Abstract
The alga Volvox carteri is one of the simplest multicellular organisms, yet it has a surprisingly complex extracellular matrix (ECM), making Volvox suitable as a model system in which to study ECM self-assembly. Here, we analyze the primary structures and post-translational modifications of two main ECM components synthesized in response to sexual induction as well as wounding. These proteins are members of the pherophorin family with as yet unknown properties. They contain polyhydroxyproline spacers as long as 500 and 2750 residues. Even the highly purified proteins retain the capacity to self-assemble and cross-link, producing an insoluble fibrous network in an apparently autocatalytic reaction. This pherophorin-based network is located within the deep zone of the ECM. A molecular genetic search for additional members of the pherophorin family indicates that at least nine different pherophorin species can be expected to serve as precursors for ECM substructures. Therefore, the highly diversified members of the pherophorin family represent region-specific morphological building blocks for ECM assembly and cross-linking.
INTRODUCTION
The volvocine algae, which range from unicellular Chlamydomonas reinhardtii to multicellular organisms in the genus Volvox, offer an attractive model in which to investigate evolutionary aspects of the transition from unicellularity to multicellularity. Molecular genetic evidence indicates that Volvox carteri has evolved from a unicellular ancestor similar to Chlamydomonas during the past 50 million years (Rausch et al., 1989). The extracellular matrix (ECM) of a multicellular organism provides a scaffolding on which to create the shape of an organism and serves as a conduit for signals passing between cells within the organism or arriving from external sources. Therefore, the development of an increasingly organized ECM from a much more simple cell wall in the unicellular ancestor (Goodenough and Heuser, 1985; Woessner and Goodenough, 1994) was one of the prerequisites necessary for the transition to multicellularity. The design of ECM proteins in plants and animals appears to follow common principles: proteins organized in a striking modular manner are able to fulfill multiple functions, and extended and usually cross-linked polypeptides with repeated sequence motifs provide tensile strength (Doolittle, 1995).
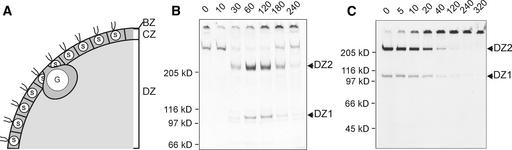
The asexually growing organism Volvox is about as simple as a multicellular organism can be. It is composed of only two cell types: 2000 to 4000 biflagellate Chlamydomonas-like somatic cells are arranged in a monolayer at the surface of a hollow sphere (Starr, 1969, 1970), and 16 much larger reproductive cells (“gonidia”) lie just below the somatic cell sheet. Volvox cells are surrounded and held together by a glycoprotein-rich ECM (for review, see Kirk et al., 1986; Sumper and Hallmann, 1998). More than 99% of a mature Volvox spheroid is ECM that is organized in a surprisingly complex manner (Kirk, 1998). It is assembled entirely from glycoproteins with a high content of Hyp (Hyp-rich glycoproteins), many of which are sulfated extensively (reviewed by Sumper and Hallmann, 1998). These Hyp-rich glycoproteins are organized into a system of highly regular fibrous layers that divide the ECM into at least three zones (Figure 1A): a boundary zone that consists of layers that coat the spheroid; a honeycomb-like array of compartments denoted the cellular zone that house the somatic cells and surround a voluminous central region; and the central region itself (the deep zone), which is filled with a network of fibers and filaments (Kirk et al., 1986).
Figure 1.
Identification of ECM Deep Zone Components.
(A) Highly stylized drawing emphasizing the main compartments of the Volvox ECM. BZ, boundary zone; CZ, cellular zone; DZ, deep zone (according to the nomenclature of Kirk et al. [1986]); G, gonidium (reproductive cell); S, somatic cells.
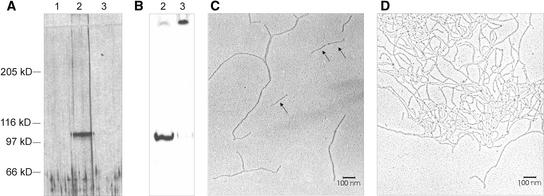
(B) Identification of DZ1 and DZ2 as ECM components secreted into the deep zone of the ECM in response to the sex-inducing pheromone. Fluorogram of a SDS–polyacrylamide gel loaded with deep zone extracts from sexually induced Volvox spheroids pulse labeled with 33P-phosphate (15 min). At time 0, sexual induction was initiated by the addition of pheromone (∼0.1 pM). A pulse-labeling experiment was initiated after the times indicated at top (minutes). At the end of the pulse-labeling period, a deep zone extract of the ECM was prepared by carefully disrupting the spheroids.
(C) In vitro polymerization of components DZ1 and DZ2. The deep zone extract obtained by 33Ppulse-labeling at 60 min after the application of the sex-inducing pheromone (lane 4 in [B]) was incubated further in vitro at 28°C for the times indicated at top (minutes), and aliquots were applied again to a SDS–polyacrylamide gel. The fluorogram of the gel is shown.
A remarkably rapid remodeling of the ECM is observed under the influence of the sex-inducing pheromone that triggers the initiation of the sexual life cycle of Volvox (Wenzl and Sumper, 1982, 1986b, 1987; Gilles et al., 1983). This pheromone is a glycoprotein that triggers the development of males and females at a concentration <10−16 M (Starr, 1970; Starr and Jaenicke, 1974; Tschochner et al., 1987; Mages et al., 1988). In particular, the synthesis of some members of the pherophorin family of ECM proteins (Wenzl and Sumper 1986b, 1987; Sumper et al., 1993; Godl et al., 1995, 1997) is induced strongly by the pheromone. Pherophorins are defined by a modular organization and by the presence of a C-terminal domain with homology with the sex-inducing pheromone.
From a subset of pherophorins (pherophorin II class), this pheromone-like domain becomes liberated proteolytically from the parent glycoproteins. It has been proposed that this modification and processing of the ECM is part of the signal amplification process required to achieve the exquisite sensitivity observed for this sexual induction system (Sumper et al., 1993). Surprisingly, most of the Volvox ECM proteins that are under the control of the sex-inducing pheromone have been shown to be induced by wounding as well (Amon et al., 1998; Ender et al., 1999). Therefore, pulse-labeling experiments performed immediately after wounding offer an alternative approach to detect ECM constituents.
Pheromone-induced remodeling of the ECM has been characterized in detail within the cellular zone (Figure 1A) of the ECM containing pherophorin I, II, and III (Wenzl and Sumper, 1982, 1986b; Ertl et al., 1989; Sumper et al., 1993; Godl et al., 1995) and to a lesser extent within the deep zone (Figure 1A) of the ECM containing pherophorin-S (Godl et al., 1997; Ender et al., 1999). The deep zone may constitute >95% of the total volume of the organism. An early study (Gilles et al., 1983) described two extracellular phosphorylated proteins (pp120 and pp240) possibly located within the deep zone that were found to be synthesized in response to the sex-inducing pheromone. However, neither biochemical nor molecular genetic data for these proteins have been published.
In this article, we analyze the primary structure and the glycosylation of these ECM proteins and demonstrate that these proteins are members of the pherophorin family with as yet unknown properties. Even the highly purified proteins retain the capacity to self-assemble and to cross-link, producing a completely insoluble fibrous network in an autocatalytic reaction. This network was visualized using recombinant DNA constructs combining one of these pherophorins with green fluorescent protein (GFP). A polymerase chain reaction–based search identified additional members of the pherophorin family that can be expected to serve as constituents of the Volvox ECM. Therefore, the highly diversified members of the pherophorin family represent the main building blocks for ECM assembly within both the cellular zone and the deep zone. Because a pherophorin-related gene was identified recently in unicellular Chlamydomonas (Rodriguez et al., 1999), diversification within this protein family appears to be part of the situation that allowed the evolution of a complex algal ECM.
RESULTS
Identification of the Deep Zone Components DZ1 and DZ2
Volvox ECM components secreted into the deep zone in response to the sex-inducing pheromone were analyzed by pulse-labeling experiments with radioactive phosphate. Deep zone material is released easily and selectively if a suspension of Volvox spheroids is forced gently through a hypodermic needle (Godl et al., 1997; Hallmann et al., 2001). This mild mechanical stress fragments the spheroids, producing hemispheres or smaller fragments of cellular sheets with the concomitant release of deep zone material. After low-speed centrifugation, the cell-free supernatant containing deep zone material was subjected to SDS-PAGE. Only ∼30 min after the application of the sex-inducing pheromone, two 33P-labeled components, with apparent molecular masses of 240 and 110 kD, become detectable in the fluorogram of the gel (Figure 1B). The same type of pulse-labeling experiment using 14C-bicarbonate produced a quite similar labeling pattern (data not shown), indicating that these components are the main products secreted into the deep zone after the application of the pheromone.
A remarkable property of the deep zone components DZ1 and DZ2 is evident during a chase experiment (Figure 1C). If a deep zone extract prepared from Volvox spheroids immediately after a 33P pulse-labeling period is incubated further at 28°C, both components disappear and become transformed into a polymer that is no longer able to penetrate the SDS gel. As shown below in more detail for DZ1, this is an inherent property of both of these proteins. Even after purification to homogeneity, DZ1 retains the capacity to polymerize to a completely insoluble and apparently covalently cross-linked network.
Purification of Component DZ1
The sulfhydryl group–modifying Ellman's reagent (Ellman, 1959), 5,5′-dithio-bis(2-nitrobenzoic acid), was shown recently to inhibit in a highly specific manner the cross-linking of ECM components in Volvox (Sumper et al., 2000). Indeed, the addition of 1 mM Ellman's reagent to a growing Volvox population inhibited the conversion of both DZ1 and DZ2 into a polymeric form, allowing the purification of these proteins in a soluble state. Thus, a deep zone extract was prepared from Volvox spheroids induced sexually in the presence of Ellman's reagent. This extract was applied first to an anion-exchange column equilibrated with 250 mM NaCl. Even at this high ionic strength, DZ1 and DZ2 bound to the ion-exchange matrix but eluted at 800 mM NaCl.
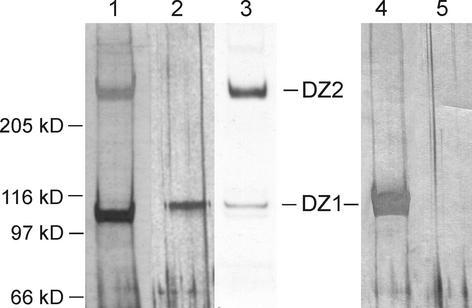
This high-salt eluate was diluted and subjected to fast protein anion-exchange chromatography. The DZ1 component eluted at 600 mM NaCl, and DZ2 required an even higher NaCl concentration (800 mM) for elution. As indicated by SDS-PAGE and silver staining (Figure 2, lane 1), DZ2 was still contaminated by a 100-kD component that was identified by peptide sequencing as pherophorin-S (Godl et al., 1997). However, DZ2 was well separated to allow the collection of amino acid sequence data by proteolytic in-gel digestion.
Figure 2.
Purification of the ECM Components DZ1 and DZ2.
Silver staining (lanes 1, 2, 4, and 5) of SDS–polyacrylamide gels loaded with different fractions from the purification procedure. Lane 1, DZ2 after anion-exchange chromatography; lane 2, DZ1 (0.5 μg) after hydroxyapatite HPLC; lane 3, fluorogram of 33P-labeled DZ1 and DZ2 applied as a reference; lane 4, to verify the homogeneity of DZ1, this gel was loaded heavily with purified DZ1 (20 μg); lane 5, only sample buffer was applied to detect staining artifacts.
DZ1 could be purified to homogeneity by an additional round of HPLC on a hydroxyapatite column (Figure 2, lane 2). Unlike typical proteins, which display dark-brown bands on a silver-stained SDS gel, DZ1 stained only weakly (staining was five times less strong compared with standard proteins) and produced a pale yellow band. Even on a heavily loaded gel (Figure 2, lane 4), no contaminating protein was seen, confirming the homogeneity of the DZ1 preparation.
Surprisingly, the amino acid sequence data obtained from the peptides isolated (Table 1) clearly indicate that DZ1 and DZ2 represent novel members of the pherophorin family, because peptide 2 (derived from DZ1) exhibited 65% identity to a corresponding sequence found in pherophorin-S (positions 116 to 135; Godl et al., 1997). In addition, peptide 4, derived from DZ2, displayed 55% identity to amino acid positions 37 to 47 of pherophorin II (Godl et al., 1995). Therefore, the names pherophorin-DZ1 and pherophorin-DZ2 were chosen to indicate their localization within the ECM.
Table 1.
Sequences of Isolated Peptides
| Peptides derived from DZ1 |
|---|
| AGTAILR |
| ATINGRPTTVGAAYDRPING |
| ATPINWSIAQASGAR |
| LLPPAFDVRPYR |
| DQTGLLPAFPFR |
| Peptides derived from DZ2 |
| FGRPV |
| LNVYPAGGPK |
| PPAIDVRPYR |
| QTGFVPNFPYR |
Pherophorin-Specific cDNA Library
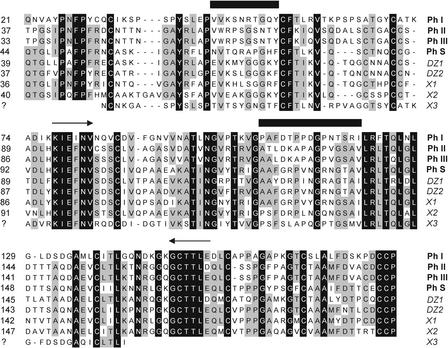
Realizing that members of the pherophorin family apparently serve as building units to establish the ECM architecture even in different ECM zones, we initiated an effort to construct a cDNA library expected to be enriched in pherophorin sequences. This approach was based on the fact that highly conserved amino acid sequence motifs are common to all known pherophorin species. Within their N-terminal domains, two elements, KIEFNV and GCTTLE, found at positions 97 and 171, respectively, in pherophorin-S (Godl et al., 1997) appear to be nearly invariant motifs. The sequence information of the latter motif was used to synthesize an antisense oligonucleotide primer to reverse transcribe mRNA isolated from both vegetatively growing and sexually induced Volvox algae. A sense primer derived from the motif KIEFNV allowed the amplification of cDNA populations of ∼240 bp. These cDNAs were cloned into the SmaI site of pUC18 vector by blunt ligation. The inserts from 25 randomly selected transformants derived from vegetatively growing Volvox populations confirmed the nearly exclusive presence of pherophorin sequences in this library.
The sequences obtained included the known sequences for pherophorin I (12 hits), pherophorin-S (3 hits), and two as yet unknown pherophorins (5 and 3 hits). Another 20 transformants were sequenced that were derived from sexually induced organisms. This produced mainly sequences encoding pherophorin II, pherophorin-S, and three novel pherophorins. The alignment of these deduced amino acid sequences (Figure 3) confirmed a high degree of homology among these genes. Remarkably, two of these novel sequences (denoted DZ1 and DZ2) encode peptides 1 and 2 obtained from DZ1 and peptide 4 derived from DZ2. Both of these cDNA sequences, therefore, were used to probe a genomic library of Volvox in the replacement vector λEMBL3 (Frischauf et al., 1983) to obtain the complete genes.
Figure 3.
Identification and Amino Acid Sequence Alignment of Members of the Pherophorin Family.
cDNA sequences were obtained from a cDNA library enriched for pherophorin (Ph) sequences. In addition to all of the known members of the family (indicated by boldface species names), five novel pherophorin species (indicated by italic species names) were found. The alignment is confined to the N-terminal domains of pherophorins preceding their central polyproline spacers. Arrows indicate highly conserved sequences used to design oligonucleotides for polymerase chain reaction amplification of pherophorin-related cDNAs. Sequences upstream of the sense primer and downstream of the antisense primer were derived from genomic clones of the corresponding pherophorins. Numbers at left indicate the amino acid position at which each alignment starts. Horizontal black bars indicate two sequence stretches that are diagnostic for a given pherophorin species.
Three positive clones were identified with the dz1 probe out of 120,000 phages screened. The 14-kb insert of one of these clones containing the complete gene was subcloned and sequenced (Figure 4). We were unable to clone a genomic DNA covering the complete dz2 gene. As detected by protein chemical analysis, pherophorin-DZ2 contains a unique polyhydroxyproline domain as long as 2750 amino acid residues (see below). Most likely, the extreme GC content (CCN is the codon for Pro) of the corresponding DNA stretch interferes with the efficient cloning of the complete dz2 gene in Escherichia coli. The GC-rich region of the dz2 clones obtained turned out to be truncated. Partial sequencing (∼200 nucleotides) confirmed the encoding of a polyproline stretch.
Figure 4.
Gene Organization and Deduced Amino Acid Sequence for Pherophorin-DZ1.
(A) Restriction map and intron/exon organization of the dz1 gene cloned from a genomic library. Black bars represent exons, and the gray bar indicates a GC-rich stretch encoding a polyproline sequence.
(B) Deduced amino acid sequence of pherophorin-DZ1. The characteristic polyproline stretch is shown on a gray background. Amino acid sequences confirmed from isolated peptides are underlined. The arrowhead marks the potential signal peptidase cleavage site. The broken line marks the C-terminal part of the polypeptide found to be homologous with the sex-inducing pheromone.
Deduced Amino Acid Sequence
A striking feature of the deduced amino acid sequence for pherophorin-DZ1 is a central domain, 502 amino acid residues long, that is composed almost exclusively (98.2%) of Pro residues (Figure 4B). Most likely, the secondary structure of this domain is a polyproline II helix that separates the N- and C-terminal domains. As expected, the N-terminal sequence (positions 1 to 23) represents a putative signal peptide to guide this polypeptide into the secretory pathway. A BLASTP search (Altschul et al., 1990) of the SWISSPROT protein sequence database confirmed significant identities to the pherophorin family from Volvox. The region of identities covers nearly the total length of the polypeptide chain. For instance, the N-terminal part of the polypeptide (amino acids 24 to 208) shows 58% identity to the N-terminal part of pherophorin-S (amino acids 17 to 200) (Godl et al., 1997), and the C-terminal part (amino acids 712 to 1009) exhibits 50% identity to the C-terminal half of pherophorin II (amino acids 182 to 484) (Sumper et al., 1993).
Glycosylation
The carbohydrate composition of pherophorin-DZ1 was determined by radio gas chromatography. Pherophorin-DZ1 purified from Volvox spheroids grown in the presence of 14C-bicarbonate was hydrolyzed, and the resulting monosaccharides were converted to the corresponding alditol acetates. Pherophorin-DZ1 contains the neutral sugars Ara and Gal in a 1:1 ratio (data not shown).
Pherophorin-DZ1 incorporated 33P-phosphate in pulse-labeling experiments. The hydrolysis of pherophorin-DZ1 in 0.5 M trifluoroacetic acid at 100°C for 2 hr quantitatively liberated bound phosphate as a low molecular mass derivatives. In earlier studies, the phosphodiester Ara-5-phospho-5′-Ara was identified as a structural element in both the ECM glycoprotein SSG 185 from Volvox (Holst et al., 1989) and in pherophorin-S (Godl et al., 1997). Proof of the existence of the same structural component in pherophorin-DZ1 was obtained by mass spectrometry as follows. Pherophorin-DZ1 (supplemented with trace amounts of 33P-labeled material) was hydrolyzed and reduced with sodium borohydride. The resulting products were separated by high-performance anion-exchange chromatography.
As expected, three radioactive peaks were obtained that corresponded to a phosphodiester, a phosphomonoester, and traces of free phosphate. The putative phosphodiester fraction was collected and analyzed by mass spectrometry. The phosphodiester produced a mass signal at a mass-to-charge ratio of 365.0. This corresponds exactly to the calculated mass for the reduced phosphodiester Ara-5-phospho-5′-Ara. The exclusive presence of arabinose in this material (obtained from 14C-labeled pherophorin-DZ1) was proven by radio gas chromatography exactly as described for pherophorin-S (Godl et al., 1997).
Characterization of the Polyproline Module
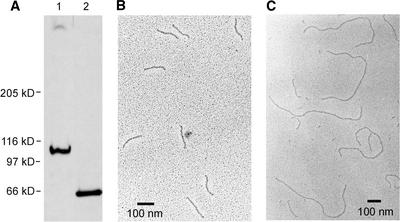
Because of its polyproline domain, pherophorin-DZ1 is partly resistant to degradation by proteases such as pronase, proteinase K, and subtilisin. The resistant core exhibits an apparent molecular mass of ∼60 kD (6% SDS-PAGE) (Figure 5A). The proteolytic degradation of 33P-labeled pherophorin-DZ1 resulted in core material that still contained all of the originally incorporated radioactivity, indicating that the phosphodiester is located within this resistant core material. To define this core material, homogenous pherophorin-DZ1 was digested with subtilisin. The 60-kD core material was purified by anion-exchange chromatography and analyzed by electron microscopy after rotary shadowing. As shown in Figure 5B, rod-like structures with an average length of 150 nm were detected. Circular dichroism spectra obtained from this material (data not shown) confirmed the presence of a polyproline II helix conformation.
Figure 5.
Characterization of the Protease-Resistant Fragment Derived from Pherophorin-DZ1 and -DZ2.
(A) Fluorogram of a SDS–polyacrylamide gel loaded with 33P-labeled pherophorin-DZ1 before (lane 1) and after (lane 2) digestion with subtilisin.
(B) Electron microscopic analysis of the resistant fragment derived from pherophorin-DZ1 after rotary shadowing.
(C) Electron microscopic analysis of the resistant fragment derived from pherophorin-DZ2 after rotary shadowing.
With a residue height in this helix of 0.3 nm (Bhatnagar and Gough, 1996), the rods should consist of ∼500 amino acid residues. Complete acid hydrolysis of this core material followed by amino acid analysis detected the presence of Hyp only (confirmed by mass spectrometry). Using exactly the same experimental procedure with pherophorin-DZ2, rods as long as 825 ± 22.5 nm were obtained (a total of 100 rods were measured), corresponding to a polyproline stretch of 2750 amino acid residues (Figure 5C). The 20-nm fragments distributed in this electron microscopy image resulted from the polyproline domain of pherophorin-S that contaminated the DZ2 preparation. The existence of this polyproline stretch in pherophorin-DZ2 was confirmed by cloning the truncated genomic clones that encoded polyproline stretches (see above).
Autocatalytic Polymer Formation
As shown above, pherophorin-DZ1 and -DZ2 in a deep zone extract did not remain in a soluble state. Rather, they were converted to polymeric and insoluble materials upon incubation at 28°C. Treatment with Ellman's reagent inhibited in a highly specific manner this assembly and cross-linking. When highly purified pherophorin-DZ1 (previously inactivated with Ellman's reagent) was treated with a thiol compound such as DTT to restore thiol groups of the protein that were converted to a mixed disulfide by Ellman's reagent, the protein displayed remarkable behavior. It polymerized and appeared to become covalently cross-linked in an autocatalytic reaction (Figures 6A and 6B). The polymerized product did not enter the gel and was no longer detectable by silver staining (Figure 6A, lane 3). Therefore, 33P-labeled DZ1 purified to homogeneity was used to detect by fluorography the polymerized material that was trapped in the well (Figure 6B, lane 3).
Figure 6.
Autocatalytic Cross-Linking of Pherophorin-DZ1.
(A) Silver staining of a SDS–polyacrylamide gel loaded with homogenous pherophorin-DZ1 before (lane 2) and after (lane 3) incubation in polymerization buffer (containing DTT). Lane 1 was loaded with SDS sample buffer only.
(B) Loading as in (A) but using homogenous 33P-labeled pherophorin-DZ1 and detection of radioactive components by fluorography.
(C) and (D) Electron microscopic images obtained from pherophorin-DZ1 before (C) and after (D) in vitro polymerization. A monomer and a dimer of pherophorin-DZ1 are marked by arrows in (C). Although the protein concentration in both preparations was the same, clotting of the cross-linked material produced a highly nonhomogenous distribution of rods in the electron microscopy image (D).
Covalent cross-links are postulated because the polymeric state of pherophorin-DZ1 cannot be released by any of the following treatments: 6 M guanidinium hydrochloride, 70% formic acid, and boiling in 1.5% SDS and 2.5% β-mercaptoethanol. Electron microscopy by rotary shadowing of pherophorin-DZ1 before and after treatment with DTT revealed additional information. Even Ellman's reagent–treated pherophorin-DZ1 assembled to form dimers and oligomeric chains (Figure 6C). However, this self-assembly occurred via noncovalent linkages, because only the 110-kD monomers were observed by SDS-PAGE of this material. Only after the restoration of the modified thiol groups did these self-assembled structures become cross-linked by autocatalysis, forming a complex fibrous network (Figure 6D). DZ1 solutions of >200 μg/mL produced a stiff gel that clotted into a visible precipitate after vigorous shaking. This material remained completely insoluble by SDS-PAGE.
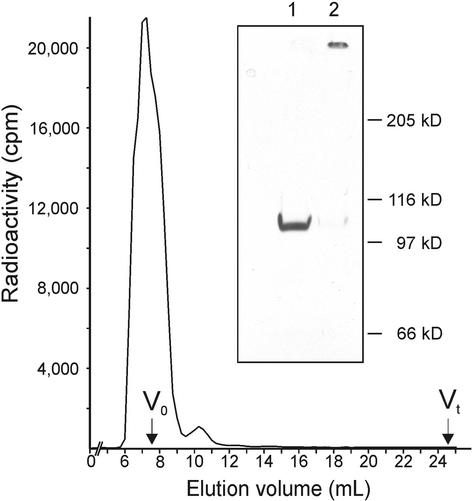
To exclude the hypothetical possibility that trace amounts of a cross-linking enzyme remain tightly bound to DZ1 even after HPLC purification steps, an additional purification step in the presence of 6 M guanidinium was included. Any tightly bound enzyme should dissociate under these conditions and should be removed by size-exclusion chromatography on Superose 12 (Mr exclusion limit, 2 × 106), because DZ1 elutes with the void volume of this column as a result of its extreme rod-shaped structure. Even after this extreme treatment, DZ1 recovered its activity to polymerize after dialysis (Figure 7). Purified pherophorin-DZ2 exhibited the same polymerization activity demonstrated for pherophorin-DZ1. By analogy, autocatalytic cross-linking appears likely, but the presence of an enzyme remains a possibility because DZ2 could not be purified to homogeneity (Figure 2, lane 1).
Figure 7.
Pherophorin-DZ1 Polymerization Assay after Size-Exclusion Chromatography in the Presence of 6 M Guanidinium Hydrochloride.
Sixty micrograms of purified 33P-labeled pherophorin-DZ1 was applied to a Superose 12 column (bed volume, 24 mL; Amersham Pharmacia Biotech) equilibrated in 6 M guanidinium hydrochloride and 100 mM sodium phosphate, pH 7.0. Pherophorin-DZ1 eluted with the void volume (V0) of 7 mL. The peak fraction was dialyzed against Volvox medium and concentrated fivefold by ultrafiltration. Twenty-microliter aliquots then were incubated for 6 hr at 28°C in the presence or absence of 1 mM DTT. Both samples were analyzed by SDS-PAGE followed by fluorography. The insert shows incubation in the absence (lane 1) or presence (lane 2) of DTT.
In Vivo Localization
For in vivo localization of pherophorin-DZ1, a chimeric gene was constructed that encodes a pherophorin-DZ1 that is linked at its C-terminal end to GFP (Figure 8A). For this construct, the genomic clone of pherophorin-dz1 that includes its own promotor was combined via a short linker region that encodes a flexible peptide sequence with a gfp sequence that has been adapted to the preferred codon usage in Chlamydomonas (Fuhrmann et al., 1999). Corresponding Volvox transformants expressed this gene product under the control of the sex-inducing pheromone and displayed strong fluorescence within the deep zone of the ECM (Figures 8B to 8E). For a more precise localization, these transformants were analyzed by confocal fluorescence microscopy (Figures 8F and 8G). Pherophorin-DZ1 was organized in a fibrous network within the deep zone and was excluded from the cellular compartments housing the large reproductive cells.
Figure 8.
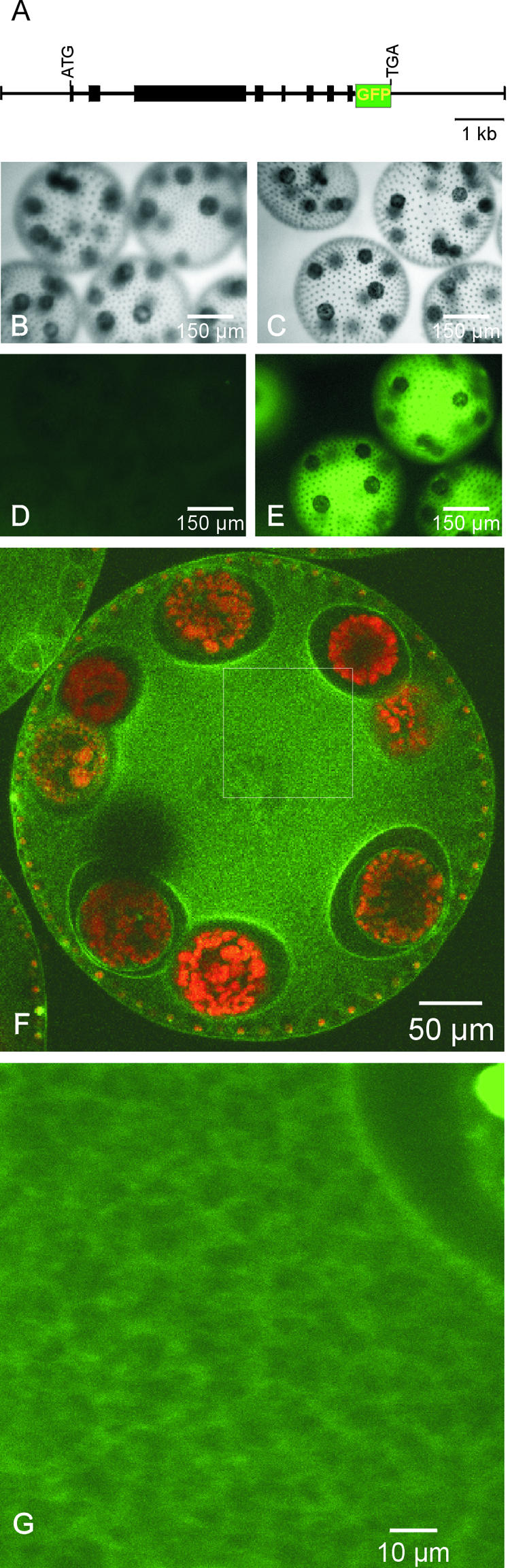
Localization within the ECM of Pherophorin-DZ1 Linked with GFP.
(A) Structure of the chimeric gene used for the transformation of Volvox containing the pherophorin-dz1 gene linked to the gfp gene.
(B) to (E) Microscopy of transformants. Volvox spheroids before (B) and after (C) sexual induction for 180 min. Fluorescence of transformants before (D) and after (E) sexual induction.
(F) and (G) Confocal fluorescence microscopy of a sexually induced transformant. (G) shows the scan of a section (white frame in [F]) at a higher magnification.
Response to Wounding
Volvox responds to the sex-inducing pheromone and to wounding (e.g., breaking up the spheroids into hemispheres) in much the same way (Amon et al., 1998; Ender et al., 1999). Both of these environmental stimuli induce changes that affect the structure of the ECM. These responses also include the induction of pherophorin-DZ1 and -DZ2 synthesis (data not shown). If Volvox spheroids are slit, the resulting hemispheres display a strong tendency to close and to restore spheres within several hours. The self-assembly of ECM components probably is involved in this healing process. This assumption is supported by the observation illustrated in Figure 9. If hemispheres are treated with Ellman's reagent, which inhibits cross-linking of pherophorins, this healing response is suppressed completely and slit spheroids are no longer able to restore a spherical structure.
Figure 9.
“Wound Healing“ in Volvox.
Spheroids were gently slit into hemispheres by passage through a hypodermic needle and incubated for another 8 hr in pure Volvox medium or medium containing 1 mM Ellman's reagent.
(A) Intact spheroids as a reference.
(B) and (C) Slit spheroids incubated in the presence (B) or absence (C) of Ellman's reagent, which inhibits cross-linking of ECM components.
DISCUSSION
The ECM accounts for >99% of the volume of a mature Volvox spheroid (Kirk, 1998), and it is the complex architecture of the ECM that places the cells into a coordinated multicellular organism. At the end of embryogenesis, the somatic cells of the embryo begin to secrete ECM material, causing each cell to move apart from its neighbors. The organism then grows in size but not in cell number. Hence, the production and deposition of ECM constituents remains a major synthetic activity of the enlarging organism. During this period, all of the ECM structures enlarge proportionally and maintain their distinctive symmetries.
Understanding how this modeling of ECM structures, partly positioned at considerable distances from the site of precursor synthesis (the somatic cells), is accomplished remains a challenging task. A major impediment to the molecular analysis of precursor molecules that create the fibrous networks within the ECM is the extensive cross-linking that makes most components completely insoluble in all conventional protein-extraction media. Recently, Ellman's reagent was recognized as an ideal tool to specifically inhibit the cross-linking of ECM components (Sumper et al., 2000), and this in turn has enabled the purification and biochemical characterization of soluble ECM precursors.
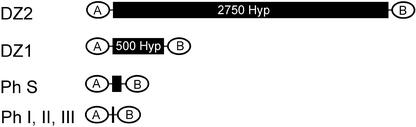
This article describes the structures and properties of two precursors of fibrous ECM structures. These monomers are novel members of the pherophorin family with striking and as yet unknown features that appear to be crucial for self-assembly and cross-linking processes. The scheme shown in Figure 10 summarizes the modular composition of known pherophorins. Unexpectedly, even after purification to homogeneity (as determined by silver staining of SDS gels), the monomers of pherophorin-DZ1 retain the capability to create in vitro a complex fibrous network. This polymeric network remains stable even in the presence of 70% formic acid or hot SDS/mercaptoethanol, indicating the formation of covalent cross-links. If this is the case, pherophorin-DZ1 protein must have the enzymatic property for autocatalytic cross-linking.
Figure 10.
Domain Organization of Pherophorins.
Black boxes represent the polyhydroxyproline (Hyp) spacers. Ph, pherophorin.
In addition, pherophorin-DZ1 and -DZ2 exhibit remarkable structural features. Their internal Hyp-rich modules (HR modules) consist of 500 (pherophorin-DZ1) and 2750 (pherophorin-DZ2) amino acid residues, producing rod-like spacers as long as 150 and 825 nm that separate the N- and C-terminal domains, which are completely devoid of Hyp residues. Most likely, these rod-shaped HR modules have a mainly structural function and serve as spacers to create different sets of defined frameworks within the ECM. Where analyzed in more detail, these HR modules were found to be targets for extensive post-translational modifications. Among the modifications found in Volvox are O-glycosylations with oligoarabinosides and attachment of saccharides containing phosphodiester bridges between Ara residues (Ertl et al., 1989; Holst et al., 1989; Godl et al., 1997). This is true as well for the HR module present in pherophorin-DZ1.
The construction of a cDNA library enriched for pherophorin-related sequences resulted in the identification of another three unknown pherophorin species. Thus, the pherophorin family consists of at least nine members in Volvox. Pherophorins are found as constituents of all major ECM compartments (cellular and deep zone) and therefore are likely to serve as building units for most if not all fibrous networks within the ECM. The identification of a pherophorin-related gene in the unicellular alga Chlamydomonas (Rodriguez et al., 1999) suggests that the divergent evolution of a pherophorin-related ancestor protein enabled the development of more and more differentiated ECM structures, because they are found within both the cellular and deep zones of multicellular algae.
The amino acid sequence alignment of the N-terminal domains reveals a high degree of homology among different pherophorins. However, there are two particular stretches, separated by ∼50 amino acids (marked in Figure 3), that are unique to a given pherophorin species. It is tempting to speculate that these motifs are responsible for self-recognition during the assembly of a specific pherophorin-based network. However, the in vitro polymerization experiments do not exclude the possibility that mixed meshworks may form in the native situation.
The experiments described here indicate that it is an inherent property of pherophorin-DZ1 monomers to create oligomers that become cross-linked into fibrous networks in an autocatalytic process. Oligomer formation and cross-linking are separate processes, as demonstrated by the fact that treatment with Ellman's reagent selectively inhibited cross-linking without affecting the formation of oligomers. The nature of the covalent cross-links is unknown at present. The participation of intermolecular disulfide bridges is highly unlikely, because the polymeric product was completely resistant to even the harshest treatments known to cleave disulfide linkages (e.g., performic acid). In contrast, anhydrous hydrogen fluoride at 0°C completely degraded pherophorin-DZ1 polymers, resulting in the same (deglycosylated) product that was produced by hydrogen fluoride treatment of DZ1 monomers (data not shown).
Anhydrous hydrogen fluoride is able to cleave glycosidic bonds as well as phosphoester bonds but leaves peptide bonds completely unaffected (Mort and Lamport, 1977). This finding indicates the participation of saccharides or phosphodiester bridges in the cross-linking reaction and excludes other types of cross-links detected in algal ECMs: products of transglutaminase reactions (Waffenschmidt et al., 1999) and peroxide-mediated oxidative cross-linking (Waffenschmidt et al., 1993). Recently, a peptide motif was identified that mediates the cross-linking of cell wall proteins from tobacco plants (Domingo et al., 1999). This motif is a Cys-rich domain. Pherophorin-DZ1 also contains Cys-rich stretches in both its N- and C-terminal domains, but these do not match the consensus sequence derived from tobacco cell wall proteins. The elucidation of the chemical nature of the cross-links in pherophorin-based networks should provide important insights into the molecular mechanism of ECM assembly and remodeling.
METHODS
Culture Conditions
The female Volvox carteri f. nagariensis strains HK10 (wild type) and 153-48 (nitA−) were grown as described previously (Godl et al.,1997).
Radioactive Labeling with 33P-Phosphate
In pulse-labeling and pulse-chase-labeling experiments with 33P-phosphate, ∼3000 Volvox spheroids were washed and suspended in 1 mL of phosphate-free Volvox medium (Provasoli and Pinter, 1959). After the addition of 50 μCi of 33P-phosphate (100 μCi/nmol), incubation under standard conditions was continued for 3 hr. Polymerization of pherophorin-DZ1 and -DZ2 was inhibited by supplementing the medium with 1 mM 5,5′-dithio-bis(2-nitrobenzoic acid) (Ellman's reagent; Sigma Aldrich) (Sumper et al., 2000).
Purification of Pherophorin-DZ1 and -DZ2
One hundred twenty liters of sexually induced Volvox spheroids (incubated for 3 hr in Volvox medium containing 1 pM pheromone and 1 mM Ellman's reagent to inhibit the polymerization of pherophorin-DZ1 and -DZ2) were harvested by filtration on a 100-μm-mesh nylon screen. The spheroids were broken up by forcing them through a 0.5-mm hypodermic needle. The disrupted spheroids were centrifuged at 16,000g for 30 min. To remove any remaining weakly charged components, the supernatant was adjusted to 20 mM Tris, pH 8.2, and 250 mM NaCl and applied to a QAE Sephadex A-25 column (volume, 100 mL) (Amersham Pharmacia Biotech) equilibrated in the same buffer. After washing the column with equilibration buffer, bound components were eluted with 20 mM Tris, pH 8.2, and 800 mM NaCl. The eluate, containing pherophorin-DZ1 and -DZ2, was diluted to 400 mM NaCl in 20 mM Tris, pH 8.2, and applied to a UNO Q-6 anion-exchange column (Bio-Rad Laboratories).
Pherophorin-DZ1
Pherophorin-DZ1 was eluted from the UNO Q-6 column by a stepwise increase of the ionic strength to 600 mM NaCl. The desalted eluate was brought to 10 mM Na-phosphate, pH 6.8, and applied to a Bio-Scale CHT5 ceramic hydroxyapatite column (Bio-Rad Laboratories) equilibrated in the same buffer. After washing the column with 100 mM Na-phosphate buffer, pH 6.8, pherophorin-DZ1 was eluted by a stepwise increase to 600 mM Na-phosphate buffer, pH 6.8. The eluate was desalted and concentrated (Centricon 10; Millipore, Bedford, MA). As determined by SDS-PAGE with subsequent silver staining, pherophorin-DZ1 was purified to homogeneity. Typically, the yield was 100 to 150 μg.
Pherophorin-DZ2
For the recovery of pherophorin-DZ2, the UNO Q-6 column was eluted further with 20 mM Tris, pH 8.2, and 800 mM NaCl. The eluate was desalted and concentrated (Centricon 30; Millipore). As verified by SDS-PAGE and subsequent silver staining, pherophorin-DZ2 was sufficiently pure for the production of peptides. Typically, the yield was 1 to 2 mg. Glycoprotein concentrations were determined according to Dubois et al. (1956).
Proteolytic Digestion and Separation of Peptides
Aliquots of 20 μg of pherophorin-DZ1 and 40 μg of enriched pherophorin-DZ2 were applied to 6% SDS-PAGE gels and stained with Alcian Blue 8GX (Sigma Aldrich). The gel slices containing pherophorin-DZ1 and -DZ2 were cut out in small pieces. Further treatment and digestion with trypsin were performed as described previously (Selmer et al., 1996). The resulting peptides were eluted from the gel with 0.2 M NH4HCO3 and 50% acetonitrile. The eluate was passed through a 0.22-μm filter (Millipore) and dried by lyophilization. The peptides were dissolved in 6 M guanidinium-HCl and 0.1% trifluoroacetic acid and separated by reverse-phase HPLC (SMART system; Amersham Pharmacia Biotech) on a 3-μm RPC C2/C18 column (Amersham Pharmacia Biotech) by applying a 50-min linear gradient of 5 to 50% acetonitrile in 0.1% trifluoroacetic acid at a flow rate of 200 μL/min. Peptides were sequenced by Edman degradation using an automated gas-phase peptide sequencer (Applied Biosystems, Foster City, CA).
Preparation of the Subtilisin Fragments of Pherophorin-DZ1 and -DZ2
Fifty micrograms of pherophorin was digested with 0.5 μg/μL subtilisin (Carlsberg type VIII; Sigma Aldrich) in 50 mM Tris, pH 8.0, and 0.5% SDS for 3 hr at 30°C and applied to a QAE Sephadex A-25 column (volume, 2 mL; Amersham Pharmacia Biotech) equilibrated in 20 mM Tris, pH 8.2, and 200 mM NaCl. The column was washed with 20 mM Tris, pH 8.2, and 400 mM NaCl. Elution of the protease-resistant fragment was performed with 20 mM Tris, pH 8.2, and 800 mM NaCl. After dialysis against water, the eluate was concentrated by lyophilization.
Amino Acid Analysis and Mass Spectrometry
Amino acid analysis was performed as described by Cohen and Strydom (1988). Molecular masses of compounds of interest were determined by electrospray mass spectrometry using an Esquire-LC apparatus (Bruker Instruments, Billerica, MA).
Preparation of the Phosphodiester of Ara
Pherophorin-DZ1 was mixed with 33P-labeled pherophorin-DZ1 and hydrolyzed in 0.5 M trifluoroacetic acid at 100°C for 2 hr. The preparation of the phosphodiester was performed as described by Godl et al. (1997).
Carbohydrate Analysis
Volvox spheroids were pulse labeled with 14C-bicarbonate as described by Wenzl et al. (1984). The neutral sugar composition of pherophorin-DZ1 was determined by radio gas chromatography of the alditol acetates as described by Wenzl and Sumper (1986a). The carbohydrate analysis of the phosphodiester was performed as described by Godl et al. (1997).
Polymerization Assay for Pherophorin-DZ1
Pherophorin-DZ1, purified from sexually induced Volvox cultures (see above), was obtained in the monomeric state. Purified, desalted, and concentrated pherophorin-DZ1 (100 to 200 μg/mL) can be transformed into the polymeric state by incubation in 1 mM DTT at 28°C for 3 hr. The kinetics of polymerization were followed by SDS-PAGE.
Stability of Polymeric Pherophorin-DZ1 in Denaturing Agents
The polymeric form of pherophorin-DZ1 sedimented quantitatively by centrifugation at 50,000g for 20 min, whereas the soluble form remained in the supernatant, as shown with 33P-labeled pherophorin-DZ1. To analyze the stability of the polymeric state, samples were dissolved in different chaotropic solutions: 6 M guanidinium-HCl, 70% formic acid, and 1.5% SDS with 2.5% β-mercaptoethanol. None of these conditions was able to convert polymeric material into the soluble form, as judged by the sedimentation assay.
Generation of a Pherophorin-Enriched cDNA Library
For reverse transcription of pherophorin mRNAs, the degenerated oligonucleotide sequence 5′-TCCAGSGTSGTGCAKCC-3′ (derived from the conserved amino acid sequence motif GCTTLE) served as a primer. This primer was biotinylated and bound to streptavidin-coated magnetic beads (Dynal Biotech, Oslo, Norway). The degenerated sense primer for cDNA amplification of pherophorin-enriched cDNA populations was 5′-AARATTGAGTTYAAYGT-3′, derived from the conserved amino acid sequence motif KIEFNV common to all pherophorins.
Cloning of the Pherophorin-DZ1 Gene
The amino acid sequence information for the peptides 5′-ATINGRPTTVGAAYDRPING-3′ and 5′-AGTAILR-3′ identified the correct cDNA clone in a collection obtained from the pherophorin-enriched cDNA library. Because of the high degree of homology among the members of pherophorin gene family, a diagnostic sequence of only 33 nucleotides (5′-CGCCTAGATCTCACACTCGCAACCGCCGCGGAC-3′) was used to probe the genomic library. The Volvox genomic library in λEMBL3 (Frischauf et al., 1983) described by Ertl et al. (1989) was used to clone the pherophorin-DZ1 gene. The screening procedure followed standard techniques (Sambrook et al., 1989).
DNA sequencing was performed by the chain-termination method (Sanger et al., 1977) or by cycle sequencing (Kim and Kim, 1994; Hengen, 1996). Subclones with stretches of an unusually high (G)C content and repetitive motifs were digested unidirectionally with exonuclease III to create targeted breakpoints for DNA sequencing (Henikoff, 1984). This was done from both sides of the subclones. The precise length of a given insert was determined by gel electrophoresis. Sequencing of (G)C-rich stretches was improved using nucleotide analogs (McConlogue et al., 1988).
To verify the intron/exon boundaries assumed by homology with other known pherophorin genes, mRNA from sexually induced (1 hr) Volvox spheroids was reverse transcribed with exon-specific pherophorin-DZ1 oligonucleotides and further amplified by polymerase chain reaction (PCR). The resulting cDNAs were ligated into the SmaI site of pUC18 and sequenced.
Construction of the Chimeric dz1-gfp Gene
A chimeric gene was constructed consisting of the complete genomic clone of dz1, which includes 1.4 kb upstream of the start codon and 2.9 kb downstream of the stop codon, and a gfp sequence, which was adapted to the codon usage of Chlamydomonas reinhardtii (Fuhrmann et al., 1999). The gfp sequence was inserted immediately in front of the stop codon of the dz1 gene. Additionally, a sequence coding for a pentaglycine spacer was introduced to separate the C-terminal amino acid of pherophorin-DZ1 from the N-terminal amino acid of green fluorescent protein. This was achieved by PCR on exon 8 of dz1 with oligonucleotide 5′-ATGGTTGCAATGATG- TGGC-3′ and the recombinant oligonucleotide 5′-CTTGGCCATGCC-ACCACCGCCGCCAGCGAAGCCGCTGCCGTA-3′, which bear the sequence coding for the pentaglycin spacer, and a MscI restriction site, which was used to fuse the dz1 and gfp sequences. The final construction was performed by standard techniques (Sambrook et al., 1989). The construct was confirmed by sequencing.
Stable Nuclear Transformation of Volvox
Volvox strain 153-48 was transformed using a particle gun to bom-bard cells with DNA-coated gold particles, as described previously (Schiedlmeier et al., 1994). Plasmids carrying the artificial gene construct were introduced into Volvox nit− strain 153-48 by cotransformation with plasmid pVcNR1 (Gruber et al., 1992; Schiedlmeier et al., 1994) containing the Volvox nitA gene. Bombarded cultures were cultivated in selective Volvox medium containing only nitrate as a nitrogen source.
Genomic PCR
Genomic PCR was used to confirm the stable transformation of Volvox. Fifty spheroids were selected using a dissecting microscope and transferred into 10 μL of sterile lysis buffer (0.1 M NaOH, 2.0 M NaCl, and 0.5% SDS). After 5 min at 95°C, 200 μL of 50 mM Tris, pH 7.5, was added. Two microliters of the resulting lysate was used for PCR (in a total volume of 100 μL). Products of PCR amplification were cloned into the pUC18 SmaI site and sequenced.
Fluorescence Microscopy
Specimens were examined with a fluorescence imaging system (Till Photonics, Gräfelfing, Germany). The green fluorescent protein fluorescence was excited with monochromatic light (15-nm half-bandwidth) with a center wavelength at 395 nm. Confocal laser scanning microscopy was performed with a LSM 5 Pascal microscope from Zeiss (Jena, Germany).
Electron Microscopy
Pherophorin-DZ1 monomers or polymers, as well as the subtilisin-resistant fragment, were rotary shadowed with platinum and visualized by electron microscopy, as described by Mörglin et al. (1988).
Software
Corel Draw 8.0 (Ottawa, Canada) and Photoshop 5.0 (Adobe Systems, Mountain View, CA) were used to process digital images.
Accession Number
The accession number for the pherophorin-DZ1 sequence is AJ429230.
Acknowledgments
We thank Dr. R. Deutzmann and E. Hochmuth for mass spectrometry and for protein sequencing, Dr. J. Engel and T. Schulthess for electron microscopy, and J. Nink for excellent technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft (SFB 521).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.000711.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Amon, P., Haas, E., and Sumper, M. (1998). The sex-inducing pheromone and wounding trigger the same set of genes in the multicellular green alga Volvox. Plant Cell 10, 781–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar, R.S., and Gough, C.A. (1996). Circular dichroism of collagen and related polypeptides. In Circular Dichroism and the Conformational Analysis of Biomolecules, G.D. Fasman, ed (New York: Plenum Press), pp. 183–199.
- Cohen, S., and Strydom, D. (1988). Amino acid analysis utilizing phenylisothiocyanate derivatives. Anal. Biochem. 174, 1–16. [DOI] [PubMed] [Google Scholar]
- Domingo, C., Sauri, A., Mansilla, E., Conejero, V., and Vera, P. (1999). Identification of a novel peptide motif that mediates cross-linking of proteins to cell walls. Plant J. 20, 563–570. [DOI] [PubMed] [Google Scholar]
- Doolittle, R.F. (1995). The multiplicity of domains in proteins. Annu. Rev. Biochem. 64, 287–314. [DOI] [PubMed] [Google Scholar]
- Dubois, M., Gilles, K.A., Hamilton, J.K., and Rebers, P.A. (1956). Colorimetric method for determination of sugars and related substances. Anal. Chem. 28, 350–356. [Google Scholar]
- Ellman, G.L. (1959). Tissue sulfhydryl groups. Arch. Biochem. Biophys. 82, 70–77. [DOI] [PubMed] [Google Scholar]
- Ender, F., Hallmann, A., Amon, P., and Sumper, M. (1999). Response to the sexual pheromone and wounding in the green alga Volvox: Induction of an extracellular glycoprotein consisting almost exclusively of hydroxyproline. J. Biol. Chem. 274, 35023–35028. [DOI] [PubMed] [Google Scholar]
- Ertl, H., Mengele, R., Wenzl, S., Engel, J., and Sumper, M. (1989). The extracellular matrix of Volvox carteri: Molecular structure of the cellular compartment. J. Cell Biol. 109, 3493–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischauf, A.-M., Lehrach, H., Poustka, A., and Murray, N. (1983). Lambda replacement vectors carrying polylinker sequences. J. Mol. Biol. 170, 827–842. [DOI] [PubMed] [Google Scholar]
- Fuhrmann, M., Oertel, W., and Hegemann, P. (1999). A synthetic gene coding for the green fluorescent protein (GFP) is a versatile reporter in Chlamydomonas reinhardtii. Plant J. 19, 353–361. [DOI] [PubMed] [Google Scholar]
- Gilles, R., Gilles, C., and Jaenicke, L. (1983). Sexual differentiation of the green alga Volvox carteri: Involvement of extracellular phosphorylated proteins. Naturwissenschaften 70, 571–572. [Google Scholar]
- Godl, K., Hallmann, A., Rappel, A., and Sumper, M. (1995). Pherophorins: A family of extracellular matrix glycoproteins from Volvox structurally related to the sex-inducing pheromone. Planta 196, 781–787. [PubMed] [Google Scholar]
- Godl, K., Hallmann, A., Wenzl, S., and Sumper, M. (1997). Differential targeting of closely related ECM glycoproteins: The pherophorin family from Volvox. EMBO J. 16, 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough, U.W., and Heuser, J.E. (1985). The Chlamydomonas cell wall and its constituent glycoproteins analyzed by the quick-freeze deep-etch technique. J. Cell Biol. 101, 1550–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber, H., Goetinck, S.D., Kirk, D.L., and Schmitt, R. (1992). The nitrate reductase-encoding gene of Volvox carteri: Map location, sequence and induction kinetics. Gene 120, 75–83. [DOI] [PubMed] [Google Scholar]
- Hallmann, A., Amon, P., Godl, K., Heitzer, M., and Sumper, M. (2001). Transcriptional activation by the sexual pheromone and by wounding: A new gene family from Volvox encoding modular proteins with (hydroxy)proline-rich and metalloproteinase homology domains. Plant J. 26, 583–593. [DOI] [PubMed] [Google Scholar]
- Hengen, P.N. (1996). Cycle sequencing through GC-rich regions. Trends Biochem. Sci. 21, 11562–11566. [PubMed] [Google Scholar]
- Henikoff, S. (1984). Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene 28, 351–359. [DOI] [PubMed] [Google Scholar]
- Holst, O., Christoffel, V., Fründ, R., Moll, H., and Sumper, M. (1989). A phosphodiester bridge between two arabinose residues as a structural element of an extracellular glycoprotein of Volvox carteri. Eur. J. Biochem. 181, 345–350. [DOI] [PubMed] [Google Scholar]
- Kim, H.J., and Kim, B.K. (1994). Simple cycle sequencing by labeling primer using Taq DNA polymerase. Biotechniques 16, 576–578, 580. [PubMed] [Google Scholar]
- Kirk, D.L. (1998). Volvox: Molecular-genetic origins of multicellularity and cellular differentiation. In Development and Cell Biology Series, J.B. Bard, P.W. Barlow, P.B. Green, and D.L. Kirk, eds (Cambridge, UK: Cambridge University Press), pp. 165–177.
- Kirk, D.L., Birchem, R., and King, N. (1986). The extracellular matrix of Volvox: A comparative study and proposed system of nomenclature. J. Cell Sci. 80, 207–231. [DOI] [PubMed] [Google Scholar]
- Mages, H.W., Tschochner, H., and Sumper, M. (1988). The sexual inducer of Volvox carteri: Primary structure deduced from cDNA sequences. FEBS Lett. 234, 407–410. [DOI] [PubMed] [Google Scholar]
- McConlogue, L., Brown, M.A., and Innis, M.A. (1988). Structure-independent DNA amplification by PCR using 7-deaza-2′-deoxyguanosine. Nucleic Acids Res. 16, 9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mörglin, M., Paulson, M., Hardingham, T.E., Heinegard, D., and Engel, J. (1988). Cartilage proteoglycans: Assembly with hyaluronate and link protein as studied by electron microscopy. Biochem. J. 253, 175–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mort, A.J., and Lamport, D.T.A. (1977). Anhydrous hydrogenfluoride deglycosylates glycoproteins. Anal. Biochem. 82, 289–309. [DOI] [PubMed] [Google Scholar]
- Provasoli, L., and Pinter, I.J. (1959). Artificial media for freshwater algae: Problems and suggestions. In The Ecology of Algae, Special Publication No. 2, C.A. Tyron and R.T. Hartman, ed (Pittsburgh, PA: Pymatuning Laboratory of Field Biology, University of Pittsburgh), pp. 84–96.
- Rausch, H., Larsen, N., and Schmitt, R. (1989). Phylogenetic relationships of the green alga Volvox carteri deduced from small-subunit ribosomal RNA comparison. J. Mol. Evol. 29, 255–265. [DOI] [PubMed] [Google Scholar]
- Rodriguez, H., Haring, M.A., and Beck, C.F. (1999). Molecular characterization of two light-induced, gamete-specific genes from Chlamydomonas reinhardtii that encode hydroxyproline-rich proteins. Mol. Gen. Genet. 261, 267–274. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sanger, F., Nicklen, S., and Coulson, A.R. (1977). DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiedlmeier, R., Schmitt, R., Müller, W., Kirk, M.M., Gruber, H., Mages, W., and Kirk, D.L. (1994). Nuclear transformation of Volvox carteri. Proc. Natl. Acad. Sci. USA 91, 5080–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer, T., Hallmann, A., Schmidt, B., Sumper, M., and von Figura, K. (1996). The evolutionary conservation of a novel protein modification, the conservation of cysteine to serinesemialdehyde in arylsulfatase from Volvox carteri. Eur. J. Biochem. 238, 341–345. [DOI] [PubMed] [Google Scholar]
- Starr, R.C. (1969). Structure, reproduction, and differentiation in Volvox carteri f. nagariensis Iyengar, strains HK9 & 10. Arch. Protistenkd. 111, 204–222. [Google Scholar]
- Starr, R.C. (1970). Control of differentiation in Volvox. Dev. Biol. Suppl 4, 59–100. [DOI] [PubMed] [Google Scholar]
- Starr, R.C., and Jaenicke, L. (1974). Purification and characterization of the hormone initiating sexual morphogenesis in Volvox carteri f. nagariensis Iyengar. Proc. Natl. Acad. Sci. USA 71, 1050–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumper, M., and Hallmann, A. (1998). Biochemistry of the extracellular matrix of Volvox. Int. Rev. Cytol. 180, 51–85. [DOI] [PubMed] [Google Scholar]
- Sumper, M., Berg, E., Wenzl, S., and Godl, K. (1993). How a sex pheromone might act at a concentration below 10−16 M. EMBO J. 12, 831–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumper, M., Nink, J., and Wenzl, S. (2000). Self-assembly and cross-linking of Volvox extracellular matrix glycoproteins are specifically inhibited by Ellman's reagent. Eur. J. Biochem. 267, 2334–2339. [DOI] [PubMed] [Google Scholar]
- Tschochner, H., Lottspeich, F., and Sumper, M. (1987). The sexual inducer of Volvox carteri: Purification, chemical characterization and identification of its gene. EMBO J. 6, 2203–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waffenschmidt, S., Woessner, J.P., Beer, K., and Goodenough, U. (1993). Isodityrosine cross-linking mediates insolubilization of cell walls in Chlamydomonas. Plant Cell 5, 809–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waffenschmidt, S., Kusch, T., and Woessner, J.P. (1999). A transglutaminase immunologically related to tissue transglutaminase catalyzes cross-linking of cell wall proteins in Chlamydomonas reinhardtii. Plant Physiol. 121, 1003–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzl, S., and Sumper, M. (1982). The occurence of different sulphated cell surface glycoproteins correlates with defined developmental events in Volvox. FEBS Lett. 143, 311–315. [Google Scholar]
- Wenzl, S., and Sumper, M. (1986. a). A novel glycosphingolipid that may participate in embryo inversion in Volvox carteri. Cell 45, 633–639. [DOI] [PubMed] [Google Scholar]
- Wenzl, S., and Sumper, M. (1986. b). Early events of sexual induction in Volvox: Chemical modification of the extracellular matrix. Dev. Biol. 115, 119–128. [Google Scholar]
- Wenzl, S., and Sumper, M. (1987). Pheromone-inducible glycoproteins of the extracellular matrix of Volvox and their possible role in sexual induction. In Algal Development: Molecular and Cellular Aspects, W. Wiessner, D.G. Robinson, and R.C. Starr, eds (Berlin: Springer-Verlag), pp. 58–65.
- Wenzl, S., Thym, D., and Sumper, M. (1984). Development-dependent modification of the extracellular matrix by a sulphated glycoprotein in Volvox carteri. EMBO J. 3, 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woessner, J.P., and Goodenough, U.W. (1994). Volvocine cell walls and their constituent glycoproteins: An evolutionary perspective. Protoplasma 181, 245–258. [Google Scholar]