Abstract
Although a large number of embryo mutants have been studied, mostly at the morphological level, the critical molecular and cellular events responsible for embryogenesis are unknown. Here, we report that using an enhancer-trap embryo mutant of Arabidopsis, we identified a gene, ROOT-SHOOT-HYPOCOTYL–DEFECTIVE (RSH), that is essential for the correct positioning of the cell plate during cytokinesis in cells of the developing embryo. We traced the earliest point of influence of RSH to the first asymmetrical division of the zygote. Homozygous rsh embryos were defective morphologically, had irregular cell shape and size, and germinated to form agravitropic-defective seedlings incapable of further development. The RSH gene encodes a Hyp-rich glycoprotein–type cell wall protein. RSH localized to the cell wall throughout the embryo and to a few well-defined postembryonic sites. Although several lines of evidence from previous work suggest that the cell wall is involved in development, the protein(s) involved remained elusive.
INTRODUCTION
Embryogenesis in flowering plants extends from the zygote to the desiccated seed. In Arabidopsis, the early part of embryogenesis is a period of rapid and precise symmetrical and asymmetrical cell divisions (Mansfield and Briarty, 1991). At the same time, cell differentiation occurs and polarity is being established, resulting in the development of a body plan with bilateral symmetry and apical-basal polarity. It is accepted generally that embryogenesis is an extremely complex process dependent on the coordination of numerous specific genetic programs as well as proper communication between cells. Many of the genes involved have essential “housekeeping” functions, whereas many more undoubtedly play critical roles in the developmental events involved in pattern formation and cell differentiation (Mayer et al., 1991). Screening of mutant banks for lethal and defective embryos has yielded large numbers of mutants (Meinke, 1994; Torres Ruiz, 1998). However, the genes identified in such screens have provided useful information relevant to plant development in general rather than to embryogenesis specifically. This is to be expected, considering that most events that occur in the embryo also occur postembryonically as the plant continues to grow and develop new organs.
Several lines of evidence suggest that the cell wall is involved in development (Berger et al., 1994; Dupree, 1996; McCabe et al., 1997; Pennell, 1998; Reinhardt et al., 1998; Braam, 1999; Belanger and Quatrano, 2000; Smith, 2001). A link between cell shape and the division plane is well established (Smith, 2001). Cell wall proteins, particularly Hyp-rich glycoprotein (HRGP) (Kieliszewski and Lamport, 1994; Cassab, 1998), are assumed to play a role in cell shape, although this has not been proven for any particular protein because of the lack of mutants. The extensins were the first HRGPs identified and are the best studied. In dicots, extensins are recognized by the repeating pentapeptide Ser-(Hyp)4 and an abundance of Tyr, Lys, His, and Val residues arranged in repetitive motifs (Kieliszewski and Lamport, 1994). Most of the Hyp and some of the Ser residues are glycosylated (Shpak et al., 1999, 2001).
Hydroxylation of the Pro residues and glycosylation occur post-translationally in the Golgi. The amino acid motifs are presumed to be important for secondary and tertiary structure, to which Ser-(Hyp)4 contributes molecular rigidity and kinks (Ferris et al., 2001), and the multiple Tyr residues are thought to allow both intramolecular and intermolecular isodityrosine cross-linking (Kieliszewski and Lamport, 1994). Four major classes of structural cell wall proteins have been described: HRGPs, Pro-rich proteins, Gly-rich proteins, and arabinogalactans (AGPs) (Showalter, 1993; Cassab, 1998). As more wall protein sequences have been identified, it has become clear that there is a continuum of common sequence domains from HRGPs through Pro-rich proteins and AGPs, with some having characteristics of more than one group (Carpita et al., 1996). Currently, HRGPs are a loosely defined superfamily of differentially expressed cell wall proteins. As the name implies, it includes glycosylated proteins rich in Hyp residues.
AGPs, a family of highly glycosylated HRGPs, are involved in determining cell fate during embryogenesis in carrot cell cultures. The AGP epitope recognized by JIM8 was shown to be distributed asymmetrically in cells about to divide and to be localized exclusively to the part of the cell that would become the basal cell (Pennell et al., 1992; Kreuger and van Holst, 1993; Egertsdotter and von Arnoldz, 1995; Marcel et al., 1997). On separation, the two cells have different fates. However, the role of the AGP in determining cell fate is unknown (McCabe et al., 1997). In the brown algae Fucus, the first asymmetrical cell division, which is critical to the establishment of the rhizoid and thallus cell fates, requires cellular polarity associated with localized secretion of cell wall components. Actin microfilaments and intact cell wall both are required for targeted vesicle secretion and to stabilize the polar axis (Shaw and Quatrano, 1996; Brownlee and Bouget, 1998; Belanger and Quatrano, 2000).
Proteins involved in secretion and cell wall synthesis have been shown to be required for normal embryo development in Arabidopsis (Scheres and Benfey, 1999; Vroemen et al., 1999). The two best-studied genes required for embryo development in Arabidopsis, GNOM (EMB30) and KNOLLE, are associated with secretion. GNOM encodes a membrane-associated GUANINE NUCLEOTIDE EXCHANGE FACTOR on ADP-RIBOSYLATION FACTOR G PROTEIN. This G protein is involved in vesicular trafficking to the cell surface (Mayer et al., 1993; Shevell et al., 1994). Data suggest that GNOM is involved in the polar localization of auxin efflux carrier proteins (Steinmann et al., 1999) and the deposition of cell wall materials (Shevell et al., 2000). Polarization of the Arabidopsis zygote requires GNOM (Vroemen et al., 1996). KNOLLE is a putative t-SNARE shown to be cell plate specific (Lukowitz et al., 1996; Lauber et al., 1997). In other systems, t-SNARES on membranes have been shown to bind to v-SNARES on arriving vesicles to facilitate vesicle fusion during cytokinesis (Jantsch-Plunger and Glotzer, 1999).
Cytokinesis is the part of cell division whereby the cytoplasm of one cell is cleaved into two (for recent reviews, see Staehelin and Hepler, 1996; Sylvester, 2000; Smith, 2001). Unlike animal cells, dividing plant cells initiate a new cell wall from within the cell, along the short axis, that grows toward the mother cell wall. The factors that determine the location and orientation of this dividing line and its site of fusion to the mother membrane and wall are largely unknown. However, it is known that before the onset of mitosis, a preprophase band of microtubules is formed when the cortically arranged interphase microtubules assemble into a band around the cell that marks the future site of cell division (Gunning and Wick, 1985; Mineyuki and Gunning, 1990). Microtubule-associated vesicles and cell wall thickening beneath the preprophase band have been observed in a variety of cell types (Packard and Stack, 1976; Galatis and Mitrakos, 1979; Galatis et al., 1982).
In the embryo, as in other plant parts, the positioning of the cell plate results in either symmetrical or asymmetrical cell divisions, with consequences for differentiation and development. Although a large number of embryo mutants have been studied at the morphological level, and some at the molecular level, a unifying framework of molecular and cellular events responsible for the critical steps in embryogenesis has not been established (Torres Ruiz, 1998). Therefore, we constructed an enhancer-trap library of Arabidopsis with the aim of identifying key embryo-expressed genes and determining their functions. Here, we report the identification of a gene, ROOT-SHOOT-HYPOCOTYL–DEFECTIVE (RSH), that encodes a HRGP, that is localized to the cell wall, and that is essential for normal embryo development. Using a knockout mutant, we show that RSH is required for normal cell shape and expansion and for correct positioning of the cell plate during cytokinesis. We trace the earliest point of influence of RSH to the first division of the zygote.
RESULTS
Embryo Mutant Identification
To discover genes with important functions in embryo development, we constructed an enhancer-trap library of Arabidopsis using an engineered transposable element (3 kb) based on the Ac/Ds system (Sundaresan et al., 1995). The element carries the neomycin phosphotransferase II (NPT) gene on a constitutive promoter for plant selection and the β-glucuronidase (GUS) reporter gene on a minimal promoter (40 bp) bordering one end of the transposon. GUS is expressed only when it is inserted in an appropriate orientation relative to plant regulatory sequences. Such a library is ideal for identifying lines with recessive mutations, in which the homozygous progeny display the mutant phenotype and the heterozygous progeny can be used to profile the expression of the gene in question. Embryo-defective mutants were identified by a two-step process. First, germination mutants were identified by a visual screen of transposant lines on agar plates on the understanding that a proportion would be caused by embryo defects. Second, the heterozygous progeny with germination mutations were screened after self-fertilization by microscopically visualizing embryos in seed of immature siliques (Figure 1G).
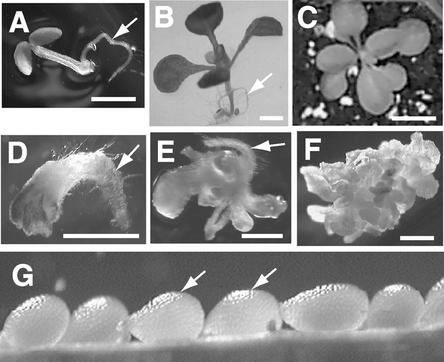
Figure 1.
Phenotypes of rsh/rsh Mutant Seedlings and Embryos Compared with the Wild Type.
(A) to (C) Phenotypes of wild-type seedlings.
(D) to (F) Phenotypes of rsh/rsh mutant seedlings.
(A) and (D) show 5-day-old seedlings; (B) and (E) show 13-day-old seedlings; and (C) and (F) show 21-day-old seedlings. All of the seedlings except the one shown in (C) were grown on sterile media; the seedling shown in (C) was transferred to soil. Arrows indicate root tissue. Bars in (A), (B), and (D) to (F) = 1 mm; bar in (C) = 1 cm.
(G) Immature silique of a self-fertilized RSH/rsh plant. Arrows indicate seed with rsh/rsh embryos.
The rsh mutant was identified as a germination-defective agravitropic mutant with severely defective root, shoot, and hypocotyl and a vitreous appearance throughout. The defective seedlings grew to a maximum length of 1 to 3 mm and were incapable of surviving for more than ∼3 weeks, even when transferred to fresh media (Figures 1D to 1F). A morphological examination of 3027 seed from 100 siliques of 20 self-pollinated heterozygous rsh plants at the bent-cotyledon stage of development showed that 753 seed (25%) had abnormally small embryos with no obvious organ differentiation. This mendelian inheritance pattern is consistent with a single recessive embryo mutation.
The rsh Mutant Is Cytokinesis Defective
A detailed examination of all stages of development of these embryos was simplified by the fact that homozygotes (rsh/rsh), heterozygotes (rsh/RSH), and wild-type (RSH/RSH) seed were present in the same silique. The wild-type zygote divides to produce a small apical cell and a larger basal cell. The small apical cell gives rise to the shoot meristem, cotyledons, and the uppermost part of the root meristem. The large basal cell gives rise to the suspensor and the hypophysis, which at the globular stage of embryogenesis produces an upper lens-shaped cell that develops into the root meristem and a lower cell that develops into the root cap.
Wild-type embryos have well-defined cell shapes in relation to their positions in the embryo and have a very precise set of cell divisions leading to cell differentiation and organ formation. In rsh mutants, a zygote with a mutant phenotype was not identified; however, the position of the cell plate at the first division of the zygote differed from that in the wild type in 25% of those observed. The plane of division was in the same orientation as that of the wild type, but its position resulted in a larger apical cell relative to the basal cell compared with the wild type. In some cases, the apical cell was equal to or larger than the basal cell. The rsh mutation also affected subsequent cell divisions in the embryo, with the plane of division occurring in apparently random directions. The protoderm layer was disorganized, the hypophysis and O-ring did not develop, and abnormally shaped cells of mutant globular-stage embryos continued to divide, but bilateral symmetry was not established (Figure 2).
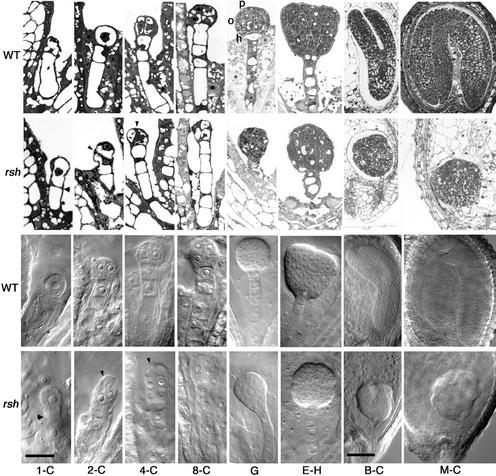
Figure 2.
Morphology of rsh/rsh Mutant Embryos Compared with Wild-Type Embryos during Development.
Stained sections (rows 1 and 2) and Nomarski images (rows 3 and 4) of wild-type (rows 1 and 3) and rsh mutant (rows 2 and 4) embryos at comparable stages of development (columns). Column 1-C, one-cell stage; column 2-C, two-cell stage; column 4-C, four-cell stage; column 8-C, eight-cell stage; column G, globule stage; column E-H, early-heart stage; column B-C, bent-cotyledon stage; column M-C, mature-cotyledon stage. Note the large apical cell in 1-C rsh, the abnormal cell shapes in 2-C and 4-C rsh (arrowheads show the dividing line in rsh), the absence of normal protoderm (p), O-ring (o), and hypophysis (h) in G rsh, and the small abnormal embryo within the normal seed coat in M-C rsh. Bar in 1-C = 20 μm for 1-C to E-H; bar in B-C = 80 μm for B-C and M-C. WT, wild type.
The rsh Mutant Has Abnormal Cell Shapes
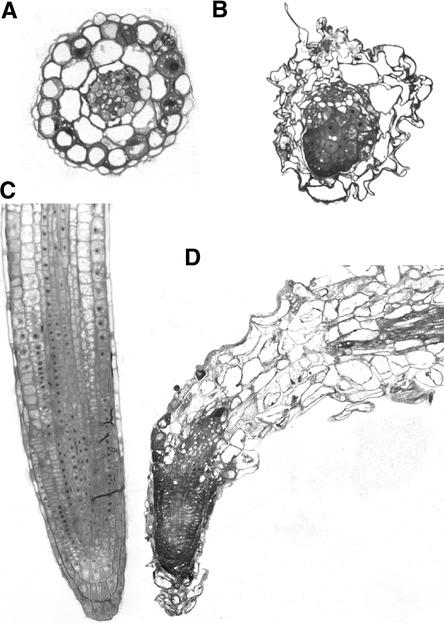
Compared with the wild type, thin sections of rsh homozygous seedling roots showed disorganized cells with severely abnormal shapes in the epidermis, ground tissue, vascular tissue, and root cap (Figure 3). The individual cell types differentiated to some degree, and polarity appeared to be established. Normal cell expansion did not occur, and cell growth appeared to be in random directions. The abnormal cell shapes made it impossible to determine the identity of the cell types. This is consistent with the abnormal cell shapes found in rsh homozygous embryos (Figure 2) and supports the claim that the RSH gene is essential for normal cell shape.
Figure 3.
Comparison of Cell Shapes in Root Sections of Young Seedlings.
(A) and (C) Wild type transverse (A) and longitudinal (C) sections.
(B) and (D) rsh homozygous mutant transverse (B) and longitudinal (C) sections.
The RSH Gene Encodes a Putative HRGP
The question of which gene (or genes) is responsible for the rsh mutant phenotype was addressed by cloning the tagged DNA contiguous to the transposon insert and using these sequences to map the insert and identify bacterial artificial chromosome clones carrying the region. A 6.5-kb EcoRI fragment of DNA was cloned from a bacterial artificial chromosome clone. Sequencing and analysis of this fragment revealed that the transposon element had inserted in chromosome I at ∼28 centimorgan and 228 bp upstream from the amino acid coding sequence of a putative 46.5-kD HRGP, which was on a 1.5-kb transcript with no detectable splice sites. A comparison of the plant DNA sequences at both ends of the transposon insert with that of the wild type showed that 4 bp was missing from the rsh sequence, eliminating the possibility that a large deletion at the insertion site could be responsible for the rsh phenotype (Figure 4).
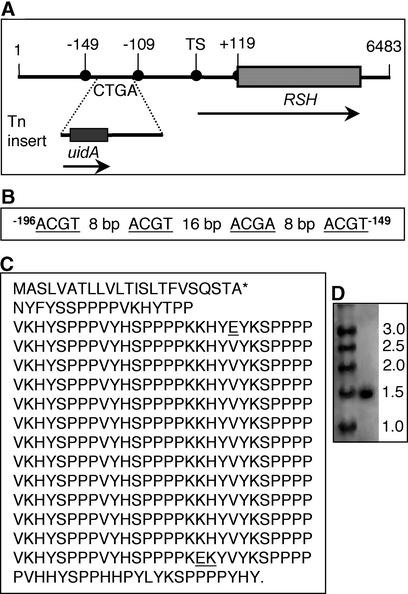
Figure 4.
Genetic Map of the Mutated Region in the rsh Mutant.
(A) Cloned 6483-bp EcoRI DNA fragment. The predicted amino acid coding sequence of RSH is from 2940 to 4220 bp. The transcription start nucleotide is marked TS (+1). The translation start nucleotide (A of ATG) is at +119. The transposon insert (Tn) mapped at −109 bp from the transcription start nucleotide. Arrows represent transcripts of RSH (1.5 kb) and uidA, which encodes the GUS reporter. CTGA are the deleted nucleotides in the rsh mutant. The scheme is not drawn to scale.
(B) Predicted bZIP response element at −149 to −196 nucleotides from the transcription start nucleotide.
(C) Amino acid sequence of RSH. The asterisk indicates the most likely cleavage site of the signal peptide (Nielsen et al., 1997). Note the 13 nearly identical repeats of 28 amino acids (nonidentical amino acids are underlined).
(D) RNA gel blot of RSH mRNA in total RNA from wild-type roots (right lane) and RNA molecular mass markers (kb) (left lane).
RSH has 13 nearly identical repeats of 28 amino acids each and an N-terminal transit peptide typical of cell wall–targeted proteins (von Heijne, 1985). The amino acid sequence of RSH has 28 Ser-(Pro)4 motifs and an abundance of Tyr, Lys, His, and Val residues, arranged mostly in repetitive motifs of Val-Lys-His-Tyr and Lys-Lys-His-Tyr-Val-Tyr-Lys, indicating that it is an extensin-type HRGP. The RSH transit peptide is identical to that of EXT3, which has 325 amino acid residues, and EXT5, which has 203 amino acid residues, both from Arabidopsis. The total amino acid sequence of RSH is related most closely to that of EXT1 (Figure 5). Experimentally, EXT1 was shown to be regulated developmentally. It is expressed normally in roots but not in leaves. Wounding reverses this expression pattern, causing expression in the leaves but not in roots. Salicylic acid, abscisic acid, and methyl jasmonate also elicit leaf expression of EXT1 (Merkouropoulos et al., 1999).
Figure 5.
Alignment of RSH and EXT1.
Pairwise alignment of the deduced amino acid sequence of EXT1 with that of RSH using the ClustalV (Higgins and Sharp, 1989) method in Megalign (DNAStar, Madison, WI) with PAM250 residue weight tables. Identical amino acids are shaded black.
The rsh Mutant Phenotype Is a Consequence of a Mutation in the RSH Gene
The rsh segregation and sequence data described above suggest that a mutation in the RSH gene is responsible for the mutant phenotype, and this was confirmed by two experiments. A genetic analysis of 200 seed from each of 29 heterozygous F6 generation rsh plants showed that the segregation ratio of kanamycin-resistant (selectable marker in the transposon insert) to kanamycin-sensitive plants was 2:1. None of the kanamycin-sensitive segregants to the F10 generation showed the rsh mutant phenotype. The 2:1 segregation ratio is indicative of a single embryo mutation, whereas cosegregation of the rsh phenotype with kanamycin resistance showed that the rsh mutation is not at a distant site to the kanamycin marker. It was confirmed further that expression of the RSH gene is required for normal embryo development by complementation of the mutation with the RSH transgene.
Heterozygous rsh plants transformed with a 6483-bp fragment of DNA carrying the RSH gene (Figure 4) with its native regulatory sequences produced both homozygous and heterozygous rsh plants, as well as wild-type plants, carrying the T-DNA in the T1 generation. The transposants were kanamycin resistant, whereas the transformed transposants were both kanamycin and Basta resistant (see Methods). The RSH genotypes of 11 independent rsh homozygous transformants were tested by polymerase chain reaction, and the results confirmed the presence of the T-DNA in a rsh homozygous background. A wild-type phenotype with no rsh mutant segregants was observed in the T3 generation in homozygous rsh plants that were homozygous for the RSH transgene, confirming that RSH is responsible for the rsh mutant phenotype. It is worth noting that wild-type plants carrying RSH on a transgene, identified as kanamycin sensitive and Basta resistant, all had wild-type phenotypes, indicating that overproduction of RSH did not result in a mutant phenotype.
Expression Profile of the RSH Gene
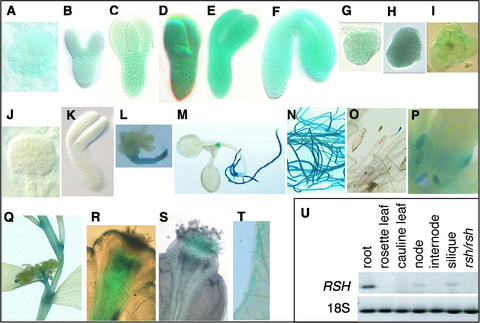
A profile of RSH expression throughout the plant was determined. In the rsh mutant, the engineered transposon had inserted so that the enhancer-trap reporter gene was in the same orientation as RSH, making it likely that the histochemical GUS assay could be used to monitor the expression of RSH. Also, a putative binding site for basic domain/Leu zipper (bZIP)–type transcription factors (Hurst, 1995) was identified 36 to 83 bp upstream from the transposon insert, suggesting that this is a trapped enhancer (Figure 4B). Reporter gene (GUS) expression was detected in cells throughout the embryo, including the suspensor, from the 32-cell stage, with expression increasing up to the torpedo stage, followed by a decrease at the bent-cotyledon stage, with little or no expression detectable in the nearly mature cotyledon stage of development. This pattern was similar for heterozygous and homozygous rsh embryos at equivalent developmental stages (Figures 6A to 6K).
Figure 6.
GUS Reporter Gene Expression Patterns during Development Viewed by Light Microscopy, and Tissue Distribution of RSH mRNA.
(A) to (K) Embryos dissected from seed coats at different stages of development.
(A) to (F) Heterozygous rsh at progressive stages of development: globular (A); heart (B); early torpedo (C); late torpedo (D); early bent cotyledon (E); and late bent cotyledon (F).
(G) to (I) Homozygous rsh at progressive stages of development: heart (G); torpedo (H); and nearly mature cotyledon (I). Compare with rsh heterozygous equivalents (B), (D), and (F), respectively.
(J) and (K) Wild-type globular and bent-cotyledons stages, respectively (negative controls). Note that blue color did not develop after the GUS assay.
(L) Homozygous rsh seedling 2 weeks after germination. Note that the root of the mutant is GUS positive.
(M) to (T) Postembryonic heterozygous rsh: seedling 2 weeks after germination (M); roots at bolting (N); roots at harvest (O); stipules (P); nodes (Q); prepollination stylar transmitting tissue (R); postpollination stigma (S); and wounded leaf edge (T). Blue color indicates GUS positive. (U) Tissue distribution of RSH mRNA and 18S rRNA in wild-type plant tissue as indicated and in 12-day-old rsh homozygous mutant seedlings (total tissue). Note that the detected RSH mRNA coincided with GUS reporter gene expression and that the rsh homozygote had no detectable RSH mRNA.
The images shown in (A) to (T) are not to scale; sizes can be estimated from Figures 1 and 2.
Although expression was not detected before the 32-cell stage, it is likely that subdetectable levels of expression occurred, because the mutant phenotype was evident at the first division of the zygote (see below). Postembryonically, RSH was expressed at the following sites: young roots, stipules, nodes, prepollination stylar transmitting tissue, and postpollination stigma, and in wound response (Figures 6L to 6T). GUS assays of all tissue from wild-type plants were negative. Reverse transcriptase–mediated polymerase chain reaction confirmed that the GUS expression profile coincided with the tissue distribution of RSH mRNA (Figure 6U). RSH mRNA was undetected in the rsh/rsh mutant, even on extended overexposure of autoradiograms, demonstrating that the transposon insert prevented expression of the RSH gene and confirming that expression of RSH is critical for normal embryo development.
The RSH Gene Product Localizes to the Cell Wall
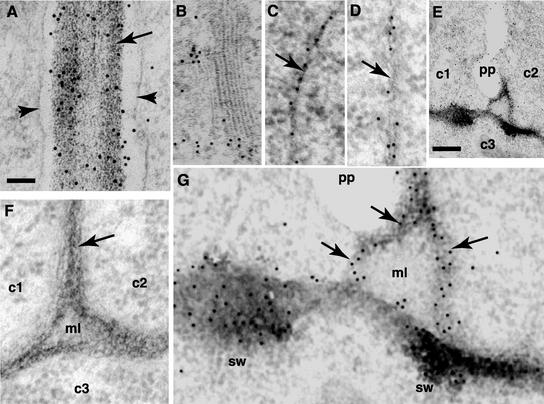
Localization of RSH to the cell wall was shown in transformed wild-type plants designed to express RSH fused to green fluorescent protein (RSH-GFP) using native RSH regulatory sequences. Because RSH is a member of a large family of glycosylated proteins, it is unlikely that antibodies specific to RSH could be obtained. The GFP tag on RSH was used to confirm the expression pattern of RSH using fluorescence microscopy and to localize RSH using immunoelectron microscopy. All 200 independent transformants examined by fluorescence microscopy in the T2 generation showed expression at varying levels, but they had the same embryonic and postembryonic expression profiles as that of the GUS reporter shown in Figure 6. To demonstrate localization at the subcellular level, two of these transformant lines were examined in the T3 generation by immunoelectron microscopy using antibodies to GFP (Figure 7). The results showed that RSH-GFP localized to the cell walls throughout the embryo and to the walls of root cells. It was concentrated at three-cell junctions in the embryo but not in the root. RSH-GFP also localized to the edges of the cell plate in embryo cells and to the Golgi and trans-Golgi network. The concentration of RSH-GFP was particularly notable in the mother cell wall at the point of contact to the connecting cell plate (Figure 7G).
Figure 7.
Electron Micrographs of RSH-GFP Localization.
(A) Localization of RSH-GFP to the cell wall of a 3-day-old root section. Note the cell wall (arrow) between two cells with a concentration of immunogold particles (sharp black spots). Arrowheads denote the plasma membrane.
(B) Golgi and trans-Golgi network in a 3-day-old root section. Note the concentration of immunogold particles.
(C) and (D) Localization of RSH-GFP to the cell walls of heart-stage embryo cells. Note the immunogold particles in the thin walls (arrows) of young, rapidly dividing cells.
(E) Fusion of the cell plate to the cell wall in a dividing heart-stage embryo cell. Two new cells (c1 and c2) with a cell plate (pp) between them. c3 indicates an adjacent cell. Note the cell wall connections between the edge of the cell plate and the swollen regions of the mother cell wall.
(F) Localization control. This embryo section was prepared like the other sections except that preimmune serum was substituted for the first antibody (anti–RSH-GFP antibody). Note the absence of immunogold particles in the three cells (c1, c2, and c3), the cell wall (arrow), and the middle lamella (ml).
(G) Enlargement of (E). Localization of RSH-GFP as seen by the concentration of immunogold particles in swollen regions (sw) of the mother wall and new connecting walls (arrows).
Bar in (A) = 100 μm for (A) to (D), (F), and (G). Bar in (E) = 25 μm.
DISCUSSION
GUS analysis of the rsh mutant, RNA gel blot analysis, and fluorescence microscopy of wild-type plants carrying RSH::GFP all showed similar patterns of expression, supporting the claim that the GUS expression profile is an accurate reflection of RSH expression. GUS was expressed in the heterozygous and homozygous mutant embryos at equivalent stages of development, confirming that the expression of RSH in the embryo is dependent on growth stage. RSH mRNA was undetectable in the homozygous rsh mutant seedling, indicating that the enhancer trap used to generate the mutant did in fact trap the regulatory element(s) responsible for the expression of RSH and caused the rsh mutant phenotype.
The RSH gene encodes a HRGP-type cell wall protein. It is accepted generally that the HRGP family of glycosylated proteins plays a role in strengthening the cell wall; hence, these proteins most likely have functional significance in determining cell shape and plant morphology (Kieliszewski and Lamport, 1994). HRGPs are known to be expressed differentially in different tissues (Keller and Lamb, 1989; Ye and Varner, 1991) and in response to wounding, pathogen attack, and elicitors (Corbin et al., 1987; Memelink et al., 1993; Parmentier et al., 1995; Wycoff et al., 1995). Our data demonstrate that a HRGP (RSH) influenced cell shape and link this shape to plant growth and development.
In carrot, the HRGP extensin-1 localizes to the cellulose layer of the cell wall (Stafstrom and Staehelin, 1988), whereas HRGP extensin-2 is limited to the expanded middle lamella at three-cell junctions (Swords and Staehelin, 1993). These authors proposed that although these two HRGPs share common carbohydrate moieties, they could have different functions, with the former being involved in cell structure and the latter playing a role in plant defense. In contrast, our data show that in Arabidopsis roots, RSH localized to the cell walls, whereas in the developing embryo, it localized to the wall and was concentrated at three-cell junctions. By analogy with the extensins in carrot, the localization patterns of RSH are consistent with a dual function, that of strengthening mature cell walls and playing a role in positioning the dividing line at cytokinesis, which would influence development.
A comparison of rsh with known embryo mutants shows that the well-studied GNOM (EMB30) mutant has the closest phenotype, in that it also has defective positioning of the cell plate at the first division of the zygote. Because GNOM encodes a protein involved in vesicular trafficking to the cell surface, it is interesting to speculate that RSH is a cargo in GNOM-associated vesicles. Subsequent to the first division of the rsh zygote, all other cell divisions produced abnormally shaped cells. This may be the consequence of the first abnormal division of the zygote or it may be attributable to the absence of RSH from cells of the developing embryo. The shape of rsh/rsh mutant cells was severely abnormal, consistent with the claim that cell shape plays a critical role in cell division (Lintilhac and Vesecky, 1984; Lynch and Lintilhac, 1997). However, the rsh homozygous zygote appeared normal in shape yet the cell plate was misplaced, indicating that shape is not the only contribution of RSH to the correct positioning of the cell plate. In the rsh/rsh zygote, an alternative, possibly a predivision cell wall protein, may be sufficient to provide shape but insufficient to position the cell plate correctly at cytokinesis.
Should RSH have a more direct function in positioning the cell plate, the following are interesting points to consider. The selection and establishment of a cortical site for cell division, and recognition of that site, are long-standing questions (Mineyuki and Gunning, 1990; Wick, 1991; Smith, 2001). We have shown that the rsh mutant either lacks, or the growing cell plate fails to recognize or bind, this cortical site. The hypothesis that RSH plays a role either directly, by being the molecule that marks the cortical site, or indirectly, by being a determining factor in marking the cortical site, for cell division now can be tested. Furthermore, the data do not preclude the possibility that RSH, within the growing cell plate, plays a role in recognizing or binding to the cortical site of cell division. The higher concentration of RSH at cell wall to plate junctions in rapidly dividing embryo cells supports the hypothesis that RSH plays a role in connecting the plate to a target site in the cell wall. Experiments now can be designed to test these hypotheses.
In conclusion, we have profiled the expression of a gene (RSH) that encodes a HRGP-type protein, shown that its product is localized to the cell wall, and shown that its expression is critical for (1) cell shape, (2) the correct positioning of the cell plate during cytokinesis, and (3) normal embryo development. The genome sequence of Arabidopsis contains a superfamily of loosely defined putative HRGPs (Kieliszewski and Lamport, 1994), some with high sequence homology with RSH. The identification and functional analysis of RSH and the availability of the rsh mutant will enable progress in defining HRGP family members and in understanding their functions.
METHODS
Mutant Library Construction
Arabidopsis thaliana Landsberg erecta starter lines CS8047 and CS8045, carrying the engineered transposons DsE and Ac, respectively, were crossed and propagated by self-pollination to generate an F3 generation enhancer-trap library of separate F1 families, as described previously (Sundaresan et al., 1995).
Microscopy
For light microscopy, seedlings and siliques were fixed (4% paraformaldehyde, 1% glutaraldehyde, and 50 mM Pipes buffer, pH 7) and embedded (LR White; London Resin Co. Ltd., London, UK), and 1-μm sections were prepared and stained with toluidine blue. For direct interference contrast microscopy, siliques were opened longitudinally, fixed at room temperature for 15 to 60 min in 50% ethanol, 5% glacial acetic acid, and 4% formaldehyde, and cleared overnight in modified Hoyer's solution (gum arabic:chloral hydrate:glycerin:water, 8:100:5:30). Samples were mounted with clearing solution under sealed cover slips and viewed using an E600 microscope (Nikon, Tokyo, Japan) with light and Nomarski optics, and images were captured with a SPOT 2 digital camera (Diagnostic Instruments, Sterling Heights, MI).
For fluorescence microscopy, fresh embryos removed from their seed coats at all stages of development, and fresh samples of other plant tissues, were mounted in PBS, pH 7.6. Green fluorescence was visualized using a Labophot-2 microscope (Nikon) with a fluorescence attachment and a filter set with excitation at 425 ± 60 nm and emission at 505 ± 40 nm (Chroma Technology, Brattleboro, VT). For subcellular localization of RSH, immunogold conjugated to secondary antibody and bound to anti–green fluorescent protein (GFP) antibody was viewed with a JOEL 100S transmission electron microscope at 80 kV acceleration.
Heart-stage embryos and 3-day-old seedling roots of wild-type Arabidopsis transformed with pQH27 (see below) expressing RSH::GFP were examined. Seed and small pieces of root were fixed (2% glutaraldehyde, 4% paraformaldehyde, 15% picric acid, and 0.1 M phosphate buffer, pH 7.2) for 2.5 hr, washed sequentially in 0.1 M phosphate buffer, pH 7.2, three times and in water two times, each for 15 min, followed by dehydration, all at 4°C. Samples were infiltrated with resin (LR White, Medium Grade; London Resin Co. Ltd.) at 4°C for 12 to 16 hr and embedded under long-wave UV light and a stream of nitrogen gas for 12 to 16 hr at 4°C. Subsequent steps were performed at room temperature.
Sections (80 nm) were collected on formvar-coated nickel grids. Samples were blocked for 30 min with 1% BSA in PBS, pH 7.2 (PBS-1%BSA), treated with a 1:50 dilution (in PBS-0.1%BSA) of primary antibody (rabbit anti-GFP antibody; Clontech, Palo Alto, CA) for 3 hr, and washed twice, each for 2 min, in PBS containing 0.1% BSA and 0.1% Tween 20 followed by two 2-min washes in PBS-0.1%BSA. Samples were treated with a 1:20 dilution (in PBS-0.1%BSA) of secondary, goat anti-rabbit, antibodies conjugated to 10-nm gold particles (Sigma) for 1 hr in darkness and washed as described above (after exposure to primary antibodies). Samples were washed twice in water, each for 2 min, stained with 2% uranyl acetate for 10 min, washed once in water, stained with 0.5% lead citrate (Fahmy, 1967) for 7 min, washed once in water, and dried. For controls, samples of root and embryo were subjected to the same treatment except that preimmune serum was substituted for the primary antibody.
Mapping and Cloning
The rsh mutation was mapped by sequencing amplified plant DNA contiguous with the DsE insert. Heterozygous rsh DNA, cut with BstYI, was amplified at the β-glucuronidase (GUS)–distal end of DsE by inverse polymerase chain reaction (PCR) using nested primers, set 1 (No. 104, 5′-GTTCGAATTCGATCGGGATAAAAC-3′; No. 105, 5′-GGTAGTCGACGAAAACGGAACGGAAAC-3′) and set 2 (No. 106; 5′-AAATCAGATCTACGATAACGGTCGG-3′; No. 107, 5′-AAACGGTACCCGGAAACGGAAACGG-3′) (Sundaresan et al., 1995). Identified sequences were mapped to bacterial artificial chromosomes (BACs) on TAMU DNA arrays (ABRC, Columbus, OH). For ease of cloning, BAC T2B3 was used to clone and sequence a 6.5-kb EcoRI fragment carrying the region of interest. Subsequently, the sequence of BAC F16F4 carrying this region was deposited in GenBank as part of the genome sequencing project. Plant DNA sequences at the GUS-proximal end of DsE were identified by sequencing a 1.3-kb fragment generated by amplifying heterologous rsh DNA with primers to the GUS gene (No. 114, 5′-CCAACGCTGATCAATTCCAC-3′) and to an upstream site in the 6.5-kb fragment (No. 123, 5′-CGATGAACATGTTACTTAATTGG-3′).
GUS Localization
Heterozygous rsh, homozygous rsh, and wild-type plants (controls) were tested. Siliques, opened longitudinally, and other plant tissues were incubated in the dark in prechilled GUS enzymatic reagent mix (Jefferson et al., 1987) on ice for 1 hr followed by 37°C overnight before clearing with several changes of 70% ethanol at 37°C. For sectioning, material was embedded in LR White (London Resin Co. Ltd.) before cutting into 4-μm sections.
RNA Analyses
Total RNA purification and RNA gel blot analysis were performed using standard protocols (Ausubel et al., 1995). In RNA gel blot analysis, the RSH message was identified using a complementary 5′ end 32P-labeled probe (No. 129, 5′-GCGGTTGATTGAGATACAAAGGTG-3′). RNA was quantified by UV light spectroscopy and confirmed by RNA gel electrophoresis. Reverse transcriptase–mediated (RT)-PCR was performed with the Access RT-PCR system (Promega) using 1 μg of total RNA purified from tissues of wild-type plants and homozygous rsh mutant seedlings, with primers for RSH (No. 129 [see above] and No. 132, 5′-GTAAGTTACCATTTTACAACGAGTGTG-5′) to generate a 185-bp product and for 18S rRNA (No. 130, 5′-AGGAATTGACGGAAGG-GCAC-3′; No. 131, 5′-GGACATCTAAGGGCATCACA-3′) to generate a 315-bp product. The 18S product is an RT-PCR qualitative control.
Equal quantities of RT-PCR products were run on 2% agarose gels and stained with ethidium bromide. The 5′ end of RSH mRNA was estimated by running a 5′ 32P end–labeled primer extension product on a sequencing gel alongside a sequence of the same fragment. The primer extension product was obtained using the primer extension system from Promega with total RNA from wild-type plants and a primer (No. 141, 5′-CAGTTTCACTATTTTCACACTCG-3′) complementary to RSH mRNA. The estimated transcription start of RSH was confirmed by the 185-bp RT-PCR fragment obtained (see above) using primers No. 129 and No. 132, because the latter is complementary to the estimated transcription start.
Transgenic Plants
For rsh rescue experiments, the wild-type genomic 6.5-kb EcoRI fragment carrying the RSH gene (Figure 4) was cloned into the EcoRI site of SLJ75515, a binary-type vector for plant transformation carrying the Basta resistance gene for plant selection (Jones et al., 1992). This construct, pQH17, was transformed into heterozygous rsh plants via Agrobacterium tumefaciens strain EHA105 (Hood et al., 1993) using the in planta procedure (Chang et al., 1994). For RSH localization experiments, the C terminus of RSH was fused to the N terminus of GFP (EGFP; Clontech). A plasmid carrying the RSH gene with its regulatory sequences on a 6.3-kb DNA fragment was used to clone a 0.75-kb DNA fragment encoding GFP.
The GFP fragment was inserted into the EcoRV site just downstream from the stop codon of RSH. The stop codon was eliminated subsequently by in vitro mutagenesis (QuikChange; Stratagene), leaving a RSH-GFP fusion with a 12–amino acid linker region (YISSLSIPSTSTVATM, where Y is the last amino acid of RSH and replaces the stop site, and M is the first amino acid of GFP). This 7.1-kb fragment was subcloned into the EcoRI-XhoI sites of SLJ6991, a binary-type vector carrying the kanamycin resistance marker for plant selection (Jones et al., 1992). This construct, pQH27, was transformed into Arabidopsis as described above.
Accession Numbers
The accession numbers for the sequences mentioned in this article are BAB20084 (EXT3), BAB20086 (EXT5), AC036104 (BAC F16F4), B96798 (EXT1), and B86346 (RSH).
Acknowledgments
We thank Frank Cannon (University of Massachusetts, Amherst) for useful discussions. We thank Lucy Yin and Dale Callaham at the Central Microscopy Facility of the University of Massachusetts for excellent technical assistance, and we acknowledge the National Science Foundation (Award NSF BBS 8714235) support of the facility. We also thank Jonathan Jones and colleagues at the Sainsbury Laboratory/Gatsby Charitable Foundation (Norwich, UK) for providing plasmids, and we acknowledge the ABRC at Ohio State University for providing Arabidopsis lines, BAC library arrays, and clones.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010477.
References
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1995). Current Protocols in Molecular Biology. (New York: John Wiley & Sons).
- Belanger, K., and Quatrano, R.S. (2000). Polarity: The role of localized secretion. Curr. Opin. Plant Biol. 3, 67–72. [DOI] [PubMed] [Google Scholar]
- Berger, F., Taylor, A., and Brownlee, C. (1994). Cell fate determination by the cell wall in early Fucus development. Science 263, 1421–1423. [DOI] [PubMed] [Google Scholar]
- Braam, J. (1999). If walls could talk. Curr. Opin. Plant Biol. 2, 521–524. [DOI] [PubMed] [Google Scholar]
- Brownlee, C., and Bouget, F. (1998). Polarity determination in Fucus: From zygote to multicellular embryo. Semin. Cell Dev. Biol. 9, 179–185. [DOI] [PubMed] [Google Scholar]
- Carpita, N.C., McCann, M., and Griffing, L.R. (1996). The plant extracellular matrix: News from the cell's frontier. Plant Cell 8, 1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassab, G.I. (1998). Plant cell wall proteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 281–309. [DOI] [PubMed] [Google Scholar]
- Chang, S.S., Park, S.K., Kim, B.C., Kang, B.J., Kim, D.U., and Nam, H.G. (1994). Stable genetic transformation of Arabidopsis thaliana by Agrobacterium inoculation in planta. Plant J. 5, 551–558. [Google Scholar]
- Corbin, D.R., Sauer, N., and Lamb, C.J. (1987). Differential regulation of a hydroxyproline-rich glycoprotein gene family in wounded and infected plants. Mol. Cell. Biol. 7, 4337–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupree, P. (1996). Cell division forms a pattern. Curr. Biol. 6, 683–685. [DOI] [PubMed] [Google Scholar]
- Egertsdotter, U., and von Arnoldz, S. (1995). Importance of arabinogalactan proteins for the development of somatic embryos of Norway spruce (Picea abies). Physiol. Plant. 93, 334–345. [Google Scholar]
- Fahmy, A. (1967). An extemporaneous lead citrate for electron microscopy. In Proceedings of the 25th Annual EMSA Meeting. (Baton Rouge, LA: Claitor's Publishing Division), p. 148.
- Ferris, P.J., Woessner, J.P., Waffenschmidt, S., Kilz, S., Drees, J., and Goodenough, U.W. (2001). Glycosylated polyproline II rods with kinks as a structural motif in plant hydroxyproline-rich glycoproteins. Biochemistry 40, 2978–2987. [DOI] [PubMed] [Google Scholar]
- Galatis, B., and Mitrakos, K. (1979). On the differential divisions and preprophase microtubule bands involved in the development of stomata of Vigna sinensis. J. Cell Sci. 37, 11–37. [DOI] [PubMed] [Google Scholar]
- Galatis, B., Apostolakos, P., Katsaros, C., and Loukaki, H. (1982). Pre-prophase microtubule band and local wall thickening in guard mother cells of some Leguminosae. Ann. Bot. 50, 779–791. [Google Scholar]
- Gunning, B.E.S., and Wick, S.M. (1985). Preprophase bands, phragmoplasts, and spatial control of cytokinesis. J. Cell Sci. 2, S157–S179. [DOI] [PubMed] [Google Scholar]
- Higgins, D.G., and Sharp, P.M. (1989). Fast and sensitive multiple sequence alignments on a microcomputer. Cabios 5, 151–153. [DOI] [PubMed] [Google Scholar]
- Hood, E.E., Gelvin, S.B., Melchers, L.S., and Hoekema, A. (1993). New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 2, 208–218. [Google Scholar]
- Hurst, H.C. (1995). Transcription factors. 1. bZIP proteins. Protein Profile 2, 105–168. [PubMed] [Google Scholar]
- Jantsch-Plunger, V., and Glotzer, M. (1999). Depletion of syntaxins in the early Caenorhabditis elegans embryo reveals a role for membrane fusion events in cytokinesis. Curr. Biol. 9, 738–747. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G., Shlumukov, L., Carland, F., English, J., Scofield, S.R., Bishop, G.J., and Harrison, K. (1992). Effective vectors for transformation, expression of heterologous genes, and assaying transposon excision in transgenic plants. Transgenic Res. 1, 285–297. [DOI] [PubMed] [Google Scholar]
- Keller, B., and Lamb, C.J. (1989). Specific expression of a novel cell wall hydroxyproline-rich glycoprotein gene in lateral root initiation. Genes Dev. 3, 1639–1646. [DOI] [PubMed] [Google Scholar]
- Kieliszewski, M.J., and Lamport, D.T.A. (1994). Extensin: Repetitive motifs, functional sites, post-translational codes and phylogeny. Plant J. 5, 157–172. [DOI] [PubMed] [Google Scholar]
- Kreuger, M., and van Holst, G. (1993). Arabinogalactan proteins are essential in somatic embryogenesis of Daucus carota L. Planta 189, 243–248. [Google Scholar]
- Lauber, M.H., Waizenegger, I., Steinmann, T., Schwarz, H., Mayer, U., Hwang, I., Lukowitz, W., and Juergens, G. (1997). The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139, 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lintilhac, P.M., and Vesecky, T.B. (1984). Stress-induced alignment of division plane in plant tissues grown in vitro. Nature 307, 363–364. [Google Scholar]
- Lukowitz, W., Mayer, U., and Juergens, G. (1996). Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 84, 61–71. [DOI] [PubMed] [Google Scholar]
- Lynch, T.M., and Lintilhac, P.M. (1997). Mechanical signals in plant development: A new method for single cell studies. Dev. Biol. 181, 246–256. [DOI] [PubMed] [Google Scholar]
- Mansfield, S.G., and Briarty, L.G. (1991). Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Can. J. Bot. 69, 461–476. [Google Scholar]
- Marcel, A., Tooner, E., Schmidt, A., and de Vries, A. (1997). Promotive and inhibitory effects of diverse arabinogalactan proteins on Daucus carota L. somatic embryogenesis. Planta 203, 188–195. [Google Scholar]
- Mayer, U., Torres-Ruis, R.A., Berleth, T., Misera, S., and Juergens, G. (1991). Mutations affecting body organization in the Arabidopsis embryo. Nature 353, 402–407. [Google Scholar]
- Mayer, U., Buttner, G., and Juergens, G. (1993). Apical-basal pattern formation in the Arabidopsis embryo: Studies on the role of the gnom gene. Development 117, 149–162. [Google Scholar]
- McCabe, P.F., Valentine, T.A., Forsberg, S., and Pennell, R.I. (1997). Soluble signals from cells identified at the cell wall establish a developmental pathway in carrot. Plant Cell 9, 2225–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke, D.W. (1994). Seed development in Arabidopsis thaliana. In ARABIDOPSIS, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 253–295.
- Memelink, J., Swords, K.M.M., de Kam, R.J., Schilperoort, R.A., Hoge, J.H.C., and Staehelin, L.A. (1993). Structure and regulation of tobacco extensin. Plant J. 4, 1011–1022. [DOI] [PubMed] [Google Scholar]
- Merkouropoulos, G., Barnett, D.C., and Shirsat, A.H. (1999). The Arabidopsis extensin gene is developmentally regulated, is induced by wounding, methyl jasmonate, abscisic and salicylic acid, and codes for a protein with unusual motifs. Planta 208, 212–219. [DOI] [PubMed] [Google Scholar]
- Mineyuki, Y., and Gunning, B.E.S. (1990). A role for preprophase bands of microtubules in maturation of new cell walls, and a general proposal on the function of preprophase band sites in cell division in higher plants. J. Cell Sci. 97, 527–537. [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6. [DOI] [PubMed] [Google Scholar]
- Packard, M.J., and Stack, S.M. (1976). The preprophase band: Possible involvement in the formation of the cell wall. J. Cell Sci. 22, 403–411. [DOI] [PubMed] [Google Scholar]
- Parmentier, Y., Durr, A., Marbach, J., Hirsinger, C., Criqui, M.-C., Fleck, J., and Jamet, E. (1995). A novel wound-inducible extensin gene. Plant Mol. Biol. 29, 279–292. [DOI] [PubMed] [Google Scholar]
- Pennell, R. (1998). Cell walls: Structures and signals. Curr. Opin. Plant Biol. 1, 504–510. [DOI] [PubMed] [Google Scholar]
- Pennell, R., Janniche, L., Scofield, G.M., Booij, H., De Vries, S.C., and Roberts, K. (1992). Identification of a transitional cell state in the developmental pathway to carrot somatic embryogenesis. J. Cell Biol. 119, 1371–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, D., Wittwer, F., Mandel, T., and Kuhlemeier, C. (1998). Localized upregulation of a new expansin gene predicts the site of leaf formation in the tomato meristem. Plant Cell 10, 1427–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres, B., and Benfey, P. (1999). Asymmetric cell division in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 505–537. [DOI] [PubMed] [Google Scholar]
- Shaw, S., and Quatrano, R. (1996). The role of targeted secretion in the establishment of cell polarity and the orientation of the division plane in Fucus zygotes. Development 122, 2623–2630. [DOI] [PubMed] [Google Scholar]
- Shevell, D.E., Leu, W.-M., Gillmor, C.S., Xia, G., Feldmann, K.A., and Chua, N.-H. (1994). EMB30 is essential for normal cell division, cell expansion, and cell adhesion in Arabidopsis and encodes a protein that has similarity to Sec7. Cell 77, 1051–1062. [DOI] [PubMed] [Google Scholar]
- Shevell, D.E., Kunkel, T., and Chua, N.-H. (2000). Cell wall alterations in the Arabidopsis emb30 mutant. Plant Cell 12, 2047–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter, A.M. (1993). Structure and function of cell wall proteins. Plant Cell 5, 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak, E., Leykan, J.F., and Kieliszewski, M.J. (1999). Synthetic genes for glycoprotein design and the elucidation of hydroxyproline-O-glycosylation codes. Proc. Natl. Acad. Sci. USA 96, 14736–14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak, E., Elisar, B., Leykan, J.F., and Kieliszewski, M.J. (2001). Contiguous hydroxyproline residues direct hydroxyproline arabinosylation in Nicotiana tabacum. J. Biol. Chem. 276, 11272–11278. [DOI] [PubMed] [Google Scholar]
- Smith, L.G. (2001). Plant cell division: Building walls in the right places. Nat. Rev. 2, 33–39. [DOI] [PubMed] [Google Scholar]
- Staehelin, L.A., and Hepler, P.K. (1996). Cytokinesis in higher plants. Cell 84, 821–824. [DOI] [PubMed] [Google Scholar]
- Stafstrom, J.P., and Staehelin, L.A. (1988). Antibody localization of extensin in carrot cell walls. Planta 174, 321–332. [DOI] [PubMed] [Google Scholar]
- Steinmann, T., Geldner, N., Grebe, M., Mangold, S., Jackson, C.L., Paris, S., Galweiler, L., Palme, K., and Juergens, G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286, 316–318. [DOI] [PubMed] [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D.G., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9, 1797–1810. [DOI] [PubMed] [Google Scholar]
- Swords, K.M.M., and Staehelin, L.A. (1993). Complementary immunolocalization patterns of cell wall hydroxyproline-rich glycoproteins studied with the use of antibodies directed against different carbohydrate epitopes. Plant Physiol. 102, 891–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester, A.W. (2000). Division decisions and spatial regulation of cytokinesis. Curr. Opin. Plant Biol. 3, 58–66. [DOI] [PubMed] [Google Scholar]
- Torres Ruiz, R.A. (1998). Embryogenesis. In Arabidopsis, M. Anderson and J.A. Roberts, ed (Boca Raton, FL: CRC Press), pp. 223–261.
- von Heijne, G. (1985). Signal sequences: The limits of variation. J. Mol. Biol. 184, 99–105. [DOI] [PubMed] [Google Scholar]
- Vroemen, C., deVries, S., and Quatrano, R. (1999). Signaling in plant embryo during the establishment of the polar axis. Semin. Cell Dev. Biol. 10, 157–164. [DOI] [PubMed] [Google Scholar]
- Vroemen, C.W., Langeveld, S., Mayer, U., Ripper, G., and Juergens, G. (1996). Pattern formation in the Arabidopsis embryo revealed by position-specific lipid transfer protein gene expression. Plant Cell 8, 783–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick, S.M. (1991). Cytoskeleton elements of the phragmosome establish the division plane in vacuolated higher plant cells. In The Cytoskeletal Basis of Plant Growth and Form, C.W. Lloyd, ed (London: Academic Press), pp. 231–244.
- Wycoff, K.L., Powell, P.A., Gonzales, R.A., Corbin, D.R., Lamb, C., and Dixon, R.A. (1995). Stress activation of a bean hydroxyproline-rich glycoprotein promoter is superimposed on a pattern of tissue-specific developmental expression. Plant Physiol. 109, 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Z.-H., and Varner, J.E. (1991). Tissue-specific expression of cell wall proteins in developing soybean tissues. Plant Cell 3, 23–37. [DOI] [PMC free article] [PubMed] [Google Scholar]