Abstract
Centromeric H3-like histones, which replace histone H3 in the centromeric chromatin of animals and fungi, have not been reported in plants. We identified a histone H3 variant from Arabidopsis thaliana that encodes a centromere-identifying protein designated HTR12. By immunological detection, HTR12 localized at centromeres in both mitotic and meiotic cells. HTR12 signal revealed tissue- and stage-specific differences in centromere morphology, including a distended bead-like structure in interphase root tip cells. The anti-HTR12 antibody also detected spherical organelles in meiotic cells. Although the antibody does not label centromeres in the closely related species Arabidopsis arenosa, HTR12 signal was found on all centromeres in allopolyploids of these two species. Comparison of the HTR12 genes of A. thaliana and A. arenosa revealed striking adaptive evolution in the N-terminal tail of the protein, similar to the pattern seen in its counterpart in Drosophila. This finding suggests that the same evolutionary forces shape centromeric chromatin in both animals and plants.
INTRODUCTION
Centromeres are the specialized chromosomal sites necessary for poleward movement during mitosis and meiosis in eukaryotes. Commonly, a centromere is evident as a prominent constriction within the heterochromatin of each metaphase chromosome. The attachment to and movement of chromosomes along the spindle is mediated by the proteinaceous kinetochores, which form at the centromeres during cell division.
Despite this highly conserved function, centromeric DNA sequences are not conserved between organisms. For example, human centromeres consist of large blocks (200 kb to several megabases) of tandemly repeated 171-bp α-satellite (Willard, 1998), but the sequences can differ from those of apes on homologous chromosomes (Haaf and Willard, 1997). Similarly, Drosophila melanogaster centromeric regions contain blocks of 5- to 12-bp satellite repeats that do not appear to be shared by homologous centromeres of sibling species (Lohe and Brutlag, 1987).
Plant centromeric regions resemble their mammalian counterparts in that both have large arrays of tandem repeats, frequently of approximately nucleosomal size. In centromere regions of maize (Alfenito and Birchler, 1993; Ananiev et al., 1998), pearl millet (Kamm et al., 1994), rice (Dong et al., 1998), sugarcane (Nagaki et al., 1998), sorghum (Zwick et al., 2000), the Australian daisy Brachycome dichromosomatica (Leach et al., 1995), rape (Xia et al., 1993), and the wild beet Beta procumbens (Gindullis et al., 2001), tandem repeats have been found that differ in sequence but that all have lengths in the range of ∼140 to 180 bp. Particular repeat arrays in cereals have been estimated to be hundreds of kilobases long (Dong et al., 1998; Kaszás and Birchler, 1998). Cereal tandem repeat arrays are interrupted by TY3/gypsy-like retrotransposons and other retrotransposons (Ananiev et al., 1998; Kumekawa et al., 2001; Nonomura and Kurata, 2001). The satellite arrays and interspersed retrotransposons of cereal centromeric regions resemble a proposed model for the structure of a centromeric region from wild beet (Gindullis et al., 2001). In this model, a central tandem array of a 160-bp satellite is flanked by arrays of a nonhomologous satellite, with both satellite arrays interrupted by TY3/gypsy-like retrotransposons.
In Arabidopsis thaliana, centromeric regions have a core consisting of large tandem arrays of 180-bp repeats (Martinez-Zapater et al., 1986; Simoens et al., 1988; Maluszynska and Heslop-Harrison, 1991). These repeat arrays are embedded in a recombination-deficient heterochromatic region that consists largely of retrotransposons and other moderately repetitive sequences (Copenhaver et al., 1999; Arabidopsis Genome Initiative, 2000; Kumekawa et al., 2000; Haupt et al., 2001). Fluorescent in situ hybridization probes to the 180-bp repeats and to the repeat 106B (Thompson et al., 1996), which is related to the heterogeneous Athila family of TY3/gypsy-like retrotransposons (Pelissier et al., 1995; Marin and Llorens, 2000), have been shown to hybridize to the leading points of metaphase chromosomes (Fransz et al., 1998, 2000; Haupt et al., 2001). This indicates that they are present in the centromere, whereas repeats in nearby clones are pericentromeric. This organization, a central domain of satellite that mediates spindle attachment flanked by pericentromeric heterochromatin, conforms to a general model of the structure of centromeric regions (Choo, 2001).
The structure of the central domain is imperfectly known. Estimates of the sizes of the central domains of 180-bp repeats range from 1.1 to 2.9 Mb (Round et al., 1997; Jackson et al., 1998; Kumekawa et al., 2000; Haupt et al., 2001). Sequencing 435 kb from the two ends of the central domain of chromosome 5 showed that they consisted of nearly equal amounts of 180-bp repeats and interspersed Athila derivatives, plus 4% other sequences (Kumekawa et al., 2000). The 180-bp repeats at one end of the core were oriented oppositely to those at the other end, similar to the organization in Schizosaccharomyces pombe centromeres (Chikashige et al., 1989; Clarke et al., 1993), in which inversely symmetrical repeat arrays flank a central core. Together, these observations suggest that centromeric sequence organizational principles derive from a common ancestor of fungi, animals, and plants.
Although their DNA sequences are not conserved, animal and fungal centromeres are characterized by special histone H3 variants, or centromeric H3s (CenH3s), which are thought to identify centromeres. The first recognized CenH3, the CENP-A protein (Earnshaw and Rothfield, 1985; Palmer et al., 1991; Sullivan et al., 1994), is present at all active human centromeres (Warburton et al., 1997; Sullivan and Willard, 1998; Tyler-Smith et al., 1999; Vafa et al., 1999). It replaces conventional H3 in centromeric nucleosomes (Shelby et al., 1997) and is associated with the nucleosome-length α-satellite repeats in phased arrays (Vafa and Sullivan, 1997). It is present constitutively on chromosomes, surviving the protamine transition in sperm that removes noncentromeric nucleosomes (Palmer et al., 1991). CenH3s also are found in other organisms: Cid in D. melanogaster (Henikoff et al., 2000), SpCENPA or Cnp1 in S. pombe (Takahashi et al., 2000), Cse4 from the “point” centromeres of Saccharomyces cerevisiae (Meluh et al., 1998), and HCP-3 from the holokinetic centromeres of Caenorhabditis elegans (Buchwitz et al., 1999). The latter two species lack the regional centromeres of other eukaryotes. Thus, CenH3s appear to be found universally at all types of functional centromeres in animals and fungi.
The parallels among animal, fungal, and plant centromeres in terms of sequence organization raise the question of whether CenH3 histones also are found in plant centromeric chromatin. Although several kinetochore proteins have been described in plants (reviewed in Yu et al., 2000), no plant CenH3 has yet been characterized. Here, we describe a gene that encodes the A. thaliana CenH3. We describe the dynamic distribution pattern of this protein, as revealed by immunofluorescence, and that of an intriguing cross-reacting meiotic organelle. Comparison of this gene, designated HTR12, between related Arabidopsis species demonstrates that the N-terminal tail of HTR12 is undergoing adaptive evolution.
RESULTS
CenH3 Identification and Analysis
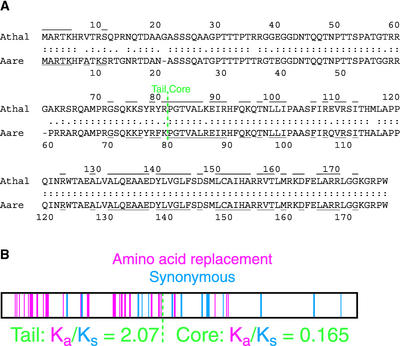
The sequence of A. thaliana histone H3 was used to search the High Throughput Genomic Sequences database (March 16, 2000) using TBLASTN (http://www.ncbi.nlm.nih.gov:80/BLAST/). A hit to part of an unordered contig of clone F6F3 from the A. thaliana genome revealed features of a CenH3 (Henikoff et al., 2000). An FGENEP (Solovyev et al., 1994) prediction of the open reading frame revealed putative N and C termini, and primers were made for amplification of the intervening region from cDNA pools of the Columbia-0 and Nossen-0 ecotypes (Henikoff and Comai, 1998). The amplified fragments from the two ecotypes were identical in sequence and revealed a coding region of 178 amino acids. The predicted protein has 58% identity to H3 in the histone core, with a longer loop 1 region and an N-terminal tail that is not alignable to H3 (Figure 1A). The histone core does not group with conventional H3 proteins in a phylogenetic tree (Malik and Henikoff, 2001).
Figure 1.
HTR12 Genes from A. thaliana and A. arenosa.
(A) Predicted protein sequences of HTR12 from A. thaliana (Athal) and A. arenosa (Aare). Amino acid identities (:) and conservative substitutions (.) are indicated. Underlines and overlines indicate amino acid identity to A. thaliana histone H3.
(B) Positions of nucleotide substitutions in the A. arenosa HTR12 gene that result in nonsynonymous (red) and synonymous (blue) codon changes relative to the A. thaliana gene.
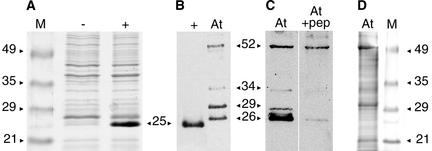
An antibody to the putative protein was raised using an octadecapeptide from the N terminus that is not encoded elsewhere in the genome. The antibody was affinity purified using the same peptide. We tested this antibody in a protein gel blot assay against a protein extract of Escherichia coli BL21(DE3)pLysS cells expressing the HTR12 cDNA under the control of an inducible T7 promoter (Figure 2A). The predicted molecular mass for the HTR12 protein is 19 kD, but it is typical for histones to run anomalously because they are highly basic. The antibody detected a single band with an apparent molecular mass of 25 kD (Figure 2B).
Figure 2.
Protein Gel Blot Analysis Using the Anti-HTR12 Antibody.
Sources of protein extracts are uninduced E. coli cells carrying the HTR12 cDNA under the control of an isopropylthio-β-galactoside–inducible T7 promoter (−), induced cells of same culture (+), and A. thaliana (At) immature inflorescences. M, molecular mass markers. Protein sizes are in kD.
(A) Coomassie blue–stained SDS-PAGE gel showing the induction of HTR12 protein expression in E. coli.
(B) Anti-HTR12 antibody detection on a protein gel blot comparing bacterially expressed HTR12 protein with proteins detected in A. thaliana.
(C) Anti-HTR12 antibody binding to identical lanes of a protein gel blot in the absence (left) or presence (right; +pep) of the octadecapeptide antigen at a peptide:antibody molar ratio of 1:2.
(D) Coomassie blue–stained SDS-PAGE gel of A. thaliana protein extract.
In protein extracts from A. thaliana immature inflorescences, the antibody detected two major bands with apparent molecular masses of 26 and 29 kD (Figure 2B), suggesting that the HTR12 protein is modified post-translationally in vivo. The intensity of the 29-kD band varied among extracts and in repeated assays of the same extract, indicating that it is relatively labile. The antibody also sometimes detected minor bands, the most consistent of which were estimated to correspond to protein sizes of 52 and 34 kD. To determine whether these bands represented antigen-specific or nonspecific binding, we used increasing concentrations of the octadecapeptide antigen to compete off antibody binding in a protein gel blot assay. Binding to the two major bands and the minor 34-kD band was reduced by the peptide, whereas the 52-kD band was affected only marginally (Figure 2C). We conclude that the latter minor band represents nonspecific binding and note that the 52-kD band may correspond to the most prominent band visible in Coomassie blue–stained protein extracts (Figure 2D).
To verify that HTR12 encodes a CenH3, we used indirect immunofluorescence with a fluorescein isothiocyanate–conjugated secondary antibody to detect anti-HTR12 antibody binding in fixed cells. We chose anthers to search for the centromeric localization of HTR12 protein because they contain both meiotic pollen mother cells (PMCs) and somatic cells such as those of the tapetum.
HTR12 in Dividing Tapetum Cells
In premeiotic anthers, interphase tapetum cells have round nuclei with large nucleoli. As PMCs progress from leptotene to pachytene, tapetum cells undergo a mitotic division without cytokinesis to generate binucleate cells. Subsequent additional rounds of mitosis or endomitosis produce subsets of cells with up to three nuclei and ploidy levels up to 8n (Weiss and Maluszynska, 2001).
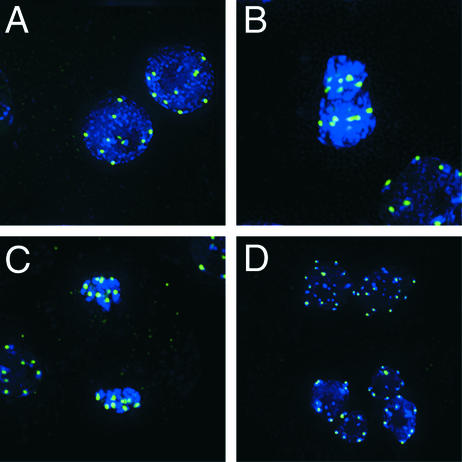
HTR12 protein was detected in all tapetum cells in premeiotic and meiotic anthers. In interphase tapetum cells from premeiotic anthers, HTR12 usually was detected in 10 spots (range, 7 to 10; Figure 3A), corresponding to the 10 centromeres of diploid cells. In interphase tapetum cells from slightly older meiotic anthers, it was apparent that the HTR12 spots were located on 4′,6-diamidino-2-phenylindole (DAPI)–bright heterochromatic regions (Figure 3B, bottom right). During metaphase, the number of spots approached 20, and spots could be seen to lie centrally on chromosomes (Figure 3B). The spots were on the leading portions of the chromosomes during anaphase (Figure 3C). This finding indicates that the protein is located at centromeres. In binucleate tapetum cells, nuclei with ∼10 or up to 20 distinct HTR12 signals were observed (Figure 3D), corresponding to diploid or tetraploid nuclei, respectively, and consistent with ploidy level determinations based on rDNA probes (Weiss and Maluszynska, 2001). We also observed HTR12 signal in trinucleate cells (data not shown). These three classes account for 99% of mature tapetum cells (Weiss and Maluszynska, 2001). The presence of signal at all mitotic stages indicates that HTR12 is a constitutive component of centromeric chromatin.
Figure 3.
HTR12 Protein in Mitotic Tapetum Cells.
Green indicates anti-HTR12; blue indicates DAPI.
(A) Mitotic interphase in a premeiotic anther.
(B) to (D) Mitotic cells in a meiotic anther: metaphase (center) and interphase (bottom right) (B); anaphase (center) and interphase (left) (C); and interphase binucleate cells (D). Note that in (D) the nuclei in the lower two cells are diploid, whereas those in the upper cell are tetraploid.
HTR12 during Meiotic Prophase
As PMCs enter meiosis, they can be distinguished from somatic cells by their larger size. When the chromosomes of PMCs became visible in leptotene, up to 10 spots of HTR12 protein were detected. The spots were distinctly larger in PMCs than in somatic cells (Figure 4A). Some of the largest spots were on clusters of centromeres, which are known to aggregate together at this stage (Ross et al., 1996). This aggregation persisted in some cells as homologs synapsed, so that by late zygotene or pachytene, between two and five large centromeric signals were visible (Figure 4B).
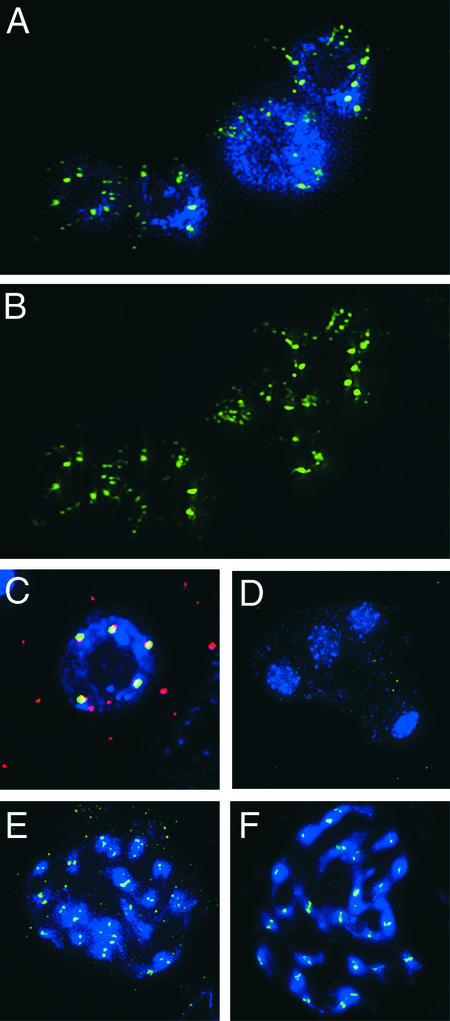
Figure 4.
Anti-HTR12 Staining in PMCs and Pollen Cells.
Green indicates anti-HTR12; blue indicates DAPI.
(A) Leptotene PMC at top left, somatic cell at bottom right.
(B) Zygotene and pachytene PMCs with somatic cells at bottom left.
(C) and (F) Pachytene chromosomes at two different pixel intensities. In (F), note the presence of signal on the euchromatic arms but its absence from the nucleolus organizers (DAPI-bright regions on the ends of the chromosomes seen at left) and the pericentric heterochromatin.
(D) and (E) Metaphase 1. Inset in (D) shows an enlargement of a niu body.
(G) Anaphase 1. There is no detectable DAPI signal under niu bodies (inset).
(H) Two cells of a meiotic tetrad. Note the location of niu bodies outside the nuclei.
(I) Binucleate tapetum cell with fluorescent vesicles and an intact pollen grain, which fluoresces in the green channel. Anti-HTR12 antibody does not penetrate the pollen wall.
(J) Crushed pollen grains and somatic cells. Strong anti-HTR12 staining in the pollen cells contrasts with smaller signals in some somatic cells (bottom left) and weak or undetectable signals in others (bottom right).
In pachytene cells, the chromosomes reach maximum synapsis and the bivalents are differentiated into chromomeres (Ross et al., 1996). The primary constrictions of the chromosomes were seen readily in the pericentric heterochromatin of these cells. HTR12 signals overlay the primary constrictions and were visibly bipartite, with the centromere of one chromosome in a bivalent distinguishable from its homolog (Figure 4C). We also detected weak anti-HTR12 signal on the euchromatic chromosome arms (Figure 4F). This is specific staining, because it was not observed in the nonchromosomal nucleoplasm or over the pericentromeric heterochromatin or nucleolus organizers. We were unable to distinguish such weak euchromatic signal from nonspecific background at other stages. The weak euchromatic signal may be specific to pachytene cells or it may be seen most easily at this stage.
Anti-HTR12 Staining from Metaphase I through Microspore Formation
Homologs desynapse in diplotene, and further chromosome condensation occurs before metaphase. Up to 10 HTR12 signals were distinguishable in metaphase I cells (Figure 4D). After alignment on the plate, the arrangement of the 10 signals in five pairs was clear (Figure 4E). On many chromosomes, each signal was visibly bipartite, indicating that sister centromeres can be distinguished at this stage. In anaphase I cells, HTR12 signal was found at the leading edge of each chromosome (Figure 4G). The signal lay just poleward to, rather than over, the DAPI staining (Figure 4G, inset). The centromeric DNA may be difficult to detect with DAPI because of the decondensation and stretching of the centromeric repeats at this stage (Fransz et al., 2000).
In addition to the centromeric signals, the anti-HTR12 antibody detected a few bright circular objects with diameters in the range of 0.5 to 0.7 μm in metaphase and anaphase cells as well as a variable number of smaller (∼0.3-μm) dots of fluorescence (Figures 4D, 4E, 4G, and 4H). We are uncertain whether the small dots represent signal, background, or a mix of both. In contrast, the larger circular objects appear to represent a unique meiotic organelle. These objects were not found in earlier meiotic stages or in mitotic cells. Typically, the objects were slightly larger and slightly brighter than centromeric signals in the same cells. They did not stain detectably with DAPI (Figure 4G, inset). They were nearly always circular or ring shaped in cross-section (Figure 4D, inset), suggesting that they are thick-walled spheres that are hollow with respect to the epitope detected by the anti-HTR12 antibody. The hollow interior could be resolved in approximately half of the objects of this size. In some of our images, the cross-sectional rings appeared to be composed of a few subunits. We refer to these objects as niu bodies, from the Hawaiian term for coconuts. To our knowledge, these meiosis-specific organelles have not been described previously. Because we do not have a second antibody to the HTR12 protein, we cannot exclude the possibility that our antibody cross-reacted to a different epitope in the niu bodies. If so, the protein apparently would be unique to this organelle.
The number and location of the niu bodies varied. One to four niu bodies were seen in metaphase I cells, sometimes located peripherally and sometimes intermixed with the chromosomes. In five of five anaphase I cells, four niu bodies were detected per cell. They were found at the poles, near the chromosomes, and at peripheral locations (Figure 4G). In postmeiotic interphase nuclei of tetrads and free microspores, five centromeric signals were visible, arranged around the nucleolus. One niu body was found outside each haploid nucleus in 20 of 26 tetrad and free microspore cells examined (Figure 4H).
Anti-HTR12 Staining during Pollen Formation
Microspores develop the specialized pollen wall, which consists of the inner intine made by the microspore and the outer patterned exine made largely from material secreted by the tapetum cells (reviewed in McCormick, 1993; Bedinger et al., 1994). The anti-HTR12 antibody did not penetrate the mature pollen wall, and most pollen grains displayed no anti-HTR12 signal, although the pollen wall itself was fluorescent in both the green (Figure 4I) and red (data not shown) channels. The fluorescent pigments were present in the exine and probably were secreted by the tapetum. A subset of tapetum cells were identified that became filled with vesicles that fluoresced in the green channel (Figure 4I) and more weakly in the red channel (data not shown). These vesicles fluoresced in both the red and green channels regardless of whether a fluorescein isothiocyanate–conjugated or a Texas Red–conjugated secondary antibody was used to detect the anti-HTR12 antibody, indicating that the fluorescence is independent of the antibody (data not shown).
Occasionally, pollen walls cracked or were ruptured in squashing, giving the anti-HTR12 antibody access to the interphase microspore nuclei. Five centromeric signals were visible in each of these nuclei (Figure 4J). These signals were larger than those in somatic cells (Figure 4J, bottom left), suggesting that the centromeres retain the chromatin structure they had throughout meiosis. We observed a niu body in only 1 of 25 pollen microspores with centromeric signals, indicating that the niu bodies disappear quickly after pollen wall formation, before the first pollen mitosis. Smaller fluorescent dots are common in pollen microspores (data not shown), suggesting that the smaller dots present in PMCs and pollen grains may represent stages in the assembly and disassembly, respectively, of niu bodies.
Although centromeric signals were visible in pollen microspores and in some somatic cells, centromeric signals in most binucleate tapetum cells became weak to undetectable at this stage (Figure 4J, bottom right). At a later stage, tapetum cells lose visible nuclei and eventually are destroyed (Owen and Makaroff, 1995). This suggests that HTR12 protein is lost as part of the developmental program in these terminally differentiated cells. A similar loss of Cid protein in terminally differentiated Drosophila cells also has been observed (J.S. Platero, personal communication).
HTR12 in Interphase Root Tips
Anther development is highly specialized, and centromere behavior in this tissue might not be general, so we also chose to examine HTR12 distribution in the rapidly dividing root apex. In interphase cells of this tissue, HTR12 signals varied from single dots of ∼0.5 μm diameter, similar to those in anther tissue, to bar shapes, to discontinuous strings of smaller bead-like dots with a total length of 1.0 to 2.0 μm (Figures 5A and 5B). There were too many of the smaller bead-like dots for each one to represent a centromere, so we conclude instead that each string of dots must represent a single centromere (or pair of sister centromeres) in a decondensed state.
Figure 5.
Anti-HTR12 Staining in Interphase Root Tips and Allotetraploids, and Colocalization with 180-bp Repeats.
(A) and (B) Anti-HTR12 staining in interphase root tip cells. DAPI staining (A) is omitted in (B) for greater visibility of the centromeric structure.
(C) A probe for the 180-bp repeat (red) and anti-HTR12 staining (green) coincide or overlap (yellow) in a pollen cell nucleus.
(D) Anti-HTR12 antibody detects only nonspecific background in a meiotic tetrad of A. arenosa.
(E) Anti-HTR12 staining in mitotic prophase cells of a synthetic allotetraploid of A. thaliana and A. arenosa.
(F) Anti-HTR12 staining in mitotic prophase cells of A. suecica.
We examined 24 root tip nuclei. Assuming a maximum of 10 centromeres or centromere pairs per nucleus, we found that in 6 nuclei the centromeres were predominantly string like (with no more than two dot-like centromeres), in 7 nuclei the centromeres were predominantly dots or bars (with two or fewer strings), and in 11 nuclei the centromeres were an approximately equal mix of dots and strings. The occurrence of nuclei with both mostly dots and mostly strings suggests that the variation in the appearance of centromeres from dots or bars to strings probably reflects different degrees of decondensation rather than structural differences among the centromeres on different chromosomes.
Colocalization of Anti-HTR12 and 180-bp Repeats
The distribution of HTR12 protein is expected to coincide with the centromeric DNA sequences, which include the 180-bp and 106B repeats (Fransz et al., 2000). We combined anti-HTR12 staining with fluorescent in situ hybridization to the 180-bp repeat. In all double-labeled cells, the signals coincided or overlapped (Figure 5C). Therefore, we conclude that HTR12 protein is found on or in close proximity to the 180-bp repeats.
The HTR12 Gene of Arabidopsis arenosa
The Centromere identifier (Cid) gene of D. melanogaster encodes a CenH3 that has been shown to be under positive adaptive selection in the N-terminal tail and the loop 1 region of the histone core (Malik and Henikoff, 2001), and such adaptive evolution has been predicted to be common in CenH3s of higher eukaryotes, including plants (Henikoff et al., 2001). To test this prediction for HTR12, we amplified and sequenced the HTR12 ortholog from the closely related autotetraploid species Arabidopsis arenosa (also known as Cardaminopsis arenosa; 2n = 4x = 32). Comparison of the sequences revealed 22 nonsynonymous base changes plus two codon deletions in the 79–amino acid A. arenosa N-terminal tail relative to the A. thaliana sequence, but only 5 synonymous substitutions (Figure 1). In contrast, silent changes exceeded replacement changes by nine to four in the histone core.
We used K-Estimator (Comeron, 1999) to estimate the average number of substitutions per synonymous site (Ks) and per nonsynonymous site (Ka) for each part of the protein. For the tail, Ks = 0.071, Ka = 0.147, and Ka/Ks = 2.07. The value of Ka exceeded that of Ks at the 99% confidence level, indicating that the tail is under positive (adaptive) selection. In contrast, for the core, Ks = 0.121, Ka = 0.020, and Ka/Ks = 0.165. The value of Ka was less than that of Ks at the 99% confidence level, indicating that the core is under negative (purifying) selection.
Because of the differences in the N-terminal tails of the A. thaliana and A. arenosa HTR12 proteins, we did not expect our anti-HTR12 antibody to cross-react to centromeres in A. arenosa. This proved to be correct, because only nonspecific background staining could be seen in repeated tests of A. arenosa anthers. Centromeric staining was not detected, even when A. arenosa anthers were on the same slide as A. thaliana anthers of a comparable developmental stage with centromeric signal. Niu bodies were not observed in A. arenosa meiotic tetrads and free microspores (Figure 5D), although they were detectable in A. thaliana tetrads and microspores on the same slide (data not shown).
HTR12 Behavior in Allotetraploids
The formation of allotetraploids is a significant mode of speciation in flowering plants (reviewed in Otto and Whitton, 2000). Because of differences in chromosome number, a new allotetraploid individual is isolated reproductively from its parents immediately. When allotetraploids are formed, the centromere-specific histones of each parent species operate in the context of a dual set of centromeric sequences. Both natural and synthetic allotetraploids of A. thaliana exist. We wondered whether the A. thaliana HTR12 protein recognized only the A. thaliana centromere sequences in these allotetraploids or whether it would recognize the centromeres from the other parent as well.
We stained for A. thaliana HTR12 protein in a recently derived synthetic allotetraploid (2n = 26) between A. thaliana and A. arenosa (Comai et al., 2000) and in Arabidopsis suecica (2n = 26), a natural allotetraploid with one genome donated by A. thaliana that may be postglacial in origin (Suominen, 1994). The other species contributing a genome to A. suecica often is thought to be A. arenosa, although a closely related species cannot be excluded (O'Kane et al., 1996). In both the natural and synthetic allotetraploids, A. thaliana HTR12 was found at the centromeres of all 26 chromosomes (Figures 5E and 5F), indicating that it interacts with both parental sets of centromeric sequences.
We sequenced the A. thaliana–derived HTR12 gene of A. suecica to determine whether any changes had occurred in response to its interaction with two distinct sets of centromeric sequences. The A. thaliana–derived HTR12 gene in A. suecica had only one silent change in the coding region and eight intronic changes relative to HTR12 from the A. thaliana Columbia ecotype. We sequenced eight additional ecotypes of A. thaliana and found that four (Landsberg erecta, Niederenz-1, Frickhofen-0, and Burghaun-20) were identical to Columbia; one (Metzudeh) had no amino acid replacements and eight silent changes; and three had G21A replacements with 8 (Bensheim-0), 9 (Köln-0), or 12 (Tacoma-0) silent changes (Table 1). The A. suecica HTR12 gene clearly falls within the range of variation seen between ecotypes, differing at only five positions from Metzudeh, and shows no evidence of either adaptive or relaxed selection since the formation of the allotetraploid.
Table 1.
Polymorphisms in the HTR12 Gene of A. thaliana
| Position | Columbia | Köln-0 | Tacoma-0 | Bensheim-0 | Metzudeh | A. suecica |
|---|---|---|---|---|---|---|
| 111 | G | G | G | G | T | G |
| 169 | C | T | T | T | C | C |
| 171 | G | C (G21A) | C (G21A) | C (G21A) | G | G |
| 226 | T | T | A | T | T | T |
| 368 | T | T | C | T | T | T |
| 428 | T | T | T | T | T | A |
| 587 | T | A | A | A | A | A |
| 598a | – a | A | A | A | A | A |
| 598b | – | A | A | A | – | – |
| 598c | – | – | A | – | – | – |
| 601 | A | C | C | C | C | C |
| 625 | T | T | T | T | T | G |
| 638 | G | T | T | T | T | T |
| 790 | G | G | G | G | G | T |
| 1024 | G | G | T | G | G | G |
| 1038 | G | T | T | T | T | T |
| 1060 | A | G | G | G | G | G |
| 1326 | C | C | C | C | G | C |
| 1519 | A | G | A | A | A | A |
Landsberg erecta, Niederenz-1, Frickhofen-0, and Burghaun-20 ecotypes are identical to Columbia. Nucleotide positions are numbered from the start of the Columbia coding sequence.
a Dash indicates a deletion relative to the Tacoma-0 sequence.
DISCUSSION
Centromere Condensation and Structure
We have shown that the HTR12 gene encodes an H3-like variant histone that is found specifically at the cytologically visible centromeres and colocalizes with the 180-bp repeats. Our anti-HTR12 antibody provides a powerful tool for the study of centromeres throughout the cell cycle. It has revealed cell- and stage-specific differences in the apparent structure and organization of centromeric chromatin, which we interpret as differences in centromere condensation.
In tapetum cells, HTR12 signals are detected as dots throughout the cell cycle, suggesting that the centromeres in these cells remain condensed between mitoses. In PMCs, on the other hand, HTR12 signals are dot like in most stages but appear distinctly larger than those in tapetum cells, suggesting that they are in a less condensed state that persists throughout meiosis and into the pollen microspores. Stage-specific changes in the condensation level of meiotic centromeres relative to surrounding pericentric regions have been documented in A. thaliana (Fransz et al., 2000). The function of such stage-specific decondensation of centromeres is unknown, but presumably it is important for normal pairing or disjunction. In hexaploid bread wheat, the Ph1 locus controls both meiotic centromere decondensation and homeologous pairing (Aragón-Alcaide et al., 1997). Meiotic condensation in the Easter lily may be regulated by meiosis-specific H1 histones (Hasenkampf et al., 2000), including a centromere-specific form (Suzuki et al., 1997).
In interphase root tip cells, at least some of the centromeres show a discontinuous bead-like distribution of HTR12 protein, which we interpret as extreme decondensation of centromeric chromatin. This pattern is quite similar to the beaded pattern of prekinetochore staining detected in interphase Indian muntjac cells (Brinkley et al., 1984) or the patterns of staining of stretched metaphase kinetochores from several species, including the African blood lily (Zinkowski et al., 1991). Our observations extend and clarify these earlier studies using anti-centromere serum from calcinosis, Raynauds phenomenon, esophogeal dysmotility, sclerodactyly, telangiectasia syndrome patients because our antibody specifically detects HTR12, implying that the discontinuous beaded pattern occurs at the level of centromeric nucleosomes (Earnshaw and Rothfield, 1985). Our observations provide support for a model of a centromere-kinetochore complex composed of repeated subunits (Zinkowski et al., 1991).
Intriguingly, the discontinuous pattern of HTR12 distribution also resembles a pattern of 180-bp repeat hybridization observed in at least one centromere in an interphase root tip cell (Figure 3f [left-most centromere] in Maluszynska and Heslop-Harrison, 1991), suggesting that a discontinuous arrangement of 180-bp repeats might underlie the discontinuous distribution of HTR12. Alternatively, HTR12 nucleosomes may be distributed discontinuously in spite of a uniform underlying sequence.
HTR12 in Euchromatin
In addition to the centromeric HTR12 signal we observed, we also detected HTR12 signal at noncentromeric sites. In pachytene chromosomes, we found weak signal over the euchromatic arms of the chromosomes, but not over heterochromatic regions. HTR12 may be incorporated randomly into euchromatin at a low level that is normally undetectable at other stages, or it may have an unknown role at specific euchromatic sequences. Alternatively, studies of human CENP-A expression suggest another explanation. The centromeric localization of CENP-A has been shown to depend on both the protein sequence and the expression pattern, with overexpression of CENP-A under its own promoter resulting in uniform nuclear localization in interphase cells (Shelby et al., 1997). This suggests that the weak euchromatic HTR12 signal may be a nonfunctional consequence of higher levels of HTR12 protein in meiotic cells.
A Novel Meiotic Organelle
The apparently high levels of HTR12 protein in pachytene cells may be the prelude to the assembly of niu bodies by metaphase. Although we cannot exclude the possibility that the anti-HTR12 signal in niu bodies is attributable to a cross-reacting protein, the absence of reactivity by our antibody to any structures at all in the closely related A. arenosa is consistent with the hypothesis that niu bodies contain HTR12 protein. This possibility is more parsimonious than assuming the simultaneous loss or gain of a second cross-reacting epitope in one of these two species. It also is intriguing that usually there are four of these bodies in a PMC and each one seems to get distributed to one of the four meiotic products. One possibility is that niu bodies contain functional HTR12 protein to interact with the spindle in some way that ensures their regular segregation into the microspores. Despite this segregation, niu bodies do not seem to be B chromosomes because they do not stain with DAPI, they have a unique structure unlike normal centromeres, and they are found outside the nuclei of the microspores.
Niu bodies may partition HTR12 protein into the microspores for later use in pollen mitosis. The generative nucleus resulting from the first pollen mitosis and its daughter sperm nuclei have condensed chromatin and are thought to have reduced transcriptional activity (McCormick, 1993). In Easter lily, the male gametic histones gH2A, gH2B, and gH3 are expressed in these cells, and they have been proposed to repress gene expression (Ueda et al., 2000). The paternal genome appears to be largely silent in early embryogenesis and endosperm formation in A. thaliana (Vielle-Caldaza et al., 2000). This suggests that HTR12 protein may be partitioned into microspores to compensate for a silenced HTR12 gene in the gametic cells. Alternatively, niu bodies may partition another essential product or a selfish element into microspores.
Adaptive Evolution of the HTR12 Gene
The satellite DNA sequences that are found at and around most eukaryotic centromeres are among the most rapidly evolving sequences known. The 180-bp repeats of A. arenosa are only 58 to 80% identical to those of A. thaliana (Kamm et al., 1995). CenH3s also evolve rapidly. The Drosophila Cid protein shows adaptive evolution in the lineages leading to D. melanogaster and Drosophila simulans (Malik and Henikoff, 2001). Adaptive changes have occurred in both the N-terminal tail and the loop 1 region of the histone core that makes DNA contacts (Luger et al., 1997), suggesting that Cid responds to changes in rapidly evolving satellite sequences. The recurrent evolution of minor groove binding motifs in the tails of CenH3s of several Drosophila species and of mammals (Malik et al., 2002) supports this interpretation.
We have shown that adaptive changes also have occurred between A. arenosa and A. thaliana in the N-terminal tail of HTR12. Although the HTR12 core as a whole is under purifying selection, our analysis lacks the power to determine whether individual amino acid replacements are adaptive. It is intriguing that two of the four nonsynonymous changes that have occurred in the core of the HTR12 gene form a dinucleotide substitution (M116A; ATG→GCG) in the loop 1 region. This suggests that the same forces that drive the adaptive evolution of Cid may affect HTR12 in both the N-terminal tail and the loop 1 region as well. Although HTR12 could be evolving in response to a cellular component other than the centromeric satellite DNA, such as a kinetochore protein, no other component of the conserved cell division apparatus is known to be evolving rapidly. If such a component were discovered, the matter of what drives its rapid evolution would remain, and the centromeric DNA would be a likely candidate.
A two-step model has been proposed to explain the rapid evolution of centromeric chromatin (Henikoff et al., 2001; Malik and Henikoff, 2001). In the first step, centromeric DNA sequence variants compete to take advantage of the asymmetric meiosis in females, in which only one of the four meiotic products contributes to the egg (Zwick et al., 1999). Any heterozygous variant that is more likely to be included in the egg, perhaps as a consequence of attracting more centromeric nucleosomes, will sweep rapidly through the population. The mechanism by which the preferential orientation of a centromere variant toward the future position of the egg is achieved during the separation of centromeres at metaphase I is unclear, but there is evidence that such nonrandom segregation occurs in other systems (Pardo-Manuel de Villena and Sapienza, 2001) and that it is under maternal control in birds (Sheldon, 1999). An analogous meiotic drive process is thought to occur in the distally located neocentromeric knobs of maize after a recombination event, although in this case it is the precocious movement of heterozygous knobs to the anaphase I poles that results in their preferential inclusion in the egg (Rhoades, 1952; Dawe and Cande, 1996).
As favored centromeric variants spread through a population, they may have deleterious consequences if inequality in the “strength” of centromeres (Novitski, 1955) in heterozygotes leads to nondisjunction and hence reduced fitness (Zwick et al., 1999). In the second step of the model, any allele of CenH3 that suppresses such inequality will be selected for and fixed, leading to the divergence of satellite/CenH3 combinations between isolated populations.
Centromere Divergence in Allotetraploids
The HTR12 protein from A. thaliana can be detected at the centromeres of all chromosomes in both our synthetic allotetraploid and A. suecica. The centromeric localization in the synthetic allotetraploid indicates that the protein is competent for localization to the A. arenosa centromeric repeats despite its divergence from the A. arenosa HTR12 protein. This may be a consequence of the formation of HTR12/H4 tetramers that include HTR12 monomers from both parental genomes and therefore are able to interact with both sets of centromeric repeats. In analogy with H3 (Luger et al., 1997), HTR12 monomers are predicted to contact each other in helix 3 of the histone fold, which is identical in A. thaliana, A. arenosa, and both genomes of A. suecica (data not shown).
The presence of A. thaliana–derived HTR12 protein at the centromeres of both parental genomes in A. suecica does not appear to be deleterious, because we found no indication of adaptive selection on the A. thaliana–derived HTR12 gene in A. suecica compared with HTR12 in A. thaliana ecotypes. The minimal divergence of these genes even in their introns is consistent with the proposed recent origin of A. suecica (Suominen, 1994).
The existence of diverged satellite-nucleosome complexes has different implications in allotetraploids and in diploid hybrids. Hybridization between diploid populations that have diverged in their centromeric satellite-nucleosome combinations is predicted to lead to reduced fitness and eventual reproductive isolation because diverged centromeres in heterozygous bivalents may be of unequal strengths and cause nondisjunction (Henikoff et al., 2001). In contrast, the formation of allotetraploids circumvents the immediate problem of unequal centromeres by providing two copies of each parental chromosome that will form a balanced bivalent. In addition, diploid hybrids segregate progeny that are homozygous for the CenH3 of one parent but retain chromosomes from both parents. Homozygous CenH3 in these progeny may not adequately recognize the centromeric satellites from the other parent. In allotetraploids, the CenH3s of both parents are retained in all progeny, and centromeric satellites cannot be segregated away from their coevolved CenH3. Allotetraploids thus avoid some of the potential problems of incompatible satellites and CenH3s encountered by diploids, which may contribute to their evolutionary success.
METHODS
Plant Materials
Growth conditions for all stocks have been described previously (Comai et al., 2000). Arabidopsis thaliana Landsberg erecta, Columbia, and Columbia erecta stocks (all 2n = 10), Arabidopsis arenosa Care-1 (WU9509; 2n = 4x = 32), and Arabidopsis suecica Sue-1 (Hanfstingl et al., 1994; 2n = 26) were used for cytological preparations. The synthetic allotetraploid line used was derived from the F1 individual 605B (Comai et al., 2000).
Gene Amplification and Sequencing
Sources of genomic DNAs and cDNA pools have been described (Henikoff and Comai, 1998). The primers 5′-ATGGCGAGAACCAAGCATCGCGTTAC-3′ and 5′-TCACCATGGTCTGCCTTTTCCTCC-3′ were used to amplify the coding region of HTR12 from cDNA pools. A second pair of primers, 5′-CAATGTAAGAGATGTGAATAGTGAGTGAGTTTTCTATCA-3′ and 5′-CAGGTGACTTTCATAATCGCAGTT-TTCGTC-3′, was used to determine the upstream and downstream sequences, including the termini of the coding region. The primers also were used to amplify A. arenosa cDNA and genomic DNA from A. arenosa and A. suecica. Amplifications were performed with Pfu DNA polymerase (Stratagene) using an annealing temperature of 58°C and 2-min extensions at 72°C for 35 cycles. Subsequent to our amplification and sequencing, the gene was designated HTR12 by the Plant Chromatin Database (http://chromdb.biosci.arizona.edu), and we have adopted this name.
Protein Gel Blot Analysis and Immunolabeling
The anti-HTR12 antibody is an affinity-purified rabbit polyclonal antibody made by Quality Controlled Biochemicals (Hopkinton, MA) against the peptide acetyl-RTKHRVTRSQPRNQTDAC-amide. Crude antibody serum gave similar results for the affinity-purified antibody in cytology and was used to detect HTR12 in anaphase cells. Preimmune serum gave no detectable signal.
For bacterial expression of the HTR12 protein, the amplified cDNA from A. thaliana was placed under a T7 promoter inducible with isopropylthio-β-galactoside using the pCRT7 TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Preparation of protein extracts from immature inflorescences and protein gel blot analysis were performed according to Dawe et al. (1999), except that the volume of extraction buffer added typically was 200 to 600 μL and cellular debris was left in suspension to avoid removing chromatin proteins. For the peptide competition experiment, five equally loaded lanes of a protein gel blot were cut apart and incubated separately with primary antibody solution to which the octadecapeptide was added at peptide-to-antibody molar ratios of 0, 0.5, 1, 5, and 10. Subsequently, the lanes were washed and treated with secondary antibody together.
For cytology, flower buds were fixed for 30 to 60 min under a vacuum in freshly made 4% paraformaldehyde in PBS with 0.2% Triton X-100. Buds were washed in PBS and then stored in PBS at 4°C. For meiotic stages, buds were selected in which the anthers were small and whitish. For pollen, slightly older buds were chosen that had some yellow color, indicating that pollen had formed. Anthers were dissected from the buds in PBS plus 0.5% Triton X-100, placed on a microscope slide, covered with a siliconized cover slip, and squashed with a 7 × 2-inch toggle clamp (Grizzly Industrial, Bellingham, WA). Slides were immersed in liquid nitrogen, the cover slips were removed, and blocking solution (PBS, 10% goat serum, and 0.5% Tween 20) was applied to the anthers for 30 min.
The anti-HTR12 antibody was diluted 1:2000 in blocking solution and applied to the anthers, which then were covered with a cover slip. The slides were incubated in a moist chamber at room temperature for 1 hr or overnight. In the latter case, the slides were sealed with rubber cement. Cover slips were removed, and the slides were washed twice with blocking solution for 2 min. The antibody was detected by applying fluorescein isothiocyanate–conjugated or Texas Red–conjugated goat anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) diluted 1:100 in blocking solution and incubated for 1 hr, followed by two washes in blocking solution for 2 min each. The slides were stained with a 0.25 μg/mL DAPI solution for 5 min, excess liquid was removed, and the anthers were mounted in Vectashield (Vector Laboratories, Burlingame, CA) and sealed with nail polish.
To verify that the lack of signal in A. arenosa anthers was not caused by slide-to-slide variation, 10 slides of A. arenosa anthers prepared on five separate days were compared with A. thaliana slides with signal prepared on the same days. In addition, seven slides bearing both types of anthers at similar developmental stages also were examined.
White root tips were taken from the edges of plants grown in 4-inch pots and were fixed and stored using the same protocol described for anthers. For immunolabeling, the root tips were digested for 25 min at 37°C with a mixture of 2.5% pectinase from Aspergillus niger (Sigma) and 2.5% cellulase Onozuka RS (Yakult Honsha Co., Tokyo, Japan) dissolved in PBS. They were washed in PBS, squashed onto slides, and detected by indirect immunofluorescence as for anthers.
Fluorescent in Situ Hybridization
The 180-bp repeat probe was made by amplifying pARR20 (Round et al., 1997) using primers 5′-CATGGTGGTAGCCAAAGTCCATA-3′ and 5′-GCTTTGAGAAGCAAGAAGAAGG-3′ and standard Taq polymerase conditions with a block preheated to 94°C and an initial denaturation of 94°C for 2 min followed by 32 cycles of 94°C for 25 sec, 57°C for 30 sec, and 72°C for 40 sec. The product was diluted to ∼100 pg/μL and then reamplified using a 1:4 ratio of Fluoro Red rhodamine-4–dUTP (Amersham Pharmacia Biotech) to deoxy thymidine triphosphate in the polymerase chain reaction. To combine HTR12 protein detection with fluorescent in situ hybridization, anthers were prepared as for immunolabeling except that after the washes following the secondary antibody, the anthers were fixed again with 4% paraformaldehyde in PBS and 0.2% Triton X-100 for 15 min.
The slides were washed with 2 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) followed by two washes in 2 × SSC in 70% formamide. The chromosomes then were denatured at 65°C for 15 min in 70% formamide followed by a dehydration series of ethanol solutions (70, 90, and 100%) and air drying. The hybridization solution, containing 50% formamide, 2 × SSC, 10% dextran sulfate, sonicated salmon sperm (0.5 mg/mL), and 1 ng/μL probe, was denatured at 90°C for 5 min and then chilled immediately on ice. Ten microliters of hybridization mixture was added to each slide and covered with a cover slip. Slides were sealed with rubber cement and incubated in a moist chamber at 37°C overnight. Posthybridization washes were in 2 × SSC (5 min, 25°C), 0.2 × SSC (10 min, 55°C), PBS plus 0.1% Triton X-100 (5 min, 25°C), and PBS (5 min, 25°C). Slides were stained with DAPI and mounted as described above.
Although both our anti-HTR12 antibody and our 180-bp repeat probe labeled all anther cells before pollen formation when these procedures were performed separately, only a fraction of anther cells were labeled by both fluorescent signals when the two procedures were combined, indicating technical limitations in the simultaneous detection of both labels using our anti-HTR12 antibody and procedure. However, in all double-labeled cells, the two signals coincided or overlapped.
Imaging
Protein gel blots and Coomassie Brilliant Blue R 250–stained gels were scanned on standard image scanners. Cytological images were collected with the DeltaVision Image Restoration Microscopy System (Applied Precision, Issaquah, WA) using a cooled charge-coupled device camera (Photometrics, Tucson, AZ), SoftWorks 2.50 software (Applied Precision), and a Zeiss Axiovert (Jena, Germany) or Olympus IX70 (Tokyo, Japan) microscope with a ×100 objective with or without ×1.6 auxiliary magnification. Stacks of deconvolved optical sections were projected into a plane for two-dimensional viewing, and measurements were taken using the tools of SoftWorks 2.50. Images were processed using Adobe Photoshop 5.5 (Mountain View, CA).
Accession Numbers
The accession numbers for the sequences described in this article are AF465800 (HTR12), AF465802 (A. arenosa cDNA), and AF465801 (A. arenosa genomic DNA).
Acknowledgments
We thank Kami Ahmad, Harmit Malik, and Danielle Vermaak for helpful discussions and technical assistance, Steve Reynolds for plant care, Jessica Johnson for genomic DNA from A. arenosa and A. suecica, and Eric Richards for pARR20. This work was supported by the Howard Hughes Medical Institute and by U.S. Department of Agriculture National Research Initiative Competitive Grants Program Grant 9901352 and National Science Foundation Plant Genome Grant 0077774.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010425.
References
- Alfenito, M.R., and Birchler, J.A. (1993). Molecular characterization of a maize B chromosome centric sequence. Genetics 135, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiev, E.V., Phillips, R.L., and Rines, H.W. (1998). Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc. Natl. Acad. Sci. USA 95, 13073–13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Aragón-Alcaide, L., Reader, S., Miller, T., and Moore, G. (1997). Centromeric behaviour in wheat with high and low homoeologous chromosomal pairing. Chromosoma 106, 327–333. [DOI] [PubMed] [Google Scholar]
- Bedinger, P.A., Hardeman, K.J., and Loukides, C.A. (1994). Travelling in style: The cell biology of pollen. Trends Cell Biol. 4, 132–138. [DOI] [PubMed] [Google Scholar]
- Brinkley, B.R., Valdivia, M.M., Tousson, A., and Brenner, S.L. (1984). Compound kinetochores of the Indian muntjac: Evolution by linear fusion of unit kinetochores. Chromosoma 91, 1–11. [DOI] [PubMed] [Google Scholar]
- Buchwitz, J.G., Ahmad, K., Moore, L.L., Roth, M.B., and Henikoff, S. (1999). A histone-H3-like protein in C. elegans. Nature 401, 547–548. [DOI] [PubMed] [Google Scholar]
- Chikashige, Y., Kinoshita, N., Nakaseko, Y., Matsumoto, T., Murakami, S., Niwa, O., and Yanagida, M. (1989). Composite motifs and repeat symmetry in S. pombe centromeres: Direct analysis by integration of NotI restriction sites. Cell 57, 739–751. [DOI] [PubMed] [Google Scholar]
- Choo, K.H.A. (2001). Domain organization at the centromere and neocentromere. Dev. Cell 1, 165–177. [DOI] [PubMed] [Google Scholar]
- Clarke, L., Baum, M., Marschall, L.G., Ngan, V.K., and Steiner, N.C. (1993). Structure and function of Schizosaccharomyces pombe centromeres. Cold Spring Harbor Symp. Quant. Biol. 58, 687–695. [DOI] [PubMed] [Google Scholar]
- Comai, L., Tyagi, A.P., Winter, K., Holmes-Davis, R., Reynolds, S.H., Stevens, Y., and Byers, B. (2000). Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell 12, 1551–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron, J.M. (1999). K-Estimator: Calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics 15, 763–764. [DOI] [PubMed] [Google Scholar]
- Copenhaver, G.P., et al. (1999). Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286, 2468–2474. [DOI] [PubMed] [Google Scholar]
- Dawe, R.K., and Cande, W.Z. (1996). Induction of centromeric activity in maize by suppressor of meiotic drive 1. Proc. Natl. Acad. Sci. USA 93, 8512–8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe, R.K., Reed, L.M., Yu, H.-G., Muszynski, M.G., and Hiatt, E.N. (1999). A maize homolog of mammalian CENPC is a constitutive component of the inner kinetochore. Plant Cell 11, 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, F., Miller, J.T., Jackson, S.A., Wang, G.L., Ronald, P.C., and Jiang, J. (1998). Rice (Oryza sativa) centromeric regions consist of complex DNA. Proc. Natl. Acad. Sci. USA 95, 8135–8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw, W.C., and Rothfield, N. (1985). Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91, 313–321. [DOI] [PubMed] [Google Scholar]
- Fransz, P.F., Armstrong, S., Alonso-Blanco, C., Fischer, T.C., Torres-Ruiz, R.A., and Jones, G. (1998). Cytogenetics for the model system Arabidopsis thaliana. Plant J. 13, 867–876. [DOI] [PubMed] [Google Scholar]
- Fransz, P.F., Armstrong, S., de Jong, J.H., Parnell, L.D., van Drunen, C., Dean, C., Zabel, P., Bisseling, T., and Jones, G.H. (2000). Integrated cytogenetic map of chromosome arm 4S of A. thaliana: Structural organization of heterochromatic knob and centromere region. Cell 100, 367–376. [DOI] [PubMed] [Google Scholar]
- Gindullis, F., Desel, C., Galasso, I., and Schmidt, T. (2001). The large-scale organization of the centromeric region in Beta species. Genome Res. 11, 253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf, T., and Willard, H.F. (1997). Chromosome-specific alpha-satellite DNA from the centromere of chimpanzee chromosome 4. Chromosoma 106, 226–232. [DOI] [PubMed] [Google Scholar]
- Hanfstingl, U., Berry, A., Kellogg, E.A., Costa, J.T., III, Rudiger, W., and Ausubel, F.M. (1994). Haplotypic divergence coupled with lack of diversity at the Arabidopsis thaliana alcohol dehydrogenase locus: Roles for both balancing and directional selection? Genetics 138, 811–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenkampf, C.A., Taylor, A.A., Siddiqui, N.U., and Riggs, C.D. (2000). meiotin-1 gene expression in normal anthers and in an-thers exhibiting prematurely condensed chromosomes. Genome 43, 604–612. [DOI] [PubMed] [Google Scholar]
- Haupt, W., Fischer, T.C., Winderl, S., Fransz, P., and Torres-Ruiz, R.A. (2001). The CENTROMERE1 (CEN1) region of Arabidopsis thaliana: Architecture and functional impact of chromatin. Plant J. 27, 285–296. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., and Comai, L. (1998). A DNA methyltransferase homolog with a chromodomain exists in multiple forms in Arabidopsis. Genetics 149, 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., Ahmad, K., Platero, J.S., and van Steensel, B. (2000). Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA 97, 716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., Ahmad, K., and Malik, H.S. (2001). The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293, 1098–1102. [DOI] [PubMed] [Google Scholar]
- Jackson, S.A., Wang, M.L., Goodman, H.M., and Jiang, J. (1998). Application of fiber-FISH in physical mapping of Arabidopsis thaliana. Genome 41, 566–572. [PubMed] [Google Scholar]
- Kamm, A., Schmidt, T., and Heslop-Harrison, J.S. (1994). Molecular and physical organization of a highly repetitive, undermethylated DNA from Pennisetum glaucum. Mol. Gen. Genet. 244, 420–425. [DOI] [PubMed] [Google Scholar]
- Kamm, A., Galasso, I., Schmidt, T., and Heslop-Harrison, J.S. (1995). Analysis of a repetitive DNA family from Arabidopsis arenosa and relationships between Arabidopsis species. Plant Mol. Biol. 27, 853–862. [DOI] [PubMed] [Google Scholar]
- Kaszás, E., and Birchler, J.A. (1998). Meiotic transmission rates correlate with physical features of rearranged centromeres in maize. Genetics 150, 1683–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumekawa, N., Hosuchi, T., Tsuruoka, H., and Kotani, H. (2000). The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 5. DNA Res. 7, 315–321. [DOI] [PubMed] [Google Scholar]
- Kumekawa, N., Ohmido, N., Fukui, K., Ohtsubo, E., and Ohtsubo, H. (2001). A new gypsy-type retrotransposon, RIRE7: Preferential insertion into the tandem repeat sequence TrsD in pericentromeric heterochromatin regions of rice chromosomes. Mol. Genet. Genomics 265, 480–488. [DOI] [PubMed] [Google Scholar]
- Leach, C.R., Donald, T.M., Franks, T.K., Spinello, S.S., Hanrahan, C.F., and Timmis, J.N. (1995). Organisation and origin of a B chromosome centromeric sequence from Brachycome dichromosomatica. Chromosoma 103, 708–714. [DOI] [PubMed] [Google Scholar]
- Lohe, A.R., and Brutlag, D.L. (1987). Identical satellite DNA sequences in sibling species of Drosophila. J. Mol. Biol. 194, 161–170. [DOI] [PubMed] [Google Scholar]
- Luger, K., Mader, A.W., Richmond, R.K., Sargent, D.F., and Richmond, T.J. (1997). Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260. [DOI] [PubMed] [Google Scholar]
- Malik, H.S., and Henikoff, S. (2001). Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157, 1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, H.S., Vermaak, D., and Henikoff, S. (2002). Recurrent evolution of DNA-binding motifs in the Drosophila centromeric histone. Proc. Natl. Acad. Sci. USA, 99, 1449–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maluszynska, J., and Heslop-Harrison, J.S. (1991). Localization of tandemly repeated DNA sequences in Arabidopsis thaliana. Plant J. 1, 159–166. [Google Scholar]
- Marin, I., and Llorens, C. (2000). Ty3/Gypsy retrotransposons: Description of new Arabidopsis thaliana elements and evolutionary perspectives derived from comparative genomic data. Mol. Biol. Evol. 17, 1040–1049. [DOI] [PubMed] [Google Scholar]
- Martinez-Zapater, J.M., Estelle, M.A., and Sommerville, C.R. (1986). A highly repeated DNA sequence in Arabidopsis thaliana. Mol. Gen. Genet. 204, 417–423. [Google Scholar]
- McCormick, S. (1993). Male gametophyte development. Plant Cell 5, 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P.B., Yang, P., Glowczewski, L., Koshland, D., and Smith, M.M. (1998). Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94, 607–613. [DOI] [PubMed] [Google Scholar]
- Nagaki, K., Tsujimoto, H., and Saskuma, T. (1998). A novel repetitive sequence of sugar cane, SCEN family, locating on centromeric regions. Chromosome Res. 6, 295–302. [DOI] [PubMed] [Google Scholar]
- Nonomura, K., and Kurata, N. (2001). The centromere composition of multiple repetitive sequences on rice chromosome 5. Chromosoma 110, 284–291. [DOI] [PubMed] [Google Scholar]
- Novitski, E. (1955). Genetic measures of centromere activity in Drosophila melanogaster. J. Cell. Comp. Physiol. 45, 151–169. [DOI] [PubMed] [Google Scholar]
- O'Kane, S.L., Schaal, B.A., and Al-Shehbaz, I.A. (1996). The origins of Arabidopsis suecica (Brassicaceae) as indicated by nuclear rDNA sequences. Syst. Bot. 21, 559–566. [Google Scholar]
- Otto, S.P., and Whitton, J. (2000). Polyploid incidence and evolution. Annu. Rev. Genet. 34, 401–437. [DOI] [PubMed] [Google Scholar]
- Owen, H., and Makaroff, C.A. (1995). Ultrastructure of microsporogenesis and microgametogenesis in Arabidopsis thaliana (L.) Heynh. ecotype Wassilewskija (Brassicaceae). Protoplasma 185, 7–21. [Google Scholar]
- Palmer, D.K., O'Day, K., Trong, H.L., Charbonneau, H., and Margolis, R.L. (1991). Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc. Natl. Acad. Sci. USA 88, 3734–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena, F., and Sapienza, C. (2001). Nonrandom segregation during meiosis: The unfairness of females. Mamm. Genome 12, 331–339. [DOI] [PubMed] [Google Scholar]
- Pelissier, T., Tutois, S., Deragon, J.M., Tourmente, S., Genestier, S., and Picard, G. (1995). Athila, a new retroelement from Arabidopsis thaliana. Plant Mol. Biol. 29, 441–452. [DOI] [PubMed] [Google Scholar]
- Rhoades, M.M. (1952). Preferential segregation in maize. In Heterosis, J.W. Gowen, ed (Ames, IA: Iowa State College Press), pp. 66–80.
- Ross, K.J., Fransz, P., and Jones, G.H. (1996). A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res. 4, 507–516. [DOI] [PubMed] [Google Scholar]
- Round, E.K., Flowers, S.K., and Richards, E.J. (1997). Arabidopsis thaliana centromere regions: Genetic map positions and repetitive DNA structure. Genome Res. 7, 1045–1053. [DOI] [PubMed] [Google Scholar]
- Shelby, R.D., Vafa, O., and Sullivan, K.F. (1997). Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 136, 501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon, B.C. (1999). At the females' whim. Curr. Biol. 9, R487–R489. [DOI] [PubMed] [Google Scholar]
- Simoens, C.R., Gielen, J., Van Montagu, M., and Inze, D. (1988). Characterization of highly repetitive sequences of Arabidopsis thaliana. Nucleic Acids Res. 16, 6753–6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev, V.V., Salmov, A.A., and Lawrence, C.B. (1994). Predicting internal exons by oligonucleotide composition and discriminant analysis of spliceable open reading frames. Nucleic Acids Res. 22, 5156–5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, B.A., and Willard, H.F. (1998). Stable dicentric X chromosomes with two functional centromeres. Nat. Genet. 20, 227–228. [DOI] [PubMed] [Google Scholar]
- Sullivan, K.F., Hechenberger, M., and Masri, K. (1994). Human CENP-A contains a histone H3 related histone fold that is required for targeting to the centromere. J. Cell Biol. 127, 581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suominen, J. (1994). Ruotsinpitkäpalon, Arabidopsis suecica, syntyseudusta. Lutukka 10, 77–84. [Google Scholar]
- Suzuki, T., Ide, N., and Tanaka, I. (1997). Immunocytochemical visualization of the centromeres during male and female meiosis in Lilium longiflorum. Chromosoma 106, 435–445. [DOI] [PubMed] [Google Scholar]
- Takahashi, K., Chen, E.S., and Yanagida, M. (2000). Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288, 2215–2219. [DOI] [PubMed] [Google Scholar]
- Thompson, H., Schmidt, R., Brandes, A., Heslop-Harrison, J.S., and Dean, C. (1996). A novel repetitive sequence associated with the centromeric regions of Arabidopsis thaliana chromosomes. Mol. Gen. Genet. 253, 247–252. [DOI] [PubMed] [Google Scholar]
- Tyler-Smith, C., Gimelli, G., Giglio, S., Floridia, G., Pandya, A., Terzoli, G., Warburton, P.E., Earnshaw, W.C., and Zuffardi, O. (1999). Transmission of a fully functional human neocentromere through three generations. Am. J. Hum. Genet. 64, 1440–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, K., Kinoshita, Y., Xu, Z.J., Ide, N., Ono, M., Akahori, Y., Tanaka, I., and Inoue, M. (2000). Unusual core histones specifically expressed in male gametic cells of Lilium longiflorum. Chromosoma 108, 491–500. [DOI] [PubMed] [Google Scholar]
- Vafa, O., and Sullivan, K.F. (1997). Chromatin containing CENP-A and alpha-satellite DNA is a major component of the inner kinetochore plate. Curr. Biol. 7, 897–900. [DOI] [PubMed] [Google Scholar]
- Vafa, O., Shelby, R.D., and Sullivan, K.F. (1999). CENP-A associated complex satellite DNA in the kinetochore of the Indian muntjac. Chromosoma 108, 367–374. [DOI] [PubMed] [Google Scholar]
- Vielle-Caldaza, J.-P., Baskar, R., and Grossniklaus, U. (2000). Delayed activation of the paternal genome during seed development. Nature 404, 91–94. [DOI] [PubMed] [Google Scholar]
- Warburton, P.E., Cooke, C.A., Bourassa, S., Vafa, O., Sullivan, B.A., Stetten, G., Gimelli, G., Warburton, D., Tyler-Smith, C., Sullivan, K.F., Poirier, G.G., and Earnshaw, W.C. (1997). Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 7, 901–904. [DOI] [PubMed] [Google Scholar]
- Weiss, H., and Maluszynska, J. (2001). Molecular cytogenetic analysis of polyploidization in the anther tapetum of diploid and autotetraploid Arabidopsis thaliana plants. Ann. Bot. 87, 729–735. [Google Scholar]
- Willard, H.F. (1998). Centromeres: The missing link in the development of human artificial chromosomes. Curr. Opin. Genet. Dev. 8, 219–225. [DOI] [PubMed] [Google Scholar]
- Xia, X., Selvaraj, G., and Bertrand, H. (1993). Structure and evolution of a highly repetitive DNA sequence from Brassica napus. Plant Mol. Biol. 21, 213–224. [DOI] [PubMed] [Google Scholar]
- Yu, H.G., Hiatt, E.N., and Dawe, R.K. (2000). The plant kinetochore. Trends Plant Sci. 5, 543–547. [DOI] [PubMed] [Google Scholar]
- Zinkowski, R.P., Meyne, J., and Brinkley, B.R. (1991). The centromere-kinetochore complex: A repeat subunit model. J. Cell Biol. 113, 1091–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick, M.E., Salstrom, J.L., and Langley, C.H. (1999). Genetic variation in rates of nondisjunction: Association of two naturally occurring polymorphisms in the chromokinesin nod with increased rates of nondisjunction in Drosophila melanogaster. Genetics 152, 1605–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick, M.S., Islam-Faridi, M.N., Zhang, H.B., Hodnett, G.L., Gomez, M.I., Kim, J.S., Price, H.J., and Stelly, D.M. (2000). Distribution and sequence analysis of the centromere-associated repetitive element CEN38 of Sorghum bicolor (Poaceae). Am. J. Bot. 87, 1757–1764. [PubMed] [Google Scholar]