Abstract
Family 3 β-d-glucan glucohydrolases are distributed widely in higher plants. The enzymes catalyze the hydrolytic removal of β-d-glucosyl residues from nonreducing termini of a range of β-d-glucans and β-d-oligoglucosides. Their broad specificity can be explained by x-ray crystallographic data obtained from a barley β-d-glucan glucohydrolase in complex with nonhydrolyzable S-glycoside substrate analogs and by molecular modeling of enzyme/substrate complexes. The glucosyl residue that occupies binding subsite −1 is locked tightly into a fixed position through extensive hydrogen bonding with six amino acid residues near the bottom of an active site pocket. In contrast, the glucosyl residue at subsite +1 is located between two Trp residues at the entrance of the pocket, where it is constrained less tightly. The relative flexibility of binding at subsite +1, coupled with the projection of the remainder of bound substrate away from the enzyme's surface, means that the overall active site can accommodate a range of substrates with variable spatial dispositions of adjacent β-d-glucosyl residues. The broad specificity for glycosidic linkage type enables the enzyme to perform diverse functions during plant development.
INTRODUCTION
β-d-Glucan glucohydrolases have been purified and characterized from barley seedlings, maize coleoptiles, soybean cultures, Acacia cells, nasturtium cells, and cultured tobacco cells (Cline and Albersheim, 1981; Nari et al., 1982; Lienart et al., 1986; Labrador and Nevins, 1989; Hrmova et al., 1996; Kotake et al., 1997; Crombie et al., 1998; Kim et al., 2000; Koizumi et al., 2000). They can hydrolyze glycosidic linkages in several β-d-glucans, in β-d-oligoglucosides containing (1→2)-, (1→3)-, (1→4)-, or (1→6)-linkages, in aryl β-d-glucosides such as 4′-nitrophenyl β-d-glucopyranoside (4NPGlc), and in some β-d-oligoxyloglucosides (Crombie et al., 1998; Hrmova and Fincher, 1998; Kim et al., 2000). The barley β-d-glucan glucohydrolases also hydrolyze cyanogenic β-d-glucosides, albeit with low activity (M. Hrmova and G.B. Fincher, unpublished data). Single Glc molecules are released from the nonreducing termini of these substrates, with retention of the anomeric configuration (Hrmova et al., 1996). Their broad substrate specificity makes it difficult to assign these higher plant β-d-glucan glucohydrolases to current Enzyme Commission classes; therefore, they have been described variously as β-d-glucan glucohydrolases, (1→3)-β-d-glucan exohydrolases, and β-d-glucosidases. Nevertheless, they can be classified according to the structural criteria of Henrissat (1991) and fall into the family 3 group of glycoside hydrolases (http://afmb.cnrs-mrs.fr/CAZY/).
The distribution of the broad-specificity β-d-glucan glucohydrolases in various tissues of monocotyledons and dicotyledons, together with the presence of expressed sequence tags in gymnosperm sequence databases (e.g., Pinus taeda), suggests that they may play a fundamental role in plant growth and development. They have been implicated in wall loosening during cell elongation, in wall remodeling, in defense reactions against fungal pathogens, in the release of Glc from wall polysaccharides as an energy source in dark-grown seedlings, and in the general recovery of Glc from different classes of polysaccharides and oligosaccharides (Hrmova and Fincher, 2001; Roulin et al., 2002).
The three-dimensional structure of the barley β-d-glucan glucohydrolase isoenzyme ExoI has been solved by x-ray crystallography to a resolution of 2.2 Å (Hrmova et al., 1998a; Varghese et al., 1999). The enzyme adopts a globular, two-domain conformation. The first domain of 357 amino acid residues has an (α/β)8 triosephosphate isomerase-barrel fold and is joined by a 16–amino acid, helix-like linker to the second domain, which consists of residues 374 to 559 arranged in an (α/β)6 sandwich. A 13-Å–deep pocket at the interface of the two domains has been identified as the active site of the enzyme. The catalytic nucleophile of the barley enzyme is amino acid residue Asp-285, and the catalytic acid/base is Glu-491 (Varghese et al., 1999; Harvey et al., 2000; Hrmova et al., 2001). The crystal structure revealed that a Glc molecule remains tightly bound in the active site pocket and is probably the product of the enzyme-catalyzed reaction that is not released after hydrolysis (Varghese et al., 1999).
The availability of procedures to crystallize the barley enzyme (Hrmova et al., 1998a), together with the previously solved three-dimensional structure (Varghese et al., 1999), allowed us to examine the structural basis for its broad specificity. In view of the structural similarities of the barley and other plant β-d-glucan glucohydrolases (Harvey et al., 2000), the barley enzyme can be used as a model to explain the broad specificity of these enzymes more generally in higher plants. In particular, a structural rationale was sought for the ability of the barley enzyme to hydrolyze the (1→2)-, (1→3)-, (1→4)-, or (1→6)-β-d–linked disaccharides sophorose, laminaribiose, cellobiose, and gentiobiose (Hrmova and Fincher, 1998).
Here, β-d-glucan glucohydrolase crystals were soaked with nonhydrolyzable S-glycoside substrate analogs of the preferred disaccharide substrate, laminaribiose, and the structure of the resulting complex was compared with that of the most slowly hydrolyzed substrate, cellobiose (Hrmova et al., 2001). The enzyme binds these analogs, but the S-glycosidic linkage is not hydrolyzed (Sulzenbacher et al., 1996; Driguez, 2001; Hrmova et al., 2001). As a result, the molecular interactions between amino acid residues on the enzyme's active site and the substrates can be defined precisely. S-Glycoside substrate analogs of sophorose and gentiobiose, the substrates that are hydrolyzed at intermediate rates, were not available for crystallography, but reliable molecular models of the corresponding enzyme/substrate complexes have been generated. In addition, a structural rationale was sought for the substrate specificity of the β-d-xylosidase–like group of family 3 enzymes from higher plants.
Substrate binding by the barley β-d-glucan glucohydrolase also was investigated by subsite-mapping techniques. The substrate binding regions of polysaccharide hydrolases are envisaged as a series of tandemly arranged subsites in which each subsite binds a single glucosyl residue of the polymeric or oligomeric substrate (Hiromi, 1970; Thoma et al., 1970). The catalytic efficiency (kcat·Km−1) values for oligoglucosides of increasing chain length have been used here to define the number of subsites and to calculate the binding affinities of individual β-d-glucosyl binding subsites in the barley β-d-glucan glucohydrolase (Hrmova et al., 1995, 1998b).
RESULTS
Catalytic Efficiencies during Hydrolysis of β-d-Oligoglucosides
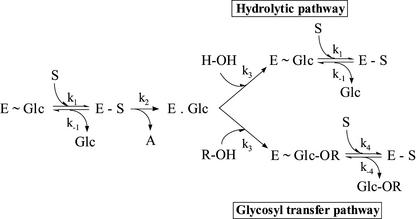
The barley β-d-glucan glucohydrolase is capable of catalyzing both hydrolytic and glycosyl transfer reactions (Figure 1), depending on substrate concentrations (Hrmova and Fincher, 1998; Kim et al., 2000). Therefore, care was exercised in steady state kinetic analyses of hydrolytic reactions to ensure that high substrate concentrations were avoided and that initial reaction rates were always measured. The kinetic parameters of hydrolysis of β-d-glucopyranosyl-(1→2)-d-glucose (sophorose or G2OG), β-d-glucopyranosyl-(1→3)-d-glucose (laminaribiose or G3OG), β-d-glucopyranosyl-(1→4)-d-glucose (cellobiose or G4OG), and β-d-glucopyranosyl-(1→6)-d-glucose (gentiobiose or G6OG) by the barley β-d-glucan glucohydrolase are shown in Table 1. The catalytic efficiency was highest for G3OG, which was identified previously as the preferred disaccharide substrate (Hrmova and Fincher, 1998), and lowest for G4OG. These values reflect earlier estimates of the relative rates of hydrolysis of the enzyme against various β-d-oligoglucosides (Hrmova and Fincher, 1998).
Figure 1.
Kinetics of Hydrolytic or Glycosyl Transfer Reactions Catalyzed by a Plant Family 3 β-d-Glucan Glucohydrolase.
After enzyme containing a noncovalently bound Glc product in the active site (E ∼ Glc) binds the first molecule of substrate (S), the Michaelis complex (E - S) is formed (k1), and the Glc product of the previous reaction is released from the active site. In the second step, the glycosidic bond is cleaved (k2), and the glycone part of the substrate becomes attached covalently to the enzyme to produce a metastable covalent glycosyl-enzyme intermediate (E . Glc). At the same time, the aglycone part of the substrate (A) is released. In the third step, the covalent glycosyl-enzyme intermediate is subjected (k3) to cleavage by a water molecule (H-OH), and a noncovalent E ∼ Glc product complex is formed, which is ready to interact (k1) with the second substrate molecule (S) to generate the next Michaelis complex (E - S), and again, the Glc molecule (Glc) is released from the active site. Alternatively, in the third step, the covalent glycosyl-enzyme intermediate (E . Glc) can be cleaved by an activated substrate molecule (R-OH), leading to a glycosyl transfer product (E ∼ Glc-OR), which remains noncovalently bound to the enzyme and is released (k4) when a second substrate molecule approaches the active site and forms the next Michaelis complex (E - S).
Table 1.
Kinetic Parameters of Barley β-d-Glucan Glucohydrolase Isoenzyme ExoI for the Hydrolysis of (1→2)-, (1→3)-, (1→4)-, and (1→6)-β-d–linked Disaccharides
| Substratea |
Km ± sd (106 M) |
kcat ± sd (sec−1) |
kcat·Km−1 (10−4 M−1· sec−1) |
ΔG‡b(kJ·mol−1) |
|---|---|---|---|---|
| G2OG | 798.7 ± 84.6 | 6.1 ± 0.6 | 0.8 | 23.1 |
| G3OG | 364.9 ± 35.0 | 11.7 ± 0.8 | 3.2 | 26.7 |
| G4OG | 2721.1 ± 179.4 | 2.4 ± 0.2 | 0.1 | 17.8 |
| G6OG | 902.9 ± 126.4 | 5.3 ± 0.6 | 0.6 | 22.4 |
α,α-Trehalose, α,β-trehalose, and β,β-trehalose were not hydrolyzed.
Calculated according to ΔG
= −RT ln [kcat·Km−1] (Fersht, 1999).
The Gibbs free energy of activation (ΔG‡) values for G4OG and G3OG were used to calculate the difference in activation energies for the glycosylation steps (Namchuk and Withers, 1995), where Δ(ΔG‡) = −RT ln [(kcat·Km−1)G3OG/(kcat·Km−1)G4OG] (Fersht, 1999). This value is ∼9 kJ·mol−1 (Table 1), which represents a small but significant difference between the two positional isomers. The nonreducing β-d-disaccharides α,α-trehalose, α,β-trehalose, and β,β-trehalose were not hydrolyzed, nor were xyloglucan or the xyloglucan oligosaccharides XXXG, with degree of polymerization 7 (DP 7), XXLG (DP 8), and XLLG (DP 9) (data not shown). Nomenclature and abbreviations for the xyloglucan oligosaccharides are as described by Fry (1995).
Glycosyl Transfer Reaction in the Presence of 4NPGlc
When incubated with 4NPGlc at concentrations greater than ∼10 mM, the barley β-d-glucan glucohydrolase (Hrmova and Fincher, 1998; Kim et al., 2000) and a putative family 3 soybean β-d-glucosidase (Nari et al., 1982) form various oligoglucosides through glycosyl transfer reactions. Here, close to 25% of products released by the enzyme after 8 hr of incubation with 100 mM 4NPGlc were oligomeric (Table 2). The major product was 4NP-β-gentiobioside, and the trisaccharide 4NP-β-gentiotrioside also was detected. The structures of these oligoglucosides were confirmed (data not shown) by electrospray ionization mass spectrometry and 13C-NMR spectroscopy (Hrmova et al., 1998b).
Table 2.
Properties of Glycosyl Transfer Products Synthesized by Barley β-d-Glucan Glucohydrolase Isoenzyme ExoI with 100 mM 4NPGlc
| Substrate or Reaction Product |
Chromatographic Mobilitya (RGlc) |
Abundance after 8 hr (%) |
|---|---|---|
| 4NPGlc | 2.2 | 51.5 |
| 4NP-β-laminaribioside | 1.8 | 2.7 |
| 4NP-β-cellobioside | 1.6 | 2.5 |
| 4NP-β-gentiobioside | 1.4 | 16.1 |
| 4NP-β-gentiotrioside | 1.1 | 2.4 |
| Glc | 1 | 21.8 |
| Disaccharides | 0.4–0.6 | 1.2 |
| Trisaccharides | 0.2–0.3 | 1.8 |
Mobilities of aryl-glycosides and other carbohydrates are relative to Glc mobility.
Subsite Mapping
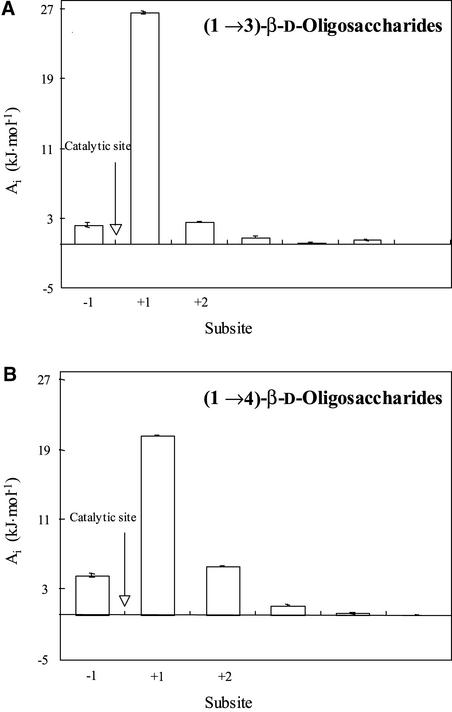
Subsite maps were determined by kinetic analysis using (1→3)-β-d-oligoglucosides and (1→4)-β-d-oligoglucosides (Table 3, Figure 2), based on the observation that the enzyme hydrolyzes G3OG at the highest rate and G4OG at the lowest rate (Table 1) (Hrmova and Fincher, 2001). In both cases, subsite binding affinities, or “transition-state interaction energies” (Malet and Planas, 1997), were highest at subsite +1, and the affinity for the penultimate glucosyl residue in (1→3)-β-d-oligoglucosides was higher than that for the same glucosyl residue in (1→4)-β-d-oligoglucosides. Binding was detected at subsite +2, particularly for the (1→4)-β-d-oligoglucosides, but binding affinities beyond subsite +2 were close to zero (Figure 2). It can be concluded from these data that there are three subsites in the active site of the barley β-d-glucan glucohydrolase.
Table 3.
Kinetic Parameters of Barley β-d-Glucan Glucohydrolase Isoenzyme ExoI for the Hydrolysis of (1→3)-β-d–Linked Oligoglucosides of DP 2 to 7 and (1→4)-β-d–Linked Oligoglucosides of DP 2 to 6
| Substrate and DP |
Km ± sd (106 M) |
kcat ± sd (sec−1) |
kcat·Km−1 ± sd (10−4 M−1·sec−1) |
|---|---|---|---|
| (1→3)-β-d–linked oligoglucosides | |||
| 2 | 364.9 ± 35.0 | 11.7 ± 0.8 | 3.2 ± 0.1 |
| 3 | 237.8 ± 29.7 | 21.1 ± 1.8 | 8.9 ± 0.4 |
| 4 | 147.4 ± 20.8 | 18.0 ± 2.3 | 12.2 ± 0.1 |
| 5 | 119.6 ± 11.8 | 15.8 ± 0.8 | 13.2 ± 0.6 |
| 6 | 93.1 ± 11.3 | 14.7 ± 1.7 | 15.8 ± 0.1 |
| 7 | 87.5 ± 12.1 | 13.7 ± 1.8 | 15.7 ± 0.1 |
| (1→4)-β-d–linked oligoglucosides | |||
| 2 | 2721.1 ± 179.4 | 2.4 ± 0.2 | 0.09 ± 0.001 |
| 3 | 343.2 ± 29.5 | 2.7 ± 0.3 | 0.8 ± 0.02 |
| 4 | 257.5 ± 37.7 | 3.1 ± 0.2 | 1.2 ± 0.1 |
| 5 | 248.0 ± 30.6 | 3.2 ± 0.2 | 1.3 ± 0.1 |
| 6 | 256.6 ± 48.8 | 3.3 ± 0.2 | 1.3 ± 0.1 |
Figure 2.
Subsite Maps of Barley β-d-Glucan Glucohydrolase Determined during Hydrolysis of (1→3)-β-d–Linked (A) and (1→4)-β-d–Linked (B) Oligoglucosides.
Arrows indicate the positions of catalytic amino acids. Subsites are labeled from −1 to +2, where the −1 subsite binds a nonreducing glucosyl residue of the substrate. Subsite affinities (Ai) were calculated using kcat·Km−1 values (Table 3) as described previously (Hrmova et al., 1998b).
Synthesis of 4-NP S-(β-d-Glucopyranosyl)-(1→3)- (3-Thio-β-d-Glucopyranosyl)-(1→3)-β-d-Glucopyranoside
The molecular and structural analyses of substrate specificity of the barley β-d-glucan glucohydrolase required oligomeric substrates that would be bound in the active site of the enzyme but that would resist hydrolysis and thus allow the collection of x-ray diffraction data of the enzyme/substrate com-plexes. A nonhydrolyzable (1→4)-β–linked substrate analog 4I,4III,4V-S-trithiocellohexaose (G4SG4OG4SG4OG4SG) was synthesized previously (Driguez, 2001) and was used to examine the S-cellobioside/enzyme complex (Hrmova et al., 2001). Here, the (1→3)-β–linked substrate analog 4-NP S-(β-d-glucopyranosyl)-(1→3)-(3-thio-β-d-glucopyranosyl)-(1→3)-β-d- glucopyranoside (4NP-G3SG3OG; Figure 3A) was synthesized using “glycosynthase” methodology (Mackenzie et al., 1998; Malet and Planas, 1998; Fort et al., 2000). A mutant barley (1→3)-β-d-glucan endohydrolase isoenzyme GII (E231G), in which the catalytic nucleophile has been altered to a Gly residue, acts as a highly efficient glycosynthase for the generation of (1→3)-β-d-glucan polymers (Hrmova et al., 2000).
Figure 3.
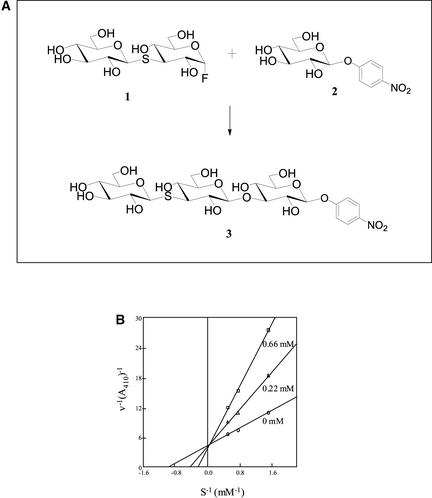
Synthesis of the Trisaccharide 4NP-G3SG3OG (Structure 3) Catalyzed by Barley (1→3)-β-d-Endohydrolase Isoenzyme GII E231G Catalytic Nucleophile Mutant Enzyme from 3-Thio-α-Laminaribiosyl Fluoride (Structure 1) and 4NPGlc (Structure 2) (A), and Double Reciprocal (Lineweaver-Burk) Plot of the Inhibition of the Barley β-d-Glucan Glucohydrolase by 4NP-G3SG3OG (B).
Using this mutant E231G (1→3)-β-d-glucan glycosynthase, thiolaminaribiosyl fluoride was condensed with 4NPGlc to generate the trisaccharide 4NP-G3SG3OG (Figure 3A) in 50% yield. The condensation reaction with thiolaminaribiosyl fluoride was less efficient than for other syntheses using glycosynthases, for which yields of 80 to 100% have been reported (Mackenzie et al., 1998; Fort et al., 2000). Nevertheless, NMR analysis of the product clearly indicated that the newly formed linkage was in the β-anomeric configuration (1H δ 4.45, J 7.5 Hz) and joined to C3 of the d-glucopyranosyl unit bearing the aromatic 4NP aglycone (13C δ 86.3) (Bock et al., 1984). Further proof of its identity was obtained by electrospray ionization mass spectrometry and by x-ray crystallography of β-d-glucan glucohydrolase crystals soaked with the synthesized 4NP-G3SG3OG.
In using these S-glycosides for structural studies, it is acknowledged that the geometries of S- and O-glycosidic linkages are not identical and that there are differences in the C1-S(O)-C3′(4′) bond angles and the C1-S(O) and C3′(C4′)-S(O) bond lengths (Perez and Vergelati, 1984). Nevertheless, it is unlikely that the glycosidic O atom of the native substrate would be placed more than 0.35 Å away from the position of the S atom in the nonhydrolyzable substrate analogs (Driguez, 2001).
Inhibition Kinetics
The compound 4NP-G3SG3OG inhibited the barley β-d-glucan glucohydrolase isoenzyme ExoI competitively, with a Ki value of 243.2 μM (Figure 3B). This value is ∼2.5 times lower than the inhibition found with G4SG4OG4SG4OG4SG (Ki = 614.6 μM) (Hrmova et al., 2001) and parallels the findings on hydrolytic preferences (Table 1). The Ki values of thiooligosaccharide inhibitors for β-d-glycosidase hydrolases vary within the micromolar (Sulzenbacher et al., 1996; Czjzek et al., 2001; Fort et al., 2001; Hrmova et al., 2001) (Figure 3B) and millimolar (Moreau et al., 1996; Reverbel-Leroy et al., 1998) ranges.
The Ki values of both inhibitors were used to calculate the difference in contributions to binding free energies Δ(ΔG‡) of the two inhibitors according to Δ(ΔG‡) = −RT ln [(Ki)G4SG4OG4SG4OG4SG/(Ki)4NP-G3SG3OG] (Fersht, 1999). The value obtained was ∼2.4 kJ·mol−1.
Crystal Structure of the S-Laminaribioside/β-d-Glucan Glucohydrolase Complex
The S-glucosyl substrate analog 4NP-G3SG3OG was diffused into crystals of the barley β-d-glucan glucohydrolase. The three-dimensional structure of the enzyme/S-laminaribioside complex was solved subsequently to 2.40 Å resolution with an R factor of 20.08%, using the rigid body refinement technique (Table 4). The coordinates have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb/; Berman et al., 2000).
Table 4.
Data Collection and Refinement Statistics of the Three-Dimensional Structure of Barley β-d-Glucan Glucohydrolase with Bound S-Laminaribioside Moiety
| Measurement | S-Laminaribioside/Complex |
|---|---|
| Resolution (Å) | 50.0–2.40 |
| Unit cell dimensions; a = b, c (Å) | 100.533; 181.959 |
| Total observations | 176,916 |
| Unique observations | 34,896 |
| Multiplicity | 5.18 |
| Rmergea (%) | 8.7; 27.4b |
| <I /σ/ (I)> | 7.8 |
| Data completeness (%) | 93.9; 85.5b |
| Rworkc (%) | 20.08 |
| Rfreec (%) | 24.96 |
| Root mean square bonds (Å); angles (o) |
0.01; 1.3 |
| Luzzati coordinate error (Å) | 0.30 |
Rmerge = 100 [Σ(Ii − <I>)2/ΣIi2] summed over all independent reflections.
For the highest resolution shell (2.46 to 2.40 Å).
Represents ∼10% of the data.
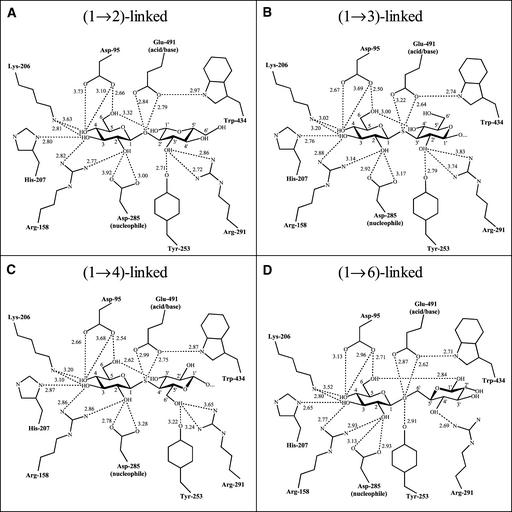
The Glc molecule that remains bound in the active site pocket of the enzyme after hydrolysis (Varghese et al., 1999) was displaced by the substrate analog, and the two nonreducing terminal (1→3)-β-glucosyl residues were defined clearly in the difference Fourier electron density map. These two residues occupy binding subsites −1 and +1. The remainder of the substrate analog molecule was disordered and therefore not visible in the electron density map (Figures 4C and 5B) because it probably projects from the surface of the enzyme without extensive molecular binding to the enzyme. For this reason, the structure is referred to as an S-laminaribioside/enzyme complex. The data showed that the enzyme was fully occupied with the S-laminaribioside moiety.
Figure 4.
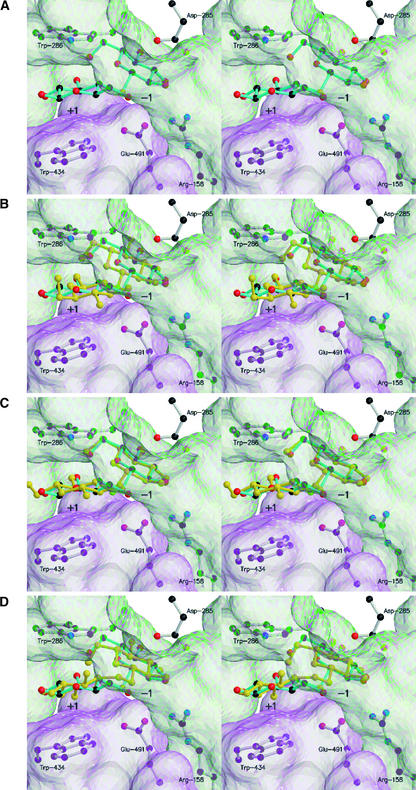
Stereo Representation of the Active Site of Barley β-d-Glucan Glucohydrolase with Bound S-Cellobioside Moiety (A), and the Superposed S-Cellobioside Moiety with G2OG (B), S-Laminaribioside Moiety (C), and G6OG (D).
A MOLSCRIPT (Kraulis, 1991) diagram of the S-cellobioside moiety is shown in cyan, and the superposed G2OG, S-laminaribioside moiety, and G6OG are shown in yellow. Fluorescent green indicates glycosidic O or S atoms in the superposed G2OG, S-laminaribioside moiety, and G6OG. Transparent green and magenta represent the molecular surfaces (Nicolls et al., 1991) of domains 1 and 2, respectively. Black, red, blue, and yellow spheres represent C, O, N, and S atoms, respectively. The structures were superposed over the Cα atoms of Asp-95, Phe-144, Arg-158, Lys-206, His-207, Glu-220, Tyr-253, Asp-285, Trp-286, Glu-287, Arg-291, Met-316, Trp-434, and Glu-491, with root mean square deviations in the Cα chain of 0.995 Å for G2OG and G4SG, 0.158 Å for G3SG and G4SG, and 1.062 Å for G6OG and G4SG. Subsites −1 and +1 are indicated. To improve the clarity of the diagrams, only amino acid residues Arg-158, Asp-285, Trp-286, Trp-434, and Glu-491 are shown. The entrance to the active site is located toward the bottom right corner. This figure is best viewed using a three-dimensional stereo viewer.
Figure 5.
Bonding Interactions of Barley β-d-Glucan Glucohydrolase with G2OG (A), S-Laminaribioside Moiety (B), S-Cellobioside Moiety (C), and G6OG (D).
Ligands are shown in the 4C1 conformation with atomic numbering of the C atoms. Dashed lines indicate hydrogen bonding interactions between the ligands and amino acid residues. All distances are expressed in angstroms and are drawn to scale where possible.
The glucopyranosyl residue at subsite −1 adopts the low-energy 4C1 conformation, without any apparent ring distortion, and is held in position by extensive hydrogen bonding interactions with six amino acid residues on the enzyme surface (Figures 4C and 5B). The C1-S-C3′ bond angle is 102.45°, and the C1-S and S-C3′ bond lengths are both 1.81 Å. The heteroglycosidic S atom is located 2.64 Å from the Oɛ1 of the catalytic acid/base Glu-491 (Figure 5B), again indicating that Glu-491 is the catalytic acid/base. The glucopyranosyl residue at subsite +1 is held by hydrophobic π-stacking interactions with Trp-286 and Trp-434 and by hydrogen bonds between C2′OH and Tyr-253 and Arg-291 (Figures 4C and 5B). More specifically, the glucopyranosyl residue at subsite +1 is clamped between the two Trp residues that form the entrance of the substrate binding pocket (Varghese et al., 1999; Hrmova et al., 2001). Furthermore, the C1-S-C3′-C4′ dihedral angle is 128.05°, compared with the dihedral angle of crystalline laminaribiose of 77.71° (Noguchi et al., 1992). Thus, the glucopyranosyl ring of the bound substrate analog at subsite +1 is rotated and translated substantially; the difference between the torsion angles is ∼50°.
Comparison of the S-Laminaribioside/ and S-Cellobioside/β-d-Glucan Glucohydrolase Complexes
The three-dimensional structure of the S-cellobioside/enzyme complex was solved previously (Hrmova et al., 2001) using the nonhydrolyzable substrate analog G4SG4OG4SG-4OG4SG (Driguez, 2001). As in the S-cellobioside/enzyme complex, only the two glucosyl residues at the nonreducing end of the analogs were visible in the electron density maps (Figure 4A). No ring distortion could be detected (Hrmova et al., 2001). It was then possible to compare directly the three-dimensional conformations of the S-laminaribioside/enzyme and S-cellobioside/enzyme complexes to provide a structural rationale for the broad specificity of the enzyme and for the differences in hydrolytic efficiencies (Table 1).
For both S-substrate analogs, the glucopyranosyl residues that are bound at subsite −1 are in almost identical positions (Figures 4C and 5B; cf. Figure 5C). The only differences are small changes in hydrogen bonding distances between the OH groups of the bound substrate analogs and two (Asp-95 and Lys-206) of the six amino acid residues involved in binding at subsite −1 (Figures 5B and 5C). These distances are shorter with the S-laminaribioside moiety than with the S-cellobioside moiety. In contrast, the glucopyranosyl residues of the S-laminaribioside and S-cellobioside moieties that occupy subsite +1 are located between the Trp-286 and Trp-434 residues, but in significantly different positions.
In the case of the S-laminaribioside moiety, the more hydrophobic β-face of the glucopyranosyl residue (apolar face) at subsite +1 is geometrically complementary with the pyrrole ring of Trp-286, whereas the more hydrophilic α-face of the glucopyranosyl residue (polar face) sits over the phenyl ring of Trp-434. In the S-cellobioside moiety, the hydrophilic α-face and the hydrophobic β-face of the glucopyranosyl residue at subsite +1 are in contact with the pyrrole ring of Trp-286 and the phenyl ring of Trp-434, respectively (Figure 6). The β-face versus α-face designation of the glucopyranosyl ring in (1→4)-β-d–linked glycoside polymers is based on a clockwise versus an anti-clockwise numbering of the carbons, respectively. Thus, the β-face of a glucopyranosyl ring is slightly more hydrophobic than the α-face (Johnston et al., 1988). Additionally, differences of ∼0.5 Å are observed in hydrogen bond lengths between NH1 of Arg-291 and C2′OH of the S-laminaribioside moiety and between NH1 of Arg-291 and C6′OH of the S-cellobioside moiety.
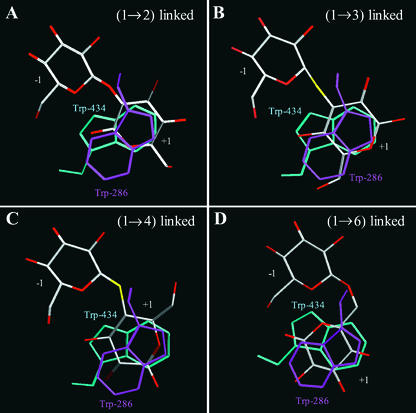
Figure 6.
Positions of G2OG (A), S-Laminaribioside Moiety (B), S-Cellobioside Moiety (C), and G6OG (D) in the Active Site of the Barley β-d-Glucan Glucohydrolase with Respect to the Two Trp Amino Acid Residues That Constitute Binding Subsite +1.
Bound carbohydrates and carbohydrate moieties are shown in atom colors, and the stacked Trp-286 (magenta) and Trp-434 (cyan) amino acid residues at subsite +1 are rotated to the positions where their pyrrole (Trp-286) and phenyl (Trp-434) portions overlap. Substrate binding subsites −1 and +1 are marked.
Another difference between the bound substrate analogs is that the interresidue hydrogen bond between C6OH and C3′OH is much shorter (2.62 Å) in the S-cellobioside/enzyme complex than between C6OH and C4′OH (3.00 Å) of the S-laminaribioside/enzyme complex (Figures 5B and 5C). These values can be compared with a C6OH-C3′OH distance of 3.12 Å in crystalline cellobiose (Chu and Jeffrey, 1968) and with a C6OH-C4′OH distance of 3.30 Å in crystalline laminaribiose (Noguchi et al., 1992). The crystallographic data indicate that the flexibility of the bound S-laminaribioside and S-cellobioside moieties could be relatively high. Furthermore, the difference in torsion angles C1-O(S)-C4′-C3′ of the two glucopyranosyl rings in the bound S-cellobioside moiety and crystalline cellobiose is only ∼7°, compared with a difference of ∼50° between the torsion angles C1-O(S)-C3′-C4′ in the S-laminaribioside moiety and crystalline laminaribiose.
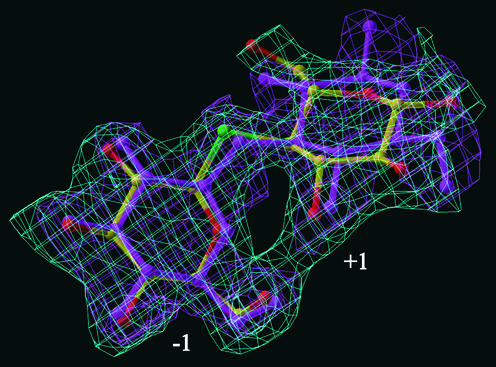
The overall differences in conformations of the glucopyranosyl rings in the bound S-laminaribioside and S-cellobioside moieties are illustrated further by superposing the two difference electron density maps (Figure 7). Again, the close coincidence of binding of the two substrate analogs at subsite −1 is evident. On the other hand, the noncoincidence of electron densities at subsite +1 indicates small, but significant, differences between the two rings at this subsite (Figure 7).
Figure 7.
Electron Density Map of the S-Laminaribioside Moiety Superposed over the −1 Subsite of the S-Cellobioside Moiety in the Active Site of the Barley β-d-Glucan Glucohydrolase.
The derived 2|Fo|-|Fc| and |Fo|-|Fc| Fourier syntheses are contoured at 1.0σ and 1.15σ for the S-cellobioside and S-laminaribioside moieties, respectively. The S-cellobioside moiety is represented in atom colors and its associated electron density map is shown in cyan, whereas the S-laminaribioside atoms and its associated electron density map are shown in magenta. The figure was prepared using “O” (Jones et al., 1991).
Molecular Modeling of G2OG and G6OG in the Active Site
The possible binding conformations of these oligoglucosides were determined by molecular modeling, based on the crystal structures of sophorose (G2OG) (Ikegami et al., 1995) and gentiobiose (G6OG) (Rohrer et al., 1980), and the structure of the barley β-d-glucan glucohydrolase (Hrmova et al., 2001). Clearly, there are interpretative limitations associated with molecular modeling based on crystal structures of the disaccharide substrates, and these were exemplified by the large difference between torsion angles observed by x-ray crystallography in the bound S-laminaribioside substrate analog and those in crystalline laminaribiose, as noted above.
Within these interpretative limitations, the modeled structures with G2OG and G6OG again indicated that the glucopyranosyl residues at subsite −1 were held in place by hydrogen bonding interactions with the six key amino acid residues and were located in almost exactly the same positions as the corresponding residues in the S-laminaribioside/enzyme and S-cellobioside/enzyme complexes (Figures 4B, 4D, 5A, and 5D). Differences of ∼1 Å were observed in the lengths of the hydrogen bonds from Oδ1 of Asp-95 to C4OH and Oδ2 of Asp-285 to C2OH for G2OG. Similar differences were observed in the lengths of hydrogen bonds from NH1 and NH2 of Arg-291 to C3′OH of G2OG and to C4′OH of G6OG. At the +1 subsite, again, the glucopyranosyl rings were located between the two hydrophobic Trp-286 and Trp-434 residues, but in slightly different positions (Figure 6).
For G2OG, the more hydrophilic α-face of the glucopyranosyl ring at subsite +1 was superposed with the pyrrole region of Trp-286, and the more hydrophobic β-face was superposed with the phenyl region of Trp-434 (Figure 6A). During the molecular modeling process, the extra rotatable bond between the two glucopyranosyl rings in the G6OG substrate resulted in the outward displacement of the subsite +1 glucopyranosyl ring from between the two Trp residues (data not shown). However, preliminary crystallographic evidence now indicates that the glucopyranosyl ring of G6OG that occupies subsite +1 is aligned between the pyrrole ring of Trp-286 and the pyrrole/phenyl region of Trp-434 (M. Hrmova, R. De Gori, J.N. Varghese, and G.B. Fincher, unpublished data), presumably because of the flexibility of the interresidue linkage in G6OG. Therefore, the glucosyl residue of G6OG at subsite +1 is aligned between the two Trp residues in Figures 4D and 6D.
Molecular Dynamics of the Binding of β-d-Glucopyranosyl-(1→3)-β-d-Glucopyranosyl- (1→3)-d-Glucose in the Active Site
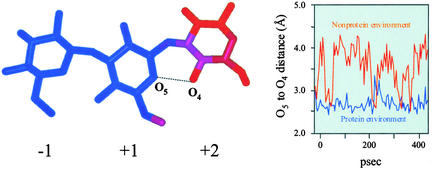
To investigate further the possible existence of subsite +2 in the active site of the barley β-d-glucan glucohydrolase (Figure 2), molecular dynamics calculations were performed. The molecular model of the β-d-glucopyranosyl-(1→3)-β-d-glucopyranosyl-(1→3)-d-glucose (G3OG3OG)/enzyme complex was calculated in a protein environment that included amino acid residues within a 10-Å radius of the bound glucosyl residue in subsite −1 of the S-laminaribioside/enzyme complex. The model of the G3OG3OG substrate was built manually, and predicted interactions with the active site were calculated. The average structure of the trisaccharide from the dynamics simulation is shown in Figure 8. The atoms are colored according to the variance in their mean positions. Atoms of the G3OG3OG substrate in subsites −1 and +1 showed low variation in their mean positions (blue), whereas atoms of the third residue at the reducing end of G3OG3OG showed significantly greater variation (red). A small difference (0.9 Å) in the distance between the O5 ring O atom of the glucopyranose in subsite +1 and the O4 hydroxyl atom of the reducing end glucopyranose ring of the G3OG3OG substrate was observed during the molecular dynamics simulation (Figure 8). This difference indicates that a hydrogen bond is formed between these two atoms, and as a result, a more constrained conformation of the trisaccharide G3OG3OG exists within the protein environment.
Figure 8.
Average Structures of the Nonhydrogen Atoms of G3OG3OG Substrate Evaluated by Molecular Dynamics Calculations.
Atoms of the putative subsite +2 show larger variation in their mean positions than those in subsites −1 and +1. Color scale: blue, <0.1 Å2; red, >0.5 Å2. The graph illustrates the variation in the distance between the O5 ring O atom of the glucopyranose in subsite +1 and the O4 hydroxyl atom of the glucopyranose in the putative subsite +2 (connected by a black dotted line) during the molecular dynamics simulation, either in a protein environment (blue) or a nonprotein environment (red). The larger variation in the O5 to O4 hydroxyl atom distance in the nonprotein environment is apparent.
On the other hand, calculations under similar conditions with the trisaccharide G3OG3OG in a water box (i.e., in a nonprotein environment) showed a much larger variation in the distance between the O5 ring O atom of the glucopyranose in subsite +1 and the O4 hydroxyl atom of the reducing end glucopyranose ring of the G3OG3OG substrate. The distance in this case varied between 2.6 and 4.3 Å (Figure 8), and the difference of 1.7 Å indicates that the hydrogen bond in the nonprotein environment would be formed only transiently.
Thus, the molecular dynamics calculations suggest that it is the protein environment that confers stability on the hydrogen bond formed between the second and third glucopyranosyl rings of the trisaccharide G3OG3OG. Furthermore, the calculations provide no evidence for direct or indirect interactions between the enzyme and the reducing end glucopyranosyl residue of the G3OG3OG substrate.
DISCUSSION
Comparative x-ray crystallographic analyses of the barley β-d-glucan glucohydrolase isoenzyme ExoI in complex with nonhydrolyzable S-glucoside substrate analogs, coupled with molecular modeling of other disaccharide/enzyme complexes, have provided a structural rationale for the broad specificity of this group of higher plant enzymes. The enzymes hydrolyze barley β-d-glucans or β-d-glucosides with (1→2)-, (1→3)-, (1→4)-, or (1→6)-linkages (Table 1) (Cline and Albersheim, 1981; Hrmova et al., 1996; Kotake et al., 1997; Hrmova and Fincher, 1998; Kim et al., 2000), despite the different relative orientations of adjacent glucopyranosyl rings imposed on the substrates by the various linkage positions. The active site pocket of the enzyme is ∼13 Å deep, which is enough to accommodate approximately two β-d-glucosyl residues, and is ∼15 Å wide at its entrance (Varghese et al., 1999). The overall kinetic schemes for both the hydrolytic and glycosyl transfer reactions catalyzed by the enzyme, together with the three-dimensional structures of key intermediates in the hydrolytic reaction pathway, have been described in detail (Figure 1) (Hrmova et al., 2001). Other glycoside hydrolases from microbial and plant sources exhibit a breadth of specificity against substrates with different glycosidic linkage positions (Frandsen and Svensson, 1998; Hashimoto et al., 1998; Stubbs et al., 1999), but no structural data are available to explain the molecular basis for this broad specificity.
The key feature of the barley β-d-glucan glucohydrolase that allows it to hydrolyze the range of (1→2)-, (1→3)-, (1→4)-, or (1→6)-linked β-d-glucosides is the presence of a relatively broad hydrophobic clamp, constituted by Trp-286 and Trp-434, which are placed ∼8 Å apart at the entrance of the active site pocket and which bind the β-d-glucosyl residue that occupies substrate binding subsite +1. Trp residues play key roles in many other types of carbohydrate–protein interactions (Quiocho, 1986; Vyas et al., 1988; Divne et al., 1998). The x-ray data presented here show that the nonreducing terminal β-d-glucosyl residue of the substrate is locked firmly in position at subsite −1 by extensive cooperative and bidentate hydrogen bonding interactions (Quiocho, 1986), with six amino acid residues at the bottom of the active site pocket. Therefore, all hydroxyl groups of the accessible hydrophilic surface area (Vyas et al., 1988) of the glucosyl residue at subsite −1 are bound to the enzyme (Figures 4 and 5). As a result, the glycosidic O atom would be held in a tightly fixed position with respect to the catalytic amino acid residues Asp-285 and Glu-491.
However, to explain the broad specificity of the enzyme for β-d-glucoside substrates, the enzyme would need to accommodate, in subsite +1, the penultimate nonreducing β-d-glucosyl residue of the various (1→2)-, (1→3)-, (1→4)-, or (1→6)-linked substrates with a significant degree of positional flexibility. The x-ray data show that this positional flexibility at subsite +1 is achieved by hydrophobic π-stacking interactions with Trp-286 and Trp-434, which sit above and below the β-d-glucosyl residue at subsite +1. Moreover, relatively few hydrogen bonds are formed between the enzyme and the β-d-glucosyl residue at subsite +1. Thus, the β-d-glucosyl residue in subsite +1 is not fixed as firmly in position as the β-d-glucosyl residue in subsite −1. The crystallographic analyses of the two S-glycoside/enzyme complexes show that the β-d-glucosyl residue in subsite +1 can be rotated and translated partially yet still remain located between the indole moieties of the two Trp residues (Figure 6). The flexibility in binding positions at subsite +1 presumably is allowed because the glucopyranosyl ring is ∼3 Å wide, whereas the indole moiety of the Trp residues is ∼5 Å wide. The Trp-286 and Trp-434 residues at the entrance of the substrate binding pocket of the barley β-d-glucan glucohydrolase also might play a role in the binding of acceptor molecules during glucosyl transfer reactions (R-OH; Figure 1).
The crystallographic studies presented here and elsewhere (Figures 4, 6, and 7) (Hrmova et al., 2001) show that glucosyl residues at subsites −1 and +1 adopt 4C1 conformations. This is in contrast to several well-characterized exo- and endo-acting β-d-glycoside hydrolases in which the subsite −1 glycosyl residues are distorted significantly (Sulzenbacher et al., 1996; Tews et al., 1996, 1997; Davies et al., 1998; Zou et al., 1999; Fort et al., 2001; Papanikolau et al., 2001).
The flexibility in substrate positioning allowed by the two relatively wide Trp residues at subsite +1, and hence the broad specificity of these family 3 enzymes for β-d-glucoside substrates, can be contrasted to the situation in a family 5 (1→3)-β-d-glucan glucohydrolase from Candida albicans. The latter enzyme also has an active site pocket that accommodates two β-d-glucosyl residues, but in this case, the penultimate β-d-glucosyl residue at subsite +1 is sandwiched between two Phe residues (Phe-144 and Phe-258) at the entrance to the pocket (Cutfield et al., 1999). One might predict, based on the central role of the Trp-286/Trp-434 clamp in allowing binding of substrates with different relative orientations of adjacent glucopyranosyl residues in the barley β-d-glucan glucohydrolase, that the relatively narrow Phe-144/Phe-258 clamp of the Candida enzyme would tighten its substrate specificity significantly. Indeed, this is the case. The relative catalytic efficiencies for the Candida (1→3)-β-d-glucan glucohydrolase during hydrolysis of G3OG, G4OG, and G6OG are 100, 0.06, and 0.14%, respectively (Stubbs et al., 1999), whereas the corresponding values for the barley enzyme are 100, 3.1, and 19%, respectively (Table 1). The catalytic efficiency value for G2OG for the Candida enzyme was not reported, but for the barley enzyme, it is 25% of the efficiency for G3OG (Table 1). Conversely, changing the Trp residues to amino acid residues with much smaller side chains might broaden the substrate specificity and alter catalytic efficiencies. Site-directed mutagenesis of the barley Trp-286 and Trp-434 residues now can be used to test further the effect of these residues on substrate specificity.
Although the structural basis for the broad specificity of the barley β-d-glucan glucohydrolase has become evident, it is not so easy to account for small differences in relative hydrolytic rates and catalytic efficiencies during hydrolysis of the dimeric β-d-oligoglucosides (Table 1). The preference for G3OG, compared with G4OG, is reflected in the generally higher subsite binding affinities for (1→3)-β–linked substrates (Figure 2), but the differences are not large. Similarly, the differences in ΔG‡ values are small, but they also parallel the differences between subsite binding energy values obtained during subsite mapping with the (1→3)-β-d-oligoglucosides and the (1→4)-β-d-oligoglucosides (Figure 2, Table 1). Additional techniques, such as time-resolved crystallography, together with site-directed mutagenesis, the use of transition-state analogs, and pre-steady state kinetic analysis, will be necessary to define the rate-limiting step during catalysis. In many cases for family 3 enzymes, the rate-limiting step is the formation or hydrolysis of the glycosyl enzyme intermediate (Legler et al., 1980; Li et al., 2001).
Additional data also will be necessary to explain the apparent discrepancy between the number of β-d-glucosyl binding subsites indicated by the subsite mapping analyses, which suggested three β-d-glucosyl binding subsites on the enzyme (Figure 2), and the x-ray crystallography and molecular dynamics predictions that there are only two subsites (Figures 4, 5, and 8). It is possible that there is weak affinity for a portion of or the entire third residue of the bound substrate beyond the +1 subsite (Stone and Svensson, 2002) and that this is responsible for the subsite mapping result. Bound substrates also might adopt unexpected curved conformations (Parsiegla et al., 2000), which would allow them to interact with other parts of the enzyme outside of the active site pocket. However, the molecular dynamics calculations provide no evidence for direct or indirect interactions between the enzyme and the third glucopyranosyl residue of the G3OG3OG; rather, they suggest that the binding energy at the +2 site observed in subsite mapping is attributable to the stabilization of intermolecular hydrogen bonding in the ligand (Figure 8). The kinetics of substrate binding now can be investigated further by isothermal titration microcalorimetry (Creagh et al., 1996) or differential scanning microcalorimetry (Creagh et al., 1998).
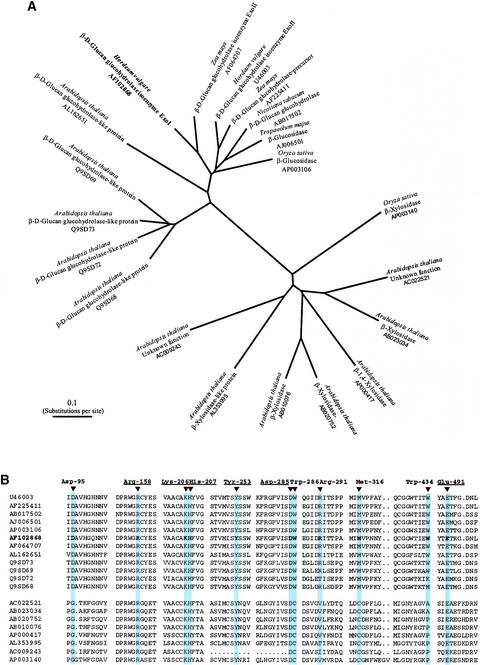
The x-ray crystallography data presented here to describe in detail the molecular interactions at the −1 and +1 subsites of the barley β-d-glucan glucohydrolase can be applied more generally to examine the structural basis for substrate specificity in other family 3 glycoside hydrolases from higher plants. There are >180 known members of this family, most of which are classified as β-d-glucosidases, β-d-xylosidases, or N-acetyl β-d-glucosaminidases and are of microbial origin (Henrissat, 1991; Harvey et al., 2000; http://afmb.cnrs-mrs.fr/CAZY/). However, when the sequences derived exclusively from higher plant enzymes are used to generate a phylogenetic tree, two quite distinct groups of enzymes become evident (Figure 9A). One group contains the β-d-glucan glucohydrolases or β-d-glucan glucohydrolase–like enzymes from plants, for which sequence identities lie in the range of 60 to 80%. The second group of higher plant enzymes from family 3 contains β-d-xylosidase–like enzymes, with sequence identities of ∼60% (data not shown).
Figure 9.
Unrooted Radial Phylogenetic Tree of Plant Family 3 Glycoside Hydrolases (A) and Partial Amino Acid Sequence Alignment of the Amino Acid Residues Involved in Substrate Binding (B).
The two major branches distinguish between the β-d-glucan glucohydrolase–like type and the β-d-xylosidase–like type, as determined with the ClustalW algorithm (Thompson et al., 1994; http://www.ebi.ac.uk/clustalw/). Branch lengths are drawn to scale. Conserved amino acid residues in the β-d-glucan glucohydrolase–like group (cyan), numbered according to the barley β-d-glucan glucohydrolase isoenzyme ExoI sequence, are compared with the amino acid residues of the β-d-xylosidase–like group of plant family 3 glycoside hydrolases. Underlined residues are totally conserved in plant members of the family. The GenBank and EMBL/SWISS-PROT accession numbers of the sequences are shown.
Although none of the higher plant β-d-xylosidase–like group in the databases have been characterized, they are assigned this specificity on the basis of their similarity to well-characterized microbial enzymes. In addition, we recently isolated cDNAs corresponding to purified family 3 β-d-xylosidases from barley seedlings, and these barley enzymes share a high level of sequence similarity to the other members of the higher plant β-d-xylosidase–like group (R.C. Lee, M. Hrmova, R.A. Burton, and G.B. Fincher, unpublished data). The clear separation of the two groups of higher plant family 3 enzymes seen in Figure 9A is reflected in sequence identities of <27% between members of the two groups (data not shown). No plant N-acetyl β-d-glucosaminidase sequences from family 3 have been reported (http://afmb.cnrs-mrs.fr/CAZY/).
In the case of the family 3 enzymes from barley, the β-d-glucan glucohydrolases do not hydrolyze β-d-xylosides (Hrmova and Fincher, 1998), and the β-d-xylosidases do not hydrolyze β-d-glucosides (R.C. Lee, M. Hrmova, R.A. Burton, and G.B. Fincher, unpublished data). The three-dimensional data presented here to explain the broad specificity of the barley β-d-glucan glucohydrolase also might explain the apparent differences in substrate specificity between the two groups (Figure 9B). The catalytic amino acid residues, corresponding to the barley Asp-285 and Glu-491, are conserved absolutely across the two groups. Similarly, amino acid residues involved in hydrogen bonding to the C2OH, C3OH, and C4OH groups (Lys-206, His-207, Arg-158, and Tyr-253) are conserved completely in both the β-d-glucan glucohydrolase–like and β-d-xylosidase–like groups (Figure 9B). However, amino acid residues corresponding to Asp-95, Arg-291, and Met-316 are replaced by other amino acid residues in the β-d-xylosidase–like group of family 3 enzymes (Figure 9B).
It is noteworthy that Asp-95 and Met-316 interact with the CH2OH group and C5 of the glucosyl residue bound at subsite −1, and Arg-291 forms a hydrogen bond with C6′OH of the glucosyl residue bound at subsite +1, at least in the case of bound G4SG. The pentose β-d-xylopyranose has no C5CH2OH group, and amino acids necessary for binding this hydroxyl group in the β-d-glucan glucohydrolase–like group could be replaced in the β-d-xylosidase–like group of family 3 enzymes. Before too many conclusions are drawn in relation to the molecular and structural basis for the differences in substrate specificity between the β-d-glucan glucohydrolase–like group and the β-d-xylosidase–like group of family 3 enzymes, it should be emphasized that members of the plant β-d-xylosidase–like group often have associated α-l-arabinofuranosidase activity (R.C. Lee, M. Hrmova, R.A. Burton, and G.B. Fincher, unpublished data). Therefore, the different sizes and conformations of the pyranosyl and furanosyl rings, together with the various orientations of projecting hydroxyl groups, need to be taken into account. In some enzymes, substrate binding amino acid residues simply adjust sterically to accommodate the binding of different groups of glycosides, as shown for a (1→4)-β-glycanase (cellulase/xylanase) from Cellulomonas fimi (White et al., 1996; Notenboom et al., 1998).
Another major difference between the higher plant β-d-glucan glucohydrolase–like and β-d-xylosidase–like groups of family 3 enzymes is that the β-d-xylosidase–like enzymes do not have the Trp-286 or Trp-434 residues that constitute the conserved hydrophobic clamp at subsite +1 in all of the β-d-glucan glucohydrolase–like enzymes (Figures 4 to 6). In the β-d-xylosidase–like group, a Cys residue replaces Trp-286 and Pro, Ala, or Met residues replace Trp-434 (Figure 9B). Whether these substitutions dramatically affect the specificity of binding at the +1 subsite of the β-d-xylosidase–like group remains to be demonstrated.
We conclude from the data presented here that the family 3 β-d-glucan glucohydrolases from higher plants can hydrolyze a range of β-d-glucans and β-d-oligoglucosides because of their ability to bind the glucosyl residues at subsite +1 in a relatively flexible manner. The β-d-xylosidase–like group of family 3 enzymes in higher plants does not have the amino acid residues necessary for hydrogen bonding to the C5 CH2OH substituent of bound substrates and also might have evolved a different mechanism for binding the glycosyl residue that occupies binding subsite +1. Clearly, the three-dimensional structure of a family 3 plant β-d-xylosidase is required to explain these possibilities further, together with the structure of the enzyme in complex with its substrates. In a more general sense, detailed structural information of the type provided here could prove useful in the functional annotation of genes discovered in genomics and genome-sequencing programs. For example, examination of sequences encoding substrate binding regions could discriminate between plant family 3 xylosidases, glucosidases, and, if they are present, N-acetyl β-d-glucosaminidases.
METHODS
Materials
The Glc diagnostic kit, 4′-nitrophenyl β-d-glucopyranoside (4NPGlc), gentiobiose, sophorose, α,α-trehalose, α,β-trehalose, β,β-trehalose, esculin, salicin, BSA, and orcinol were purchased from Sigma (St. Louis, MO). Microcon microconcentrators were from Amicon (Beverly, MA), Sep-Pak Plus cartridges were from Waters (Milford, MA), Kieselgel 60 thin layer plates and sodium 2,2-dimethyl-2-silapentane-5-sulfonic acid were from Merck (Darmstadt, Germany), chromatography paper (3 MM Chr) was from Whatman (Maidstone, Kent, UK), and (1→3)-β-d-oligoglucosides of degree of polymerization (DP) 2 to 7 and (1→4)-β-d-oligoglucosides of DP 2 to 6 were from Seikagaku Kogyo (Tokyo, Japan). Tamarind (Tamarindus indica) xyloglucan and the xyloglucan oligosaccharides XXXG (DP 7), XXLG (DP 8), and XLLG (DP 9), were provided by Vladimir Farkas (Institute of Chemistry, Bratislava, Slovak Republic).
Enzyme Isolation and Purity
Barley (Hordeum vulgare) β-d-glucan glucohydrolase isoenzyme ExoI was purified from a homogenate of 8-day-old seedlings as described by Hrmova et al. (1996). The purity of the enzyme was assessed by SDS-PAGE, in which a single protein band was detected at high protein loadings, and by N-terminal amino acid sequence analysis (Hrmova et al., 1996).
Steady State Kinetic Analyses
Kinetic parameters on (1→3)-β-d-oligoglucosides of DP 2 to 7 and (1→4)-β-d-oligoglucosides of DP 2 to 6 were determined at 37°C by incubating 1 to 3 pmol of the purified β-d-glucan glucohydrolase isoenzyme ExoI in 100 mM sodium acetate buffer, pH 5.25, containing 160 μg/mL BSA. Initial rates of hydrolysis were determined in triplicate at substrate concentrations ranging from 0.3 to 3 times the Km values. Glc released was measured by the Glc oxidase method as described previously (Hrmova et al., 1998b). Kinetic data were processed by a proportional weighted fit using a nonlinear regression analysis program based on Michaelis-Menten model equations (Perella, 1988). The initial enzyme concentrations were [E]0<<[S]0, where [E]0 and [S]0 represent enzyme and substrate concentrations at time 0; therefore, initial reaction rates were measured in all cases. One unit of activity is defined as the amount of enzyme required to release 1 μmol of Glc from β-d-glucosides per minute and corresponds to 16.67 nanokatals.
Subsite Mapping and Evaluation of Subsite Binding Affinities
Subsite affinities (Hiromi, 1970), or transition-state interaction energies (Malet and Planas, 1997; Fersht, 1999), of β-d-glucan glucohydrolase isoenzyme ExoI were calculated using Km values and catalytic rate constants or catalytic center activities (kcat) during the hydrolysis of (1→3)-β-d-oligoglucosides of DP 2 to 7 and (1→4)-β-d-oligoglucosides of DP 2 to 6. The calculation procedures, using equations for the subsite analysis of the exo-acting enzymes, are summarized in Hrmova et al. (1998b). The values of kint for (1→3)- and (1→4)-β-d-oligoglucosides were 25.1 and 3.4 sec−1, respectively. The validity of the subsite maps was confirmed by comparing the experimental and theoretical values.
Glycosyl Transfer Reactions
A reaction mixture containing 100 mM freshly prepared and temperature-equilibrated 4NPGlc in 20 mM sodium acetate buffer, pH 5.25, was incubated with 6 to 7 pmol of the purified β-d-glucan glucohydrolase isoenzyme ExoI for up to 96 hr at 37°C. The reaction products were subjected to quantitative and qualitative analyses as described below.
Quantitative Product Analysis
Aliquots of the reaction mixture were separated by thin layer chromatography on Kieselgel 60 thin layer plates and developed in ethyl acetate:acetic acid:water (3:2:1, v/v/v). The compounds were excised and eluted from the chromatogram with water, and their relative abundances were determined spectrophotometrically at 300 nm. In addition, the thin layer plates containing the compounds were probed for reducing sugars with the orcinol reagent (Hrmova and Fincher, 1993).
Qualitative Product Analysis
Preparative amounts of the aryl-glycosides were synthesized, separated by descending paper chromatography on Whatman 3 MM Chr paper, and developed in ethyl acetate:acetic acid:water (3:2:1, v/v/v) at ambient temperature. The compounds were excised from the chromatogram, eluted with water, concentrated under reduced pressure, and subjected to analyses by electrospray ionization mass spectrometry and 13C-NMR spectroscopy, as described previously (Hrmova et al., 1998b).
Preparation of 4-NP S-(β-d-Glucopyranosyl)-(1→3)-(3-Thio- β-d-Glucopyranosyl)-(1→3)-β-d-Glucopyranoside
(1→3)-β-d-Glucan endohydrolase mutant E231G (0.01 mg) was added to a solution of 3-thio-α-laminaribiosyl fluoride (Hrmova et al., 2000) (70 mg, 1 equivalent) and 4NPGlc (234 mg, 4 equivalents) in phosphate buffer (4 mL of 0.25 M, pH 7.0), and the solution was incubated for 2 hr at 37°C. The mixture was separated by chromatography on a reverse-phase C18 silica gel column using a methanol/water gradient (0 to 20%) to yield, after evaporation and lyophilization, 4-NP S-(β-d-glucopyranosyl)-(1→3)-(3-thio-β-d-glucopyranosyl)-(1→3)-β-d-glucopyranoside (4NP-G3SG3OG) as a colorless powder (62 mg, 50%). The sample was dissolved in d6-DMSO/D2O and lyophilized three times before NMR analysis. 1H-NMR (d6-DMSO; 300 MHz, 303 K) 8.22 to 8.15, 7.25 to 7.20 (m, 4H, aromatic H), 5.22 [d, J(H-1I, H-2I) = 7.3 Hz, H-1I], 4.54 [d, J(H-1III, H-2III) = 9.7 Hz, H-1III], 4.45 [d, J(H-1II, H-2II) = 7.5 Hz, H-2II], 2.97 to 3.74 (m, 17H, H-2I-III, H-3I,III, H-4I-III, H-5I-III, H-6I-III), 2.78 [t, J(H-2II, H-3II) = J(H-3II, H-4II) = 10.2 Hz, H-3II]. 13C-NMR (D2O; 300 MHz) δ 162.04, 143.02, 126.76, 117.78, aromatic; 105.69, 100.92, C1I,II; 86.39, 86.33, C1III, 3I; 81.62, 80.17, 79.02, 77.68, 74.38, 74.23, 74.00, 70.97, 69.47, 69.22, C2I-III, 3III, 4I-III, 5I-III; 62.54, 62.36, 62.08, C6I-III; 56.03, C3II. Electrospray ionization mass spectrometry calculated for C24H35NO17S (M+Na)+ 664.6; found m/z, 664.2. Electrospray ionization mass spectrometry calculated for C24H35NO17S (M+K)+ 680.7; found m/z, 680.2. All other experimental details have been described previously (Fort et al., 2000; Driguez, 2001).
Inactivation of β-d-Glucan Glucohydrolase by 4NP-G3SG3OG
Inactivation of β-d-glucan glucohydrolase isoenzyme ExoI by 4NP-G3SG3OG was monitored at 37°C by incubating 13 pmol of the purified enzyme in 100 mM sodium acetate buffer, pH 5.25, containing 0.2% (w/v) 4NPGlc, 160 μg/mL BSA, and 0 to 660 μM 4NP-G3SG3OG. Residual enzyme activity was monitored spectrophotometrically at 410 nm as specified above. The Ki constant was determined by a proportional weighted fit using a nonlinear regression analysis program (Perella, 1988) as described previously (Hrmova et al., 1998b, 2001).
Molecular Modeling of β-d-Glucopyranosyl-(1→2)-d-Glucose and β-d-Glucopyranosyl-(1→6)-d-Glucose in the Active Site of β-d-Glucan Glucohydrolase
The molecular models of the barley β-d-glucan glucohydrolase with bound β-d-glucopyranosyl-(1→2)-d-glucose (sophorose or G2OG) and β-d-glucopyranosyl-(1→6)-d-glucose (gentiobiose or G6OG) were constructed manually with the program SYBYL 6.62 (Blundell et al., 1988) and involved two stages. The first stage represented energy minimization of the β-d-glucan glucohydrolase structure (Hrmova et al., 2001) on amino acid residues within a 15-Å radius of the active site. This initial three-dimensional structure excluded water molecules but contained several conserved amino acid residues and a bound S-cellobioside moiety. Optimization of the structure was performed with SYBYL without imposing charges on the protein, using the Powell method (Powell, 1977) and the TRIPOS force-field parameters (Clark et al., 1989). The resultant protein model was used as a template for docking of all disaccharides.
The second stage involved the building of the disaccharides for docking. The starting three-dimensional structures of the G2OG (Ikegami et al., 1995), with two rotatable bonds (C1-O and O-C2′) around the glycosidic linkage, and G6OG (Rohrer et al., 1980), with three rotatable bonds (C1-O, O-C6′, and C6′-C5′) around the glycosidic linkage, were obtained from the Cambridge Crystallographic Data Centre (http://www.ccdc.cam.ac.uk/). The two low molecular mass carbohydrates were reoptimized in SYBYL and allowed to relax by a round of molecular mechanics calculations. The optimized coordinates of G2OG and G6OG were used in subsequent investigations with β-d-glucan glucohydrolase isoenzyme ExoI. This involved superposing the nonreducing ends of G2OG and G6OG within the −1 subsite of the G4SG moiety, which was removed subsequently from the active site to yield a starting position for the docking procedure.
The next round of docking was performed manually by positioning the disaccharide atoms against the protein template and simultaneous energy minimization of the steric overlaps and clashes between the disaccharide and the enzyme active site. From this preliminary model, a number of key interactions were identified, and these were restrained during refinement (Blundell et al., 1988). Finally, an exhaustive search of the possible orientations of the G2OG and G6OG moieties in the active site was undertaken, using information on the −1 subsite of the G4SG moiety of the structure of the S-cellobioside/enzyme complex.
For each G2OG/ and G6OG/β-d-glucan glucohydrolase complex, the bonds in the glycosidic linkage were subjected to an angular search and were oscillated by 10° steps to determine the most favorable position in the active site and to remove the bias introduced by the superposition of their nonreducing ends during the early stages of model building. For each solution of the enzyme/disaccharide complex, several steps of energy minimization were performed. No restraints were applied, and TRIPOS force-field parameters established for protein/carbohydrate complexes were applied (Perez et al., 1995).
Molecular Dynamics of Binding of G3OG3OG
An active site model of the protein and the trisaccharide G3OG3OG was generated using Insight II molecular modeling software (Accelrys, Burlington, MA). All residues outside a radius of 10 Å from the ligand in the −1 subsite of the x-ray structure of the S-laminaribioside/β-d-glucan glucohydrolase complex were excluded. All crystallographic water molecules within this radius were retained. The Glc in the putative +2 site was built manually, and the S atom was replaced with O. H atoms were added to fill unsatisfied valences. The standard protonation state of ionizable residues at pH 7.0 was adopted, with the exception of the catalytic acid/base Glu-491 that was modeled in its neutral form. The active site model was placed in a cubic water box of 40 Å (side dimensions). The positions of all H atoms, atoms of the trisaccharide, and the solvent were minimized, that is, nonhydrogen atoms of the protein were fixed at their experimental positions, with periodic boundary conditions, using the Discover program, version 2.98 (Accelrys).
Force-field parameters for the protein, trisaccharide, and solvent were taken from the consistent valence forcefield. The method of steepest descents was applied initially until the gradient decreased to <10.0 kcal·mol−1·Å−1, after which further minimization using the method of conjugate gradients was applied until the gradient decreased to <0.01 kcal·mol−1·Å−1. The dielectric constant was unscaled (i.e., set to 1.0), and a nonbond interaction cutoff distance of 15 Å, with a 1-Å neighbor-list buffer region and a 1.5-Å switch distance, was applied. Molecular dynamics calculations at 300 K were performed under the same conditions. An initial equilibration of 1 psec was followed by molecular dynamics of 425 psec, with integration time steps of 1 fsec. Molecular dynamics calculations of the trisaccharide G3OG3OG in a 25-Å cubic water box were performed under similar conditions.
X-Ray Crystallography of the S-Laminaribioside/β-d-Glucan Glucohydrolase Complex
Crystals of β-d-glucan glucohydrolase isoenzyme ExoI (Hrmova et al., 1998a) with a c axis of ∼200 μm were transferred into a solution of 100 mM Hepes-NaOH buffer, pH 7.0, containing 1.2% (w/v) polyethylene glycol 400, 0.8 M ammonium sulfate (solution A), and 10 mM 4NP-G3SG3OG. After ∼2 hr of soaking at 4 ± 2°C, the crystal was transferred into solution A containing 30% (v/v) glycerol as a cryoprotectant. The crystal was mounted subsequently on a goniometer and flash frozen to −283°C in a stream of nitrogen gas (Oxford Instruments, Oxford, UK). The data were collected to 2.40 Å resolution using a MAC SRA M18XH1 rotating copper anode x-ray generator (MacScience, Yokohama, Japan) operating at 40 kV and 50 mA and fitted with focusing mirrors for Kα radiation. A total of 120° data frames were collected using 1° oscillations and 1-hr exposure times on an R-axis II detector (Rigaku/MSC, Woodlands, TX) with a crystal-to-film set distance of 120 mm.
The diffraction data were integrated, scaled, and reduced by HKL (Otwinowski and Minor, 1996). Autoindexing determined that the primitive tetragonal crystals belong to the space group P43212. Model refinement was performed with the Crystallographic and NMR System (Brünger et al., 1998) using the Glc/β-d-glucan glucohydrolase isoenzyme ExoI three-dimensional structure (Hrmova et al., 2001) as a search model. Atomic B-factor values for the protein, water molecules, and carbohydrates were reset to an overall thermal parameter of 30 Å2. The residues within a 10-Å radius of the active site, excluding the bound Glc, were set to zero occupancy. The initial model used to locate the position of the protein in the unit cell was built with a restrained rigid body refinement technique from 15 to 3.0 Å. After a final round of rigid body refinement, the crystallographic factor Rwork was 24.15%, Rfree was 24.51%, and the overall B value was 25.45 Å2. Geometrical positional refinement followed by individual B factor refinement was calculated subsequently using the β-d-glucan glucohydrolase isoenzyme ExoI model with a bulk solvent correction applied and a maximum likelihood method implemented.
The electron density map was calculated from the observed structure factors and phases using the β-d-glucan glucohydrolase isoenzyme ExoI structure and MAPMAN (Kleywegt and Jones, 1994). The thioglycoside moiety was located in the active site pocket by examining a difference Fourier electron density map at >4σ, which showed peaks for the two sugars of the S-laminaribioside moiety, which were built manually in the electron density map using “O” (Jones et al., 1991) and refined. The PROCHECK program (Laskowski et al., 1993) was used to assess the geometric quality of the structure, and the structure was found not to deviate from ideality. The data collection and final refinement statistics are summarized in Table 4. The three-dimensional structures of the search model and the Glc/β-d-glucan glucohydrolase complex were superposed over the Cα atoms of Asp-95, Phe-144, Arg-158, Lys-206, His-207, Glu-220, Tyr-253, Asp-285, Trp-286, Glu-287, Arg-291, Met-316, Trp-434, and Glu-491, and the two three-dimensional structures showed a root mean square deviation of 0.099 Å. The atomic coordinates of the refined structure of the S-laminaribioside/β-d-glucan glucohydrolase were deposited with the Protein Data Bank (http://www.rcsb.org/pdb/; Berman et al., 2000).
Accession Numbers
The accession numbers for the sequences mentioned in this article are AW754572 (Pinus taeda expressed sequence tag), 1J8V (the three-dimensional structure of the S-laminaribioside/enzyme complex), 1IEX (the three-dimensional structure of the S-cellobioside/enzyme complex), 1IEQ (the three-dimensional structure of Glc/β-d-glucan glucohydrolase isoenzyme ExoI), and AF102868 (barley β-d-glucan glucohydrolase isoenzyme ExoI).
Acknowledgments
We are grateful to Dr. Peter Biely and Jelle Lahnstein for expert assistance, to Dr. Vladimir Farkas for providing the xyloglucan and the xyloglucan oligosaccharides, to Professor Bruce Stone for invaluable advice, and to Professor Peter Colman for ongoing support. This work was supported by grants from the Australian Research Council and the Grains Research and Development Corporation of Australia (to G.B.F.), from the Centre National de la Recherche Scientifique (to H.D.), and from the Australian National Beam Line Facility (to J.N.V.).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010442.
References
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., and Bourne, P.E. (2000). The Protein Data Bank. Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell, T., Carney, D., Gardner, S., Hayes, F., Howlin, B., Hubbard, T., Overington, J., Singh, D.A., Sibanda, B.L., and Sutcliffe, M. (1988). 18th Sir Hans Krebs Lecture: Knowledge-based protein modelling and design. Eur. J. Biochem. 172, 513–520. [DOI] [PubMed] [Google Scholar]
- Bock, K., Pedersen, C., and Pedersen, H. (1984). Carbon-13 nuclear magnetic resonance data for oligosaccharides. Adv. Carbohydr. Chem. Biochem. 42, 193–225. [Google Scholar]
- Brünger, A.T., Adams, P.D., Clorc, G.M., DeLano, W.L., Gros, P., Grosse-Kunsteve, W., and Warren, G.L. (1998). Crystallographic and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Chu, S.S.C., and Jeffrey, G.A. (1968). The refinement of the crystal structures of β-d-glucose and cellobiose. Acta Crystallogr. Sect. B 24, 830–838. [Google Scholar]
- Clark, M., Cramer, R.D.I., and Opdenbosch, V.D. (1989). Validation of the general purpose Tripos 5.2 force field. J. Comp. Chem. 8, 982–1012. [Google Scholar]
- Cline, K., and Albersheim, P. (1981). Host-pathogen interactions. XVI. Purification and characterization of a glucosyl hydrolase/transferase present in the walls of soybean cells. Plant Physiol. 68, 207–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creagh, A.L., Ong, E., Jervis, E., Kilburn, D.G., and Haynes, C.A. (1996). Binding of the cellulose-binding domain of exoglucanase Cex from Cellulomonas fimi to insoluble microcrystalline cellulose is entropically driven. Proc. Natl. Acad. Sci. USA 93, 12229–12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creagh, A.L., Koska, J., Johnson, P.E., Tomme, P., Joshi, M.D., McIntosh, L.P., Kilburn, D.G., and Haynes, C.A. (1998). Stability of oligosaccharide binding of the N1 cellulose-binding domain of Cellulomonas fimi endoglucanase CenC. Biochemistry 37, 3529–3537. [DOI] [PubMed] [Google Scholar]
- Crombie, H.J., Chengappa, S., Hellyer, A., and Reid, J.S.G. (1998). A xyloglucan oligosaccharide-active, transglycosylating β-d-glucosidase from the cotyledons of nasturtium (Tropaeolum majus L) seedlings: Purification, properties and characterization of a cDNA clone. Plant J. 15, 27–38. [DOI] [PubMed] [Google Scholar]
- Cutfield, S.M., Davies, G.J., Marshudov, G., Anderson, B.F., Moody, P.C.E., Sullivan, P.A., and Cutfield, J.F. (1999). The structure of the exo-β-(1,3)-glucanase from Candida albicans in native and bound forms: Relationship between a pocket and groove in family 5 glycosyl hydrolases. J. Mol. Biol. 294, 771–783. [DOI] [PubMed] [Google Scholar]
- Czjzek, M., Cicek, M., Zamboni, V., Burmeister, W.P., Bevan, D.R., Henrissat, B., and Esen, A. (2001). Crystal structure of a monocotyledon (maize ZMGlu1) β-glucosidase and a model of its complex with p-nitrophenyl β-d-thioglucoside. Biochem. J. 354, 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, G.J., Mackenzie, L., Varrot, A., Dauter, M., Brzozowski, A.M., Schülein, M., and Withers, S.G. (1998). Snapshots along an enzymatic reaction coordinate: Analysis of a retaining β-glycoside hydrolase. Biochemistry 37, 11707–11713. [DOI] [PubMed] [Google Scholar]
- Divne, C., Stahlberg, J., Teeri, T.T., and Jones, T.A. (1998). High resolution crystal structures reveal how a cellulose chain is bound in the 50 Å long tunnel of cellobiohydrolase I from Trichoderma reesei. J. Mol. Biol. 275, 309–325. [DOI] [PubMed] [Google Scholar]
- Driguez, H. (2001). Thiooligosaccharides as tools for structural biology. Chembiochem. Eur. J. Chem. Biol. 2, 311–318. [DOI] [PubMed] [Google Scholar]
- Fersht, A. (1999). Enzyme-substrate complementarity and the use of binding energy in catalysis. In Structure and Mechanism in Protein Science. (New York: W.H. Freeman), pp. 349–376.
- Fort, S., Boyer, V., Greffe, L., Cottaz, S., Davies, G.J., Moroz, O., Christiansen, L., Schülein, M., and Driguez, H. (2000). Highly efficient synthesis of β(1→4)-oligo- and polysaccharides using a mutant cellulase. J. Am. Chem. Soc. 122, 5429–5437. [Google Scholar]
- Fort, S., Varrot, A., Schülein, M., Cottaz, S., Driguez, H., and Davies, G.J. (2001). Mixed-linkage cellooligosaccharides: A new class of glycoside hydrolase inhibitors. Chembiochem. Eur. J. Chem. Biol. 2, 319–325. [DOI] [PubMed] [Google Scholar]
- Frandsen, T.P., and Svensson, B. (1998). Plant α-glucosidases of the glycoside hydrolase family 31: Molecular properties, substrate specificity, reaction mechanism, and comparison with family members of different origin. Plant Mol. Biol. 37, 1–13. [DOI] [PubMed] [Google Scholar]
- Fry, S.C. (1995). Polysaccharide-modifying enzymes in the plant cell wall. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 497–520. [Google Scholar]
- Harvey, A.J., Hrmova, M., De Gori, R., Varghese, J.N., and Fincher, G.B. (2000). Comparative modeling of the three-dimensional structures of family 3 glycoside hydrolases. Proteins Struct. Funct. Genet. 41, 257–269. [DOI] [PubMed] [Google Scholar]
- Hashimoto, W., Miki, H., Nankai, H., Sato, N., Kawai, S., and Murata, K. (1998). Molecular cloning of two genes of β-d-glucosidase in Bacillus sp. GL1 and identification of one as a gellan-degrading enzyme. Arch. Biochem. Biophys. 360, 1–9. [DOI] [PubMed] [Google Scholar]
- Henrissat, B. (1991). A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280, 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiromi, K. (1970). Interpretation of dependency of rate parameters on the degree of polymerization of substrate in enzyme-catalyzed reactions: Evaluation of subsite affinities of exo-enzyme. Biochem. Biophys. Res. Commun. 40, 1–6. [DOI] [PubMed] [Google Scholar]
- Hrmova, M., and Fincher, G.B. (1993). Purification and properties of three (1→3)-β-d-glucanase isoenzymes from young leaves of barley (Hordeum vulgare). Biochem. J. 289, 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrmova, M., and Fincher, G.B. (1998). Barley β-d-glucan exohydrolases: Substrate specificity and kinetic properties. Carbohydr. Res. 305, 209–221. [Google Scholar]
- Hrmova, M., and Fincher, G.B. (2001). Structure-function relationship of β-d-glucan exohydrolases from higher plants. Plant Mol. Biol. 47, 73–91. [PubMed] [Google Scholar]
- Hrmova, M., Garrett, T.P.J., and Fincher, G.B. (1995). Subsite affinities and disposition of catalytic amino acids in the substrate-binding region of barley 1,3-β-d-glucanases: Implications in plant-pathogen interactions. J. Biol. Chem. 270, 14556–14563. [DOI] [PubMed] [Google Scholar]
- Hrmova, M., Harvey, A.J., Wang, J., Shirley, N.J., Jones, G.P., Stone, B.A., Hoj, P.B., and Fincher, G.B. (1996). Barley β-d-glucan exohydrolases with β-d-glucosidase activity: Purification, characterization, and determination of primary structure from a cDNA clone. J. Biol. Chem. 271, 5277–5286. [DOI] [PubMed] [Google Scholar]
- Hrmova, M., Varghese, J.N., Høj, P.B., and Fincher, G.B. (1998. a). Crystallization and preliminary X-ray analysis of β-d-glucan exohydrolase isoenzyme ExoI from barley (Hordeum vulgare). Acta Crystallogr. Sect. D 54, 687–689. [DOI] [PubMed] [Google Scholar]
- Hrmova, M., MacGregor, A.E., Biely, P., Stewart, R.J., and Fincher, G.B. (1998. b). Substrate binding and catalytic mechanism of a barley β-d-glucosidase/(1,4)-β-d-glucan exohydrolase. J. Biol. Chem. 273, 11134–11143. [DOI] [PubMed] [Google Scholar]
- Hrmova, M., Rutten, S.J., Fairweather, J.K., Fincher, G.B., and Driguez, H. (2000). Wild-type and mutant retaining (1→3)-β-glucan endohydrolases from barley are efficient tools in the synthesis of (1→3)-β-linked oligo- and polysaccharides. In 20th International Carbohydrate Symposium, J. Thiem, ed (Hamburg: LCI), p. 148.
- Hrmova, M., Varghese, J.N., De Gori, R., Smith, B.J., Driguez, H., and Fincher, G.B. (2001). Catalytic mechanisms and reaction intermediates along the hydrolytic pathway of a plant β-d-glucan glucohydrolase. Structure 9, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Ikegami, M., Sato, T., Suzuki, K., Noguchi, K., Okuyama, K., Kitamura, S., Takeo, K., and Ohno, S. (1995). Molecular and crystal structures of 2,3,4,6,1′,3′,4′,6′-octa-O-acetyl-β-sophorose, methyl 2,3,4,6,1′,3′,4′,6′-hepta-O-acetyl-β-sophoroside, and methyl 2,3,4,6,1′,3′,4′-hexa-O-acetyl-6′-deoxy-β-sophoroside. Carbohydr. Res. 271, 137–150. [Google Scholar]
- Johnston, L.N., Cheetham, J., McLaughlin, P.J., Acharya, K.R., Barford, D., and Phillips, D.C. (1988). Protein-oligosaccharide interactions: Lysozyme, phosphorylase, amylases. Curr. Top. Microbiol. Immunol. 139, 81–134. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. (1991). Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kim, J.B., Olek, A.T., and Carpita, N.C. (2000). Cell wall and membrane-associated exo-β-d-glucanases from developing maize seedlings. Plant Physiol. 123, 471–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt, G.J., and Jones, T.A. (1994). Halloween masks and bones. In From the First Map to Final Model (CCP4), S. Bailey, R. Hubbard, and D. Waller, eds (Daresbury, UK: SERC Daresbury Laboratory), pp. 59–66.
- Koizumi, N., Okushima, Y., and Sano, H. (2000). Isolation, characterization and molecular cloning of β-d-glucan exohydrolase from cultured tobacco cells. J. Plant Physiol. 157, 691–698. [Google Scholar]
- Kotake, T., Nakagawa, N., Takeda, K., and Sakurai, N. (1997). Purification and characterization of wall-bound exo-1,3-β-d-glucanase from barley (Hordeum vulgare L) seedlings. Plant Cell Physiol. 38, 194–200. [DOI] [PubMed] [Google Scholar]
- Kraulis, P. (1991). MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950. [Google Scholar]
- Labrador, E., and Nevins, D.J. (1989). An exo-β-d-glucanase derived from Zea coleoptile walls with a capacity to elicit cell elongation. Physiol. Plant. 77, 479–486. [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. (1993). PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- Legler, G., Sinnott, M.L., and Withers, S.G. (1980). Catalysis by β-glucosidase A3 of Aspergillus wentii. J. Chem. Soc. Perkin Trans. 2, 1376–1383. [Google Scholar]
- Li, Y.K., Chir, J., and Chen, F.Y. (2001). Catalytic mechanism of a family 3 β-glucosidase and mutagenesis study on residue Asp-247. Biochem. J. 355, 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienart, Y., Comtat, J., and Barnoud, F. (1986). A wall-bound exo-β-d-glucanase from Acacia cultured cells. Biochim. Biophys. Acta 883, 353–360. [Google Scholar]
- Mackenzie, L.F., Wang, Q., Warren, R.A.J., and Withers, S.G. (1998). Glycosynthases: Mutant glycosidases for oligosaccharide synthesis. J. Am. Chem. Soc. 120, 5583–5584. [Google Scholar]
- Malet, C., and Planas, A. (1997). Mechanism of Bacillus 1,3:1,4-β-d-glucan 4-glucanohydrolases: Kinetic and pH studies with 4-methylumbelliferyl β-d-oligosaccharides. Biochemistry 36, 13838–13848. [DOI] [PubMed] [Google Scholar]
- Malet, C., and Planas, A. (1998). From β-glucanase to β-glucansynthase: Glycosyl transfer to α-glycosyl fluorides catalyzed by a mutant endoglucanase lacking its catalytic nucleophile. FEBS Lett. 440, 208–212. [DOI] [PubMed] [Google Scholar]
- Moreau, V., Viladot, J.L., Samain, E., Planas, A., and Driguez, H. (1996). Design and chemoenzymatic synthesis of thiooligosaccharide inhibitors of 1,3:1,4-β-d-glucanases. Bioorg. Med. Chem. 4, 1849–1855. [DOI] [PubMed] [Google Scholar]
- Namchuk, M.N., and Withers, S.G. (1995). Mechanism of Agrobacterium β-glucosidase: Kinetic analysis of the role of noncovalent enzyme/substrate interactions. Biochemistry 34, 16194–16202. [DOI] [PubMed] [Google Scholar]
- Nari, J., Noat, G., Ricard, J., Franchini, E., and Moustacas, A.-M. (1982). Catalytic properties and tentative function of a cell wall β-glucosyltransferase from soybean cells cultured in vitro. Plant Sci. Lett. 28, 313–320. [Google Scholar]
- Nicolls, A., Sharp, K., and Hönig, B. (1991). Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 4, 281–296. [DOI] [PubMed] [Google Scholar]
- Noguchi, K., Okuyama, K., Kitamura, S., and Takeo, K. (1992). Crystal structure of methyl 3-O-β-d-glucopyranosyl-β-d-glucopyranoside (methyl β-d-laminaribioside) monohydrate. Carbohydr. Res. 237, 33–43. [DOI] [PubMed] [Google Scholar]
- Notenboom, V., Birsan, C., Warren, R.A.J., Withers, S.G., and Rose, D.R. (1998). Exploring the cellulose/xylan specificity of the β-1,4-glycanase Cex from Cellulomonas fimi through crystallography and mutation. Biochemistry 37, 4751–4758. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z., and Minor, W. (1996). A processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Papanikolau, Y., Prag, G., Tavlas, G., Vorgias, C.E., Oppenheim, A.B., and Petratos, K. (2001). High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into mechanism of catalysis. Biochemistry 40, 11338–11343. [DOI] [PubMed] [Google Scholar]
- Parsiegla, G., Reverbel-Leroy, C., Tardif, C., Belaich, J.O., Driguez, H., and Haser, R. (2000). Crystal structures of the cellulase Cel48F in complex with inhibitors and substrates give insights into its processive action. Biochemistry 39, 11238–11246. [DOI] [PubMed] [Google Scholar]
- Perella, F. (1988). EZ-FIT: A practical curve-fitting microcomputer program for the analysis of the enzyme kinetic data on IBM-PC compatible computers. Anal. Biochem. 174, 437–447. [DOI] [PubMed] [Google Scholar]
- Perez, S., Meyer, C., and Imberty, A. (1995). Practical tools for accurate modeling of complex carbohydrates and their interactions with proteins. In Modeling of Biomolecular Structures and Mechanisms, A. Pullman, J. Jortner, and B. Pullman, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp 425–454.
- Perez, V., and Vergelati, C. (1984). Structure and conformational analysis of methyl α-thiomaltoside, C13H24O10S. Acta Crystallogr. Sect. B 40, 294.–299. [Google Scholar]
- Powell, M.J.D. (1977). Restart procedures for the conjugate gradient method. Math. Progr. 12, 241–254. [Google Scholar]
- Quiocho, F.A. (1986). Carbohydrate-binding proteins: Tertiary structures and protein-sugar interactions. Annu. Rev. Biochem. 55, 287–315. [DOI] [PubMed] [Google Scholar]
- Reverbel-Leroy, C., Parsiegla, G., Moreau, V., Juy, M., Tardif, C., Driguez, H., Belaich, J.P., and Haser, R. (1998). Crystallization and the catalytic domain of Clostridium cellolyticum CeIF cellulase in the presence of a newly synthesized cellulase inhibitor. Acta Crystallogr. Sect. D 54, 114–118. [DOI] [PubMed] [Google Scholar]
- Rohrer, D.C., Sarko, A., Bluhm, T.L., and Lee, Y.N. (1980). The structure of gentiobiose. Acta Crystallogr. Sect. B 36, 650.–654. [Google Scholar]
- Roulin, S., Buchala, A.J., and Fincher, G.B. (2002). Induction of (1→3,1→4)-β-d-glucan hydrolases in leaves of dark-incubated barley seedlings. Planta, in press. [DOI] [PubMed]
- Stone, B.A., and Svensson, B. (2002). Enzymic depolymerisation and synthesis of polysaccharides. In Glycoscience: Chemistry and Chemical Biology, B. Fraser-Reid, K. Tatsuta, and J. Thiem, eds (Berlin: Springer-Verlag), pp. 1–86.
- Stubbs, H.J., Brasch, D.J., Emerson, G.W., and Sullivan, P.A. (1999). Hydrolase and transferase activities of the β-1,3-exoglucanase of Candida albicans. Eur. J. Biochem. 263, 889–895. [DOI] [PubMed] [Google Scholar]
- Sulzenbacher, G., Driguez, H., Henrissat, B., Schülein, M., and Davies, G.J. (1996). Structure of the Fusarium oxysporum endoglucanase I with a nonhydrolyzable substrate analogue: Substrate distortion gives rise to the preferred axial orientation for the leaving group. Biochemistry 35, 15280–15287. [DOI] [PubMed] [Google Scholar]
- Tews, I., Perrakis, A., Oppenheim, A., Dauter, Z., Wilson, K.S., and Vorgias, C.E. (1996). Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease. Nat. Struct. Biol. 3, 638–648. [DOI] [PubMed] [Google Scholar]
- Tews, I., Terwisscha, A.C., Perrakis, A., Wilson, K.S., and Dijkstra, B.W. (1997). Substrate-assisted catalysis unifies two families of chitinolytic enzymes. J. Am. Chem. Soc. 119, 7954–7959. [Google Scholar]
- Thoma, J.A., Brothers, C., and Spradlin, J. (1970). Subsite mapping of enzymes: Studies on Bacillus subtilis amylase. Biochemistry 9, 1768–1775. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). ClustalW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese, J.N., Hrmova, M., and Fincher, G.B. (1999). Three-dimensional structure of a barley β-d-glucan exohydrolase, a family 3 glycosyl hydrolase. Structure 7, 179–190. [DOI] [PubMed] [Google Scholar]
- Vyas, N.K., Vyas, M.N., and Quiocho, F.A. (1988). Sugar and signal transducer binding sites of the Escherichia coli galactose chemoreceptor protein. Science 242, 1290–1295. [DOI] [PubMed] [Google Scholar]
- White, A., Tull, D., Johns, K., Withers, S.G., and Rose, D.R. (1996). Crystallographic observation of a covalent catalytic intermediate in a β-glycosidase. Nat. Struct. Biol. 3, 149–154. [DOI] [PubMed] [Google Scholar]
- Zou, J.Y., Kleywegt, G.J., Stahlberg, J., Driguez, H., Nerinckx, W., Claeyssens, M., Koivula, A., Teeri, T.T., and Jones, T.A. (1999). Crystallographic evidence for substrate distortion and protein conformational changes during catalysis in cellobiohydrolases Cel6A from Trichoderma reesei. Structure 7, 1035–1045. [DOI] [PubMed] [Google Scholar]