Abstract
Previous studies demonstrated that in somatic plant cells, homologous recombination (HR) is several orders of magnitude less efficient than nonhomologous end joining and that HR is little used for genomic double-strand break (DSB) repair. Here, we provide evidence that if genomic DSBs are induced in close proximity to homologous repeats, they can be repaired in up to one-third of cases by HR in transgenic tobacco. Our findings are relevant for the evolution of plant genomes because they indicate that sequences containing direct repeats such as retroelements might be less stable in plants that harbor active mobile elements than anticipated previously. Furthermore, our experimental setup enabled us to demonstrate that transgenic sequences flanked by sites of a rare cutting restriction enzyme can be excised efficiently from the genome of a higher eukaryote by HR as well as by nonhomologous end joining. This makes DSB-induced recombination an attractive alternative to the currently applied sequence-specific recombination systems used for genome manipulations, such as marker gene excision.
INTRODUCTION
Until now, it was generally accepted that homologous recombination (HR) is a minor recombination pathway in somatic plant cells under all circumstances. It has been reported that HR proceeds at frequencies that are several orders of magnitude lower than nonhomologous end joining (NHEJ) (for reviews, see Puchta and Hohn, 1996; Mengiste et al., 1999; Vergunst and Hooykaas, 1999). This assumption was based on several lines of evidence, such as the lack of a feasible gene-targeting technique for somatic cells (Puchta, 1998a, 2002; Gallego et al., 1999; Reiss et al., 2000) and the low rates of intrachromosomal recombination, both spontaneous and after application of genotoxic stress (Tovar and Lichtenstein, 1992; Lebel et al., 1993; Puchta et al., 1995a; Kovalchuk et al., 1998; Ries et al., 2000).
Rare cutting restriction enzymes or transposable elements have been used to induce a double-strand break (DSB) at a specific location in the plant genome and to study its repair (for review, see Gorbunova and Levy, 1999). These studies indicated that HR can be enhanced drastically by the induction of a DSB. However, under the experimental conditions applied, the break was repaired by HR in only a small fraction of the cases. At most, one out of 100 DSBs was repaired by the use of homology of an incoming T-DNA (Puchta et al., 1996) and one out of 10,000 DSBs was repaired by the use of an ectopic homology (Shalev and Levy, 1997; Puchta, 1999a). It also was reported that DSB induction enhances intrachromosomal recombination by only one order of magnitude (Chiurazzi et al., 1996), although a recent report suggested a much stronger induction (Xiao and Peterson, 2000). However, it could not be excluded that this finding was attributable to a transposon-specific effect.
To reevaluate the role of HR in DSB repair between closely linked sequences in plants, we set up an assay system in tobacco based on interrupted overlapping halves of a β-glucuronidase (GUS) gene (Swoboda et al., 1994). A negative selectable marker gene flanked by two I-SceI sites was inserted between the overlaps in the transgene. As a consequence of I-SceI expression, the negative selectable marker is excised and the resulting genomic break can be repaired either by NHEJ or HR. Our results show that loss of the negative selectable marker involves HR in up to one-third of the cases. The implications of this finding for our understanding of plant genome evolution and its putative application for controlled alterations of plant genomes are discussed.
RESULTS
Experimental Setup and Induction of Recombination
The binary vector pGU.C.USB was constructed for the analysis of the efficiency of homologous DSB repair in the presence of nearby homology (for details, see Methods and Figure 1). A negative selectable marker gene (cytosine deaminase; codA [Stougaard, 1993]) was flanked by two I-SceI sites in direct orientation. Two halves of a GUS gene harboring an overlap of 557 bp (Tinland et al., 1994) were positioned next to the I-SceI sites. DSBs were induced at the borders of the codA expression cassette by expression of the restriction enzyme I-SceI, thus excising the intervening codA sequence from the genomic locus. The DSB then was repaired either by NHEJ or by HR, in the latter case resulting in the restoration of the GUS gene. Because the two I-SceI sites were cloned in direct orientation, a religation reaction between the two sites also was possible. Thus, plant cells that became resistant to 5-fluorouracil (5-FC) and expressed GUS were deficient in codA activity as a result of HR. Cells that were resistant to 5-FC but did not express GUS were derived from NHEJ events.
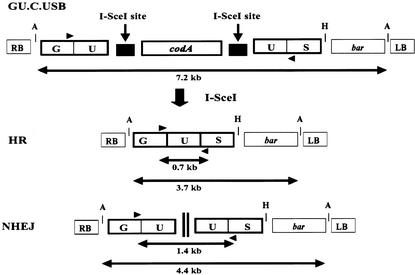
Figure 1.
Scheme of the T-DNA from the Binary Plasmid pGU.C.USB.
A codA gene flanked by two I-SceI sites was integrated between overlapping halves of a GUS gene. The T-DNA used for transformation also contained the bar gene as a transformation marker. Possible outcomes of the excision reaction of the codA marker gene are depicted. Triangles represent the primers used for PCR amplification of the recombined junctions. A 0.7-kb fragment is indicative of HR, and a 1.4-kb fragment is indicative of NHEJ. Restriction sites used for DNA gel blot analysis were Acc65I (A) and HindIII (H) (see Figure 4). A 3.7-kb Acc65I GUS-specific fragment is indicative of HR, and a 4.4-kb fragment is indicative of NHEJ. LB, left border; RB, right border.
Tobacco seedlings were transformed by Agrobacterium tumefaciens with the binary vector pGU.C.USB. Plants were regenerated, and lines carrying the transgenic sequences at a single locus were identified by segregation analysis and characterized by DNA gel blot analysis. Three lines were chosen for further analysis: GU.C.USB 1, containing a single copy of the transgene, and GU.C.USB 3 and 7, containing two closely linked copies of the transgene. Detailed restriction analysis indicated that in both lines the copies were not joined directly in tandem to one another but were separated by nontransgenic sequences of at least several kilobases. The lines with the two copies were chosen to test whether individual DSB repair events at a transgenic locus might influence each other, as has been reported previously for intrachromosomal recombination in tobacco (Peterhans et al., 1990).
Seedlings of the F1 generation of all three lines were inoculated with an Agrobacterium strain that harbored on its T-DNA an I-SceI open reading frame under the control of the 35S promoter of Cauliflower mosaic virus to achieve transient expression of the enzyme (Puchta, 1999b). After 3 days, the seedlings were put on callus-inducing medium that contained 5-FC and phosphinotricin for selection of DSB-induced deletion events. The resulting calli were cut into two pieces. Part of the callus was used for histochemical staining with 5-bromo-4-chloro-3-indolyl-β-d-glucuronide to determine whether the GUS gene was restored (Figure 2). From the remaining part of the callus, either DNA was extracted or plants were regenerated. The results are shown in Table 1. We found surprisingly high numbers of 5-FC–resistant calli that expressed GUS. Depending on the line, a GUS gene was restored in ∼20 to 40% of the cases. In control experiments, no 5-FC–resistant calli could be obtained from similar numbers of uninoculated seedlings. This finding indicates that the isolated calli arose as a result of DSB induction.
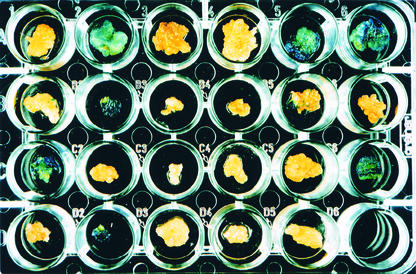
Figure 2.
GUS Assay of 5-FU–Resistant Calli.
Transformants in which the marker gene was removed by HR result in blue staining of the whole callus. The small sectors visible in some of the other calli are caused by secondary recombination events that occurred during callus growth. The calli tested and the experiment numbers are as follows: GU.C.USB 3-1 (A1), 3-3 (A2), 3-5 (A3), 3-6 (A4), 3-7 (A5), and 3-18 (A6); GU.C.USB 7-14 (B1), 7-34 (B2), 7-58 (B3), 7-96 (B4), and 7-102 (B5); GU.C.USB 1-83 (C1), 1-61 (C2), 1-60 (C3), 1-55 (C4), and 1-23 (C5); and GU.C.USB 3-10 (D1), 3-18 (D2), 3-19 (D3), 3-21 (D4), and 3-65 (D5). As a positive control, callus of pBG5 (transgenic tobacco with a functional GUS gene [Puchta et al., 1995b]) was used (C6); as a negative control, callus of the nontransformed tobacco line SR1 was used (B6). In accord with the histochemical staining, the molecular analysis of lines GU.C.USB 1-83, GU.C.USB 3-3, 3-7, and 3-18, and GU.C.USB 7-34 by PCR and DNA gel blot analysis revealed the restoration of the GUS gene by HR (see also Figure 3). PCR and DNA gel blot analysis of lines GU.C.USB 1-23, 1-55, and 1-61, GU.C.USB 3-1, 3-5, 3-6, 3-10, 3-19, 3-21, and 3-65, and GU.C.USB 7-14 and 7-102 demonstrated that the elimination of the codA gene occurred by NHEJ (see also Figure 3). PCR analysis of lines GU.C.USB 1-60 and GU.C.USB 7-58 and 7-96 resulted in the 1.4-kb band indicative of NHEJ, whereas in the case of line GU.C.USB 3-65, the lack of a PCR product indicated a major rearrangement of the transgene locus.
Table 1.
Compilation of 5-FC–Resistant Tobacco Calli Obtained after Induction of DSBs by Transient Expression of I-SceI in Lines GU.C.USB 1, 3, and 7
| Line | No. of Seedlings | Resistant Calli | GUS Positive | HR (%) |
|---|---|---|---|---|
| 1 | 290 | 56 | 22 | 39 |
| 3 | 490 | 90 | 24 | 27 |
| 7 | 370 | 59 | 11 | 19 |
Molecular Analysis of the Recombination Products of Line GU.C.USB 1
The calli were analyzed further depending on the number of transgene copies in the original lines. Polymerase chain reaction (PCR) analysis was performed on a large number of calli from the single-copy line GU.C.USB 1. For amplification, oligonucleotides were designed that allowed the amplification of a 0.7-kb fragment if the GUS gene was restored by HR. A 1.4-kb fragment was indicative of a NHEJ event between the genomic ends of the two I-SceI sites (Figure 1). The results of this analysis are shown in Table 2. Three different classes of events could be distinguished: events resulting in a 0.7-kb band, events resulting in a 1.4-kb band, and events yielding no amplification product.
Table 2.
Results of PCR Analysis of 5-FC–Resistant Tobacco Calli Obtained after Induction of DSBs by Transient Expression of I-SceI with the Single-Copy Line GU.C.USB 1
Histochemical staining of all 10 calli demonstrated the functional restoration of the GUS gene. Blue staining was not detected for the other calli analyzed in this experiment.
For a number of 5-FC–resistant calli, no PCR fragment was amplified under the conditions used. The most likely explanation is that in these calli, the DSB caused a deletion that included at least part of the primer binding sites. Alternatively, insertions too large to be amplified by PCR or other major rearrangements might be responsible for the lack of a PCR fragment.
The 0.7-kb band is indicative of a HR reaction within the GUS gene, resulting in the restoration of the marker. This was proven further by the fact that the tested calli containing this band turned blue after histochemical staining with 5-bromo-4-chloro-3-indolyl-β-d-glucuronide. Three 0.7-kb bands were sequenced, and the normal sequence of the GUS gene was detected, as expected for a HR event between the overlaps. DNA gel blot analysis of representative lines confirmed the PCR results. To discriminate unambiguously between HR and NHEJ, a digest with Acc65I was especially useful, because the indicative bands could be discriminated easily by size: a 4.4-kb band indicated NHEJ, and a 3.7-kb band indicated HR (Figure 1). As shown for the representative line GU.C.USB 1-83, the expected 3.7-kb band was detected with a GUS-specific probe (Figure 3C, lane 3). The codA-specific sequences were removed completely (disappearance of the transgene-specific bands in lane 3 of Figures 3A and 3C after rehybridization of the blot with a codA-specific probe, lane 3 of Figures 3B and 3D).
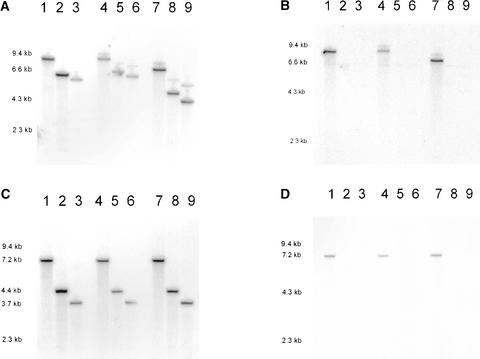
Figure 3.
DNA Gel Blot Analysis with Restriction Digested DNA of Plant Lines GU.C.USB 1, 3, and 7 and Some Recombinants.
Lane 1, GU.C.USB 1; lane 2, GU.C.USB 1-61; lane 3, GU.C.USB 1-83; lane 4, GU.C.USB 3; lane 5, GU.C.USB 3-1; lane 6, GU.C.USB 3-3; lane 7, GU.C.USB 7; lane 8, GU.C.USB 7-14; lane 9, GU.C.USB 7-34. The faint band at ∼6 kb in all nine lanes in (B) is attributable to a HindIII-specific fragment present in wild-type tobacco that cross-hybridizes weakly with the codA-specific probe.
(A) HindIII-restricted DNA hybridized with a GUS-specific probe.
(B) HindIII-restricted DNA from (A) washed to remove the GUS probe and rehybridized with a codA-specific probe.
(C) Acc65I-restricted DNA hybridized with a GUS-specific probe.
(D) Acc65I-restricted DNA from (C) washed to remove the GUS probe and rehybridized with a codA-specific probe.
Because the 1.4-kb band originated from a NHEJ process, we expected that individual products would differ in their sequences at the rejoined break sites, as has been shown previously (Gorbunova and Levy, 1997; Salomon and Puchta, 1998; Kirik et al., 2000). The sequence was determined from five junctions (Figure 4). Deletions at the newly formed junctions constituted between 1 and 9 bp. In one case, the I-SceI site was restored, indicating that the break also could be repaired by simple ligation. The restoration of the I-SceI sites also was confirmed by restriction digest of the PCR fragment. DNA gel blot analysis of representative lines confirmed the PCR analysis. As shown for the representative line GU.C.USB 1-61, the expected 4.4-kb band was detected with a GUS-specific probe (Figure 3C, lane 2). The codA-specific sequences were removed completely (disappearance of the transgene-specific bands in lane 2 of Figures 3A and 3C after rehybridization of the blot with a codA-specific probe, lane 2 of Figures 3B and 3D).
Figure 4.
Compilation of Five Junctions Originating from GU.C.USB 1 in Which End Joining between the Two I-SceI Sites Had Occurred.
Nucleotides of the I-SceI recognition sequence are highlighted in red. The 5′ 13 nucleotides of the recognition site originate from the proximal I-SceI site, and the remaining five nucleotides originate from the distal I-SceI site, of pGU.C.USB.
Molecular Analysis of the Recombination Products of Lines GU.C.USB 3 and 7
Because both lines GU.C.USB 3 and 7 harbored two closely linked copies of the transgene, a simple PCR analysis was not appropriate to evaluate the outcome of the recombination reactions that were necessary to remove both functional codA genes. Plants were regenerated from the calli, DNA was prepared, and DNA gel blot analysis was performed. The number of transgene copies could be determined directly by digestion with HindIII and the use of a GUS- or a codA-specific probe (Figure 1). Whereas for line pGU.C.USB 1, a single band was indicative of a single copy, in lines pGU.C.USB 3 and 7, two bands represented two copies that hybridized with the GUS-specific probe as well as with the codA-specific probe (Figures 3A and 3B, cf. lane 1 with lanes 4 and 7). Because Acc65I sites are present at both ends of the T-DNA of GU.C.USB (Figure 1), digestion of DNA of all three lines resulted in the same type of pattern in the DNA gel blot. This result demonstrates the integrity of the integrated T-DNAs (Figures 3C and 3D, cf. lane 1 with lanes 4 and 7).
After elimination of the codA expression cassette by NHEJ reaction between the two I-SceI sites, the HindIII fragment should be reduced by 2.8 kb. If the codA gene is eliminated by HR, the fragment should be smaller by another 0.7 kb (the length of the overlapping region). In Figure 3A (which shows a DNA gel blot of HindIII-restricted DNA hybridized with a GUS-specific probe), the changes in size of each of the two individual transgene copies are documented for representative recombinant lines of GU.C.USB 3 and 7 (Figure 3A, lanes 5 and 8 [NHEJ of lines GU.C.USB 3-1 and GU.C.USB 7-14] and lanes 6 and 9 [HR of lines GU.C.USB 3-3 and GU.C.USB 7-34]). The absence of any transgene-specific signal on the blot probed with codA indicates that the codA expression cassette was eliminated, as predicted in all cases from both copies (Figure 3B, lanes 5, 6, 8, and 9). Because lines GU.C.USB 3 and 7 harbor two transgene copies, probing of Acc65I-restricted DNA with a GUS-specific probe was especially informative for the discrimination of HR and NHEJ events. The simultaneous presence of a 4.4-kb band and a 3.7-kb band indicated the repair of one copy by HR and the other by NHEJ, whereas the appearance of only a 4.4-kb band indicated the repair of both copies by NHEJ and the appearance of only a 3.7-kb band indicated the repair of both copies by HR.
A number of transgenic lines were analyzed this way, and the results are shown in Table 3. Surprisingly, we were unable to detect cells harboring both fragments. If one copy recombined by HR, the second copy was coconverted (Figure 3C, lanes 6 and 9). We amplified 0.7-kb bands by PCR in cases in which the DNA gel blots indicated that HR occurred. As expected, the sequence analysis of four PCR products (two for each line) revealed the functional GUS sequence resulting from a HR event between the overlaps within pGU.C.USB. DNA gel blot analysis also clearly demonstrated that the recombination reactions occurred within the transgene copies and not between them, resulting in a loss of intervening sequences. In the latter case, a single GUS-specific HindIII fragment would have been visible after recombination on the DNA gel blot. However, in all lines tested in this study, two GUS-specific HindIII fragments were clearly visible after the recombination reaction (Figure 3A). Compared with the parent lines, both fragments were reduced by the same size, as expected from intratransgenic recombination events.
Table 3.
Compilation of DNA Gel Blot Analysis of Plants of Lines GU.C.USB 3 and 7 Regenerated from 5-FC–Resistant Tobacco Calli Obtained after Induction of DSBs by Transient Expression of I-SceI
| GUS-Specific Acc65I Fragment (kb)
|
||||
|---|---|---|---|---|
| Line | 3.7 (HR) |
4.4 (NHEJ) |
3.7 and 4.4 (HR and NHEJ) |
Othersa |
| 3 | 6 | 18 | 0 | 15 |
| 7 | 2 | 5 | 0 | 7 |
The DNA gel blot pattern resulted in several cases in the presence of one or more GUS-specific fragments smaller than 3.0 kb, indicating the deletion of parts of the transgene locus. In two cases, larger bands indicative of further rearrangements were detected. However, no codA-specific sequences were detected in any line tested.
The third class of recombination events harbored mainly deletions within the transgene. Bands smaller than 3 kb were detected with a GUS-specific probe in several cases, indicating deletions of various sizes within the transgene locus. In two cases, further rearrangements occurred, resulting in several novel transgene-specific bands. In all cases tested, these deletions or rearrangements were accompanied by the complete loss of the codA-specific sequences (data not shown).
DISCUSSION
The Role of HR and NHEJ in Genomic DSB Repair in Plants
DSBs are critical lesions in genomes. Therefore, efficient repair of DSBs is important for the survival of all organisms. In principle, DSBs can be repaired by illegitimate recombination or by HR. A major goal of recent studies was to elucidate under which conditions each pathway is used preferentially. For this purpose, rare cutting restriction endonucleases were applied for the induction of breaks at defined loci within eukaryotic genomes (for review, see Paques and Haber, 1999; Jasin, 2000), a technique that was used for plants as well (Puchta et al., 1993, 1996; Chiurazzi et al., 1996; Puchta, 1998b, 1999a; Salomon and Puchta, 1998; Kirik et al., 2000).
Because we could not measure the relation between HR and NHEJ in planta directly, we devised an experimental system in which, after the excision of a negative selectable marker gene, a single genomic DSB had to be repaired. Because the marker gene was lost, all resulting repair events (independent of their nature) could be selected for. Important for the evaluation of the results was that both I-SceI sites were cut simultaneously by the transiently expressed restriction enzyme. If in most cases only one site were cut, the single DSB itself could induce the elimination of the codA gene by HR, resulting in a biased picture and thus an overestimation of the frequency of HR events.
In previous experiments conducted in tobacco, we demonstrated that the induction of a DSB within the codA gene leads to the production of a large number of deletions of various sizes that still contained nonfunctional parts of the codA gene (Salomon and Puchta, 1998; Kirik et al., 2000). Because we were unable to detect a single case of a truncated codA gene (including all deletions or rearrangements) in the current experiments, we are convinced that in most, if not all, transformed cells, both sites were cut during transient expression of the I-SceI gene. Moreover, the relation between the number of 5-FU–resistant calli and the inoculated seedlings did not differ significantly between the two lines that harbor two copies of the transgene (and thus four I-SceI sites) and line GU.C.USB 1, with only one copy and two I-SceI sites. This finding indicates that in these lines the cutting of the I-SceI sites was not rate limiting, supporting our hypothesis that HR occurs at high frequencies when DSBs are induced between directly repeated sequences.
Efficient Repair of Genomic DSBs by Single-Strand Annealing of Homologous Sequences
Whereas in bacteria such as Escherichia coli and in lower eukaryotes such as yeast, HR generally is the prominent pathway of DSB repair, initial studies indicated that the opposite might be true for higher eukaryotes. In these studies, the experimental conditions clearly differed from those used in this report. The homologies were supplied either in trans at an ectopic site in the genome (Puchta, 1999a) or by an incoming T-DNA (Puchta et al., 1996). It also has been reported that DSB induction enhances intrachromosomal recombination by only one order of magnitude (Chiurazzi et al., 1996). However, in that study, the recombination substrate used contained an inverted repeat to specifically detect gene conversion events. In all of these cases, the marker genes had to be restored by a synthesis-dependent strand-annealing type of recombination mechanism (Chiurazzi et al., 1996; Puchta, 1998a; Gorbunova and Levy, 1999).
The synthesis-dependent strand-annealing model describes the directed transfer of information from a homologous donor sequence to the break site. This process does not result in the loss of genomic sequences because the donor locus is conserved. This is in contrast to the experimental situation applied in the present study. Recombination between direct repeated sequences is described best by the single-strand annealing (SSA) model of recombination. The SSA model was suggested first for extrachromosomal recombination between plasmids in mammalian cells (Lin et al., 1984, 1990). After induction of a DSB, the free double-stranded ends are resected by a single-strand–specific exonuclease, leaving behind 3′ single-stranded overhangs. These single strands can anneal with each other at regions of complementarity, overhanging nonhomologous sequences are digested, single-stranded gaps are refilled by repair synthesis, and in a last step, the double strand is restored by religation of the remaining nicks. As a result, a deletion occurs and the sequence information between the repeated sequences is lost.
This model has been used to describe extrachromosomal recombination in Xenopus laevis oocytes (Maryon and Carroll, 1991a, 1991b) and yeast (Fishman-Lobell et al., 1992). In plants, three different approaches unambiguously demonstrated that extrachromosomal recombination proceeds efficiently by SSA (Puchta and Hohn, 1991; Bilang et al., 1992; De Groot et al., 1992; for review, see Puchta and Meyer, 1994). Interestingly, a similar mechanism is used for NHEJ: the majority of junctions contain small patches of homologous nucleotides between reaction partners, which is best explained by the operation of a “SSA-like” mechanism (Lehman et al., 1994; Nicolas et al., 1995; Mason et al., 1996; Gorbunova and Levy, 1997; Salomon and Puchta, 1998; for review, see Gorbunova and Levy, 1999). Thus, SSA and SSA-like mechanisms might be the most prominent mode for the rejoining of broken DNA molecules in higher eukaryotes. If there are homologous sequences in close proximity to the break, they are used regularly for the annealing reaction; if they are not available, short patches of homology, which are present abundantly, are used.
We are convinced that the repair of genomic DSBs using directly repeated sequences is an efficient pathway in eukaryotes in general and not a phenomenon restricted to plants or even tobacco. This hypothesis is sustained by two recent studies in mammalian cells (Liang et al., 1998; Pipiras et al., 1998). We also assume that the high frequencies of homologous recombination in Arabidopsis reported from a construct carrying a transposable element between repeated GUS sequences (Xiao and Peterson, 2000) are caused by the transient DSB induced during the reaction rather than from a transposon-specific effect. Both possibilities were suggested by the authors (Xiao and Peterson, 2000). The transgenes used in both studies were derived from the same recombination substrate, pGU.US (Tinland et al., 1994), which includes an overlap of 557 bp of GUS sequences. Whereas in our study, the negative selectable marker gene, flanked by I-SceI sites, was cut out of the transgene after I-SceI expression, in the former study, a Ds transposon was excised after Ac transposase expression, leaving a similar kind of reaction intermediate behind.
Using lines that harbor two copies of the T-DNA, we were surprised to find in all cases that both copies were changed by HR, similar to the high coconversion frequencies reported for several copies of an intrachromosomal recombination substrate integrated at a single locus in tobacco (Peterhans et al., 1990). This could be attributable either to the fact that the cells were in a “HR-prone” state at the time of transformation or to the proximity of the DSBs at the specific locus. We favor the second hypothesis and suggest that, as a result of the close proximity of the transgenes, the HR machinery processing the DSBs might have excluded the presence of a NHEJ mechanism. Notably, we were unable to detect a recombination event in which only one recombined copy of the overlapping sequence was left behind in the genome in these two lines. However, because of our experimental setup, we cannot draw final conclusions from this finding. In both lines, the two transgene copies were in close proximity (a 3:1 segregation of the hygromycin marker), but they were not attached directly to one another (no common restriction fragment was found using restriction enzymes that cut 6-mer sequences; no PCR product with the intervening sequences could be obtained). Therefore, we were unable to determine whether both copies are located in the same orientation on the chromosome. Only this kind of configuration would allow a SSA reaction between the outer overlaps of both copies, resulting in the loss of all sequences between them.
Consequences for Genome Evolution
The demonstration that in plants DSBs can be repaired efficiently by the use of nearby homologies also seems to be relevant for plant genome evolution. In barley, a 7- to 42-fold excess of single long terminal repeats (LTRs) over internal regions was found for the retroelement BARE-1 (Vicient et al., 1999; Kalendar et al., 2000; Shirasu et al., 2000). Because intrachromosomal recombination had been demonstrated with model substrates to be quite infrequent in plants (Puchta and Hohn, 1996), it was surprising to find such a high incidence of single LTRs. These could have arisen only by a homology-dependent deletion of the internal region of the retrotransposon. BARE-1 is active in barley (Kalendar et al., 2000; Vicient et al., 2001), and during their spread, retroelements integrate regularly into one another (San Miguel et al., 1996).
If one assumes that transient breaks within the internal part of BARE-1 are induced either by the activity of transposons (Xiao et al., 2000) or by aberrant integration of retroelements, a DSB-induced HR event between the two flanking LTRs may yield single LTRs. Thus, it is tempting to speculate that the accumulation of multiple single LTRs in plant genomes is linked to the presence of active mobile genetic elements that are able to induce breaks during their life cycles. One might argue that in our experimental setup, the DSB was induced directly adjacent to the direct repeats and that intervening sequences of several kilobases, as is the case for retrotransposons, might hinder an efficient SSA reaction. However, a study in yeast clearly demonstrated that SSA between repeats occurs with high efficiency in the presence of intervening sequences of 4.4 kb: the overall rate of deletion formation was reduced by only one-third compared with that in the absence of an intervening sequence (Fishman-Lobell et al., 1992).
Biotechnological Application: Marker Gene Excision
Our results clearly demonstrate that a marker gene can be eliminated from the plant genome by NHEJ as well as by HR after induction of DSBs by a highly specific restriction endonuclease. The technique described may have potential for biotechnological applications. Site-specific recombinases have been used efficiently for the excision of selectable marker genes (for review, see Vergunst and Hooykaas, 1999; Hohn et al., 2001; Ow, 2002). Using this approach, recognition sites of the recombinases are left behind in the genome. Thus, for the performance of multiple genomic changes, a combination of different site-specific recombination systems has to be applied.
The development of alternative approaches for site-specific alterations of genomes is of great interest in biotechnology. A very promising approach would involve the combined elimination of the transgene sequences and their respective recognition sequences. Therefore, the use of a highly specific restriction endonuclease to remove transgenic sequences from the plant genome might be a useful, irreversible alternative to the established site-specific techniques. In the current study, we demonstrated that a marker gene can be eliminated efficiently from the plant genome by either HR or NHEJ (including religation) by inducing DSBs with a rare cutting restriction enzyme. Thus, any sequence flanked by restriction sites could be eliminated from genomes of plants and probably also from other eukaryotic organisms. The sequence in our case was the codA gene, which was used simultaneously as a negative selectable marker. In practice, other sequences to be removed also could be included adjacent to this marker. For example, a selectable marker gene could be included in the I-SceI cassette of the transgene. Other rare cutting endonucleases also might be useful for this purpose.
PCR analysis of resistant calli in the single-copy line GU.C.USB 1 showed precise excision of the marker gene in more than two-thirds of the cases (in 22 of 29 calli). Taking all isolated recombination events of lines GU.C.USB 1, 3, and 7 into account (Table 1), we obtained 57 homologous recombinants in 205 5-FC–resistant calli (>27%) and conclude that almost every third excision event of the marker is attributable to HR.
Our findings also might help explain recent results on the HR-mediated elimination of a selectable marker gene from the tobacco genome (Zubko et al., 2000). The selectable marker gene, placed between two homologous, 352-bp attP sites of bacteriophage λ, was excised efficiently in two of 11 transgenic lines, although no sequence-specific recombinase was expressed. Thus, transgenes with the desired rearranged structure were obtained at a much higher frequency by intrachromosomal recombination than was anticipated previously (Puchta and Hohn, 1996). It was suggested that this effect might be caused by a certain instability at these two loci and might be linked to a nonspecific DSB induction mechanism (Puchta, 2000).
By means of controlled induction of DSBs by a restriction endonuclease, we now demonstrate that even higher frequencies of HR can be obtained in all of our lines. The obvious advantage of the controlled induction of DSBs is that the marker gene is removed efficiently independent of its location in the genome. No indications for the instability of the transgenic loci in the absence of I-SceI expression, an important prerequisite for bringing transgenic plants into the field, were found. We were unable to isolate deletion events without DSB induction. However, we were able to determine the overall rate of spontaneous homologous recombination by performing histochemical staining of 4-week-old tobacco seedlings. The obtained frequencies were 10−6 or lower (per genome; data not shown), similar to that reported in previous studies (Puchta et al., 1995a, 1995b). Because under our experimental conditions the induction and repair of the DSB takes place in tobacco seedlings before callus induction (the elimination of the negative selectable marker must occur as a prerequisite), we believe that DSBs can stimulate intrachromosomal recombination by as much as five orders of magnitude.
METHODS
DNA Constructs
The binary plasmid pGU.US was used for construction of the recombination substrate. The plasmid contains on its T-DNA two halves of the β-glucuronidase (GUS) gene with an overlap of 557 bp (Tinland et al., 1994). A hygromycin gene is inserted in a unique XbaI site between the GUS sequences. In the first step, a gluphosinate-selectable marker gene cassette harboring the open reading frame (ORF) under the control of the 35S promoter of Cauliflower mosaic virus (CaMV) and the CaMV terminator (isolated as a HindIII fragment from pRC [Puchta et al., 1996]) was inserted in the unique HindIII site of pGU.US within the T-DNA behind the GUS-specific sequences. By means of polymerase chain reaction (PCR), the ORF containing the negative selectable marker gene cytosine deaminase (codA) under the control of the CaMV 35S promoter and a nopaline synthase terminator was amplified from plasmid pNE3 (Stougaard, 1993) from which an internal XbaI site had been removed by a Klenow filling-in reaction.
The oligonucleotides used in the PCR reaction were 5′-pCGGCTCTAGAGCGGCCGCCTAGGGATAACAGGGTAATAGAATCCCACAAAAATCTGAGCTTAACAG-3′ and 5′-pCGGCTCTAGACTATTACCCTGTTATCCCTAGGCCCGATCTAGTAACATAGATGA-CACCGCGCGCG-3′. The PCR product carrying I-SceI sites at its ends was digested by XbaI-NotI and exchanged in pGU.US with the XbaI-NotI fragment carrying the hygromycin gene, resulting in the binary plasmid pGU.C.USB (Figure 1). In this binary plasmid, the codA gene is flanked by two I-SceI sites in direct orientation. The I-SceI expression vector pCISceI (Puchta et al., 1996) contains a synthetic I-SceI ORF under the control of the CaMV 35S promoter (Puchta et al., 1993) between T-DNA borders.
Plant Transformation
Tobacco plants (Nicotiana tabacum cv Petite Havana line SR1) were transformed with an Agrobacterium tumefaciens strain harboring the binary vector pGU.C.USB. Vacuum infiltration of tobacco seedlings and plant regeneration was performed as described previously (Salomon and Puchta, 1998). Segregation of the selfed transformants was tested by germination of the seed on Murashige and Skoog (1962) medium supplemented with 300 μg of phosphinotricin and 500 μg of 5-fluorouracil (5-FC) per milliliter.
In a second series of experiments, F1 seedlings of the transgenic lines GU.C.USB 1, 3, and 7 were inoculated with an Agrobacterium strain harboring the binary plasmid pCISceI as described by Puchta (1999b). Double selection using 100 μg of 5-FC and 100 μg of phosphinotricin per milliliter of medium was applied. In most cases, surviving calli were cut into two pieces after an additional 6 weeks. One piece was used for shoot regeneration or DNA isolation, and the other piece was used for GUS staining.
GUS assays were performed as described by Swoboda et al. (1994).
Plant DNA Extraction and DNA Gel Blot Analysis
DNA extraction from leaf tissues and calli was performed as described previously (Salomon and Puchta, 1998). DNA gel blot analysis using the hybridization membrane Hybond N (Amersham, Little Chalfont, UK) was performed as described by Salomon and Puchta (1998). The DNA probes were labeled using a random priming labeling kit (Megaprime DNA labeling system RPN1607; Amersham) and α-32P-dATP (Amersham). For identification of plants with single-copy integration, the codA coding region was used as a hybridization probe.
PCR and Sequence Analysis
Genomic DNA was analyzed by PCR using primers GUSH1 (5′-CGGAAGCTTCGTCACCAATCCCAATTCGATCTAC-3′) and GUSR1 (5′-CGGAAGCTTCCACTTGCAAAGTCCCGCTAGTGCC-3′). PCR and the direct sequencing of the amplification products were performed as described by Hartung and Puchta (2000).
Acknowledgments
We thank Charles White and Ingo Schubert for useful criticism of the manuscript and Frank Hartung, Brigit Gisler, and Fabian Heitzeberg for help and discussions. This study was funded partly by a grant from the biotechnology program of the Kultusministerium of Sachsen-Anhalt.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.001727.
References
- Bilang, R., Peterhans, A., Bogucki, A., and Paszkowski, J. (1992). Single-stranded DNA as recombination substrate in plants assessed by stable and transient expression. Mol. Cell. Biol. 12, 329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiurazzi, M., Ray, A., Viret, J.-F., Perera, R., Wang, X.-H., Lloyd, A., and Signer, E.R. (1996). Enhancement of somatic intrachromosomal homologous recombination in Arabidopsis by HO-endonuclease. Plant Cell 8, 2057–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groot, M.J.A., Offringa, R., Does, M.P., Hooykaas, P.J.J., and van den Elzen, P.J.M. (1992). Mechanisms of intermolecular homologous recombination in plants as studied with single- and double-stranded DNA molecules. Nucleic Acids Res. 20, 2785–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman-Lobell, J., Rudin, N., and Haber, J.E. (1992). Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol. Cell. Biol. 12, 1292–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego, M.E., Sirand-Pugnet, P., and White, C.I. (1999). Positive-negative selection and T-DNA stability in Arabidopsis transformation. Plant Mol. Biol. 39, 83–93. [DOI] [PubMed] [Google Scholar]
- Gorbunova, V., and Levy, A.A. (1997). Non-homologous DNA end joining in plant cells is associated with deletions and filler DNA insertions. Nucleic Acids Res. 25, 4650–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova, V., and Levy, A.A. (1999). How plants make ends meet: DNA double-strand break repair. Trends Plant Sci. 4, 263–269. [DOI] [PubMed] [Google Scholar]
- Hartung, F., and Puchta, H. (2000). Molecular characterization of two paralogous SPO11 homologues in Arabidopsis thaliana. Nucleic Acids Res. 28, 1548–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn, B., Levy, A., and Puchta, H. (2001). Elimination of selection markers from transgenic plants. Curr. Opin. Biotechnol. 12, 139–143. [DOI] [PubMed] [Google Scholar]
- Jasin, M. (2000). Chromosome breaks and genomic instability. Cancer Invest. 18, 78–86. [DOI] [PubMed] [Google Scholar]
- Kalendar, R., Tanskanen, J., Immonen, S., Nevo, E., and Schulman, A.H. (2000). Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc. Natl. Acad. Sci. USA 97, 6603–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik, A., Salomon, S., and Puchta, H. (2000). Species-specific double-strand break repair and genome evolution in plants. EMBO J. 19, 5562–5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk, I., Kovalchuk, O., Arkhipov, A., and Hohn, B. (1998). Transgenic plants are sensitive bioindicators of nuclear pollution caused by the Chernobyl accident. Nat. Biotechnol. 11, 1054–1059. [DOI] [PubMed] [Google Scholar]
- Lebel, E.G., Masson, J., Bogucki, A., and Paszkowski, J. (1993). Stress-induced intrachromosomal recombination in plant somatic cells. Proc. Natl. Acad. Sci. USA 90, 422–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman, C.W., Trautman, J.K., and Carroll, D. (1994). Illegitimate recombination in Xenopus: Characterization of end-joined junctions. Nucleic Acids Res. 22, 434–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, F., Han, M., Romanienko, P.J., and Jasin, M. (1998). Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl. Acad. Sci. USA 95, 5172–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, F.-L., Sperle, K., and Sternberg, N. (1984). Model for homologous recombination during transfer of DNA into mouse L cells: Role for DNA ends in the recombination process. Mol. Cell. Biol. 4, 1020–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, F.-L., Sperle, K., and Sternberg, N. (1990). Intermolecular recombination between DNAs introduced into mouse L cells is mediated by a nonconservative pathway that leads to crossover products. Mol. Cell. Biol. 10, 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryon, E., and Carroll, D. (1991. a). Involvement of single-stranded tails in homologous recombination of DNA injected into Xenopus laevis oocyte nuclei. Mol. Cell. Biol. 11, 3268–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryon, E., and Carroll, D. (1991. b). Characterization of recombination intermediates from DNA injected into Xenopus laevis oocytes: Evidence for a nonconservative mechanism of homologous recombination. Mol. Cell. Biol. 11, 3278–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason, R.M., Thacker, J., and Fairman, M.P. (1996). The joining of non-complementary DNA double-strand breaks by mammalian extracts. Nucleic Acids Res. 24, 4946–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste, T., Revenkova, E., Bechtold, N., and Paszkowski, J. (1999). An SMC-like protein is required for efficient homologous recombination in Arabidopsis. EMBO J. 18, 4505–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nicolas, A.L., Munz, P.L., and Young, C.S. (1995). A modified single-strand annealing model best explains the joining of DNA double-strand breaks in mammalian cells and cell extracts. Nucleic Acids Res. 23, 1036–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow, D. (2002). Recombination-directed plant transformation for the post-genomic era. Plant Mol. Biol. 48, 183–200. [PubMed] [Google Scholar]
- Paques, F., and Haber, J.E. (1999). Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterhans, A., Schlüpmann, H., Basse, C., and Paszkowski, J. (1990). Intrachromosomal recombination in plants. EMBO J. 9, 3437–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipiras, E., Coquelle, A., Bieth, A., and Debatisse, M. (1998). Interstitial deletions and intrachromosomal amplification initiated from a double-strand break targeted to a mammalian chromosome. EMBO J. 17, 325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta, H. (1998. a). Repair of genomic double-strand breaks in somatic plant cells by one-sided invasion of homologous sequences. Plant J. 13, 331–339. [Google Scholar]
- Puchta, H. (1998. b). Towards targeted transformation in plants. Trends Plant Sci. 3, 77–78. [Google Scholar]
- Puchta, H. (1999. a). DSB-induced recombination between ectopic homologous sequences in somatic plant cells. Genetics 152, 1173–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta, H. (1999. b). Use of I-SceI to induce double-strand breaks in Nicotiana. Methods Mol. Biol. 113, 447–451. [DOI] [PubMed] [Google Scholar]
- Puchta, H. (2000). Removing selectable marker genes: Taking the shortcut. Trends Plant Sci. 5, 273–274. [DOI] [PubMed] [Google Scholar]
- Puchta, H. (2002). Gene replacement by homologous recombination in plants. Plant Mol. Biol. 48, 173–182. [PubMed] [Google Scholar]
- Puchta, H., and Hohn, B. (1991). The mechanism of extrachromosomal homologous DNA recombination in plant cells. Mol. Gen. Genet. 230, 1–7. [DOI] [PubMed] [Google Scholar]
- Puchta, H., and Hohn, B. (1996). From centimorgans to basepairs: Homologous recombination in plants. Trends Plant Sci. 1, 340–348. [Google Scholar]
- Puchta, H., and Meyer, P. (1994). Substrate specificity of plant recombinases determined in extrachromosomal recombination systems. In Homologous Recombination and Gene Silencing in Plants, J. Paszkowski, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 123–155.
- Puchta, H., Dujon, B., and Hohn, B. (1993). Homologous recombination in plant cells is enhanced by in vivo induction of double-strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res. 21, 5034–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta, H., Swoboda, P., and Hohn, B. (1995. a). Induction of intrachromosomal homologous recombination in whole plants. Plant J. 7, 203–210. [Google Scholar]
- Puchta, H., Swoboda, P., Gal, S., Blot, M., and Hohn, B. (1995. b). Intrachromosomal homologous recombination events in populations of plant siblings. Plant Mol. Biol. 28, 281–292. [DOI] [PubMed] [Google Scholar]
- Puchta, H., Dujon, B., and Hohn, B. (1996). Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc. Natl. Acad. Sci. USA 93, 5055–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss, B., Schubert, I., Köpchen, K., Wendeler, E., Schell, J., and Puchta, H. (2000). RecA stimulates sister chromatid exchange and the fidelity of double-strand break repair, but not gene targeting, in plants transformed by Agrobacterium. Proc. Natl. Acad. Sci. USA 97, 3358–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries, G., Heller, W., Puchta, H., Sandermann, H.J., Seidlitz, H.K., and Hohn, B. (2000). Elevated UV-B radiation reduces genome stability in plants. Nature 406, 98–101. [DOI] [PubMed] [Google Scholar]
- Salomon, S., and Puchta, H. (1998). Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J. 17, 6086–6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel, P., Tikhonov, A., Jin, Y.K., Motchoulskaia, N., Zakharov, D., Melake-Berhan, A., Springer, P.S., Edwards, K.J., Lee, M., Avramova, Z., and Bennetzen, J.L. (1996). Nested retrotransposons in the intergenic regions of the maize genome. Science 274, 765–768. [DOI] [PubMed] [Google Scholar]
- Shalev, G., and Levy, A.A. (1997). The maize transposable element Ac induces recombination between the donor site and an homologous ectopic sequence. Genetics 146, 1143–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu, K., Schulman, A.H., Lahaye, T., and Schulze-Lefert, P. (2000). A contiguous 66-kb barley DNA sequence provides evidence for reversible genome expansion. Genome Res. 10, 908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stougaard, J. (1993). Substrate-dependent negative selection in plants using a bacterial cytosine deaminase gene. Plant J. 3, 755–761. [Google Scholar]
- Swoboda, P., Gal, S., Hohn, B., and Puchta, H. (1994). Intrachromosomal homologous recombination in whole plants. EMBO J. 13, 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinland, B., Hohn, B., and Puchta, H. (1994). Agrobacterium tumefaciens transfers single stranded T-DNA into the plant cell nucleus. Proc. Natl. Acad. Sci. USA 91, 8000–8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar, J., and Lichtenstein, C. (1992). Somatic and meiotic chromosomal recombination between inverted duplications in transgenic tobacco plants. Plant Cell 4, 319–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergunst, A.C., and Hooykaas, P.J.J. (1999). Recombination in the plant genome and its application in biotechnology. Crit. Rev. Plant Sci. 18, 1–31. [Google Scholar]
- Vicient, C.M., Suoniemi, A., Anamthawat-Jonsson, K., Tanskanen, J., Beharav, A., Nevo, E., and Schulman, A.H. (1999). Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. Plant Cell 11, 1769–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient, C.M., Jaaskelainen, M.J., Kalendar, R., and Schulman, A.H. (2001). Active retrotransposons are a common feature of grass genomes. Plant Physiol. 125, 1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Y.L., and Peterson, T. (2000). Intrachromosomal homologous recombination in Arabidopsis induced by a maize transposon. Mol. Gen. Genet. 263, 22–29. [DOI] [PubMed] [Google Scholar]
- Xiao, Y.L., Li, X., and Peterson, T. (2000). Ac insertion site affects the frequency of transposon-induced homologous recombination at the maize p1 locus. Genetics 156, 2007–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko, E., Scutt, C., and Meyer, P. (2000). Intrachromosomal recombination between attP regions as a tool to remove selectable marker genes from tobacco transgenes. Nat. Biotechnol. 18, 442–445. [DOI] [PubMed] [Google Scholar]