Abstract
The signal transduction pathways that control cytokinesis in plants are largely uncharacterized. Here, we provide genetic evidence that mitogen-activated protein kinase kinase kinases (MAPKKKs) play a role in the control of plant cell division. Using a reverse-genetic approach, we isolated plants carrying knockout alleles of the Arabidopsis MAPKKK genes ANP1, ANP2, and ANP3. The resulting single-mutant plants displayed no obvious abnormal phenotypes; two of the three double-mutant combinations displayed defects in cell division and growth; and the triple-mutant combination was not transmitted through either male or female gametes. The molecular and structural phenotypes displayed by the double mutants support a model in which the ANP family of MAPKKKs positively regulates cell division and growth and may negatively regulate stress responses.
INTRODUCTION
Work with yeast and animal systems has demonstrated that mitogen-activated protein (MAP) kinase cascades are critical components of the signaling mechanisms that allow many extracellular inputs to be transduced into intracellular responses. A MAP kinase cascade is composed of three components: a MAP kinase, a MAP kinase kinase, and a MAP kinase kinase kinase (MAPKKK). Activation of the MAPKKK by upstream signals causes a phosphorylation cascade that culminates in the activation of the MAP kinase, which then phosphorylates various downstream targets. Specific MAP kinase cascades are known to be involved in the regulation of growth, development, and stress responses in many eukaryotes (Gustin et al., 1998; Widmann et al., 1999).
Sequence analysis indicates that there are at least 25 MAPKKK genes present in the Arabidopsis genome (Tena et al., 2001). Two of these MAPKKK genes, CTR1 and EDR1, have emerged from forward-genetic screens in Arabidopsis (Kieber et al., 1993; Frye et al., 2001). CTR1 is responsible for the negative regulation of ethylene-induced gene expression, and EDR1 negatively regulates defense responses. In the present study, we used a reverse-genetic approach to systematically investigate the functions of the remaining genetically uncharacterized Arabidopsis MAPKKKs. To this end, we isolated individual plants carrying T-DNA insertions within three members of the Arabidopsis MAPKKK gene family: ANP1, ANP2, and ANP3 (Nishihama et al., 1997). This group of genes was chosen because it constitutes a distinct branch of the MAPKKK phylogenetic tree (Jouannic et al., 1999).
The Arabidopsis ANP genes were isolated originally as a result of their homology with the tobacco NPK1 gene (Banno et al., 1993; Nishihama et al., 1997; Machida et al., 1998; Nakashima et al., 1998). Work by Nishihama et al. (2001) has demonstrated that the NPK1 gene is involved in the regulation of cytokinesis in tobacco. In their study, it was shown that the NPK1 protein localizes to the phragmoplast during cytokinesis. In addition, they found that the expression of a kinase-negative version of NPK1 interfered with the formation of the cell plate and led to failed cytokinesis. Additional insight into the function of the ANP genes has come from the work of Kovtun et al. (1998)(2000), whose experiments involving the transient overexpression of constitutively active ANPs in protoplast cell cultures have implicated the ANPs in both oxidative stress– and auxin-related signaling pathways. In this study, we investigated directly the in planta function of the ANP genes by studying Arabidopsis plants in which the ANPs have been disrupted by T-DNA insertional mutagenesis.
RESULTS
Genetic Analysis
Insertional mutations within ANP1, ANP2, and ANP3 were recovered from populations of T-DNA–transformed Arabidopsis plants using a polymerase chain reaction (PCR)–based screening strategy (Figure 1) (Krysan et al., 1996, 1999). No obvious abnormal phenotypes were displayed by any of the single-mutant plants homozygous for T-DNA insertions within ANP1, ANP2, or ANP3. This result raised the possibility that functional redundancy was masking the appearance of a mutant phenotype in these plants. Therefore, we sought to create the various double- and triple-mutant combinations. To this end, we used genetic crosses to create a plant that was heterozygous for mutations at all three loci. This plant was allowed to self-fertilize, and 359 individuals from the resulting population of plants segregating all three mutant loci were analyzed by PCR to determine their genotypes (Table 1). All three of the possible homozygous double-mutant combinations were identified in this population; however, no homozygous triple mutants were found. We also observed that the homozygous single mutants appeared at a rate lower than the expected 25% mendelian ratio, which may reflect a reduction in the competitive ability of mutant gametes caused by defects in cellular growth, as described below.
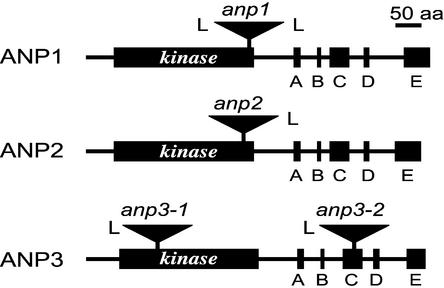
Figure 1.
The ANP Gene Family.
Locations of the T-DNA insertions with respect to the amino acid sequences of the ANP1, ANP2, and ANP3 proteins. The kinase domains of the proteins are indicated. All four T-DNA insertions occurred within predicted introns. A to E refer to five kinase-unrelated sequence domains that are conserved between the Arabidopsis ANP gene family and the tobacco NPK1 gene (Nishihama et al., 1997). L indicates the T-DNA left border. Bar = 50 amino acids (aa).
Table 1.
Progeny of an anp1/ANP1;anp2/ANP2;anp3/ANP3 Parenta
| Genotype | Number Observed | Number Expectedb |
|---|---|---|
| anp1; ____; ____ | 78 (18%) | – |
| ____; anp2; ____ | 53 (15%) | – |
| ____; ____; anp3-2 | 62 (16%) | – |
| anp1; anp2; ____ | 7 (2%) | 10 (3%) |
| anp1; ____; anp3-2 | 7 (2%) | 10 (3%) |
| ____; anp2; anp3-2 | 2 (0.5%) | 9 (2%) |
| anp1; anp2; anp3-2 | 0 (0%) | 2 (0.4%) |
a Genotypes of 359 progeny were determined using PCR. A ____ indicates that the genotype is either heterozygous or wild type for that locus. anp1, anp2, and anp3-2 indicate that the genotype is a homozygous mutant for that locus. Progeny lacking a homozygous mutant allele are not included on this table. Percentages indicate the number observed divided by 359.
Expected values for the double- and triple-mutant combinations were calculated using the observed allele frequencies for this experiment.
To improve our chances of identifying a homozygous triple mutant, we collected seed from a plant that was homozygous for the anp1 mutation and heterozygous at both ANP2 and ANP3. A total of 179 individuals from this segregating population were genotyped using PCR, and again, we failed to recover a homozygous triple mutant (Table 2). In addition, it was found that every plant homozygous at two of the mutant loci was homozygous wild type at the third locus (Table 2). No plant homozygous for two mutations and heterozygous for the third mutation was identified. Because these loci are not linked genetically, this result suggested that ANP1, ANP2, and ANP3 may be “collectively essential,” meaning that haploid gametes carrying all three mutant loci are not viable. To test this hypothesis directly, we performed reciprocal crosses in which one parent plant was anp1/anp1;anp2/ANP2;anp3/ANP3 and the other parent was fully wild type (Table 3). No triple-mutant gametes were observed from either the male or the female parent, indicating that the triple-mutant combination is not viable.
Table 2.
Progeny of an anp1/anp1;anp2/ANP2;anp3/ANP3 Parenta
| Genotype | No. Observed | No. Expected |
|---|---|---|
| WT; WT | 31 | 41 |
| WT; Het | 87 | 78 |
| Het; Het | 35 | 34 |
| Homo; WT | 26 | 13 |
| Homo; Het | 0 | 11 |
| Homo; Homo | 0 | 1 |
a Genotypes of 179 progeny were determined by PCR. Homozygous (Hom), heterozygous (Het), and wild type (WT) refer to the genotypes at the ANP2 and ANP3 loci. For example, “Homo; WT” indicates that the plant was homozygous for one of the mutations and wild type at the other locus. Expected values for the given genotype combinations were calculated using the observed allele frequencies for this experiment.
Table 3.
Reciprocal Crosses between anp1/anp1;anp2/ANP2;anp3/ANP3 and Wild Type a
| Gamete Frequency | |||||
|---|---|---|---|---|---|
|
anp1; ANP2;ANP3 |
anp1; anp2;ANP3 |
anp1; ANP2;anp3 |
anp1; anp2;anp3 |
||
| Cross | P Valueb | ||||
| Female mutant | |||||
| Observed | 35 | 40 | 36 | 0 | – |
| Expected 1 | 28 | 28 | 28 | 28 | <0.0001* |
| Expected 2 | 37 | 37 | 37 | 0 | 0.95 |
| Male mutant | |||||
| Observed | 94 | 52 | 71 | 0 | – |
| Expected 1 | 54 | 54 | 54 | 54 | <0.0001* |
| Expected 2 | 72 | 72 | 72 | 0 | 0.007* |
Genotypes of 328 progeny from reciprocal crosses were determined by PCR and used to infer the genotype of the gamete contributed by the mutant parent in each cross. The mutant parent was anp1/anp1;anp2/ANP2;anp3/ANP3 and the other parent was fully wild type (ANP1/ANP1;ANP2/ANP2;ANP3/ANP3). The raw number of gametes observed for each genotype is shown. Expected 1 indicates the number of gametes expected when the four possible gametes are equally viable. Expected 2 indicates the number of gametes expected when the anp1;anp2;anp3 gametes are not viable. The chi-squared test was used to determine if the observed data were significantly different from each of these expected distributions.
Significant P values are indicated with an asterisk.
The observed frequency distribution of female gamete genotypes indicated that the anp1 anp2 and anp1 anp3 double-mutant genotypes do not have any negative effect on female gametogenesis. By contrast, we observed that these double-mutant genotypes were underrepresented when transmitted by the male, indicating that these double-mutant combinations may have a negative effect on pollen competitiveness, especially the anp1 anp2 combination (Table 3).
Reduced Plant Size
To gain insight into the in planta function of these collectively essential genes, we analyzed the homozygous double-mutant plants described above. The most severe phenotypic changes were observed with the anp2 anp3 double mutants, which displayed an overall reduction in total plant size (Figure 2). Measurement of the fresh weights of 18-day-old plants grown in soil under constant light indicated that the anp2 anp3 double mutants weighed approximately threefold less than wild-type plants (Figure 3). The fresh weights of the single mutants and the remaining double mutants were not significantly different from those of the wild-type.
Figure 2.
Reduced-Size Phenotype of the anp2 anp3 Double Mutant.
(A) Single-mutant and double-mutant plants homozygous for the indicated mutations were grown for 4 weeks in soil under constant white light. Ws, Wassilewskija (wild type).
(B) Phenotypic rescue via genetic complementation. The seven small plants are untransformed anp2 anp3 plants that display the characteristic reduced-size phenotype. The large plant in the center also is homozygous for anp2 and anp3-1, but it has been fully restored to the wild-type phenotype as a result of transformation by a T-DNA vector carrying an ∼6-kb fragment of Arabidopsis genomic DNA encompassing the ANP2 gene.
Figure 3.
Growth Characteristics of the Different Mutant Combinations.
(A) Hypocotyl length after 3 days of growth in the dark on vertical agar plates (n = 48 to 74).
(B) Fresh weight of the aerial tissue from plants grown in soil for 18 days under constant light (n = 23 to 28). Plants are homozygous for the indicated mutations. Ws, Wassilewskija (wild type).
The graphs display the mean for each sample, and error bars indicate the 95% confidence limits of each mean. In addition, the t test was used to compare each mutant genotype with the wild-type control. Samples that are significantly different from the wild type (P < 0.05) are indicated with asterisks. Triple asterisks indicate P < 0.001.
We also measured the lengths of 3-day-old dark-grown hypocotyls for the various mutant combinations. After 3 days of growth in the dark, the anp2 anp3 double-mutant hypocotyls were ∼40% shorter than those of the wild type (Figure 3). The other mutant genotypes were indistinguishable from the wild type, with the exception of the anp2 single mutants and the anp1 anp2 double mutants. For these two genotypes, we observed a small but statistically significant increase in the hypocotyl length compared with the wild type (8% for anp2 and 15% for anp1 anp2). Although this subtle increase in hypocotyl length may reflect an important function of the ANP genes, further study will be required to determine whether this mild quantitative effect is in fact attributable to the ANP mutations. For the present study, we focused on the more pronounced phenotype displayed by the anp2 anp3 double-mutant plants.
Genetic Complementation and Additional Alleles
To confirm that the mutant phenotype of the anp2 anp3 plants was caused by the MAPKKK mutations, a plant carrying a second mutant allele of ANP3 (anp3-2) was isolated and crossed with an anp2 plant (Figure 1). Analysis of the progeny segregating anp2 and anp3-2 confirmed that the mutant phenotype was dependent on a mutation of ANP3: the mutant phenotype was exclusive to the homozygous double mutants, and all double mutants displayed the phenotype.
Because no additional mutant alleles of ANP2 were found in our T-DNA–transformed populations, we used genetic complementation to prove that mutation of ANP2 is required for the appearance of the mutant phenotype. A 6-kb fragment of genomic DNA from the ANP2 locus, including ∼450 bp upstream and downstream of the transcribed region, was amplified by PCR, cloned into a T-DNA binary vector, and transferred into anp2 anp3-1 plants using Agrobacterium tumefaciens transformation. Thirty-two transformed plants were recovered, and they all displayed a fully wild-type phenotype (Figure 2).
We used PCR amplification to confirm that the rescued plants were still homozygous for the anp2 and anp3-1 mutations. To distinguish between the wild-type chromosomal ANP2 locus and the ANP2 gene carried on our T-DNA construct, we used a PCR primer specific for the chromosomal region immediately adjacent to, but not contained on, our ANP2 T-DNA vector. Such a primer allows for the amplification of the wild-type chromosomal ANP2 locus, but it does not amplify the ANP2 gene carried on the T-DNA. In addition, we collected seed from two of the anp2 anp3-1 plants that had been rescued with the wild-type ANP2 construct and analyzed their progeny. We observed that the reduced-size phenotype was segregating in the progeny of these rescued plants and that the mutant phenotype appeared only in plants lacking the wild-type ANP2 rescuing construct.
These genetic complementation experiments, in conjunction with the results obtained using the two independent alleles of anp3, demonstrate conclusively that the mutant phenotype displayed by anp2 anp3 plants is caused by mutations of both ANP2 and ANP3.
Defective Cell Growth
The anp2 anp3 combination was the only double mutant that had a substantial effect on plant growth at the macroscopic level. To determine if any microscopic changes had occurred, we used scanning electron microscopy to analyze the epidermis of dark-grown hypocotyls. In the wild type, the epidermal cells were arranged as orderly rows of uniformly shaped cells (Figure 4). By contrast, anp2 anp3 plants displayed a number of irregularly shaped cells characterized by reduced elongation and radial swelling. The average epidermal cell length in 3-day-old dark-grown anp2 anp3 hypocotyls was 190 μm ± 66 μm, compared with 403 μm ± 102 μm for the wild type. The hypocotyls of the remaining double-mutant combinations and each of the single mutants were indistinguishable from those of the wild type.
Figure 4.
Cellular Defects in the Epidermis of Hypocotyls and Flowers.
(A) Scanning electron microscopy of 2-day-old dark-grown hypocotyls. Magnification ×120.
(B) Flowers, featuring mainly sepals. Magnification ×70.
(C) Petal surface. Magnification ×500.
Genotype notations indicate that the plants were homozygous for the indicated mutations. Ws, Wassilewskija (wild type).
The floral organs of anp2 anp3 and anp1 anp3 plants displayed cellular defects similar to those seen in the anp2 anp3 hypocotyls (Figure 4). This result was particularly interesting in light of the observation that the hypocotyl cells and all other aspects of the anp1 anp3 plants are phenotypically normal. By contrast, the anp1 anp2 plants appeared wild type in all of the tissues examined. Together, these results demonstrate that functional redundancy within the Arabidopsis ANP gene family is not an all-or-nothing phenomenon. Instead, there are complex patterns of redundancy such that the different double-mutant combinations manifest developmentally distinct phenotypic consequences. In particular, anp1 anp2 plants appear normal throughout development; anp1 anp3 plants show defects in their floral organs but are normal otherwise; and anp2 anp3 plants are affected in all of the stages of development analyzed.
Defective Cell Division
To better characterize the cellular defects described above, we used transmission electron microscopy to observe the cells of dark-grown anp2 anp3 hypocotyls (Figure 5). Numerous binucleate cells and cell wall stubs were revealed by this analysis. In addition, light microscopy of longitudinal thin sections also showed the presence of cell wall stubs in dark-grown anp2 anp3 hypocotyls (Figure 5). Although the anp2 anp3 mutants have reduced hypocotyl length, these double mutants have substantially wider hypocotyls than the wild type. This combination of reduced length and increased radial swelling has been documented for other Arabidopsis mutations that affect cell growth and division (Assaad et al., 1996; Arioli et al., 1998; Nicol et al., 1998). We also analyzed longitudinal thin sections from anp2 anp3 embryos using light microscopy and again observed the presence of cell wall stubs and binucleate cells (Figure 5). Cell wall stubs or two nuclei were observed in ∼6% of the cells in anp2 anp3 embryos (83 of 1409 cells), compared with <1% of the cells in wild-type embryos (1 of 333 cells). These results demonstrate that cytokinesis is disturbed in anp2 anp3 plants.
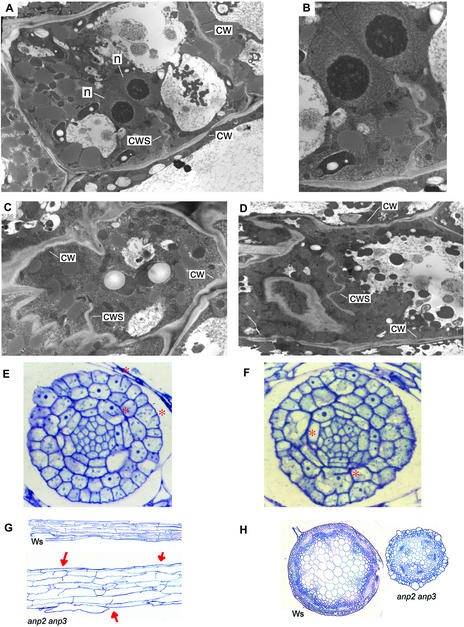
Figure 5.
Cell Division Is Disrupted in anp2 anp3 Plants.
(A) to (D) Transmission electron microscopy of hypocotyl cells of 2-day-old dark-grown anp2 anp3 plants. cw, cell wall; cws, cell wall stub; n, nucleus.
(A) Cell with two nuclei and a cell wall stub.
(B) Higher magnification view of (A).
(C) Cell with a cell wall stub.
(D) Incomplete cell wall in the middle of a cell.
(E) and (F) Longitudinal sections of anp2 anp3 embryos. Red asterisks indicate the locations of cell wall stubs. The lower asterisk in (F) indicates a binucleate cell.
(G) Longitudinal sections of 3-day-old dark-grown hypocotyls. Both sections are shown at the same magnification. Arrows indicate cell wall stubs.
(H) Transverse sections through inflorescence bolts. Both sections are shown at the same magnification.
We next analyzed transverse sections through the inflorescence bolts of 25-day-old soil-grown plants. It was found that the epidermal layer of the anp2 anp3 bolts featured numerous unusually large, swollen cells, in contrast with the uniformly small-diameter cells of the wild-type epidermis (Figure 5). In addition, the total cell number for the anp2 anp3 inflorescence bolts was approximately half that of the wild type. The reduced diameter of the anp2 anp3 bolts, therefore, is attributable to a reduction in the total number of cells present in the bolt rather than to a reduction in the size of the cells. Similarly, loss-of-function alleles of AINTEGUMENTA reduce the size of Arabidopsis shoot organs by reducing the total number of cell divisions that occur during development (Mizukami and Fischer, 2000). The levels of ANP1 protein present in anp2 anp3 plants may be sufficient to limit the number of cell divisions that occur, resulting in thinner bolts.
Hormone Sensitivity
The results described above demonstrate that the ANP genes are involved in the control of cellular growth and division. Therefore, we tested the sensitivity of the anp2 anp3 plants to the phytohormones abscisic acid, auxin, brassinosteroids, cytokinin, ethylene, and gibberellin using agar plate assays. Vertically oriented plates were grown either in the dark for 3 days or in constant light for 1 week. During growth in the light, the plants were observed daily. None of these exogenous hormone treatments “rescued” the mutant phenotype of the double mutants. In addition, all of the mutants displayed a level of sensitivity to each hormone treatment that was equivalent to that of the wild type (i.e., the degree of growth inhibition or stimulation was the same; data not shown). Furthermore, we observed no qualitative changes in growth that were unique to the mutant genotypes under the various hormone treatments.
Genome-Wide Gene Expression Analysis
To gain additional insight into the signaling pathways affected by the ANP mutations, we used the Affymetrix Arabidopsis Gene Chip to compare the RNA levels for 8200 genes in anp2 anp3 double-mutant plants and wild-type plants. Plants were grown in soil for 11 days under constant light, and the aerial tissue then was harvested and used for RNA analysis. Overall, RNA levels increased threefold or greater for 211 genes, whereas levels decreased threefold or greater for 30 genes (see supplemental material at www.biotech.wisc.edu/krysan/). The most striking result from the gene chip experiment was that a number of pathogen- and stress-related genes were upregulated in the anp2 anp3 plants. These genes include numerous disease-resistance genes, chitinases, glucanases, peroxidases, glutathione S-transferases, and several heat shock–related genes (Table 4).
Table 4.
Upregulation of Stress-Related Genes in the anp2;anp3 Double Mutant
| Accession Numbers | Fold Change
|
Abundancea
|
||||
|---|---|---|---|---|---|---|
| Descriptive Names | Mean | Exp. 1b | Exp. 2 | Exp. 1 | Exp. 2 | |
| Resistance Genes | ||||||
| At2g32680 | Putative disease resistance proteinc | 37.5 | 21.4 | 53.5 | 9,277 | 8,640 |
| At2g24160 | Putative disease resistance protein | 9.5 | 10.4 | 8.6 | 2,279 | 2,228 |
| M90508 | Pathogenesis-related 1-like protein | 9.0 | 10.7 | 7.3 | 33,940 | 27,893 |
| At2g40000 | Putative nematode resistance protein | 4.7 | 4.5 | 4.9 | 4,025 | 2,548 |
| At4g13900 | Putative disease resistance protein | 4.1 | 3.6 | 4.5 | 7,312 | 7,234 |
| At4g33300 | Similar to NBS/LRR disease resistance proteine | 3.3 | 3.3 | 3.2 | 4,197 | 4,036 |
| AF098963 | Disease resistance protein RPP1-WsBf | 3.0 | 3.2 | 2.8 | 2,242 | 2,253 |
| Pathogen Response | ||||||
| A71590 | Berberine bridge/anti-fungal gene | ∼12.2 | ∼11.1 | ∼13.3 | 1,874 | 1,687 |
| AJ243377 | EST Athsr4 hypersensitive response | ∼8.6 | ∼7.3 | ∼9.9 | 726 | 556 |
| AF188329 | Phytoalexin-deficient 4 lipase (PAD4) | 8.2 | 7.5 | 8.9 | 7,902 | 9,175 |
| M90510 | Thaumatin-like mRNA | 7.4 | 8.2 | 6.5 | 44,930 | 42,938 |
| AF128407 | EDS1 lipase homolog | 3.1 | 3.2 | 3.1 | 3,522 | 2,444 |
| At2g43570 | Putative endochitinase | 34.5 | 37.4 | 31.5 | 2,103 | 24,342 |
| Y14590 | Class IV chitinase | ∼11.4 | 8.6 | ∼14.2 | 1,545 | 1,766 |
| AB023448 | Basic endochitinase | ∼11.2 | ∼11.3 | ∼11.0 | 1,536 | 1,139 |
| At2g43620 | Putative endochitinase | ∼9.4 | ∼7.6 | ∼11.2 | 1,157 | 1,376 |
| At2g43590 | Putative endochitinase | 5.0 | 4.1 | 5.8 | 2,463 | 1,932 |
| At4g01700 | Similar to class I chitinases | 4.2 | 3.9 | 4.5 | 1,968 | 1,981 |
| M38240 | Basic chitinase gene | 3.2 | 3.8 | 2.5 | 1,471 | 1,347 |
| At4g16260 | β-1,3-Glucanase class I precursor | ∼22.8 | ∼25.0 | 20.6 | 3,774 | 2,978 |
| M90509 | β-1,3-Glucanase | 13.6 | 15.5 | 11.7 | 54,947 | 38,994 |
| M58464 | β-1,3-Glucanase (BG3) | 8.6 | 7.2 | 10.0 | 2,159 | 2,050 |
| Oxidative Stress | ||||||
| At4g10500 | Iron/ascorbate family oxidoreductase | ∼65.7 | 57.9 | ∼73.4 | 9,447 | 9,265 |
| At2g29460 | Putative glutathione S-transferase | ∼21.9 | ∼22.5 | 21.3 | 3,361 | 3,302 |
| Y14251 | Glutathione S-transferase GST11 | 12.2 | 14.7 | 9.6 | 34,338 | 31,074 |
| Y18227 | Blue copper binding-like protein | 9.2 | 8.5 | 9.9 | 24,296 | 26,922 |
| AJ006960 | p9a gene peroxidase | 6.8 | 5.2 | 8.3 | 4,115 | 3,423 |
| At2g02930 | Putative glutathione S-transferase | 3.8 | 4.4 | 3.1 | 28,819 | 31,217 |
| X71794 | prxCb mRNA for peroxidase | 3.5 | 3.8 | 3.2 | 11,189 | 1,104 |
| At2g37130 | Putative peroxidase ATP2a | 3.2 | 3.4 | 3.0 | 1,333 | 1,253 |
| Heat Shock | ||||||
| U68017 | Heat shock transcription factor 4 | ∼23.9 | 14.8 | ∼32.9 | 4,762 | 4,281 |
| X77199 | Heat shock cognate 70-1 | 7.3 | 8.5 | 6.1 | 5,460 | 4,855 |
| U68561 | Heat shock transcription factor 21 | 5.0 | 4.5 | 5.5 | 1,930 | 1,862 |
| M62984 | Heat shock protein 83 | 3.7 | 5.0 | 2.4 | 4,703 | 4,009 |
| Y08892 | Heat shock cognate 70-G8 protein | 3.5 | 3.9 | 3.0 | 8,417 | 8,482 |
a Abundance indicates the average difference value from the Gene Chip experiment, which is a direct measure of RNA abundance.
b Exp., experiment.
c Descriptive names are preceded by database accession numbers for each gene.
d ∼, approximate fold change.
e NBS/LRR is nucleotide-binding site plus leucine-rich repeat.
f RPP1-WsB is recognition of Peronospora parasitica-Wassilewskija.
We next used reverse transcriptase–mediated (RT) PCR to confirm the gene chip results (Figure 6). For RT-PCR analysis, we chose six genes that showed an induction in the gene chip experiment of greater than ninefold as well as a histone H2B gene that showed no change in expression with the gene chip (see Methods). Each RT-PCR gel contained one primer for the H2B histone gene as an internal control, one primer for a differentially expressed gene, and one universal primer specific for the poly(A) end of every transcript. Various numbers of PCR cycles were tested to ensure that the reaction had not plateaued.
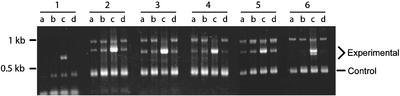
Figure 6.
RT-PCR Confirms the Expression Chip Data.
RT-PCR products visualized on an agarose gel from selected genes that were found to be strongly upregulated in the anp2 anp3 mutants in the gene chip experiment. Each PCR cycle simultaneously monitors the RNA level of a control gene and the gene of interest (see Methods). Four genotypes were compared in this experiment: a, anp2; b, anp3; c, anp2 anp3; and d, the wild type. Products attributable to the histone control gene and the various experimental genes are seen at the indicated positions on the gel. Primers used for PCR were as follows: 1, RT-1; 2, RT-2; 3, RT-3; 4, RT-4; 5, RT-5; and 6, RT-8.
The reliability of our RT-PCR assay was confirmed by performing RT-PCR using the exact cDNA populations used for the gene chip experiments. These controls confirmed that the differential gene expression detected by the gene chip could be detected by our RT-PCR assay. We next used this RT-PCR assay to determine RNA levels in independently grown samples of anp1, anp2, anp1 anp2, and wild-type plants (Figure 6). These experiments confirmed the results of the gene chip analysis and indicated that the upregulation of these genes occurred only in the anp2 anp3 double mutant and not in either of the single-mutant lines.
DISCUSSION
In the present study, we have demonstrated the effectiveness of a purely reverse-genetic approach to understanding MAPKKK function in a multicellular eukaryote, even in the face of functional redundancy. The three Arabidopsis ANP genes that we studied constitute a distinct branch of the MAPKKK gene family tree (Jouannic et al., 1999). This structural grouping parallels the functional grouping that was revealed by the phenotypes of the single, double, and triple mutants. As a whole, these three genes were found to be collectively essential, meaning that triple-mutant gametes are not viable. By contrast, any single-gene mutation was phenotypically silent, presumably as a result of the functional redundancy provided by the remaining two isoforms. In the double mutants, the incomplete functional redundancy that resulted in mutant phenotypes provided us with insight into the in planta functions of these genes. Because the majority of genes in any multicellular eukaryote are known only by their DNA sequences, our demonstration that reverse-genetic analysis guided only by DNA sequence information is able to reveal functional associations is significant. It remains to be seen, however, how precise the clustering of function and sequence will be within the context of the entire group of at least 25 MAPKKK genes found in Arabidopsis.
Our experiments documented two main consequences associated with mutating the ANP genes. First, we demonstrated significant structural changes in the mutant plants that indicate aberrant control of cell division and elongation (Nacry et al., 2000). Second, we measured dramatic molecular changes in the form of increased RNA levels for a number of stress-related genes in the mutants. Our next challenge will be to determine the precise relationships between the ANP mutations and these structural and molecular phenotypes.
The main structural defects that we observed in the anp2 anp3 double-mutant plants were the presence of cell wall stubs, binucleate cells, enlarged cells, and disorganized cell geometry. This combination of mutant phenotypes has been observed for other Arabidopsis mutants known to affect cytokinesis, such as knolle, keule, cyd1, and tso1 (Assaad et al., 1996, 2001; Lukowitz et al., 1996; Liu et al., 1997; Yang et al., 1999; Waizenegger et al., 2000; Assaad, 2001). knolle and keule have been shown to be involved in vesicle trafficking at the phragmoplast during cell plate maturation (Waizenegger et al., 2000; Assaad, 2001). The biochemical functions of cyd1 and tso1 are not known.
The localization of a protein to the phragmoplast is one indication that that protein might be involved in the cell division process. Therefore, it is significant that the tobacco NPK1 protein has been shown to localize to the phragmoplast in tobacco cells (Nishihama et al., 2001). Because the ANP genes are the closest Arabidopsis homologs of NPK1, it is possible that the ANP proteins will show subcellular localization patterns similar to those of NPK1. Further evidence for the presence of MAP kinase signaling components at the phragmoplast can be found in the study of Bogre et al. (1999), who demonstrated that the Medicago MAP Kinase3 protein is localized to the plane of cell division in alfalfa cells.
The functional parallels between the Arabidopsis ANP genes and the tobacco NPK1 gene also are evident in the phenotypes displayed when these gene functions are disrupted in their respective organisms. Nishihama et al. (2001) demonstrated that the expression of a kinase-negative version of NPK1 in tobacco caused defects in cytokinesis, and we have demonstrated similar cytokinesis defects in the anp2 anp3 double mutants in Arabidopsis. Our study used insertional mutagenesis to create knockout mutant plants in which the ANP gene product is absent from the plant.
Although this would seem to be a straightforward means of creating a loss-of-function situation for a given gene product, the presence of multigene families and functional redundancy raises the possibility that an observed phenotype could be caused by something more complex than a simple loss of function (Bent, 2001). For instance, the absence of the ANP proteins might be compensated for by the activity of a more distantly related MAPKKK that inappropriately activates or represses a given pathway. In such a situation, however, the observed phenotype might report accurately on the downstream pathways normally controlled by the ANP genes. Whether a given pathway is inactivated by the absence of ANP proteins or stimulated by the opportunistic activity of the wrong MAPKKK does not change the identity of that downstream pathway and the processes under its control.
By comparison, Nishihama et al. (2001) used the overexpression of a kinase-inactive allele of NPK1 to create a loss-of-function situation for NPK1 activity in tobacco cells. This strategy also is subject to potentially confounding effects attributable to the existence of multigene families and functional redundancy. In particular, the kinase-inactive protein could interact with MAPKKK signaling complexes other than the intended NPK1 complexes. Therefore, an observed phenotype could be caused by the inappropriate inhibition of these unrelated pathways. In such a situation, it is possible that the phenotypes induced by a dominant-negative allele of one MAPKKK could provide misleading information concerning the signaling pathways that are controlled normally by that kinase.
It is clear that any single method for interrogating gene function will have both strengths and weaknesses. In light of this situation, it is significant that our work with the ANP genes and the work of Nishihama et al. (2001) with NPK1 both suggest a role for this group of MAPKKKs in the control of cell division. The use of distinct methods to inactivate the MAPKKK pathways in each of our studies strengthens the argument that the in planta role of these kinases is to control cell division.
In addition to the cytokinesis defects discussed above, we also observed the upregulation of numerous stress-related genes in the anp2 anp3 double mutants. It is not clear how direct the linkage is between the disrupted ANP kinases and the upregulation of these stress genes. It is possible that the ANP genes in wild-type plants normally function to downregulate stress responses and that in the mutants this negative regulation is removed, resulting in the increased expression of stress-related genes. This model is analogous to the type of control exerted by the other characterized Arabidopsis MAPKKK genes, CTR1, which negatively regulates ethylene responses (Kieber et al., 1993), and EDR1, which negatively regulates pathogen responses (Frye et al., 2001).
Alternatively, it is possible that the upregulation of stress-related genes in the ANP mutants is a more indirect effect whereby the observed defects in cell division lead to a general stress response in the plants. Further experiments will be needed to determine the relevance of the stress responses activated in the anp2 anp3 double mutants. In any case, we have clearly demonstrated that mutating a group of MAPKKK genes does result, one way or another, in the upregulation of a large number of stress response genes.
Using a transient expression system, Kovtun et al. (2000) demonstrated that the ANP genes can positively regulate oxidative stress–responsive genes and can negatively regulate auxin-induced genes. Therefore, one might have predicted that knockout alleles of the ANP genes would lead to upregulation of auxin-induced genes and reduced induction of oxidative stress responses. We found, however, that the knockout mutations led to upregulation of stress responses and no apparent change in auxin-regulated gene expression. The structural and temporal differences between the two experimental systems could account for the seemingly contradictory effects on gene expression; that is, our experiments dealt with intact plants that had grown for many cell generations with a fixed genetic program, whereas the transient assay system monitored gene expression on a time scale of hours after the introduction of a new gene into individual protoplasts. It is also possible that the stress response pathways activated in the anp2 anp3 plants are distinct from the pathways activated in the protoplast experiments.
In this study, we have demonstrated a direct link between mutation of the ANP genes and the disruption of cell division. Therefore, upstream signals that stimulate cell division are likely to be transduced to the ANPs, which in turn may orchestrate many of the biochemical events that take place at the phragmoplast during cytokinesis. In addition to these direct effects on cell division, it is possible that the ANPs may negatively regulate stress responses. Thus, the ANP genes may participate in more than one signaling pathway in the cell. Future studies will attempt to characterize these putative pathways by identifying the various upstream and downstream components that use the ANP proteins as part of their signaling networks.
METHODS
T-DNA Screening
Ordered populations of Arabidopsis thaliana were screened for the presence of T-DNA insertions within the ANP genes using polymerase chain reaction (PCR) primers specific for each gene in combination with primers specific for either the T-DNA left border sequence or the T-DNA sequence encoding the BAR gene: ANP1, 5′-ACAGAGACTACGATTGTAACTGGGACTTG; ANP1, 3′-AGTTGCTTTCTT-CCTTCGTTTCTCAGTTC; ANP2, 5′-AATCACTTCCTGTATCCGCAA-ATCAATGG; ANP2, 3′-CACTCAATATGCACCAACATTCTCACATC; ANP3, 5′-CTCTACACATTCACCGTCGGCTTCTCAAA; ANP3, 3′-ATA-CAGTTTCTCTTTAGCCGTTCACTATC; T-DNA left border, CATTTT-ATAATAACGCTGCGGACATCTAC; T-DNA BAR gene, AGCACGGGA-ACTGGCATGAC. anp2 and anp3-1 were recovered from a population of 60,480 kanamycin-resistant lines transformed with a derivative of the pD991 vector (Krysan et al., 1999). anp3-2 was recovered from a population of 72,960 Basta-resistant lines transformed with the pSKI15 vector (Kardailsky et al., 1999). DNA sequencing of the genomic DNA flanking the T-DNA insertion sites confirmed the identity of each gene and the precise locations of the insertions. All of the T-DNA mutants and wild-type plants in this study were of ecotype Wassilewskija.
Genetic Complementation
The PCR primers 5′-AAAAAGAGCTCAACGTGCATTTTCTGGAAG-3′ and 5′-AAAAAGGATCCCAGATAAATACACCGAATC-3′ were used to amplify an ∼6-kb fragment of genomic DNA that included the ANP2 gene. The resulting PCR product was digested with SacI and BamHI (enzyme sites are present in the PCR primers) and ligated into pCAMBIA3300S (kindly provided by Scott Michaels, Department of Biochemistry, University of Wisconsin–Madison), a spectinomycin-resistant derivative of pCAMBIA3300 (http://www.cambia.org.au/main/r_et_camvec.htm). Plasmids containing the ANP2 gene were introduced into Arabidopsis using the Agrobacterium tumefaciens floral dip method (Clough and Bent, 1998). Transformed plants were selected in soil in the next generation by spraying seedlings at 3, 5, 8, and 11 days after germination with the herbicide Basta.
Hypocotyl Cell Measurements
Three-day-old dark-grown hypocotyls were observed using Nomarski optics and a digital video system. Individual hypocotyl cells were measured using captured images. Starting at the base of the hypocotyl, cells at positions 1 through 17 were measured for 10 wild-type hypocotyls and 10 anp2 anp3 hypocotyls, for a total of 170 measurements per genotype.
Scanning Electron Microscopy
Tissue samples were fixed in 4% (v/v) glutaraldehyde in 0.05 M K-phosphate buffer, pH 7, dehydrated with ethanol, critical point dried, and sputter coated with gold before viewing by scanning electron microscopy.
Light Microscopy
Tissue samples were fixed overnight at 4°C in 4% (v/v) glutaraldehyde, 0.5% (w/v) paraformaldehyde, 0.001% igepal CA-630 (Sigma Chemical, St. Louis, MO), and 0.05 M sodium cacodylate and treated with 2% (w/v) osmium tetroxide in 0.05 M sodium cacodylate overnight at 4°C. They were then dehydrated, infiltrated with London Resin White (Ted Pella, Redding, CA), and polymerized, and 5-μm sections were taken using a glass blade. Sections were stained with toluidine blue and viewed with bright-field illumination.
Transmission Electron Microscopy
Seedlings were fixed and embedded in London Resin White as described above for light microscopy. Sections (0.1 μm) were cut using a diamond knife, and staining was performed using uranyl acetate and lead citrate.
Hormone Sensitivity Assays
One percent (w/v) agar plates, pH 5.8, containing 0.5× Murashige and Skoog (1962) salts, 1% (w/v) Suc, and one of the following additives were prepared: 10 μM abscisic acid; 10 μM 1-aminocyclopropane-1-carboxylic acid; 1 or 10 μM epibrassinolide; 10 μM gibberellic acid; 0.1, 1, or 10 μM indole-3-acetic acid; and 10 or 50 μM kinetin. Seed were surface-sterilized and sown on agar plates, kept in the dark at 4°C for 3 days, exposed to light for 30 min, and then incubated at room temperature in either darkness or constant light. Plants were observed daily to determine if differences in hormone sensitivity could be detected between the mutants and the wild-type control for each treatment. No significant differences were observed.
Genome-Wide Expression Analysis
The Affymetrix Arabidopsis Genome Array Gene Chip (Affymetrix, Santa Clara, CA) was used. Plants were grown for 11 days in soil under constant light, and the aboveground parts were harvested and submerged immediately in liquid nitrogen. Two separate batches of Wassilewskija wild type and anp2 anp3 were harvested and processed independently. Total RNA was isolated using a Qiagen plant RNAeasy kit (Valencia, CA). cDNA synthesis and probe preparation were performed according to the Affymetrix guidelines using total RNA as the starting material. Complete gene chip data are available as an Excel file at www.biotech.wisc.edu/krysan/.
Reverse Transcriptase–Mediated PCR
The following primers were used for the reverse transcriptase–mediated (RT) PCR experiments: RT-1, 5′-TAGACTCCATTGCTTTCC-CATAG-3′ (putative Fe[II]/ascorbate oxidase SRG1 protein); RT-2, 5′-AAGTCCTTACTCTTCGCTCAAAC-3′ (putative disease resistance protein); RT-3, 5′-TTGCAGCAAATATGGCTACTGTG-3′ (endochitinase isolog); RT-4, 5′-TTAGCGATCTTATGGACAAAGTG-3′ (putative cytochrome P450); RT-5, 5′-TCAGCTCAACACTTACGGATTTC-3′ (heat shock transcription factor HSF4); RT-8, 5′-AGTTGTACTGGACACAGCGATAC-3′ (Ser/Thr kinase–like protein KI domain–interacting kinase 1); H2B, 5′-GAGAAACTTGCTGGTGAGTCTTC-3′ (histone H2B–like protein); and T7-2, 5′-GGCCAGTGAATTGTAATACGACT-3′ (universal primer that recognizes the 3′ end of cDNAs synthesized using the Affymetrix T7-dT24 primer). The thermal cycling parameters used for the RT-PCR were 94°C for 15 sec, 60°C for 30 sec, and 72°C for 30 sec repeated for a total of 30 cycles. Products were analyzed by agarose gel electrophoresis.
Accession Numbers
Accession numbers are listed in Table 4.
Acknowledgments
We thank Adam Durski, Laura Katers, Sean Monson, Jason Young, Rick Amasino, and Sandra Austin-Phillips for their work with the T-DNA–transformed populations; Sandra Splinter-BonDurant for assistance with the gene chip experiment; Sarah Patterson and Martin Garment for assistance with the scanning electron microscope; Randall Massey for assistance with the transmission electron microscope; and James Krysan for expert photographic assistance.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.001164.
References
- Arioli, T., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279, 717–720. [DOI] [PubMed] [Google Scholar]
- Assaad, F.F. (2001). Plant cytokinesis: Exploring the links. Plant Physiol. 126, 509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad, F.F., Mayer, U., Wanner, G., and Jurgens, G. (1996). The KEULE gene is involved in cytokinesis in Arabidopsis. Mol. Gen. Genet. 253, 267–277. [DOI] [PubMed] [Google Scholar]
- Assaad, F.F., Huet, Y., Mayer, U., and Jurgens, G. (2001). The cytokinesis gene KEULE encodes a Sec1 protein that binds the syntaxin KNOLLE. J. Cell Biol. 152, 531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banno, H., Hirano, K., Nakamura, T., Irie, K., Nomoto, S., Matsumoto, K., and Machida, Y. (1993). NPK1, a tobacco gene that encodes a protein with a domain homologous to yeast BCK1, STE11, and Byr2 protein kinases. Mol. Cell. Biol. 13, 4745–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent, A.F. (2001). Plant mitogen-activated protein kinase cascades: Negative regulatory roles turn out positive. Proc. Natl. Acad. Sci. USA 98, 784–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogre, L., Calderini, O., Binarova, P., Mattauch, M., Till, S., Kiegerl, S., Jonak, C., Pollaschek, C., Barker, P., Huskisson, N.S., Hirt, H., and Heberle-Bors, E. (1999). A MAP kinase is activated late in plant mitosis and becomes localized to the plane of cell division. Plant Cell 11, 101–114. [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Frye, C.A., Tang, D., and Innes, R.W. (2001). Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl. Acad. Sci. USA 98, 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin, M.C., Albertyn, J., Alexander, M., and Davenport, K. (1998). MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62, 1264–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouannic, S., Hamal, A., Leprince, A.S., Tregear, J.W., Kreis, M., and Henry, Y. (1999). Plant MAP kinase kinase kinases structure, classification and evolution. Gene 233, 1–11. [DOI] [PubMed] [Google Scholar]
- Kardailsky, I., Shukla, V.K., Ahn, J.H., Dagenais, N., Christensen, S.K., Nguyen, J.T., Chory, J., Harrison, M.J., and Weigel, D. (1999). Activation tagging of the floral inducer FT. Science 286, 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kieber, J.J., Rothenberg, M., Roman, G., Feldmann, K.A., and Ecker, J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72, 427–441. [DOI] [PubMed] [Google Scholar]
- Kovtun, Y., Chiu, W.L., Zeng, W., and Sheen, J. (1998). Suppression of auxin signal transduction by a MAPK cascade in higher plants. Nature 395, 716–720. [DOI] [PubMed] [Google Scholar]
- Kovtun, Y., Chiu, W.L., Tena, G., and Sheen, J. (2000). Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 97, 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., Tax, F., and Sussman, M.R. (1996). Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc. Natl. Acad. Sci. USA 93, 8145–8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., and Sussman, M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11, 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., Running, M.P., and Meyerowitz, E.M. (1997). TSO1 functions in cell division during Arabidopsis flower development. Development 124, 665–672. [DOI] [PubMed] [Google Scholar]
- Lukowitz, W., Mayer, U., and Jurgens, G. (1996). Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 84, 61–71. [DOI] [PubMed] [Google Scholar]
- Machida, Y., Nakashima, M., Morikiyo, K., Banno, H., Ishikawa, M., Soyano, T., and Nishihama, R. (1998). MAPKKK-related protein kinase NPK1: Regulation of the M phase of plant cell cycle. J. Plant Res. 111, 243–246. [Google Scholar]
- Mizukami, Y., and Fischer, R.L. (2000). Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. USA 97, 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nacry, P., Mayer, U., and Jurgens, G. (2000). Genetic dissection of cytokinesis. Plant Mol. Biol. 43, 719–733. [DOI] [PubMed] [Google Scholar]
- Nakashima, M., Hirano, K., Nakashima, S., Banno, H., Nishihama, R., and Machida, Y. (1998). The expression pattern of the gene for NPK1 protein kinase related to mitogen-activated protein kinase kinase kinase (MAPKKK) in a tobacco plant: Correlation with cell proliferation. Plant Cell Physiol. 39, 690–700. [DOI] [PubMed] [Google Scholar]
- Nicol, F., His, I., Jauneau, A., Vernhettes, S., Canut, H., and Hofte, H. (1998). A plasma membrane-bound putative endo-1,4-beta-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17, 5563–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihama, R., Banno, H., Kawahara, E., Irie, K., and Machida, Y. (1997). Possible involvement of differential splicing in regulation of the activity of Arabidopsis ANP1 that is related to mitogen-activated protein kinase kinase kinases (MAPKKKs). Plant J. 12, 39–48. [DOI] [PubMed] [Google Scholar]
- Nishihama, R., Ishikawa, M., Araki, S., Soyano, T., Asada, T., and Machida, Y. (2001). The NPK1 mitogen-activated protein kinase kinase kinase is a regulator of cell-plate formation in plant cytokinesis. Genes Dev. 15, 352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena, G., Asai, T., Chiu, W., and Sheen, J. (2001). Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 4, 392–400. [DOI] [PubMed] [Google Scholar]
- Waizenegger, I., Lukowitz, W., Assaad, F., Schwarz, H., Jurgens, G., and Mayer, U. (2000). The Arabidopsis KNOLLE and KEULE genes interact to promote vesicle fusion during cytokinesis. Curr. Biol. 10, 1371–1374. [DOI] [PubMed] [Google Scholar]
- Widmann, C., Gibson, S., Jarpe, M.B., and Johnson, G.L. (1999). Mitogen-activated protein kinase: Conservation of a three-kinase module from yeast to human. Physiol. Rev. 79, 143–180. [DOI] [PubMed] [Google Scholar]
- Yang, M., Nadeau, J.A., Zhao, L., and Sack, F.D. (1999). Characterization of a cytokinesis defective (cyd1) mutant of Arabidopsis. J. Exp. Bot. 50, 1437–1446. [DOI] [PubMed] [Google Scholar]