Abstract
Hepatitis C virus (HCV) induces microtubule aggregates in infected hepatocytes. To determine if cytoskeletal elements are important for HCV RNA synthesis, we examined the effect of cytoskeleton inhibitors on HCV replicon transcription in Huh7 cells. The data demonstrate that HCV replication complex-mediated RNA synthesis requires microtubule and actin polymerization.
Hepatitis C virus (HCV) infection is the leading cause of liver transplantation in the United States, with sequelae including liver fibrosis, cirrhosis, and hepatocellular carcinoma (reviewed in reference 19). Identified in 1989 as a plus-strand RNA virus (4), HCV causes an infection that was originally distinguished by its characteristic induction of microtubule (MT) aggregates in infected hepatocytes, implicating MTs in HCV-associated disease (2, 11, 23, 28, 29, 32, 36). Studies with a related flavivirus (Kunjin virus) have demonstrated that MT aggregates similar to those induced by HCV are also important for Kunjin RNA synthesis (15, 22). Moreover, MT paracrystals have recently been detected in cultured cells transfected with an HCV subgenomic replicon (21). Taken together, these studies suggest that cytoskeletal elements may be required for HCV replication. Yet, despite the historical emphasis on MTs and HCV from a clinical perspective, the role of MTs in the HCV life cycle at a molecular level remains poorly understood.
To determine if HCV RNA synthesis requires functional actin or MT networks, we examined the effects of cytoskeleton inhibitors on the efficiency of HCV RNA synthesis in the HCV replicon cell system (Fig. 1) (18). In this system, Huh7 cells stably transfected with an HCV replicon RNA are used to mimic the RNA synthesis that occurs in an ongoing, persistent infection with HCV. Because the replicon construct encodes a neomycin resistance gene for G418 (Geneticin) selection, HCV RNA synthesis in the replicon cells can be detected by quantitative PCR using a primer-probe set specific for the neomycin sequence. We have validated this approach by demonstrating a dose-dependent decrease in replicon RNA levels upon alpha interferon treatment (data not shown), using alpha interferon concentrations similar to those cited previously by other laboratories (1, 10, 18). In addition, we have confirmed that transcription of the replicon in our HCV replicon cells is resistant to actinomycin D, as originally demonstrated by Lohmann et al. (18), verifying that transcription of replicon RNA is specific to the RNA-dependent RNA polymerase activity of the HCV replication complex. A single HCV replicon cell line—a serially passaged line originally generated by stable transfection of Huh7 cells with a replicon with a sequence identical to that used by Lohmann et al. (18) (Fig. 1)—was used for all experiments.
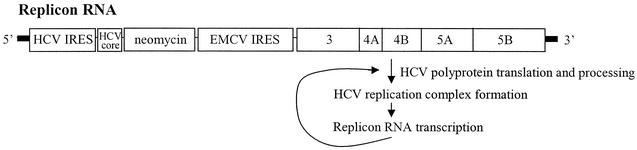
FIG. 1.
Schematic of the HCV subgenomic replicon. Translation of the neomycin resistance sequence (neomycin) is initiated at the HCV IRES, whereas expression of the polyprotein containing the HCV nonstructural proteins 3 (protease and helicase), 4A, 4B, 5A, and 5B (RNA-dependent RNA polymerase) is initiated at the EMCV IRES. An amino-terminal 17-amino-acid portion of the HCV core coding sequence (HCV core) is included immediately downstream of the HCV IRES. Processing of the viral polyprotein by the viral proteinase results in formation of the membrane-associated HCV replication complex. The complex mediates transcription of the replicon RNA from the authentic HCV 3′ and 5′ untranslated regions (bold lines) (19).
Because vinblastine sulfate (VS) is a well-characterized inhibitor of MT polymerization and has been shown to alter Kunjin virus replication (15, 22), we first determined the effect of VS on HCV replicon RNA synthesis. One day prior to addition of the inhibitor, HCV replicon cells were plated at 12,000 cells per well in a 96-well plate in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) and 0.5 mg of G418 per ml. The following day, the growth medium was replaced with DMEM containing 10% FBS and either 11 nM, 1 nM, or 0.5 nM VS in the absence of G418. Quadruplicate wells were incubated with each concentration of the inhibitor for 16 or 24 h. As a control for potential effects of the solvent on HCV RNA synthesis, four additional wells for each time point were incubated in DMEM-10% fetal bovine serum containing a volume of solvent (methanol) equivalent to the 11 nM VS concentration. At 16 and 24 h after inhibitor addition, the cells were photographed using a 10× Nikon objective. Cells were then immediately lysed in ABI lysis buffer as indicated by the manufacturer. Total RNA for each sample was isolated by use of an ABI Prism 6700 automated nucleic acid workstation. Equivalent RNA volumes were subsequently analyzed on an ABI Prism 7900HT sequence detection system for quantitative PCR, with one primer-probe set specific for the neomycin (neo) sequence and a second set specific for 18S rRNA (18S) to monitor cell number. To calculate the percentage of HCV replicon RNA remaining in the presence of each concentration of inhibitor in a standardized number of cells (% replicon RNA remaining), the mean neo RNA levels for the quadruplicate wells of each sample type were standardized to the mean 18S RNA level of the solvent-only control wells for the relevant time point. The % replicon RNA remaining was then calculated using the previously described comparative threshold cycle (CT) method (ABI Prism 7700 Sequence Detection System bulletin no. 2, PE Applied Biosystems, Foster City, Calif.).
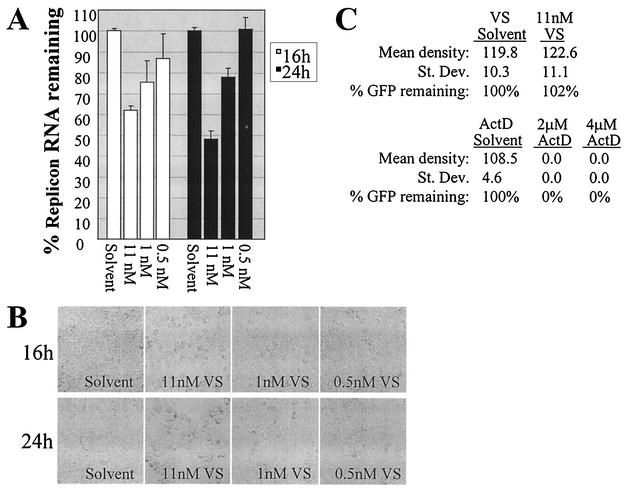
As indicated in Fig. 2A, VS decreased HCV RNA levels in the replicon cells in a dose-dependent manner. The observed cell rounding (Fig. 2B) was consistent with previously published reports of VS-induced MT depolymerization at similar concentrations in other cell types. Notably, since the data were standardized to the 18S RNA level of solvent-treated cells, the calculated % replicon RNA remaining corresponded to the level of HCV replicon RNA in a uniform number of cells. Hence, decreases in replicon RNA levels could not be attributed to cell loss.
FIG. 2.
Inhibition of HCV RNA synthesis by the MT inhibitor VS. (A) Dose-dependent decrease in HCV replicon RNA levels at 16 and 24 h of incubation with VS (11, 1, or 0.5 nM) compared to incubation with methanol equivalent to 11 nM VS (solvent). (B) Morphology of the HCV replicon cells in the presence of VS or solvent. (C) Quantification of GFP in HCV replicon cells transfected with the pCMV-EMCV IRES-GFP reporter DNA in the presence of actinomycin D (ActD), ethanol equivalent to 4 μM actinomycin D (ActD solvent), VS, or methanol equivalent to 11 nM VS (VS solvent). The mean density as well as the standard deviation of the density (St. Dev.) of the GFP band from the anti-GFP Western blot is shown for each sample. The percentage of GFP in the inhibitor-treated cells (% GFP remaining) is also indicated, with the quantity of GFP in the relevant solvent only-treated cells set at 100%. The % GFP remaining for the inhibitor-treated cells thus indicates the quantity of GFP in the inhibitor-treated cells compared to that in the solvent-treated cells.
The effect of VS also could not be explained by loss of the replicon from the cells upon removal of G418 selection, since the solvent- and VS-treated cells were identically maintained in the absence of G418 during the 16- or 24-h window of the experiment. Nevertheless, because the appearance of an inhibitory effect of VS on HCV RNA synthesis would be exaggerated in cells in which a significant amount of replicon RNA was lost upon removal of G418 selection, we examined the total amount of replicon RNA in parallel wells of cells in the presence or absence of G418. During this experiment, the variation in replicon RNA levels attributable to removal of G418 was within acceptable parameters (13% ± 8% at 16 h and 24% ± 2% at 24 h). The data therefore suggested that MT inhibition blocked the formation and/or RNA synthesis ability of the HCV replication complex. However, it remained possible that the decrease in replicon RNA was a result of the inhibition of translation mediated by the encephalomyocarditis virus (EMCV) internal ribosomal entry site (IRES). In this case, the decreased replicon RNA levels would merely represent decreased translation of the HCV nonstructural proteins. To confirm that the decrease in HCV replicon levels was due to the lack of a functional HCV replication complex rather than the lack of nonstructural protein expression, we determined the effect of 24 h of VS exposure on EMCV IRES translation in HCV replicon cells transfected with an EMCV IRES-green fluorescent protein (GFP) reporter construct whose transcription was driven by a cytomegalovirus (CMV) promoter. Eight hours after Lipofectamine PLUS-mediated transfection (Invitrogen), the transfection medium was replaced with phenol red-free DMEM containing 10% FBS and either 11 nM VS, VS solvent equivalent to that in the 11 nM VS, or 4 or 2 μM actinomycin D (to inhibit DNA-dependent RNA polymerase activity). At 24 h after the addition of the inhibitor (32 h posttransfection), whole-cell lysates were harvested in buffer containing 150 mM NaCl, 10 mM Tris (pH 7.4), 2 mM EDTA, 1% NP-40, 0.1% sodium dodecyl sulfate, and 1× protease inhibitor (Roche). A 50-μg aliquot of each cell lysate was then subjected to anti-GFP Western blotting. Equivalent protein loading was confirmed by Ponceau staining (Sigma; data not shown). The GFP signal from each of the samples on the Western blot was subsequently quantified using NIH Image software. For each sample, the mean density of the GFP band and the standard deviation of the band's density were determined. The amount of GFP remaining in cells treated with the inhibitor was calculated as the percentage of GFP in the solvent-treated cells. As demonstrated in Fig. 2C, VS did not alter EMCV IRES translation, even at the highest dose of this MT inhibitor. In contrast, actinomycin D abolished GFP expression, as expected for CMV promoter-driven transcription (Fig. 2C). We also confirmed that no GFP was detectable by fluorescence microscopy at the time of inhibitor addition, indicating that the level of GFP detected after the 24-h VS treatment was from ongoing GFP expression and not from long-term stability of GFP protein expressed prior to addition of the MT inhibitor (data not shown). Thus, the decrease in HCV replicon levels in the VS-treated cells was not due to inhibition of EMCV IRES-mediated translation by VS but rather was suggestive of inhibition of the HCV replication complex.
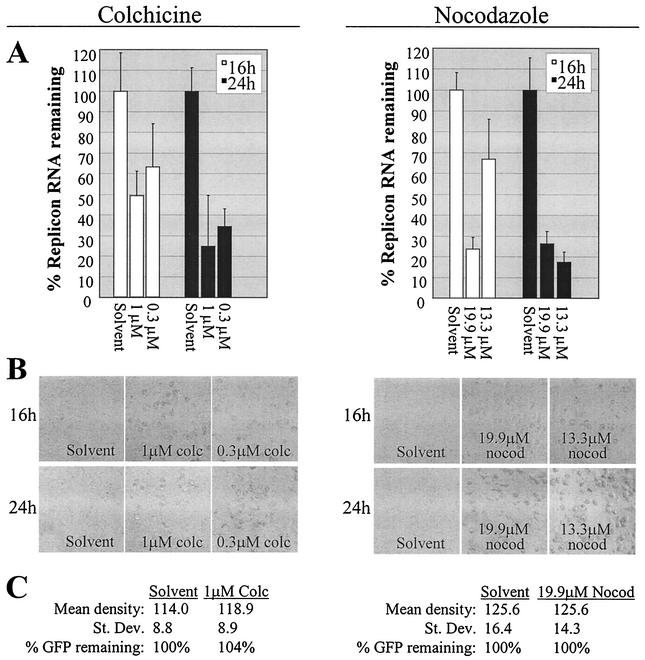
To confirm the dependency of HCV RNA synthesis on MTs, we also determined the effect of the MT depolymerizing agents colchicine and nocodazole. Two concentrations of each drug were chosen based on accepted dose ranges (7, 13, 22). By use of methods identical to those used for the VS experiment (including standardization of the data to 18S RNA of the solvent-treated cells to account for differences in cell number), colchicine and nocodazole were shown to decrease HCV replicon levels in a dose-dependent manner by 16 h of treatment (Fig. 3A). Additionally, replicon levels were generally further decreased by 24 h. As for the VS-treated cells, the addition of colchicine or nocodazole induced cell rounding indicative of MT inhibition (Fig. 3B). The extensive cell rounding at 24 h in the presence of 13.3 μM nocodazole revealed that this concentration was functionally equivalent to the higher concentration of the drug (19.9 μM), consistent with the very similar percentages of replicon RNA inhibition at the 24-h time point (Fig. 3A). Neither colchicine nor nocodazole inhibited EMCV IRES-mediated translation as determined with the CMV promoter-driven GFP reporter construct (Fig. 3C). The data are therefore consistent with an important role for MTs in HCV RNA synthesis. Furthermore, because MTs mediate intracellular membrane trafficking between the endoplasmic reticulum (ER) and the Golgi (5, 12, 26, 33), and also because HCV replication complexes are known to be anchored in ER or ER-like membranes (3, 8, 9, 16, 17, 20, 21, 24, 25, 27, 30, 31, 35), our data suggest that HCV RNA synthesis may be dependent on MT-mediated delivery of membranes required to establish the functional HCV replication complex.
FIG. 3.
Inhibition of HCV RNA synthesis by the MT inhibitors colchicine and nocodazole. Effects of colchicine (colc) and nocodazole (nocod) are shown on the left and right panels of the figure, respectively. (A) Decreased HCV replicon RNA in cells treated with colchicine (1 or 0.3 μM) compared to those treated with ethanol equivalent to 1 μM colchicine (solvent) or in cells treated with nocodazole (19.9 or 13.3 μM) compared to those treated with DMSO equivalent to 19.9 μM nocodazole (solvent). (B) Morphology of the HCV replicon cells corresponding to those harvested for RNA detection for panel A. (C) Quantification of GFP in HCV replicon cells transfected with the pCMV-EMCV IRES-GFP reporter DNA in the presence of colchicine or nocodazole or the respective solvent control described for panel A. The figure design is identical to that described for Fig. 2C.
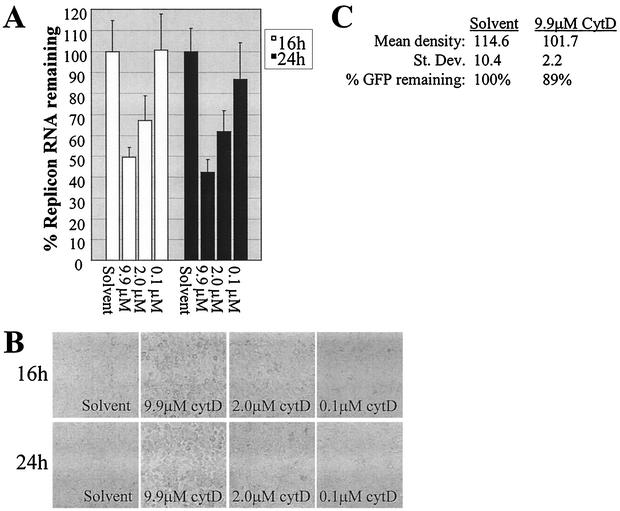
Since MTs and actin often have parallel or cooperative functions in orchestrating membrane trafficking (reviewed in reference 6), we next sought to determine if actin polymerization was important for HCV RNA synthesis. When replicon cells were treated with increasing concentrations of the actin inhibitor cytochalasin D, HCV replicon RNA levels were decreased accordingly (Fig. 4A). Actin inhibition was verified by the expected cellular morphology (Fig. 4B). Furthermore, the effect on HCV RNA was not accounted for by inhibition of EMCV IRES-mediated translation (Fig. 4C). These combined data indicated that HCV's RNA-dependent RNA polymerase activity could be blocked by inhibiting either the actin or the MT network.
FIG. 4.
Inhibition of HCV RNA synthesis by the actin inhibitor cytochalasin D (cytD). (A) Effect of cytochalasin D (9.9, 2.0, or 0.1 μM) on HCV replicon RNA levels compared to the effect of DMSO equivalent to 9.9 μM cytochalasin D (solvent). (B) Dose-dependent cell rounding induced by actin inhibition in replicon cells treated with cytochalasin D. (C) Quantification of GFP in HCV replicon cells transfected with the pCMV-EMCV IRES-GFP reporter DNA in the presence of 9.9 μM cytochalasin D or DMSO equivalent to 9.9 μM cytochalasin D (solvent). See the legend for Fig. 2C for further description of the figure design.
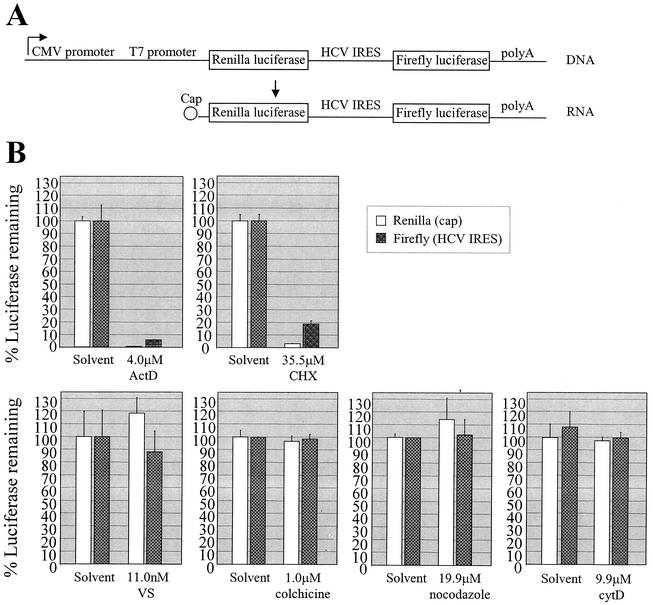
As described above, the GFP reporter data demonstrated that the actin and MT inhibitors did not block EMCV IRES-mediated translation in the HCV replicon. By extension, we reasoned that actin- and MT-mediated membrane trafficking would not likely be required for HCV IRES translation. Thus, in the context of an actual HCV infection, cytoskeleton-dependent membrane transport would be expected to be required not for providing access of the viral genome to translation factors but rather for construction, maintenance, and/or functioning of the HCV replication complex following genome translation. To determine if HCV IRES translation was dependent on actin and/or MT polymerization, we transfected the HCV replicon cells with a dual luciferase reporter encoding a Renilla luciferase gene to be expressed by cap-dependent translation as well as a firefly luciferase gene to be expressed from the same transcript by HCV IRES-dependent translation (Fig. 5A) (14). The dual system allowed us to examine potential differences in cytoskeletal requirements for HCV IRES-mediated translation versus the control cap-mediated translation. The HCV replicon cells were plated in 96-well plates in G418-containing medium with 10% FBS the day before transfection. Cells were then identically transfected with the dual luciferase reporter DNA according to the Lipofectamine PLUS manufacturer's protocol (Invitrogen). Seven hours after transfection, the medium was changed to G418-free DMEM containing 10% FBS and either the inhibitors or corresponding solvents at the concentrations indicated in Fig. 5B. Each sample condition was performed in triplicate wells. After a 21-h incubation at 37°C, the cells were prepared for luciferase detection using the Promega Dual-Luciferase reporter assay system. The mean of the triplicate luciferase values for each solvent-treated sample was calculated and designated 100% luciferase remaining, indicating the quantity of luciferase remaining after the 21-h incubation in the presence of the solvent-containing medium. The mean of the triplicate luciferase values for each corresponding inhibitor-treated sample was then calculated, and the % luciferase remaining in the presence of the inhibitor after the 21-h incubation was calculated as a percentage of the relevant solvent control. As expected, both cap- and HCV IRES-dependent translation were inhibited by actinomycin D and cycloheximide, verifying that the luciferase signal was specific to the transcription and translation, respectively, of the reporter construct (Fig. 5B). In contrast, expression of either Renilla or firefly luciferase was not significantly altered by VS, colchicine, nocodazole, or cytochalasin D (Fig. 5B). Thus, actin and MT polymerization were not required for efficient HCV IRES translation.
FIG. 5.
Dual luciferase reporter assay. (A) Schematic of the dual luciferase reporter (15). (B) Effect of actinomycin D (ActD), cycloheximide (CHX), VS, colchicine, nocodazole, or cytochalasin D (cytD) or relevant solvent-only control media (containing ethanol, ethanol, methanol, ethanol, DMSO, and DMSO, respectively) on cap-dependent translation (Renilla luciferase) and HCV IRES translation (firefly luciferase) in cells transfected with the dual luciferase reporter.
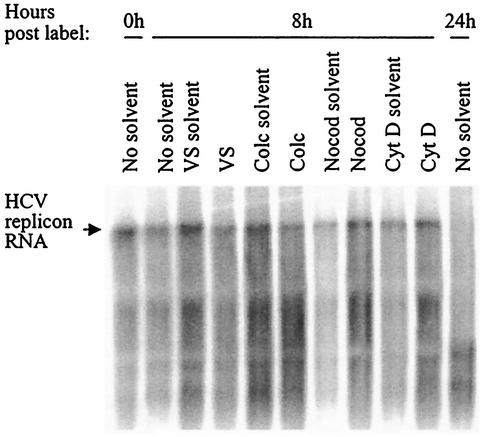
Having demonstrated that actin and/or MT inhibition reduced levels of HCV replicon RNA, we next sought to distinguish between two possible mechanisms of inhibition: (i) inhibitor-induced enhancement of replicon RNA degradation (thereby blocking HCV RNA synthesis at the level of destroying the RNA template) or (ii) inhibition of HCV RNA synthesis without altering RNA stability, such as by preventing replication complex formation or inhibiting the replication complex's RNA synthesis abilities. To determine if HCV RNA degradation was induced by the addition of VS, colchicine, nocodazole, cytochalasin D, and/or their respective solvents (methanol, ethanol, dimethyl sulfoxide [DMSO], DMSO), a pulse-chase RNA labeling experiment was performed. HCV replicon cells were plated at a density of 5 × 105 cells per 60-mm-diameter dish and incubated at 37°C for 24 h prior to a 2-h incubation in DMEM containing 4 μg of actinomycin D per ml. Cells were then washed in phosphate-free DMEM (Invitrogen), followed by a 16-h incubation at 37°C in phosphate-free medium containing 4 μg of actinomycin D per ml and 200 μCi of 32P radionuclide (NEN). To optimize the length of the chase period for maximum replicon RNA detectability and maximum time postlabeling, parallel plates of cells were incubated in labeling medium without inhibitor or solvent and were harvested at time zero postlabeling (immediately after the 16-h labeling period) as well as at 8 and 24 h postlabeling (8 or 24 h after replacing the labeling medium with fresh DMEM). The last time points were selected based on estimates of HCV RNA stability or to correspond to the 24-h time point of the inhibitor studies shown in previous figures, respectively. Total RNA was isolated from each plate by adding 600 μl of RNeasy reagent (Qiagen). Equivalent volumes of the eluted RNA were then heated at 65°C for 15 min, loaded on a formaldehyde-1% agarose gel, dried, and subjected to phosphorimager analysis.
HCV replicon RNA was readily detectable at zero time postlabeling, confirming good 32P incorporation during the 16-h labeling period (Fig. 6). As expected, migration of the primary RNA species on the gel was similar to that of in vitro-transcribed HCV replicon RNA, as expected (data not shown). The HCV replicon RNA from the cells remained detectable after an 8-h chase period but was largely degraded by 24 h postlabeling (Fig. 6). A 16-h labeling period and an 8-h chase were therefore chosen for determining the effect of the actin and MT inhibitors on HCV RNA degradation in the replicon cells. To this end, replicon cells were plated and labeled as described above. Following the 16-h labeling period, cells were washed with DMEM and were incubated at 37°C for 8 h in DMEM containing 11 nM VS, 1 μM colchicine, 19.9 μM nocodazole, 9.9 μM cytochalasin D, or equivalent concentrations of the relevant solvents. At the end of the 8-h chase period, total RNA was harvested. To ensure collection of all RNA, floating cells were gathered from the growth supernatant by centrifugation, and each plate of cells was scraped after the addition of the RNeasy reagent. Equivalent volumes of extracted RNA were then subjected to agarose gel analysis as described above. Replicon RNA levels at 8 h postlabeling were not diminished in the cells treated with solvent only compared to cells incubated in media lacking solvent for 8 h (Fig. 6). VS treatment of cells resulted in only a minimal reduction of HCV replicon RNA at 8 h postlabeling compared to solvent only-treated cells and was not indicative of substantial VS-induced RNA degradation (Fig. 6). In addition, HCV replicon RNA levels in cells treated with either colchicine, nocodazole, or cytochalasin D were consistent with the levels in cells treated with the relevant solvent alone (Fig. 6). The data therefore indicate that neither the solvents nor the cytoskeleton inhibitors induce significant HCV RNA degradation in the replicon cells. Thus, the reduction of HCV RNA levels in cells treated with the inhibitors (Fig. 2, 3, and 4) is not due to rapid degradation of template replicon RNAs but rather to inhibition of the processes required for RNA synthesis from existing template RNAs.
FIG. 6.
Stability of HCV replicon RNA in cells treated with cytoskeleton inhibitors or with solvent alone. The time of cell harvest postlabeling is indicated at the top of the figure as the number of hours after completion of the 16-h labeling period and removal of the 32P medium. The incubation conditions during the chase period are noted above each well. The bands representing the HCV replicon RNA are indicated by the arrow to the left of the gel. No solvent, cells incubated in DMEM alone; VS solvent, methanol equivalent to 11 nM VS; VS, 11 nM VS; Colc solvent, ethanol equivalent to 1 μM colchicine; Colc, 1 μM colchicine; Nocod solvent, DMSO equivalent to 19.9 μM nocodazole; Nocod, 19.9 μM nocodazole; Cyt D solvent, DMSO equivalent to 9.9 μM cytochalasin D; Cyt D, 9.9 μM cytochalasin D.
Together, the data in this report demonstrate a cytoskeletal requirement for HCV RNA synthesis. Although the precise mechanism for this requirement remains to be determined, several possibilities exist. For example, it is not yet clear whether HCV RNA synthesis occurs in a stationary complex whose formation requires MTs and actin or whether the migration of the replication complex between cellular organelles (which may be altered by the replication process itself) is necessary for generating nascent RNAs. The role of the ER stress response in this process also remains undetermined. Among the options for mechanisms, we favor a model of actin- and MT-mediated formation and/or maintenance of the vesicles which serve as the membrane anchoring sites for the HCV replication complex. This model is especially attractive in light of recent electron micrographs of HCV replicon cells demonstrating the proximity of ER membrane-associated HCV nonstructural proteins with paracrystalline structures reminiscent of the MT aggregates detected in HCV-infected chimpanzee hepatocytes or near sites of viral RNA synthesis in Kunjin virus-infected cells (21, 28, 29, 34). Notably, the model allows for but does not require alterations in MT or ER structure. For example, a requirement for MT-mediated vesicular maintenance during early stages of HCV RNA synthesis would neither require nor preclude the subsequent alteration of MT morphology (such as the development of paracrystalline arrays). In addition, since an intact MT network could presumably serve equally well as a “railroad” for vesicles during normal ER-related trafficking or during ER rearrangement, a cytoskeletal requirement for HCV RNA synthesis would not be contingent on a particular ER structure. Thus, a role for cytoskeletal elements in HCV RNA synthesis is consistent with the development of paracrystalline structures and rough ER fragmentation in some HCV replicon-containing cells (21) but not in others (24). The development of a stable tissue culture system capable of producing infectious HCV particles will be important for determining if such cytoskeletal alterations, which may be initiated by early steps in HCV replication, are enhanced during the late stages of the HCV life cycle. An infectious HCV tissue culture system will also enable experiments to determine if MT and actin polymerization are important for HCV assembly and maturation, as our data indicate for HCV RNA synthesis.
Acknowledgments
We gratefully acknowledge the technical expertise of Angel Lai and Xiaolan Lu, the statistical analysis of Mark Farmen, and the review of the manuscript by Joseph Colacino.
REFERENCES
- 1.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 2.Bradley, D. W., K. A. McCaustland, E. H. Cook, C. A. Schable, J. W. Ebert, and J. E. Maynard. 1985. Posttransfusion non-A, non-B hepatitis in chimpanzees. Physicochemical evidence that the tubule-forming agent is a small, enveloped virus. Gastroenterology 88:773-779. [PubMed] [Google Scholar]
- 3.Brass, V., E. Bieck, R. Montserret, B. Wolk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130-8139. [DOI] [PubMed] [Google Scholar]
- 4.Choo, Q.-L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 5.Cole, N. B., N. Sciaky, A. Marotta, J. Song, and J. Lippincott-Schwartz. 1996. Golgi dispersal during microtubule disruption: regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol. Biol. Cell 7:631-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DePina, A. S., and G. M. Langford. 1999. Vesicle transport: the role of actin filaments and myosin motors. Microsc. Res. Tech. 47:93-106. [DOI] [PubMed] [Google Scholar]
- 7.Duvet, S., L. Cocquerel, A. Pillez, R. Cacan, A. Verbert, D. Moradpour, C. Wychowski, and J. Dubuisson. 1998. Hepatitis C virus glycoprotein complex localization in the endoplasmic reticulum involves a determinant for retention and not retrieval. J. Biol. Chem. 273:32088-32093. [DOI] [PubMed] [Google Scholar]
- 8.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fipaldini, C., B. Bellei, and N. La Monica. 1999. Expression of hepatitis C virus cDNA in human hepatoma cell line mediated by a hybrid baculovirus-HCV vector. Virology 255:302-311. [DOI] [PubMed] [Google Scholar]
- 10.Frese, M., T. Pietschmann, D. Moradpour, O. Haller, and R. Bartenschlager. 2001. Interferon-alpha inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J. Gen. Virol. 82:723-733. [DOI] [PubMed] [Google Scholar]
- 11.Gudat, F., G. Eder, C. Eder, L. Bianchi, E. Stocklin, G. Krey, U. Durmuller, and H. P. Spichtin. 1983. Experimental non-A, non-B hepatitis in chimpanzees: light, electron and immune microscopical observations. Liver 3:110-121. [DOI] [PubMed] [Google Scholar]
- 12.Hammond, A. T., and B. S. Glick. 2000. Dynamics of transitional endoplasmic reticulum sites in vertebrate cells. Mol. Biol. Cell 11:3013-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harada, M., S. Sakisaka, K. Terada, R. Kimura, T. Kawaguchi, H. Koga, E. Taniguchi, K. Sasatomi, N. Miura, T. Suganuma, H. Fujita, K. Furata, K. Tanikawa, T. Sugiyama, and M. Sata. 2000. Role of ATP7B in biliary copper excretion in a human hepatoma cell line and normal rat hepatocytes. Gastroenterology 118:921-928. [DOI] [PubMed] [Google Scholar]
- 14.Honda, M., S. Kaneko, E. Matsushita, K. Kobayashi, G. A. Abell, and S. M. Lemon. 2000. Cell cycle regulation of hepatitis C virus internal ribosomal entry site-directed translation. Gastroenterology 118:152-162. [DOI] [PubMed] [Google Scholar]
- 15.Hong, S. S., and M. L. Ng. 1987. Involvement of microtubules in Kunjin virus replication. Arch. Virol. 97:115-121. [DOI] [PubMed] [Google Scholar]
- 16.Hugle, T., F. Fehrmann, E. Bieck, M. Kohara, H. G. Krausslich, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 284:70-81. [DOI] [PubMed] [Google Scholar]
- 17.Kim, J. E., W. K. Song, K. M. Chung, S. H. Back, and S. K. Jang. 1999. Subcellular localization of hepatitis C viral proteins in mammalian cells. Arch. Virol. 144:329-343. [DOI] [PubMed] [Google Scholar]
- 18.Lohmann, V., F. Korner, J.-O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 19.Moradpour, D., A. Cerny, M. H. Heim, and H. E. Blum. 2001. Hepatitis C: an update. Swiss Med. Wkly. 131:291-298. [DOI] [PubMed] [Google Scholar]
- 20.Moradpour, D., P. Kary, C. M. Rice, and H. E. Blum. 1998. Continuous human cell lines inducibly expressing hepatitis C virus structural and nonstructural proteins. Hepatology 28:192-201. [DOI] [PubMed] [Google Scholar]
- 21.Mottola, G., G. Cardinali, A. Ceccacci, C. Trozzi, L. Bartholomew, M. R. Torrisi, E. Pedrazzini, S. Bonatti, and G. Migliaccio. 2002. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology 293:31-43. [DOI] [PubMed] [Google Scholar]
- 22.Ng, M. L., J. S. Pedersen, B. H. Tob, and E. G. Westaway. 1983. Immunofluorescent sites in Vero cells infected with the flavivirus Kunjin. Arch. Virol. 78:177-190. [DOI] [PubMed] [Google Scholar]
- 23.Pfeifer, U., R. Thomssen, K. Legler, U. Bottcher, W. Gerlich, E. Weinmann, and O. Klinge. 1980. Experimental non-A, non-B hepatitis: four types of cytoplasmic alteration in hepatocytes of infected chimpanzees. Virchows Arch. B Cell. Pathol. Incl. Mol. Pathol. 33:233-243. [DOI] [PubMed] [Google Scholar]
- 24.Pietschmann, T., V. Lohmann, G. Rutter, K. Kurpanek, and R. Bartenschlager. 2001. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 75:1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polyak, S. J., D. M. Paschal, S. McArdle, M. J. J. Gale, D. Moradpour, and D. R. Gretch. 1999. Characterization of the effects of hepatitis C virus nonstructural 5A protein expression in human cell lines and on interferon-sensitive virus replication. Hepatology 29:1262-1271. [DOI] [PubMed] [Google Scholar]
- 26.Presley, J. F., N. B. Cole, T. A. Schroer, K. Hirschberg, K. J. Zaal, and J. Lippincott-Schwartz. 1997. ER-to-Golgi transport visualized in living cells. Nature 389:81-85. [DOI] [PubMed] [Google Scholar]
- 27.Santolini, E., L. Pacini, C. Fipaldini, G. Migliaccio, and N. Monica. 1995. The NS2 protein of hepatitis C virus is a transmembrane polypeptide. J. Virol. 69:7461-7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaff, Z., G. Eder, C. Eder, and K. Lapis. 1990. Intracytoplasmic crystalline inclusions in the hepatocytes of humans and chimpanzees. Ultrastruct. Pathol. 14:303-309. [DOI] [PubMed] [Google Scholar]
- 29.Schaff, Z., G. Eder, C. Eder, and K. Lapis. 1992. Ultrastructure of normal and hepatitis virus infected human and chimpanzee liver: similarities and differences. Acta Morphol. Hung. 40:203-214. [PubMed] [Google Scholar]
- 30.Schmidt-Mende, J., E. Bieck, T. Hugle, F. Penin, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent-RNA polymerase. J. Biol. Chem. 276:44052-44063. [DOI] [PubMed] [Google Scholar]
- 31.Shi, S. T., S. J. Polyak, H. Tu, D. R. Taylor, D. R. Gretch, and M. M. Lai. 2002. Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology 292:198-210. [DOI] [PubMed] [Google Scholar]
- 32.Spichtin, H. P., F. Gudat, M. Shmid, M. Pirovino, J. Altorfer, and L. Bianchi. 1982. Microtubular aggregates in human chronic non-A, non-B hepatitis with bridging hepatic necrosis and multinucleated hepatocytic giant cells. Liver 2:355-360. [DOI] [PubMed] [Google Scholar]
- 33.Thyberg, J., and S. Moskalewski. 1999. Role of microtubules in the organization of the Golgi complex. Exp. Cell Res. 246:263-279. [DOI] [PubMed] [Google Scholar]
- 34.Westaway, E. G., J. M. Mackenzie, M. T. Kenny, M. K. Jones, and A. A. Khromykh. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 71:6650-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolk, B., D. Sansonno, H. G. Krausslich, F. Dammacco, C. M. Rice, H. E. Blum, and D. Moradpour. 2000. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J. Virol. 74:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshizawa, H., Y. Itoh, S. Iwakiri, K. Kitajima, Y. Noguchi, T. Tachibana, Y. Miyakawa, and M. Mayumi. 1984. Beta-propiolactone for the inactivation of non-A/non-B type hepatitis virus capable of inducing cytoplasmic tubular ultrastructures in chimpanzees. Vox Sang. 46:86-91. [DOI] [PubMed] [Google Scholar]