; Schrick et al., 2000). We compared the concentrations of the three major sterols in hyd1 and hyd2/fk seedlings (Table 1). Both hyd1 and hyd2/fk have much reduced levels of sitosterol and campesterol and increased levels of stigmasterol, similar to other fk alleles. hyd1-1 and hyd1-2 mutants showed no significant differences. Sitosterol and campesterol are the most abundant sterols in Arabidopsis, each being essential in ordering acyl chains in the membrane lipid bilayer and in regulating membrane permeability; stigmasterol cannot functionally replace either sterol (Schuler et al., 1991; Hartmann, 1998). These results demonstrate the requirement for a functional Δ8-Δ7 sterol isomerase activity for sterol composition in Arabidopsis and indicate that mutations in sterol isomerase (in hyd1) and in C14 reductase (in hyd2/fk), respectively, result in similar gross changes in sterol profiles.
Table 1.
Mean Sterol Concentrations in the Wild Type and hyd1 and hyd2/fk Mutants
| Sterol | Wild Type (μg/g fresh weight) |
hyd1 (% of wild type) |
hyd2/fk (% of wild type) |
|---|---|---|---|
| Campesterol | 4.20 ± 0.02 | 12 | 0 |
| Sitosterol | 19.51 ± 0.40 | 2 | 4 |
| Stigmasterol | 1.62 ± 0.00 | 182 | 322 |
Pooled seedlings at day 10 after germination were analyzed for the three major sterols, and the means of triplicate assays ± sem are presented.
To determine whether the exogenous application of sterols can rescue the mutants, seedlings were grown in the presence of 1, 5, 10, 50, and 100 μM sitosterol, campesterol, or stigmasterol. In no case was any evidence of phenotypic rescue seen. We showed previously that the hyd mutants are not rescued by BR application (Topping et al., 1997).
hyd1 and hyd2/fk Are Defective in Ethylene and Auxin Signaling Pathways
The pleiotropic phenotypes of hyd1 and hyd2/fk suggest that correct sterol profiles are required for diverse plant signaling pathways to function correctly. For example, the root hair phenotype of hyd2/fk suggests defective ethylene signaling, because ethylene is a positive regulator of hair formation (Tanimoto et al., 1995), and defective vascular patterning suggests defective auxin transport or signaling (Mattsson et al., 1999). It also is possible that any failure of the hyd/fk mutants to regulate auxin or ethylene signaling, and to respond to exogenous BRs, may be attributable to defective membrane-dependent processes, such as receptor activity, or to defects in permeability leading to the incorrect tissue distribution of signaling molecules. To determine whether specific known signaling pathways are altered in the mutants, we focused on cellular processes in which auxin and ethylene play important roles.
Ethylene Signaling
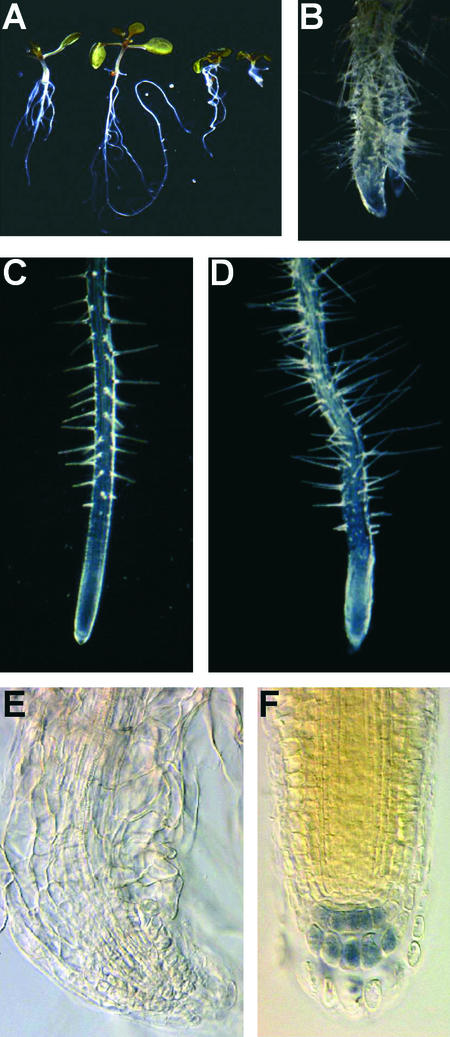
Both hyd1 and hyd2/fk roots have root hairs closer to the root tip than is seen in the wild type, with very abnormal patterning evident in hyd2/fk (Figure 2). To investigate whether ethylene signaling is abnormal in the mutant roots, hyd1 and hyd2 mutants were crossed with the ethylene-resistant mutant etr1-1 (Figure 4). The double mutants showed a restored wild-type pattern of root hair formation (Figures 4B and 4D) and meristem patterning and activity (Figures 4E and 4F) with sustained growth. Similarly, when hyd1 and hyd2 mutants were grown in the presence of 10 μM silver ions, which inhibit ethylene action by binding to the receptor (Beyer, 1976), a similar rescue of root structure and growth was observed. These results indicate either that hyd2/fk mutants accumulate relatively high levels of ethylene or that the ethylene receptor ETR1 is altered constitutively in activity to promote ethylene signaling and that this is responsible for the observed root hair and defective meristem phenotypes.
Figure 4.
hyd Root Phenotype Is Rescued Partially by Inhibiting Ethylene Action.
(A) Wild-type (left), etr1-1 (second from left), hyd2 etr1-1 (third from left), and hyd2 (right) seedlings showing increased root length in the double mutant compared with the hyd2 mutant.
(B) to (D) Roots of hyd2 (B), the wild type (C), and hyd2 etr1-1 (D) showing root hairs. Magnification ×10.
(E) hyd2 root 14 days after germination stained with Lugol to reveal that the columella is lacking. Magnification ×200.
(F) hyd2 etr101 root 14 days after germination stained with Lugol. Magnification ×200.
To determine whether the hyd1 and/or hyd2/fk mutants produce abnormal levels of ethylene, seedlings were grown in vitro for 7 days after germination, and evolved ethylene levels were assayed. In two independent sets of experiments, the hyd1 mutants (which are in a C24 background) produced ethylene at mean rates of 3.8 and 21.5 nL·g−1 fresh weight·hr−1, respectively, compared with mean rates of 11.9 and 10.8 nL·g−1 fresh weight·hr−1 for the C24 wild type. The hyd2 mutant (Wassilewskija background) produced ethylene at mean rates of 15.5 and 6.3 nL·g−1 fresh weight·hr−1, compared with 16.7 and 12.0 nL·g−1 fresh weight·hr−1 for the wild type. These findings indicate that the mutants produce variable levels of ethylene but do not produce dramatically different mean levels of ethylene compared with the wild type.
Treatment of mutant seedlings with the ethylene biosynthesis inhibitor aminoethoxyvinylglycine (0.5 μM) did not rescue the root or shoot phenotypes in hyd2/fk, consistent with evidence from the ethylene assays that abnormally high levels of ethylene biosynthesis are not responsible for the observed phenotypes; in contrast, hyd1 seedlings showed some phenotypic rescue of root growth by aminoethoxyvinylglycine (data not shown). These data suggest that the hyd2/fk mutants are defective in ethylene signal transduction rather than ethylene biosynthesis, although we cannot exclude the possibility that the mutants produce locally high ethylene levels in specific tissues that are not detectable in the assays.
Auxin Signaling
To investigate possible defects in auxin signaling, we analyzed the expression of GUS fusion genes driven by gene promoters previously demonstrated to be responsive to auxin. DR5::GUS and ACS1::GUS, each of which marks auxin-responsive cells in the shoot and hypocotyl, were introduced into both the hyd1 and the hyd2/fk mutant backgrounds by crossing. In the wild-type seedling, ACS1::GUS was expressed in developing leaves and in the endodermal layer from the hypocotyl to the root expansion zone (Figures 5A and 5B) and at the site of lateral root initiation (Rodrigues-Pousada et al., 1999). ACS1::GUS was upregulated by auxin in the shoot and upregulated by ethylene and downregulated by auxin in the root. Expression did not extend into the meristem and the root tip. DR5::GUS expression in the shoot was seen in the hydathodes and developing vasculature only and showed a “maximum” in the root meristem region in the columella initials (Sabatini et al., 1999).
Figure 5.
hyd Mutants Show Altered Gene Expression.
(A) to (E) ACS1::GUS expression in the wild type 3 days after germination ([A] and [B]), in hyd2 3 days after germination (C), in hyd2 6 days after germination (D), and in hyd2 9 days after germination (E). Magnification ×5 (A) and ×200 (B) to (E).
(F) to (I) DR5::GUS expression in the wild type 3 days after germination (F), in hyd2 3 days after germination (G), in hyd2 6 days after germination (H), and in hyd2 9 days after germination (I). Magnification ×100.
GUS activity in homozygous hyd mutants containing the gene fusions was localized by histochemistry up to 20 days after germination. Expression was abnormal in a developmentally regulated way, with the most dramatic changes seen in hyd2/fk. In hyd2, ACS1::GUS expression was normal in the root until day 6 after germination, when strong GUS activity was seen throughout the entire root, in all radial layers and extending into the meristem and the root tip (Figure 5D). By day 9, this ectopic expression was downregulated, and it was normal for the rest of development (Figure 5E). In hyd1, ACS1::GUS was expressed in a pattern similar to that seen in the wild type, although there was more expression in the shoot. Similarly, in hyd1, the expression of DR5::GUS was normal in the root throughout development. hyd2 showed normal expression until days 12 to 14, but after this stage, the expression was reduced and shifted distally to the meristem; by day 18, there was no expression in the primary root tip (Figure 5). These observations are consistent with altered auxin distribution, altered distribution of auxin-perceptive cells, and/or altered ethylene signaling in the mutants.
To determine whether the hyd mutants exhibit altered responses to auxin, mutant and wild-type seedlings were grown in the presence of exogenous 2,4-D (100 nM and 1 μM) and naphthylacetic acid (1 μM). It was found that both hyd1 and hyd2/fk mutants underwent a callusing response at these concentrations of both auxins within 3 to 5 days (Figures 6B and 6C), whereas the wild-type seedlings produced more lateral roots at these concentrations (Figure 6A) but showed evidence of callusing only at 10- to 50-fold higher auxin concentrations. In hyd1 and hyd2/fk mutants containing DR5::GUS and grown similarly in the presence of auxins, the entire plant/callus stained very strongly for GUS (Figure 6H). In contrast, auxin-treated wild-type seedlings showed no dramatic upregulation of GUS activity, except specifically in emerging lateral roots at the higher 2,4-D concentration (Figure 6E). The observed callusing response and enhanced DR5::GUS activity of the mutants suggest that the mutants either are more sensitive to auxins or take up more exogenous auxins than the wild type.
Figure 6.
hyd Mutants Show Altered Responses to Auxin.
(A) to (C) Response of wild-type (A), hyd1 (B), and hyd2 (C) seedlings on 1 μM naphthylacetic acid.
(D) Wild-type seedlings transgenic for DR5::GUS treated with 1 μM 2,4-D (left) and 1 μM 2,4-D plus 10 μM 1-NOA (right).
(E) and (F) Roots of wild-type seedlings transgenic for DR5::GUS treated with 1 μM 2,4-D (E) and 1 μM 2,4-D plus 10 μM 1-NOA (F).
(G) to (I) hyd2 (G) and hyd2::DR5-GUS transgenic plants treated with 1 μM 2,4-D (H) and 1 μM 2,4-D plus 10 μM 1-NOA ([I]).
(J) hyd2 axr3 root 18 days after germination showing lack of root hairs and less defective cellular organization.
(K) Cleared hyd1 axr1-12 leaves showing rescue of petiole length, morphology, and vascular pattern (cf. with Figure 1E).
Magnification ×4 (A), ×10 ([B] and [C]), ×6 (D), ×16 ([E] and [F]), ×15 ([G] and [I]), ×18 (H), ×40 (J), and ×10 (K).
To determine whether the observed response of the hyd/fk mutants to auxin might be caused by increased cell permeability, mutant and wild-type seedlings containing DR5:: GUS were grown in the presence of 2,4-D in the presence and absence of the auxin transport inhibitor 1-naphthoxyacetic acid (1-NOA). 2,4-D uptake requires an influx carrier (Delbarre et al., 1996), and 1-NOA inhibits intracellular uptake of 2,4-D, most likely by inhibiting AUX1 function, and has been found to inhibit cellular responses to 2,4-D (Parry et al., 2001) (Figure 6D). Seedlings were grown for 3 days after germination in the presence of 10 μM 1-NOA and then transferred to 10 μM 1-NOA plus 1 μM 2,4-D for another 3 days. In both the presence and absence of 1-NOA, the hyd1 and hyd2/fk mutants exhibited a callusing response (Figures 6G to 6I), demonstrating sensitivity to the hormone and also an upregulation of GUS activity that was similar to the upregulation when grown in the presence of 2,4-D alone.
On the other hand, wild-type seedlings showed reduced DR5::GUS activity when grown on 1-NOA plus 2,4-D (Figure 6F) compared with 2,4-D alone (Figure 6E). This finding suggests either that 2,4-D can bypass the 1-NOA–sensitive auxin influx carrier in the hyd/fk mutants, but not in the wild type, or that a low level of inward leakage of 2,4-D through the plasma membrane is sufficient to trigger auxin responses in the mutants but not in wild-type seedlings. These data suggest that some aspects of the hyd mutants, such as altered vascular patterning or defective root development, might be the result of altered (e.g., ectopic) auxin distribution or signaling.
To investigate whether we could rescue the mutants by blocking auxin action, we performed double mutant analysis between hyd1 and hyd2/fk and the auxin-resistant mutants axr1-12 and axr3-1, which are defective in the correct response to auxins (Lincoln et al., 1990; Rouse et al., 1998). Both axr1-12 and axr3-1 have a reduced frequency of root hairs. All double mutants had longer roots and much reduced root hair production compared with the hyd1 and hyd2/fk single mutants (Figure 6J). The hyd1 axr1-12 and hyd2 axr1-12 double mutants showed partial rescue of leaf development, with longer petioles, less disrupted vascular patterning, and a smoother leaf surface (Figure 6K), and were relatively long-lived. There was no obvious effect of the axr3-1 mutation on the development of the hyd1 and hyd2 mutant shoots. These results indicate that (1) introducing an auxin-resistant axr1-12 background into the hyd mutants ameliorates the abnormal leaf phenotype, and (2) both axr1-12– and axr3-1–dependent auxin resistance can reduce the root defects in the hyd mutants.
hyd/fk Mutants Exhibit Correct Trafficking of PIN Proteins
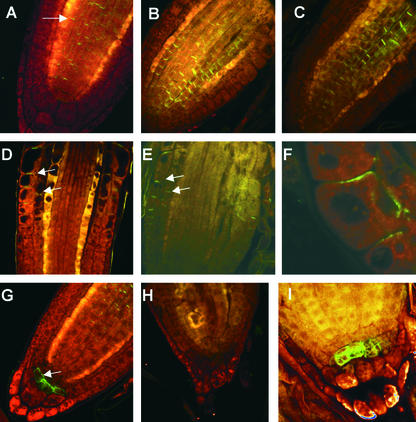
It is possible that the observed defective sterol composition in the hyd1 and hyd2/fk mutants could interfere with correct membrane trafficking to translocate regulatory (e.g., receptor or transport) proteins to the plasma membrane or other cellular compartments. For example, the PIN proteins are predicted components of the auxin efflux carrier system and are localized precisely to mediate polar auxin transport. As a model to study vesicle trafficking, we determined whether the mutants are defective in the trafficking of PIN1, PIN2, and PIN3 by immunolocalization. In the wild-type Arabidopsis root, PIN 1 localized to the basal membranes of vascular cells (Gälweiler et al., 1998) (Figure 7A), whereas PIN2 localized to the apical membranes in the root cortex and epidermis (Muller et al., 1998) (Figure 7D). PIN3 was localized to the first tier of columella cells distal to the columella initials at all faces of the plasma membrane (J. Friml and K. Palme, unpublished data) (Figure 7G).
Figure 7.
PIN Localization Is Unaffected in hyd Mutants.
(A) to (C) PIN1 localization (arrow) in wild-type (A), hyd1 (B), and hyd2 (C) roots 6 days after germination.
(D) to (F) PIN2 localization (arrows) in wild-type (D), hyd1 (E), and hyd2 (F) roots 6 days after germination.
(G) to (I) PIN3 localization (arrow) in wild-type (G), hyd2 ([H]; 14 days after germination), and hyd2 etr1-1 ([I]; 14 days after germination) roots 6 days after germination.
Magnification ×200.
It was observed that PIN1 and PIN2 proteins were localized normally in hyd1 and hyd2/fk roots (Figures 7A to 7F). Similarly, PIN3 localization was identical to that in the wild type (Figure 7G) until approximately day 9 after germination (data not shown). However, between days 9 and 12, PIN3 localization shifted to the columella initials, and by day 14 after germination, it disappeared in parallel with the disorganization of the root meristem and columella (Figure 7H). PIN3 localization was rescued in seedlings treated with silver or in hyd2 etr1 double mutants, as demonstrated by the fact that the root meristem and columella patterning was rescued (Figure 7I).
DISCUSSION
Sterols Are Required for Correct Plant Morphogenesis
Sterols are known to contribute to two biosynthetic pathways in plants: the BRs and the bulk sterols (Hartmann, 1998; Clouse, 2000). A number of mutants in plant sterol biosynthetic genes have been identified, most of which have been found to encode enzymes required for the synthesis of BRs. Such mutants typically are dwarfed and can be rescued phenotypically by exogenous BR application (Szekeres et al., 1996; Choe et al., 1999a, 1999b). However, recent studies identified a sterol mutant, fk, that encodes a defective sterol C14 reductase and is embryonic defective (Jang et al., 2000; Schrick et al., 2000). Interestingly, fk mutants are deficient in BRs but cannot be rescued by their exogenous application. Seedlings homozygous for mutant alleles of this gene are very similar phenotypically to the previously described hyd1 and hyd2 mutants, which also cannot be rescued by BR application (Topping et al., 1997). Here, we show that the hyd2 mutant is allelic to fk.
To further strengthen the view that sterols are required for plant development, we demonstrate here that the HYD1 gene encodes the sterol biosynthetic enzyme Δ8-Δ7 sterol isomerase, which is found one step downstream of the C14 reductase (Grebenok et al., 1998). This proximity of the enzymes in the pathway presumably accounts for the similarities of sterol profiles in the hyd1 and hyd2/fk mutants (Table 1) and the similar phenotypes. The subtle differences in sterols may account for the observed developmental differences between the mutants. HYD1 occurs upstream of SMT2, cosuppression of which leads to high campesterol and low sitosterol contents as well as reduced plant growth and fertility, which also cannot be rescued by BR application (Schaeffer et al., 2001).
The HYD1 sterol isomerase has been cloned previously and demonstrated to have sterol isomerase enzyme activity (Grebenok et al., 1998). No corresponding developmental mutant phenotypes have been identified elsewhere in yeast, plants, or animals. It exhibits regions of strong homology with mammalian EBP, a membrane-bound σ1 factor receptor with orthologs identified previously in mouse (Silve et al., 1996a), human (Hanner et al., 1995; Jbilo et al., 1997), and guinea pig (Hanner et al., 1995). EBP is resident in the endoplasmic reticulum and the nuclear envelope (Jbilo et al., 1997; Dussossoy et al., 1999).
Both the Arabidopsis and mammalian sterol isomerase genes complement the yeast erg2 mutant defective in this enzyme (Silve et al., 1996a; Grebenok et al., 1998), although EBP has no significant structural homology with yeast sterol isomerase and was thought previously to be unique to mammals. However, EBP is now believed to be the mammalian Δ8-Δ7 sterol isomerase. The HYD1 protein, in common with EBP, has four predicted membrane-spanning domains and a consensus endoplasmic reticulum retrieval domain at the C terminus, K(X)KXX. There are several other conserved sequence domains, with one being very highly conserved (60% identical and 91% similar over amino acids 94 to 115). The activity of the HYD1 promoter in transgenic embryos is consistent with the embryonic phenotype of the hyd1 mutant.
EBP has high affinity for emopamil, a phenylalkylamine Ca2+ antagonist (Hanner et al., 1995), and for trifluoperazine, an inhibitor of calcium signaling pathways in animals and plants. It has been proposed that the effect on calcium signaling is mediated via alterations to membrane sterol composition that affect correct calcium transport across membranes or, possibly, calmodulin action at the membrane (Silve et al., 1996a). The hypersensitivity of the smt1 mutant of Arabidopsis to calcium, which has reduced levels of sitosterol and other C24 sterols (Diener et al., 2000), correlates with defective root growth and may similarly reflect altered membrane permeability to, or perception of, this ion.
Pharmacological inhibition of the yeast C-14 sterol reductase ERG24 (Marcireau et al., 1990) and mammalian Δ8-Δ7 sterol isomerase in recombinant yeast and in mammalian cells (Silve et al., 1996b) leads to cell cycle arrest. In contrast to the essential yeast ERG24, ERG2 (yeast sterol isomerase) has been found to be functionally redundant (Ashman et al., 1991; Lees et al., 1995). Therefore, the hyd1 mutation provides the first direct evidence of a requirement for Δ8-Δ7 sterol isomerase in eukaryotic morphogenesis.
Molecular Basis for hyd/fk Defects
A key question is how defects in sterol biosynthesis are linked to the pleiotropic developmental defects seen in the hyd1 and hyd2/fk mutants. A number of possibilities can be considered. It is possible that BRs, or sterols with hormone-like activities missing in the mutants, are required during development. For example, specific sterols that are reduced in the mutants may have signaling roles directly or indirectly, such as through interaction with sterol binding domains in proteins such as PHABULOSA, which is known to play an important role in Arabidopsis development (McConnell et al., 2001).
Furthermore, fk mutants have been shown to accumulate low levels of BRs, which could contribute to the dwarfed fk phenotype (Jang et al., 2000; Schrick et al., 2000). However, neither hyd1 nor hyd2/fk mutants can be rescued by the exogenous application of BRs, although mutants defective in enzymes closer to the branch of the pathway that is committed specifically to BR biosynthesis can. This suggests that the hyd/fk mutants, which are defective in bulk sterol biosynthesis, with low campesterol as well as low sitosterol and high stigmasterol, are defective in the perception or signal transduction of exogenous BRs, a process mediated by the membrane-bound receptor kinase BRI1 (Li and Chory, 1997).
Additionally, it was not possible to rescue hyd/fk mutants by the exogenous application of bulk sterols. Although the limited uptake or transport of sterols could prevent rescue, this may be unlikely because exogenous BRs can rescue BR mutants (Clouse, 2000). It also is possible that the embryonic defects would not be corrected by sterol application at the seedling stage, although HYD1 is expressed in seedlings (J. Topping, M. Pullen, X.-L. Zhang, and K. Lindsey, unpublished data), indicating a requirement for sterol biosynthesis after germination. However, we found no evidence of any alteration in the development of the mutant seedlings by such treatments.
Sterols could influence the function of transport vesicles and thus the trafficking of important signaling or other proteins to membrane compartments, and this process could be disrupted in sterol mutants. However, analysis of PIN protein localization indicates that their targeting to the plasma membrane is not affected detectably in either the hyd1 or the hyd2/fk mutant. This finding suggests that the defective sterol composition of the mutants does not interfere with correct vesicle transport to the plasma membrane; therefore, the observed spatial defects in auxin-mediated gene expression patterns are not caused by altered PIN localization.
We have shown that both hyd1 and hyd2/fk mutants are defective in ethylene- and auxin-mediated gene expression and cell differentiation. Both hyd1 and hyd2/fk mutants callus at lower auxin concentrations than do wild-type seedlings. The observation that 1-NOA, an inhibitor of auxin influx, can reduce 2,4-D responses in the wild type but not in hyd mutants suggests that either the NOA-sensitive component of auxin influx, likely AUX1 (Delbarre et al., 1996; Yamamoto and Yamamoto, 1998; Parry et al., 2001), is bypassed or the hyd mutants are altered in auxin sensitivity rather than increased in membrane permeability to auxin. Because 2,4-D can bypass the influx carrier, albeit at low efficiency, it is possible that in NOA-treated mutants, sufficient auxin enters the cell to elicit the callusing response because of enhanced auxin perception or signal transduction. This view is supported by the increased sensitivity of hyd mutants to naphthylacetic acid, which does not require AUX1-mediated import. The partial rescue of hyd mutant leaf morphology by the axr1 mutation and of the root defects by the axr1 and axr3 mutations, both of which lead to an inability to elicit some auxin responses (Leyser, 1998), further supports the view that the mutants exhibit ectopic auxin action.
The root hair phenotype of the mutants also suggests altered ethylene and/or auxin signaling. Interestingly, GL2:: green fluorescent protein expression in the hyd/fk mutants showed ectopic expression in hair-producing cell files (Figure 2F). Because GL2 is a transcription factor that is expressed normally in atrichoblasts of the root epidermis and is considered a negative regulator of hair formation (Masucci et al., 1996), it would be expected that its ectopic expression pattern would result in fewer, rather than more, hair-producing cell files. Because the GL2 protein has a predicted sterol binding domain (Ponting and Aravind, 1999; McConnell et al., 2001), it is possible that its activity is suppressed by a lack of correct sterol ligands in the hyd/fk mutants, resulting in a glabrous root phenotype. However, it is known that the hormones auxin and ethylene act downstream of GL2 to regulate hair formation (Masucci and Schiefelbein, 1996), and it is possible that the observed auxin and ethylene response defects in the mutants bypass the ectopic GL2 expression to promote the glabrous phenotype. The observed rescue of hyd/fk root cell patterning by the inhibition of auxin and ethylene action supports this view.
The ethylene assays show that the mutants do not detectably overproduce ethylene, and the ethylene biosynthesis inhibitor aminoethoxyvinylglycine fails to rescue the hyd2/fk mutant phenotype, with only partial rescue of hyd1 roots. We conclude that the mutants either overproduce ethylene at low levels, such as in a tissue-specific way not detectable by the assays, or exhibit altered ethylene signaling, perhaps as a result of a conformational change in the ethylene receptor(s), which is rescued by the etr1-1 mutation (which is dominant; Hall et al., 1999), or by silver, which also may modify ETR1 conformation or the signal propagation to the kinase domain of the receptor.
In summary, our studies provide strong evidence that defects in plant sterol profiles can lead to defective hormone signaling. The results of the auxin/1-NOA, aminoethoxyvinylglycine, and double mutant experiments in particular support the view that the hyd/fk mutants are defective in membrane-bound protein activity (i.e., defective auxin/ethylene receptor function) or membrane permeability to auxin. The lack of response to exogenous BRs similarly suggests defective signaling mediated by the membrane-bound BRI1. It has been shown previously that membrane sterol content can modulate the activity of membrane-bound proteins, such as the plasma membrane H+-ATPase from maize roots, a protein required for ion and auxin transport, cell wall acidification, and cell expansion (Grandmougin-Ferjani et al., 1997). Furthermore, sterols can affect ATPase activity through the modulation of fatty acyl chains and membrane fluidity (Cooke and Burden, 1990). In animal cells, cholesterol can reorganize lipid packing in the bilayer and create domains of varying lipid composition, leading to altered enzyme activity (Carruthers and Melchior, 1986).
One interesting question is why mutants defective relatively early in sterol biosynthesis (hyd1, hyd2/fk, and smt1) have severely aberrant embryogenesis, whereas mutants that affect enzymes closer to BR biosynthesis (dwf1 and dwf7) (Choe et al., 1999a, 1999b) do not. It is possible that the DWF1 and DWF7 genes are not required during embryogenesis or are redundant during that phase of development, whereas HYD1, HYD2/FK, and SMT1 are functional and nonredundant. It also is possible that, as alluded to above, hyd1, hyd2/fk, and smt1 lack sterol-based signaling molecules, or sterols required for membrane-bound signaling protein function, that are functionally distinct from BRs, and that these are essential for correct embryogenesis. The role of sterols in plant cell signaling likely will be a fruitful area of future investigation.
METHODS
Mutants and Marker Lines
The hyd1 and hyd2 mutants were identified in a screen of transgenic lines as described previously (Topping et al., 1997). Genetic crosses were performed with a stereomicroscope, using fine watchmaker's forceps to dissect floral buds, remove immature anthers, and transfer mature pollen from the male parent to the stigma. For in vitro growth studies, Arabidopsis thaliana seed were vernalized, surface-sterilized, and plated on growth medium (half-strength Murashige and Skoog [1962] medium [1/2MS10; Sigma], 1% Suc, and 3.25 g/L phytagel agar [Sigma]) as described (Topping et al., 1997). For hormone/inhibitor application experiments, seed were germinated aseptically on growth medium containing various concentrations of hormones and assayed according to the particular experiment. Tissue localization of β-glucuronidase enzyme activity was performed as described (Topping et al., 1997).
Cloning of the HYD1 Gene
The HYD1 gene was identified as being putatively T-DNA tagged by cosegregation of the mutant phenotype and T-DNA from the vector 3850:1003 (Errampalli et al., 1991). The T-DNA vector contains a NPT-II gene conferring kanamycin resistance to seedlings, which were plated on 1/2MS10 supplemented with 25 mg/L kanamycin sulfate. Sixty of 60 (100%) hyd1 seedlings were demonstrated to be kanamycin resistant, and 124 of 205 (61%) phenotypically wild-type seedlings, progeny from a selfed hyd heterozygote, were kanamycin resistant. This finding indicated that the T-DNA was present at a single genetic locus. DNA gel blot analysis on the progeny of a hyd1-2 heterozygote demonstrated that a single copy of the T-DNA was present in 25 of 25 of the heterozygotes and 0 of 10 of the segregating wild types. T-DNA flanking sequence was cloned by plasmid rescue (Behringer and Medford, 1992). Genomic DNA from an individual hyd1-2 heterozygote plant was digested with SalI to allow left border rescues.
DNA fragments were cloned in DH5α Escherichia coli cells, and a left border clone, containing ∼2.6 kb of flanking plant genomic DNA, was identified by colony hybridization. The genomic DNA was amplified by direct polymerase chain reaction using primers located in the pBR322 origin of the replication region and the left border of the T-DNA and cloned directly into the pCRII vector (Invitrogen, Carlsbad, CA). Nucleotide and deduced protein sequences were used to search for homologies in the National Center for Biotechnology Information nucleotide and protein sequence databases using the BLAST network service (Altschul et al., 1990). A corresponding Δ8-Δ7 sterol isomerase expressed sequence tag was obtained from the ABRC (Columbus, OH) and demonstrated to be a full-length cDNA. The position of the T-DNA in the gene was determined by sequence analysis of the plasmid-rescued T-DNA–genome junctions.
The full-length sterol isomerase cDNA was cloned as a transcriptional fusion under the control of either the 35S promoter of Cauliflower mosaic virus or its own promoter in the T-DNA binary vector pCIRCE (a gift from M. Bevan, John Innes Centre, Norwich, UK) containing either the NPT-II gene for kanamycin selection or the phosphomannose isomerase gene (Joersbo et al., 1998) for retransformation of kanamycin-resistant mutants. Seed transformed with constructs containing the phosphomannose isomerase gene were selected on 1/2MS10 plates supplemented with 4 g/L d-Man.
Wild-type Columbia and hyd1 heterozygotes were transformed by the floral dip method (Clough and Bent, 1998) using Agrobacterium tumefaciens strain C58C3 (Dale et al., 1989) and also by the in vitro root transformation method using Agrobacterium strain LBA4404 (Clarke et al., 1992). The morphologies of the resulting transformants were identical in either transformation procedure. hyd1 mutants were transformed in vitro using a modified version of the cotyledon explant method described by Schmidt and Willmitzer (1991). Whole hyd1 mutants including the roots were grown on 1/2MS10 for 14 days, chopped into ∼1-mm2 pieces, and cultured on callus-inducing medium plates without a feeder layer.
After preculture, the explants were incubated with Agrobacterium strain LBA4404 harboring either the 35S Cauliflower mosaic virus–driven sterol isomerase cDNA or 2 kb of the genomic sterol isomerase promoter region driving the sterol isomerase cDNA. Agrobacteria were grown in Luria-Bertani medium supplemented with 10 mg/L acetosyringone. After Agrobacterium inoculation, the hyd1 explants were transferred to shoot-inducing medium (Schmidt and Willmitzer, 1991) without Man/hygromycin selection. Selection was omitted to promote regeneration of the transformed tissue; noninfected hyd1 explants were used as a control.
For sterol feeding experiments, seedlings were germinated and grown for 21 days on 1/2MS10 supplemented with 0 to 100 μM campesterol, stigmasterol, and sitosterol (Sigma) dissolved in 1% dimethyl formamide. Sterol assays in extracts of hyd mutants and wild-type seedlings (10 days after germination) were performed by gas chromatography in triplicate pooled samples by the commercial Lipid Analytical Service at the Scottish Crops Research Institute (Dundee, UK). For ethylene assays, wild-type and mutant seedlings were germinated and grown for 7 days on 1/2MS10 and then transferred for 5 hr to sealed vials, where air was sampled and ethylene evolution was assayed, as described (Llop-Tous et al., 2000). Assays were performed on populations of five seedlings per vial, and between two and four vials were assayed for each mutant/wild-type pair. These experiments were performed in duplicate.
Immunolocalization and Microscopy
Both stained and unstained tissues were fixed for 15 min in a solution of 0.24 N HCl and 20% methanol at 57°C. This was replaced with a solution of 7% NaOH and 60% ethanol and incubated for 15 min at room temperature. Samples then were rehydrated for 5 min each in 40, 20, and 10% ethanol and infiltrated for 15 min in 5% ethanol and 25% glycerol. Samples were mounted in 50% glycerol and viewed with Zeiss Axioskop (Jena, Germany), Nikon Optiphot (Tokyo, Japan; differential interference contrast optics), or Olympus SZH10 (Tokyo, Japan) microscopes. Images were captured on Ektachrome 160T film (Kodak, Rochester, NY) and processed in Photoshop 4.0 (Adobe Systems, Mountain View, CA).
For immunolocalization of PIN proteins, tissue was fixed in 4% paraformaldehyde/MTSB (50 mM Pipes, 5 mM EGTA, 5 mM MgSO4, pH 6.9 to 7.0) under vacuum for 1 hr and washed with MTSB/Triton X-100 (0.1%) five times and then with water/Triton X-100 (0.1%) five times. Tissue was permeabilized in a 2% Driselase/MTSB solution for 30 min at room temperature and then washed with MTSB/Triton X-100 (0.1%) five times. Samples were incubated for 1 hr in 10% DMSO/Nonidet P-40 (0.5%)/MTSB, washed with MTSB/Triton X-100 (0.1%) five times, and incubated for 1 hr in 3% BSA/MTSB.
Tissue was incubated with anti-PIN primary antibody in 3% BSA/MTSB for 4 hr, washed five times in MTSB/Triton X-100 (0.1%), and then incubated with fluorescein isothiocyanate–labeled secondary antibody in 3% BSA/MTSB for 3 hr. Samples were washed five times with MTSB/Triton X-100 (0.1%) and then five times with water. Samples were mounted on slides in a glycerol-based Slowfade antifade agent (Molecular Probes, Eugene, OR) and viewed on a Leica DMIRBE TCS 4D confocal microscope (Wetzlar, Germany), with fluorescein isothiocyanate–specific detection at 530 ± 15 nm and autofluorescence-specific detection at 580 ± 15 nm. Images were captured and exported to Photoshop 4.0, in which the two images were overlaid.
For Lugol staining of roots, tissues were incubated in Lugol solution (Sigma) for 5 to 10 min and mounted in chloral hydrate (Topping and Lindsey, 1997) or 20% glycerol for microscopic analysis.
Acknowledgments
We thank Dr. Ken Feldmann for providing prospective hyd alleles, Dr. Jane Murfett for providing DR5::GUS seed, Dr. D. Van Der Straeten for providing ACS1::GUS seed, Dr. John Schiefelbein for providing GL2::GFP seed, and Dr. Ottoline Leyser for axr1-12 and axr3-1 seed. etr1 and fk seed was obtained from the Nottingham Arabidopsis Stock Centre. This work was supported by a Biotechnology and Biological Science Research Council research studentship to M.S., a Durham University studentship to M.P., and Biotechnology and Biological Science Research Council Grant 12/P02330 to J.T.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.001248.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Ashman, W.H., Barbuch, R.J., Ulbright, C.E., Jarrett, H.W., and Bard, M. (1991). Cloning and disruption of the yeast C-8 sterol isomerase. Lipids 26, 628–632. [DOI] [PubMed] [Google Scholar]
- Behringer, F.J., and Medford, J.I. (1992). A plasmid rescue technique for the recovery of plant DNA disrupted by T-DNA insertion. Plant Mol. Biol. Rep. 10, 190–198. [Google Scholar]
- Beyer, E.M.J. (1976). A potent inhibitor of ethylene action in plants. Plant Physiol. 58, 268–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carayon, P., Bouaboula, M., Loubet, J.F., Bourrie, B., Petitpretre, G., Le Fur, G., and Casellas, P. (1995). The sigma ligand 31747 prevents the development of acute graft-versus-host disease in mice by blocking IFN-gamma and GM-CSF mRNA expression. Int. J. Immunopharmacol. 17, 753–761. [DOI] [PubMed] [Google Scholar]
- Carruthers, A., and Melchior, D.L. (1986). How bilayer lipids affect membrane-protein activity. Trends Biochem. Sci. 11, 331–335. [Google Scholar]
- Choe, S., Dilkes, B.P., Gregory, B.D., Ross, A.S., Yuan, H., Noguchi, T., Fujioka, S., Takatsuto, S., Tanaka, A., Yoshida, S., Tax, F.E., and Feldmann, K.A. (1999. a). The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol. 119, 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe, S., Noguchi, T., Fujioka, S., Takatsuto, S., Tissier, C.P., Gregory, B.D., Ross, A.S., Tanaka, A., Yoshida, S., Tax, F.E., and Feldmann, K.A. (1999. b). The Arabidopsis dwf7/ste1 mutant is defective in the Δ7 sterol C-5 desaturation step leading to brassinosteroid synthesis. Plant Cell 11, 207–221. [PMC free article] [PubMed] [Google Scholar]
- Clarke, M.C., Wei, W., and Lindsey, K. (1992). High frequency transformation of Arabidopsis thaliana by Agrobacterium tumefaciens. Plant Mol. Biol. Rep. 10, 178–189. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Clouse, S.D. (2000). Plant development: A role for sterols in embryogenesis. Curr. Biol. 10, R601–R604. [DOI] [PubMed] [Google Scholar]
- Cooke, D.J., and Burden, R.S. (1990). Lipid modulation of plasma-membrane-bound ATPases. Physiol. Plant. 78, 153–159. [Google Scholar]
- Dale, P.J., Marks, M.S., Brown, M.M., Woolston, C.J., Gunn, H.V., Mullineaux, P.M., Lewis, D.M., Kemp, J.M., and Chen, D. (1989). Agroinfection of wheat: Inoculation of in vitro grown seedlings and embryos. Plant Sci. 63, 237–245. [Google Scholar]
- Delbarre, A., Muller, P., Imhoff, V., and Guern, J. (1996). Comparison of mechanisms controlling uptake and accumulation of 2,4-dichlorophenoxy acetic acid, naphthalene-1-acetic acid, and indole-3-acetic acid in suspension-cultured tobacco cells. Planta 198, 532–541. [DOI] [PubMed] [Google Scholar]
- Derocq, J.M., Bourrie, B., Segui, M., Le Fur, G., and Casellas, P. (1995). In vivo inhibition of endotoxin-induced pro-inflammatory cytokines production by the sigma ligand SR 31747. J. Pharmacol. Exp. Ther. 272, 224–230. [PubMed] [Google Scholar]
- Diener, A.C., Li, H., Zhou, W.-x., Whoriskey, W.J., Nes, W.D., and Fink, G.R. (2000). STEROL METHYLTRANSFERASE 1 controls the level of cholesterol in plants. Plant Cell 12, 853–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussossoy, D., Carayon, P., Belugou, S., Feraut, D., Bord, A., Goubet, C., Roque, C., Vidal, H., Combes, T., Loison, G., and Casellas, P. (1999). Colocalization of sterol isomerase and sigma1 receptor at endoplasmic reticulum and nuclear envelope level. Eur. J. Biochem. 263, 377–385. [DOI] [PubMed] [Google Scholar]
- Errampalli, D., Patton, D., Castle, L., Mickelson, L., Schnall, J., Feldmann, K., and Meinke, D. (1991). Embryonic lethals and T-DNA insertional mutagenesis in Arabidopsis. Plant Cell 3, 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachotte, D., Husselstein, T., Bard, M., Lacroute, F., and Benveniste, P. (1996). Isolation and characterization of an Arabidopsis thaliana cDNA encoding a Δ7-sterol-C-5-desaturase by functional complementation of a defective yeast mutant. Plant J. 9, 391–398. [DOI] [PubMed] [Google Scholar]
- Gälweiler, L., Guan, C., Muller, A., Wisman, E., Mendgen, K., Yephremov, A., and Palme, K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230. [DOI] [PubMed] [Google Scholar]
- Grandmougin-Ferjani, A., Schuler-Muller, I., and Hartmann, M.-A. (1997). Sterol modulation of the plasma membrane H+-ATPase activity from corn roots reconstituted into soybean lipids. Plant Physiol. 113, 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebenok, R.J., Ohnmeiss, T.E., Yamamoto, A., Huntley, E.D., Galbraith, D.W., and Della Penna, D. (1998). Isolation and characterization of an Arabidopsis thaliana C-8,7 sterol isomerase: Functional and structural similarities to mammalian C-8,7 sterol isomerase/emopamil-binding protein. Plant Mol. Biol. 38, 807–815. [DOI] [PubMed] [Google Scholar]
- Hall, A.E., Chen, Q.H.G., Findell, J.L., Schaller, G.E., and Bleecker, A.B. (1999). The relationship between ethylene binding and dominant insensitivity conferred by mutant forms of the ETR1 ethylene receptor. Plant Physiol. 121, 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanner, M., Moebius, F.F., Webers, F., Grabner, M., Striessnig, J., and Glossman, H. (1995). Phenylalkylamine Ca2+ antagonist binding protein. J. Biol. Chem. 270, 7551–7557. [DOI] [PubMed] [Google Scholar]
- Hartmann, M.-A. (1998). Plant sterols and the membrane environment. Trends Plant Sci. 3, 170–175. [Google Scholar]
- Hauser, M.-T., and Bauer, E. (2000). Histochemical analysis of root meristem activity in Arabidopsis thaliana using a cyclin:GUS (β-glucuronidase) marker line. Plant Soil 226, 1–10. [Google Scholar]
- Jang, J.-C., Fujioka, S., Tasaka, M., Seto, H., Takatsuto, S., Ishii, A., Aida, M., Yoshida, S., and Sheen, J. (2000). A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev. 14, 1485–1497. [PMC free article] [PubMed] [Google Scholar]
- Jbilo, O., et al. (1997). Purification and characterization of the human SR 31747A-binding protein: A nuclear membrane protein related to yeast sterol isomerase. J. Biol. Chem. 272, 27107–27115. [DOI] [PubMed] [Google Scholar]
- Joersbo, M., Donaldson, I., Kreiberg, J., Guldager Peterson, S., Brunstedt, J., and Okkels, F.T. (1998). Analysis of mannose selection used for transformation of sugar beet. Mol. Breeding 4, 111–117. [Google Scholar]
- Kallen, C.B., Billheimer, J.T., Summers, S.A., Stayrook, S.E., Lewis, M., and Strauss, J.F. (1998). Steroidogenic acute regulatory protein (StAR) is a sterol transfer protein. J. Biol. Chem. 273, 26285–26288. [DOI] [PubMed] [Google Scholar]
- Lees, N.D., Skaggs, B., Kirsch, D.R., and Bard, M. (1995). Cloning of the late genes in the ergosterol biosynthetic pathway of Saccharomyces cerevisiae: A review. Lipids 30, 221–226. [DOI] [PubMed] [Google Scholar]
- Leyser, O. (1998). Auxin signalling: Protein stability as a versatile control target. Curr. Biol. 8, R305–R307. [DOI] [PubMed] [Google Scholar]
- Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. [DOI] [PubMed] [Google Scholar]
- Lincoln, C., Britton, J.H., and Estelle, M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2, 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llop-Tous, I., Barry, C.S., and Grierson, D. (2000). Regulation of ethylene biosynthesis in response to pollination in tomato flowers. Plant Physiol. 123, 971–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcireau, C., Guilloton, M., and Karst, F. (1990). In vivo effects of fenpropimorph on the yeast Saccharomyces cerevisiae and determination of the molecular basis of the antifungal property. Antimicrob. Agents Chemother. 34, 989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci, J.D., and Schiefelbein, J.W. (1996). Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell 8, 1505–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci, J.D., Rerie, W.G., Foreman, D.R., Zhang, M., Galway, M.E., Marks, M.D., and Schiefelbein, J.W. (1996). The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122, 1253–1260. [DOI] [PubMed] [Google Scholar]
- Mattsson, J., Sung, Z.R., and Berleth, T. (1999). Responses of plant vascular systems to auxin transport inhibition. Development 126, 2979–2991. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. [DOI] [PubMed] [Google Scholar]
- Muller, A., Guan, C.H., Galweiler, L., Tanzler, P., Huijser, P., Marchant, A., Parry, G., Bennett, M., Wisman, E., and Palme, K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17, 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Parry, G., Delbarre, A., Marchant, A., Swarup, R., Perrot-Rechenmann, C., and Bennett, M. (2001). Physiological characterization of a novel class of auxin influx carrier inhibitors. Plant J. 25, 399–406. [DOI] [PubMed] [Google Scholar]
- Ponting, C.P., and Aravind, L. (1999). START: A lipid binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem. Sci. 24, 130–132. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Pousada, R., Van Caeneghem, W., Chauvaux, N., Van Onekelen, H., Van Montagu, M., and Van Der Straeten, D. (1999). Hormonal cross-talk regulates the Arabidopsis thaliana 1-aminocyclopropane-1-carboxylate synthase gene 1 in a developmental and tissue-dependent manner. Physiol. Plant. 105, 312–320. [Google Scholar]
- Rouse, D., Mackay, P., Stirnberg, P., Estelle, M., and Leyser, O. (1998). Changes in auxin response from mutations in an AUX/IAA gene. Science 279, 1371–1373. [DOI] [PubMed] [Google Scholar]
- Sabatini, S., Beis, D., Wolkenfelt, H., Murfett, J., Guilfoyle, T., Malamy, J., Benfey, P., Leyser, O., Bechtold, N., Weisbeek, P., and Scheres, B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99, 463–472. [DOI] [PubMed] [Google Scholar]
- Schaeffer, A., Bronner, R., Benveniste, P., and Schaller, H. (2001). The ratio of campesterol to sitosterol that modulates growth in Arabidopsis is controlled by STEROL METHYLTRANSFERASE 2;1. Plant J. 25, 605–615. [DOI] [PubMed] [Google Scholar]
- Schmidt, R., and Willmitzer, L. (1991). Arabidopsis regeneration and transformation (leaf and cotyledon explant system). In Plant Tissue Culture Manual: Fundamentals and Applications, Vol. A6, K. Lindsey, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–17.
- Schrick, K., Mayer, U., Horrichs, A., Kuhnt, C., Bellini, C., Dangl, J., Schmidt, J., and Jürgens, G. (2000). FACKEL is a sterol C-14 reductase required for organized cell expansion in Arabidopsis embryogenesis. Genes Dev. 14, 1471–1484. [PMC free article] [PubMed] [Google Scholar]
- Schuler, I., Milon, A., Nakatini, Y., Ourisson, G., Albrecht, A.M., Beneniste, P., and Hartmann, M.-A. (1991). Differential effects of plant sterols on water permeability and on acyl chain ordering of soybean phosphatidylcholine bilayers. Proc. Natl. Acad. Sci. USA 88, 6926–6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silve, S., Dupuy, P.H., Labit-Lebouteiller, C., Kaghad, M., Chalon, P., Rahier, A., Taton, M., Lupker, J., Shire, D., and Loison, G. (1996. a). Emopamil-binding protein, a mammalian protein that binds a series of structurally diverse neuroprotective agents, exhibits a Δ8-Δ7 sterol isomerase activity in yeast. J. Biol. Chem. 271, 22434–22440. [DOI] [PubMed] [Google Scholar]
- Silve, S., et al. (1996. b). The immunosuppressant SR 31747 blocks cell proliferation by inhibiting a steroid isomerase in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 2719–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres, M., Nemeth, K., Koncz-Kalman, Z., Mathur, J., Kauschmann, A., Altmann, T., Redei, G.P., Nagy, F., Schell, J., and Koncz, C. (1996). Brassinosteroids rescue the deficiency of CYp90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182. [DOI] [PubMed] [Google Scholar]
- Tanimoto, M., Roberts, K., and Dolan, L. (1995). Ethylene is a positive regulator of root hair development in Arabidopsis thaliana. Plant J. 8, 943–948. [DOI] [PubMed] [Google Scholar]
- Topping, J.F., and Lindsey, K. (1997). Promoter trap markers differentiate structural and positional components of polar development in Arabidopsis. Plant Cell 9, 1713–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topping, J.F., May, V.J., Muskett, P.R., and Lindsey, K. (1997). Mutations in the HYDRA1 gene of Arabidopsis perturb cell shape and disrupt embryonic and seedling morphogenesis. Development 124, 4415–4424. [DOI] [PubMed] [Google Scholar]
- Voytas, D.F., Konieczny, A., Cummings, M.P., and Ausubel, F.M. (1990). The structure, distribution and evolution of the TA1 retrotransposable element family of Arabidopsis thaliana. Genetics 126, 713–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, M., and Yamamoto, K. (1998). Differential effects of 1-naphthaleneacetic acid, indole-3-acetic acid and 2,4-dichlorophenoxyacetic acid on the gravitropic response of roots in an auxin resistant mutant of Arabidopsis, aux1. Plant Cell Physiol. 39, 660–664. [DOI] [PubMed] [Google Scholar]