Abstract
In spite of intensive and increasingly successful attempts to determine the multiple steps involved in colorectal carcinogenesis, the mechanisms responsible for metastasis of colorectal tumors to the liver remain to be clarified. To identify genes that are candidates for involvement in the metastatic process, we analyzed genome-wide expression profiles of 10 primary colorectal cancers and their corresponding metastatic lesions by means of a cDNA microarray consisting of 9121 human genes. This analysis identified 40 genes whose expression was commonly upregulated in metastatic lesions, and 7 that were commonly downregulated. The upregulated genes encoded proteins involved in cell adhesion, or remodeling of the actin cytoskeleton. Investigation of the functions of more of the altered genes should improve our understanding of metastasis and may identify diagnostic markers and/or novel molecular targets for prevention or therapy of metastatic lesions.
Keywords: metastasis, cDNA microarray, microdissection, colorectal cancer (CRC), T7-based RNA amplification
Introduction
Liver metastasis is a major cause of death among patients with colorectal cancer (CRC). Despite progress that has been achieved with therapeutic approaches, a complete cure awaits more effective strategies. Prevention or effective treatment of liver metastasis will save the lives of thousands of patients. We have attempted to identify genes involved in metastasis of CRC to the liver in order to learn more about the underlying mechanisms, to identify clinically applicable diagnostic markers and preventive approaches, and to find molecules that might be useful targets for therapy.
Liver metastasis occurs in multiple steps that include release of cancer cells from the primary site, intravasation to neighboring vessels, transport to the site of metastasis through blood flow, extravasation and/or infarction to the distant organ, and re-growth of the invading cells with acquisition of nutrition in the new environment. Therefore, multiple genes must play roles. Although many investigators have been working on this clinically important issue, the precise mechanisms or identification of the critical genes remains to be clarified. A number of molecules associated with liver metastasis have been reported, but as most studies have focused on one or a few molecules, the question of the importance of each gene in the complex process has not been addressed.
Because microarray technology permits analysis of expression levels of thousands of genes in a single experiment, new classifications of cancer types can now be proposed on the basis of altered expression of multiple genes in tumor tissues [1,2]. Moreover, analyses of gene expression profiles have disclosed specific patterns that may reflect progression of tumor cells [3,4]. Recently, two groups detected genes responsible for metastasis of malignant melanomas, using cDNA microarrays. One group compared expression profiles of highly metastatic melanoma cells with less metastatic cells established from the same cell lines [5]; the other analyzed expression profiles among various melanoma cell lines and primary melanomas [6].
Here we report identification of 47 genes whose expression was commonly altered in metastatic CRC cells to the liver by comparing expression profiles of primary and metastatic CRCs on a cDNA microarray containing 9121 genes. This information will be useful for clarifying the mechanisms of liver metastasis and for discovering new molecular targets for its diagnosis, treatment, or prevention.
Materials and Methods
Tissue Samples and Laser Capture Microdissection (LCM)
Primary CRC tissues and corresponding metastatic foci from liver were obtained with informed consent from 10 patients who underwent colectomy and hepatectomy in the same operation. All of the samples were imbedded in TissueTek OCT medium (Sakura, Tokyo, Japan) and frozen at -80°C. Later, the frozen sections were fixed in 70% ethanol for 45 seconds, stained with hematoxylin and eosin, and dehydrated in 70:30, 50:50, and 30:70 of ethanol:xylene for 30 seconds at each step, followed by a final dehydration in 100% xylene for 2 minutes. Once air-dried, the stained tissues were microdissected using a PixCell LCM system (Arcturus Engineering, Mountain View, CA) according to the manufacturer's protocols. Cancerous cells from the primary lesions and from liver metastases were selectively microdissected (∼2x104 cells from each sample).
RNA Extraction and T7-Based RNA Amplification
Total RNA was extracted from each sample of laser-captured cells into 350 µl of RLT lysis buffer (QIAGEN, Hilden, Germany). The extracted RNA was treated for 1 hour at 37°C with 10 U of DNase I (Roche, Basel, Switzerland) in the presence of 1 U of RNase inhibitor (TOYOBO, Osaka, Japan) to remove any contaminating genomic DNA. After inactivation at 70°C for 10 minutes, the RNA was purified with an RNeasy Mini Kit (QIAGEN) according to the manufacturer's recommendations. All DNase I-treated RNA was subjected to T7-based amplification as described previously [7]. Three rounds of amplification yielded 8 to 70 µg of amplified RNA (aRNA) from each sample.
Construction and Analysis of the cDNA Microarray
We had selected 9121 independent cDNA, including some ESTs, from the UniGene database of the National Center for Biotechnology Information. The cDNA spotted on the microarray slides was prepared by reverse transcription polymerase chain reaction (RT-PCR) using sets of genespecific primers and a mixture of commercially provided poly A RNA (Clontech, Palo Alto, CA) as a template [7]. The products were applied to electrophoresis on agarose gels and those showing a single band of expected size were utilized for spotting. Further sequence analyses of randomly selected 2485 (27.2%) products from 9121 genes corroborated the complete concordance of their cDNA sequences.
Duplicate sets of cDNA spots were used for each analysis of expression profiles to reduce experimental fluctuation. Three-microgram aliquots of each aRNA from primary CRCs and metastatic lesions were labeled with Cy5-dCTP or Cy3-dCTP, respectively (Amersham Pharmacia Biotech, Buckinghamshire, England). Equal amounts of Cy3- and Cy5-labeled probes were co-hybridized onto the microarray slides. Hybridization, washing, and scanning were performed as described previously [7].
Data Analysis
The intensity of each duplicated signal was evaluated photometrically by the Array Vision computer program (Imaging Research, St. Catharines, Ontario, Canada) and normalized so that the averaged Cy3/Cy5 ratio of 52 housekeeping genes was 1.0. [3,7] (Figure 1). After the normalization of signal intensity of each spot, we excluded data if both Cy3 and Cy5 signals were less than those of either the Arabidopsis thaliana putative chlorophyll synthetase (G4) gene or pBluescript II SK(+) vector DNA, which had been spotted on the microarray slides as negative controls. The Cy3/Cy5 ratio for each gene was calculated by averaging duplicate spots [3,7]. Genes whose Cy3/Cy5 ratios were greater than 2 were considered to be upregulated in the metastatic tissues, whereas those with ratios less than 0.5 were considered to be downregulated. Finally, genes that showed increased expression in six or more of the 10 cases examined were selected as “frequently upregulated” genes, and those that showed decreases in five or more cases were selected as “frequently downregulated.” We evaluated the significance of altered expression of the selected genes by calculating the P values using a matched pair permutation test as described previously [3].
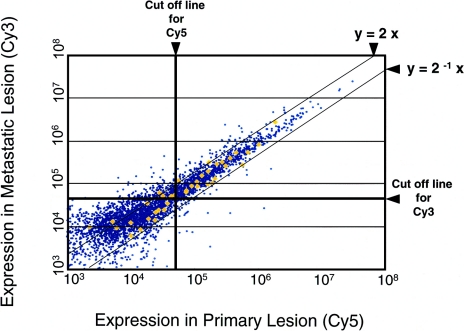
Figure 1.
Representative scatter plot (normalized) from the cDNA microarray analysis. Amplified RNA from the primary (Cy5-labeled) and metastatic (Cy3-labeled) lesions from case no. 101 were hybridized to the microarray slides. The y=2x line represents the cut-off border for upregulated genes, and y=2-1x is the border for downregulated genes. The horizontal line (Cy3) and the vertical line (Cy5) are cut-off borders for reliable signal intensity. Yellow diamonds indicate housekeeping genes, which were used for normalization of data.
Quantitative RT-PCR (TaqMan-PCR)
To verify the data obtained from microarray, we carried out quantitative RT-PCR (TaqMan-PCR) with additional eight pairs of CRCs according to the manufacturer's recommendations (Applied Biosystems, Foster City, CA). Total RNA extracted from microdissected tissues was subjected to one round of T7-based RNA amplification followed by cDNA synthesis using 0.5 µg of random hexamer (Roche) and SuperScript II according to the supplier's protocol (Life Technologies, Gaithersburg, MD). The synthesized cDNA was diluted and used as template for amplification on ABI Prism 7700 Sequence Detection System (Applied Biosystems). A set of primers and a TaqMan probe for a housekeeping gene, β2-microglobulin (Applied Biosystems), were used as endogenous control. Sequences of primers and probes of genes selected for verification were as follows: microtubule-associated protein 2 (MAP2) (forward) 5′-TCGAATCTCCTCAGCTTGCC-3′, (reverse) 5′-CACAAGCCCTGCTTAGCGA-3′, (probe) 5′-Fam-CTTTGGCTGAGGATGTCACTGCTGCA-Tamra-3′; N-WASP (forward) 5′-ACTGTTAGACCAGATACGACAGGGT-3′, (reverse) 5′-TGCAGGTGTTGGTGGTGTAGA-3′, (probe) 5′-Fam-TCCAACTAAAATCTGTGGCTGATGGCCA-Tamra-3′; neural cell adhesion molecule L1 (NCAM-L1) (forward) 5′-GTCTGAACAGTTGTCTTCCTCAGC-3′, (reverse) 5′-CAAACTTTCAAAGTCACGGTGTATTT-3′, (probe) 5′-Fam-TCCTCCCGCCCCCACTTGG-Tamra-3′; S3 ribosomal protein (forward) 5′-TGAGCATTGTGGAACCCAAA-3′, (reverse) 5′-GCTTCCCACCCTTCTGTTCTG-3′, (probe) 5′-Fam-ATGAGATACTGCCCACCACCCCCATC-Tamra-3′; caltractin (forward) 5′-AGCACCTGCCATTTTGCC-3′, (reverse) 5′-GTTCTGTGTTTGTGGATATGTGGAA-3′, (probe) 5′-Fam-TGCATCGTTTCCCTCGTCATGCA-Tamra-3′; EST (BC005071) (forward) 5′-TCAGTTATCTCCAGAGCCATTTCA-3′, (reverse) 5′-GACTGGATAATAAACCACCTCTGACA-3′, (probe) 5′-Fam-CCTTTAGAGTGAGTCACATGCAGGGAGTGTG-Tamra-3′. Comparative CT Method was employed to determine the expression ratio of each gene (Applied Biosystems).
Results
Isolation of Primary CRCs and Corresponding Metastatic Lesions by LCM
To obtain precise expression profiles of primary versus metastatic cancer cells, it was essential to collect pure populations of each type. Therefore, we employed LCM to avoid contamination by non-cancerous cells. The proportion of cancer cells that selected by this procedure was estimated to be >95%, as determined by microscopic visualization (data not shown).
To investigate further the amount of hepatocyte contamination in the microdissected metastatic lesions, we examined the signal intensity of the albumin gene, which is not expressed in colon cancer cells. When hepatocytes were selectively dissected from a non-cancerous liver specimen, the ratio of the average signal intensity of the albumin gene to that of the GAPDH gene was approximately 53.7, whereas the ratio in microdisssected metastatic cells was 0.14. Consequently, the proportion of contaminating hepatocytes among the dissected cancer cells we analyzed was estimated to be less than 0.3%. Because the cut-off value we applied in this study was higher than the signal intensity of the albumin gene, we considered that contamination by hepatocytes would have little effect on our analysis of expression profiles.
Identification of Genes Expressed Differently in Metastasized Versus Primary CRCs
The numbers of selected genes in each expression category, as judged by their ratios of signal intensity of metastatic to primary cancer tissues, are summarized in Table 1; a representative scatter plot is shown in Figure 1. Given the inevitable diversity among individual tumors and the variety of factors that could affect gene expression, we selected for further study only the genes whose expression in metastatic tissue was elevated more than two-fold in six or more, or reduced to less than half in five or more, of the 10 cases examined. In this manner, we identified 40 genes that commonly showed elevated expression in the metastatic lesions and seven genes, two of them ESTs, that revealed reduced expression (Tables 2 and 3). These commonly altered genes corresponded to about 0.44% and 0.08%, respectively, of the 9121 genes analyzed. The selected group included genes known to be involved in cell adhesion, or remodeling of the actin cytoskeleton. Among them, three [MAP 2, T lymphocyte-specific adaptor protein (SH2D2A), and G protein-coupled receptor STRL33.1 (STRL33)] were upregulated in 8 of 10 cases. Among the genes showing reduced expression, the HNK-1 gene was downregulated in six cases, while others, including genes encoding autonomously replicating sequence (ARS), caltractin, S3 ribosomal protein, and 40S ribosomal protein S19, were downregulated in five cases.
Table 1.
Number of Selected Genes.
| Case ID | 71 | 95 | 97 | 100 | 101 | 103 | 145 | 147 | 158 | 159 |
| Up | 427 | 286 | 136 | 560 | 515 | 311 | 279 | 492 | 528 | 583 |
| Unchanged | 899 | 475 | 1521 | 556 | 1941 | 1860 | 571 | 1016 | 700 | 1356 |
| Down | 19 | 51 | 270 | 220 | 51 | 60 | 48 | 62 | 50 | 41 |
| Total | 1345 | 812 | 1927 | 1336 | 2511 | 2231 | 898 | 1570 | 1278 | 1980 |
Table 2.
Upregulated Genes.
| Frequency | Average Ratio±SD | P Value | Accession Number | UniGene Cluster | Definition |
| 8 | 2.8±0.9 | 1.56E-04 | U01828 | 167 | Human microtubule-associated protein 2 (MAP2) mRNA; complete cds |
| 8 | 5.7±4.1 | 1.10E-04 | AJ000553 | 103 527 | Homo sapiens mRNA for T lymphocyte-specific adaptor protein (SH2D2A) |
| 8 | 5.7±3.7 | 2.24E-05 | U73529 | 34 526 | Human G protein-coupled receptor STRL33.1 (STRL33) mRNA; complete cds |
| 7 | 4.7±2.0 | 5.33E-04 | M26682 | 1149 | Human T cell translocation gene 1 (Ttg-1) mRNA; complete cds |
| 7 | 3.0±1.0 | 4.38E-05 | M24594 | 20 315 | Human interferon-inducible 56 kDa protein mRNA; complete cds |
| 7 | 4.5±1.7 | 1.59E-05 | D14520 | 84 728 | Human mRNA for GC box binding protein BTEB2; complete cds |
| 7 | 4.4±2.1 | 1.36E-05 | J04759 | 183 994 | Human protein phosphatase I alpha subunit (PPPIA) mRNA; 3′ end |
| 7 | 5.0± 3.8 | 1.36E-03 | M55172 | 2159 | Human large aggregating cartilage proteoglycan core protein mRNA; complete cds |
| 7 | 6.6±4.7 | 5.87E-05 | Y00816 | 193 716 | Human mRNA for complement receptor type 1 (CR1, C3b/C4b receptor, CD35) |
| 7 | 4.4±3.0 | 4.36E-04 | AB014539 | 15 711 | H. sapiens mRNA for KIAA0639 protein; partial cds |
| 7 | 5.1±2.7 | 1.05E-04 | Z29373 | 1757 | H. sapiens gene for neural cell adhesion molecule L1 |
| 7 | 3.7±1.4 | 1.92E-04 | AF116238 | 23 723 | H. sapiens pseudouridine synthase 1 (PUS 1) mRNA; partial cds |
| 6 | 3.7±1.7 | 1.81E-04 | L09749 | 112 494 | H. sapiens (clone F4) transmembrane protein mRNA sequence |
| 6 | 4.2±2.3 | 3.24E-03 | M17254 | 45 514 | Human erg2 gene encoding erg2 protein; complete cds |
| 6 | 2.5± 0.5 | 3.70E-03 | U77086 | 117 367 | Human organic cation transporter 1 (hOCT1) mRNA; complete cds |
| 6 | 4.7±2.8 | 5.24E-04 | U97519 | 16 426 | H. sapiens podocalyxin-like protein mRNA; complete cds |
| 6 | 4.4±2.1 | 2.82E-05 | D88460 | 288 830 | H. sapiens mRNA for N-WASP; complete cds |
| 6 | 5.1±3.2 | 2.94E-05 | AF062075 | 49 587 | H. sapiens leupaxin mRNA; complete cds |
| 6 | 4.8±2.9 | 5.18E-05 | X77197 | 199 250 | H. sapiens mRNA for chloride channel |
| 6 | 4.4±3.0 | 1.55E-04 | X12830 | 193 400 | Interleukin 6 receptor |
| 6 | 4.8±3.0 | 1.46E-03 | X71355 | 100 001 | H. sapiens mRNA for sodium phosphate transport system 1 |
| 6 | 5.2±3.4 | 5.11E-04 | L20431 | 79 391 | H. sapiens Huntington disease-associated protein (HD) mRNA; complete cds |
| 6 | 2.5±0.3 | 1.39E-04 | L41351 | 75 799 | H. sapiens prostasin mRNA; complete cds |
| 6 | 3.3±1.1 | 1.38E-03 | U90441 | 3622 | Human prolyl 4-hydroxylase alpha (II) subunit mRNA; complete cds |
| 6 | 4.0±1.4 | 1.89E-04 | AF007216 | 5462 | H. sapiens sodium bicarbonate cotransporter (HNBC1) mRNA; complete cds |
| 6 | 3.1±0.7 | 3.28E-05 | U10689 | 37 108 | Human MAGE-5a antigen (MAGE5a) gene; complete cds |
| 6 | 5.4± 1.7 | 1.13E-04 | M35296 | 121 521 | Human tyrosine kinase arg gene mRNA |
| 6 | 3.7±1.9 | 3.07E-06 | M86752 | 75 612 | Human transformation-sensitive protein (IEF SSP 3521) mRNA; complete cds |
| 6 | 5.0± 2.3 | 1.06E-04 | X73502 | 84 905 | H. sapiens mRNA for cytokeratin 20 |
| 6 | 3.5±1.3 | 1.06E-04 | U68723 | 211 773 | Human checkpoint suppressor 1 mRNA; complete cds |
| 6 | 3.5±1.5 | 1.14E-04 | X52151 | 88 251 | H. sapiens arylsulphatase A mRNA; complete cds |
| 6 | 5.0±2.8 | 1.23E-03 | AF056929 | 50 550 | H. sapiens sarcosin mRNA; complete cds |
| 6 | 3.0±1.3 | 3.60E-06 | X98801 | 74 617 | H. sapiens mRNA for dynactin |
| 6 | 4.5±2.2 | 2.66E-05 | W25099 | 41 997 | ESTs, highly similar to alpha-1B-glycoprotein (H. sapiens) |
| 6 | 4.1±2.2 | 2.53E-04 | AF151351 | 274 287 | H. sapiens pyrroline 5-carboxylate reductase isoform (P5CR2) mRNA; complete cds |
| 6 | 3.5±1.1 | 8.38E-05 | X99688 | 154 658 | H. sapiens mRNA from TYL gene |
| 6 | 3.3±1.0 | 1.95E-04 | AF044953 | 31 547 | H. sapiens NADH:ubiquinone oxidoreductase PGIV subunit mRNA; nuclear gene encoding mitochondrial protein |
| 6 | 3.3±1.0 | 1.22E-05 | AL133558 | 55 209 | H. sapiens mRNA; cDNA DKFZp434K0514 (from clone DKFZp434K0514); partial cds |
| 6 | 4.6±2.4 | 1.39E-04 | M20543 | 1288 | Human skeletal alpha-actin gene; complete cds |
| 6 | 3.7±1.5 | 1.70E-04 | AL133068 | 62 880 | H. sapiens mRNA; cDNA DKFZp434B0717 (from clone DKFZp434B0717) |
Table 3.
Downregulated Genes.
| Frequency | Average Ratio±SD | P Value | Accession Number | UniGene Cluster | Definition |
| 6 | 0.3±0.2 | .0134 | AF033827 | 155 553 | Homo sapiens HNK-1 sulfotransferase mRNA; complete cds |
| 6 | 0.3±0.1 | .0142 | BC005071 | 332 382 | ESTs |
| 5 | 0.3±0.2 | .0135 | S42658 | 252 259 | S3 ribosomal protein (human, colon, mRNA, 826 nt) |
| 5 | 0.3±0.2 | .0061 | X72964 | 82 794 | H. sapiens mRNA for caltractin |
| 5 | 0.4±0.1 | .0142 | X99977 | 103 505 | Human autonomously replicating sequence (ARS) mRNA |
| 5 | 0.3±0.1 | .0227 | M81757 | 298 262 | 40S ribosomal protein S19 |
| 5 | 0.4±0.1 | .0318 | N54851 | 108 222 | ESTs |
Validation of Expression Differences by Quantitative PCR
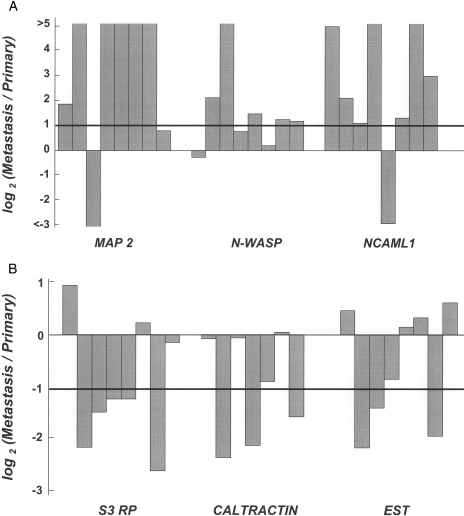
To verify the result of microarray, we selected three upregulated genes (MAP 2, N-WASP, and NCAML1) and three downregulated genes [EST (BC005071), S3 ribosomal protein, and caltractin], and performed quantitative RTPCR using additional eight pairs of CRCs. As shown in Figure 2, the frequencies of elevated expression of upregulated genes (6/8 cases of MAP 2, 5/8 of N-WASP, and 7/8 of NCAML1), and those of reduced expression of downregulated genes (5/8 cases of S3 ribosomal protein, 3/8 of caltractin, and 3/8 of the EST) were almost in good agreement with the data of microarray (8/10 cases of MAP 2, 6/10 of N-WASP, 7/10 of NCAML1, 5/10 of S3 ribosomal protein, 5/10 of caltractin, 6/10 of the EST).
Figure 2.
Quantitative RT-PCR analysis of three upregulated genes (A) and three downregulated genes (B). Relative expression in liver metastasis to primary lesion is indicated as log2 (Metastasis/Primary).
Discussion
Many molecular events, some involving acquisition of malignant properties, are involved in carcinogenesis. Among the various steps in the progression of cancer, metastasis is the one that critically affects the prognosis of individual patients. It is clear that multiple gene products must play essential roles. Consistent with that view, our comparison of gene expression profiles between primary CRCs and corresponding metastatic lesions disclosed altered expression of at least 47 genes among the 9121 genes examined.
A similar study, using oligonucleotide microarrays to screen melanoma cells representing different metastatic phenotypes but established from the same cell lines, disclosed a number of genes that might be associated with metastasis [5]. However, because the experiments involved injection of melanoma cells into the murine spleen and resection of the resulting tumors, genes involved specifically in invasion or intravasation theoretically would not be detected; the genes identified in the study were in fact much fewer than we detected in ours. A separate group also analyzed expression of various melanoma cell lines and tissues by microarray, and their data demonstrated expression profiles associated with different malignant properties such as growth, local invasion, and, of course, metastasis [6]. We chose a combined strategy; i.e., a direct comparison between primary cancer tissue and a metastatic lesion from the same patient, based on the hypothesis that the properties of primary and metastatic cells are different and/or that the proportion of cells with high metastatic potential is increased in metastatic foci. However, the genes selected in our study may include not only those representing the nature of cancer cells but also genes that were affected by metastasis as secondary events, e.g., by responding to changes in the local environment (liver versus colon). Investigations are ongoing to clarify the importance of the candidate genes and their actual roles, if any, in liver metastasis.
The commonly upregulated genes identified in our experiments included several whose products are associated with the cytoskeleton, e.g., MAP2, N-WASP, and dynactin. N-WASP, being tightly regulated by multiple signals including CDC42 and phosphatidylinositol (4,5)-biphosphate (PIP2), plays a role in the remodeling of the actin cytoskeleton [8]; dynactin, as a complex with cytoplasmic dynein, interacts directly with microtubules to form transporter vesicles and organelles [9]. These data suggest that microtubules and molecules involved in remodeling of the cytoskeleton may play crucial roles in metastatic events by changing cellular morphology during intra- and extravasation.
In our experiments, genes associated with cellular adhesion such as NCAML1, podocalyxin-like protein, and leupaxin were also upregulated; others have reported a significant increase in NCAM expression in aggressive melanomas with high metastatic potential [10]. Leupaxin contains three LD motifs, which have been implicated in binding to cytoplasmic protein tyrosine kinase and vinculin [11]; this protein may be engaged in a signal transduction pathway associated with cell adhesion. The combined data underscore the importance of adhesion molecules in the process of metastasis. Moreover, genes with elevated expression may be good candidates to serve as metastatic markers for clinical diagnosis and are potential molecular targets for treatment and/or prevention of metastases.
Genes that were commonly downregulated in the metastatic lesions studied here included HNK-1 and caltractin. The HNK-1 epitope, a cell surface carbohydrate, has been implicated in cell-to-cell and cell-to-extracellular matrix adhesion [12]; its expression was suppressed in liver metastases of melanomas [10]. Moreover, HNK-1 expression has shown a positive correlation with differentiation of prostate tumors and survival of patients with prostate cancer [13]. These data suggest an antimetastasizing potential of HNK-1. Caltractin, for its part, shows a strong similarity in amino acid sequence to CDC31 — a protein that is required for proper duplication and segregation of the centrosome, a major microtubuleorganizing center [14]. Therefore, reduced expression of the caltractin gene may contribute to the malignant nature of cancer cells by abrogating duplication.
In addition to known and well characterized genes, the lists reported here contain various ESTs and uncharacterized genes. Investigation of their functions may shed light on the detailed mechanisms of liver metastasis and yield clues for identification of more reliable and sensitive diagnostic markers and more effective therapeutic modalities.
Acknowledgements
We are grateful to Norihiko Shiraishi, Hideaki Ogasawara, Kenji Hirotani, Akihiko Saito-Hisaminato, Hiroko Bando, Noriko Nemoto, and Noriko Sudo for fabrication of the CDNA microarray; to Tae Makino for preparation of samples by cryostat; to Marcelo Eidi Nita, Seiji Sato, and Hiroshi Okabe for surgical and pathological advice; to Toyokazu Seki for technical advice; to Hideyuki Suzuki and Jun-ichi Okutsu for technological advice; and to Meiko Takahashi for preparation of the manuscript.
Abbreviations
- aRNA
amplified RNA
- EST
expressed sequence tag
- CRC
colorectal cancer
- LCM
laser capture microdissection
Footnotes
Supported, in part, by Research for the Future Program grant 00L01402 from the Japan Society for the Promotion of Science.
References
- 1.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 2.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 3.Kitahara O, Furukawa Y, Tanaka T, Kihara C, Ono K, Yanagawa R, Nita ME, Takagi T, Nakamura Y, Tsunoda T. Alterations of gene expression during colorectal carcinogenesis revealed by CDNA microarrays after laser capture microdissection of tumor tissues and normal epithelia. Cancer Res. 2001;61:3544–3549. [PubMed] [Google Scholar]
- 4.Okabe H, Satoh S, Kato T, Kitahara O, Yanagawa R, Yamaoka Y, Tsunoda T, Furukawa Y, Nakamura Y. Genome-wide analysis of gene expression in human hepatocellular carcinomas using CDNA microarray: identification of genes involved in viral carcinogenesis and tumor progression. Cancer Res. 2001;61:2129–2137. [PubMed] [Google Scholar]
- 5.Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–535. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 6.Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, et al. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406:536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 7.Ono K, Tanaka T, Tsunoda T, Kitahara O, Kihara C, Okamoto A, Ochiai K, Takagi T, Nakamura Y. Identification by CDNA microarray of genes involved in ovarian carcinogenesis. Cancer Res. 2000;60:5007–5011. [PubMed] [Google Scholar]
- 8.Prehoda KE, Scott JA, Dyche Mullins R, Lim WA. Integration of multiple signals through cooperative regulation of the N-WASP-Arp2/3 complex. Science. 2000;290:801–806. doi: 10.1126/science.290.5492.801. [DOI] [PubMed] [Google Scholar]
- 9.Holzbaur EL, Tokito MK. Localization of the DCTN1 gene encoding p150Glued to human chromosome 2p13 by fluorescence in situ hybridization. Genomics. 1996;31:398–399. doi: 10.1006/geno.1996.0068. [DOI] [PubMed] [Google Scholar]
- 10.Mooy CM, Luyten GP, de Jong PT, Jensen OA, Luider TM, van der Ham F, Bosman FT. Neural cell adhesion molecule distribution in primary and metastatic uveal melanoma. Hum Pathol. 1995;26:1185–1190. doi: 10.1016/0046-8177(95)90191-4. [DOI] [PubMed] [Google Scholar]
- 11.Lipsky BP, Beals CR, Staunton DE. Leupaxin is a novel LIM domain protein that forms a complex with PYK2. J Biol Chem. 1998;273:11709–11713. doi: 10.1074/jbc.273.19.11709. [DOI] [PubMed] [Google Scholar]
- 12.Ong E, Yeh JC, Ding Y, Hindsgaul O, Fukuda M. Expression cloning of a human sulfotransferase that directs the synthesis of the HNK-1 glycan on the neural cell adhesion molecule and glycolipids. J Biol Chem. 1998;273:5190–5195. doi: 10.1074/jbc.273.9.5190. [DOI] [PubMed] [Google Scholar]
- 13.Liu XH, Yoshiki T, Kokuho M, Okada Y, Tomoyoshi T, Higuchi K. The prognostic value of the HNK-1 (Leu-7) antigen in prostatic cancer — an immunohistochemical study. Hinyokika Kiyo. 1993;39:439–444. [PubMed] [Google Scholar]
- 14.Lee VD, Huang B. Molecular cloning and centrosomal localization of human caltractin. Proc Natl Acad Sci USA. 1993;90:11039–11043. doi: 10.1073/pnas.90.23.11039. [DOI] [PMC free article] [PubMed] [Google Scholar]