Abstract
Checkpoint kinase 1 (Chk1) is a checkpoint gene that is activated after DNA damage. It phosphorylates and inactivates the Cdc2 activating phosphatase Cdc25C. This in turn inactivates Cdc2, which leads to G2/M arrest. We report that blocking Chk1 expression by antisense or ribozymes in mammalian cells induces apoptosis and interferes with the G2/M arrest induced by adriamycin. The Chk1 inhibitor UCN-01 also blocks the G2 arrest after DNA damage and renders cells more susceptible to adriamycin. These results indicate that Chk1 is an essential gene for the checkpoint mechanism during normal cell proliferation as well as in the DNA damage response.
Keywords: Chk1, antisense, ribozyme, checkpoint, chemotherapy sensitization
Introduction
Checkpoint kinase 1 (Chk1), initially identified in Schizosaccharomyces pombe [1], is a G2/M checkpoint gene that is conserved throughout the eukaryotic kingdom. Checkpoint genes are activated after DNA damage. The sensors of DNA damage send out two signals. The first signal activates checkpoint pathways that halt the cell cycle in either G1 or G2. The second signal leads to the activation of DNA repair [2].
In S. pombe or mammalian cells the cyclin B-Cdc2 kinase complex controls cell cycle progression through the G2/M boundary. Cdc2 is inactivated through phosphorylation at Tyr-15 by either Wee1 or Myt1. The phosphatases Cdc25 (S. pombe) or Cdc25C (mammalian) can remove the inhibitory phosphate from Tyr-15 on Cdc2 and permit cell cycle progression [3–8].
The function of Chk1 has been studied in both S. pombe and mammalian cells and found to be roughly analogous. In S. pombe, after DNA damage, Chk1 induces cell cycle arrest but is not involved in the activation of DNA repair. The Δchk1 strain of S. pombe is viable under normal conditions but is not viable when DNA is damaged. This strain is also defective in delaying mitosis following γ-irradiation in a wee1-50/Δmik1 background (Mik1 phosphorylate Cdc2 at Tyr-15 and inactivates it in S. pombe) [9]. S. pombe Chk1 is phosphorylated in a Rad3-dependent manner shortly after DNA damage [10]. Active Chk1 phosphorylates Cdc25, which provides a binding site for the 14-3-3 family protein Rad24. Once bound by Rad24, Cdc25 is exported out of the nucleus [11]. However, the nuclear exclusion of Cdc25 is not required for the DNA damage response and direct inhibition of Cdc25 phosphatase activity by Chk1 may be sufficient for the cell cycle arrest [12,13]. As a result, Cdc2 is kept inactive by Wee1 and cells are arrested in G2. Chk1 has also been shown to phosphorylate Wee1 in vitro, implying that Chk1 may also facilitate G2 arrest through Wee1 [14,15].
An analogous checkpoint pathway involving ATM/ATR, Chk1, and Cdc25C has been identified in mammalian cells [16–18]. In Xenopus, ATR is required for Chk1 phosphorylation in the SQ/TQ motifs. This phosphorylation is required for DNA damage response [19]. After DNA damage, human Chk1 phosphorylates Cdc25C at Ser-216. As a result, Cdc2 is not activated and cells are arrested at G2. Hypersensitivity to radiotherapy is observed in ataxia telangiectasia patients (ATM deficient). Cells from these patients (A–T cells) are also hypersensitive to γ-irradiation in vitro. Expression of S. pombe Chk1 in A–T cells complements the defects in the G2 DNA damage checkpoint and improves the survival rate of these cells after γ-irradiation [20]. Recently, Chk1 has been reported to stabilize Cdc25A and p53 during the DNA damage response [21,22]. Therefore, Chk1 could also reinforce the G1 checkpoint.
A second mechanism, distinct from Chk1, which can phosphorylate Cdc25/Cdc25C has been identified in yeast and mammals. The S. pombe Cds1/S. cerevisiae Rad53/human Chk2 play roles similar to Chk1 downstream of MEC1/Rad3/ATM in G2 checkpoint control. [23–25]. Cdc25C can also be phosphorylated by c-TAK1, the physiological function of which is not known [26].
Cdc25C phosphorylation at serine 216 is tightly regulated and controls the timing of mitosis during the normal cell cycle. During interphase, Cdc25C is phosphorylated, bound by 14-3-3 proteins, and exported out of the nucleus. In mitosis, dephosphorylated Cdc25C remains in the nucleus where it activates Cdc2.
Recent reports have shown that CHK1-/- mouse embryos or ES cells exhibit a defect in the G2 checkpoint mechanism. [18,27]. In this report, we blocked Chk1 expression and examined the phenotype in human cell tumor cell lines. We used several approaches including full-length antisense cDNA, ribozymes, and antisense oligonucleotides to block Chk1 expression. Full-length Chk1 antisense and one of the ribozymes were the most effective in blocking Chk1 expression in human cell lines. Cells with reduced levels of Chk1 were found to be more prone to apoptosis. Furthermore, when treated with DNA-damaging agents such as adriamycin or etoposide, cells harboring low levels of Chk1 protein exhibited defects in the G2/M checkpoint. We also observed an additive cell-killing effect when treating cells with Chk1 antisense/ribozyme together with adriamycin. In addition, we showed that potent Chk1 inhibitor UCN01 was able to override the adriamycininduced G2 arrest. Furthermore, we found that UCN01 sensitized the human lung carcinoma cell line H1299 to adriamycin.
Materials and Methods
Ribozyme Design and Plasmid Construction
Ribozymes are enzymatic RNA molecules that cleave the targeted mRNA [28]. The Chk1 mRNA secondary structure was analyzed using the MFold program/GCG software [Wisconsin Package Version 10.0, Genetics Computer Group (GCG), Madison, WI]. Ribozymes were designed to hybridize to the putative single-stranded regions. The sequences of these ribozymes are: ribo910: AG-CTTCTTCTCCActgatgaggccgaaaggccgaaaGGCACCTTT; ribo1112: AGCTTTCTTCAGTctgatgaggccgaaaggccgaaaCTCTATTCT; ribo1314: AGCTTATCCCTGTctgatgaggccgaaaggccgaaaGTTATTCCT; ribo1516: AGCTTCTGAGATTTTctgatgaggccgaaaggccgaaaGGTTATCCT. The uppercase letters represent the regions that hybridize to the Chk1 coding region. The lowercase letters represent the ribozyme catalytic sequence. The ribozymes, full-length Chk1 sense cDNA and antisense cDNA were cloned into a pCMV vector between HindIII and XbaI sites. Clones were confirmed by sequencing.
Cell Lines, Chemicals and Reagents
NCI-H1299, HCT116, and Hela cells were obtained from ATCC. UCN-01 was obtained from the Synthetic Chemistry Department at the National Cancer Institute, Bethesda, MD. Nocodazole was purchased from Calbiochem, San Diego, CA. Unless otherwise stated all other chemicals were purchased from Sigma. The following rabbit polyclonal antibodies were purchased from Santa Cruz (La Jolla, CA): Chk1 (C-16; Cat#: sc-7234); actin (I-19; Cat#: sc-1616).
Transient Transfection
H1299 cells were grown at 37°C in a 5% CO2 atmosphere in RPMI 1640 supplemented with 10% fetal bovine serum, 1 mM sodium pyruvate, 1 mM Hepes, 1 mM glutamine, and 4 g l-1 glucose. Transfection with LipofectAMINE PLUS DNA (1.5 µg ml-1) was mixed with LipofectAMINE PLUS (Life Technology, Rockville, MA, Cat#: 10964-013) for 15 minutes at room temperature. This mixture was then mixed with LipofectAMINE reagent and incubated for 15 minutes at room temperature to form DNA complexes. The DNA complexes were added to cells in RPMI media without serum. After a 4-hour incubation at 37°C, the transfection mixture was replaced with normal growth media. Transfection with LipofectAMINE 2000. The appropriate DNA (1.5 µg ml-1 unless otherwise specified) was mixed with LipofectAMINE 2000 (Life Technology, Cat#: 11668-027) for 15 minutes at room temperature to form DNA complexes. The DNA complexes were added to each tissue-culture plate with the complete media. After a 6-hour incubation at 37°C, the transfection mixture was replaced with normal growth media.
Western Blotting
Cells were lysed in lysis buffer [20 mM Tris-Cl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 1% Na deoxycholate, 0.1% SDS, 1 mM DTT, 0.25 mM PMSF, 1x protease inhibitor cocktail (Boehringer Mannheim, Germany, Cat# 1697498)]. After brief sonication, insoluble debris were pelleted (14,000 rpm, 30 minutes, 4°C) and the protein concentration of the resulting supernatant was determined using the Bradford method. Total cell lysates (100 µg protein) were loaded onto SDS-gradient gel (Novex, San Diego, CA), electrophoresed, and electrotransferred to 0.2 µ nitrocellulose membranes (Novex). Antibody detecting was carried out according to the manufacturer's instructions (Amersham Pharmacia Biotech, IL).
Apoptosis Assay
Both the floating and adherent cells were harvested, washed, and resuspended in 1.0 ml of ice-cold phosphatebuffered saline (PBS) containing 2 µg ml-1 4′,6-diamidino-2-phenylindole (DAPI). The cells were rocked for 1 hour at room temperature in the dark and then washed two times with PBS after staining. The resuspended sample (15 µl) was dropped onto a glass slide, covered with a glass coverslip and apoptotic cells were scored. For each sample, at least 600 cells were randomly counted by fluorescence microscopy and the percentage of apoptotic cells was determined based on evidence of nuclear fragmentation and abnormal chromosome condensation.
Alamar Blue Assay
Twenty-four hours after transfection, the transfected cells were trypsinized and re-plated onto a 96-well tissue culture dish at 7500 cells per well. Cells were allowed to grow in complete media at 37°C for 48 hours. The cells were then gently washed with 200 µl of PBS. Alamar Blue reagent (Biosource International, Camarillo, CA; Cat#: DAL1100) was diluted 1/10 in normal growth media. The diluted Alamar Blue reagent (100 µl) was added to each well on the 96-well plate and incubated until the reaction was complete as per manufacturer's instructions. Analysis was performed using an fmax Fluorescence Microplate Reader (Molecular Devices, Sunnyvale, CA), set at the excitation wavelength of 544 nm and emission wavelength of 595 nm. Data was analyzed using SOFTmax PRO software provided by the manufacturer.
Flow Cytometry
Cells were harvested, washed, and resuspended in 500 µl of ice-cold PBS. Five milliliters of 100% ethanol was added to fix cells at 4°C overnight. Cells were then washed once with ice-cold PBS plus 0.1% bovine serum albumin (BSA) and resuspended in 800 µl of ice-cold PBS+0.1% BSA. Resuspended cells were treated with RNase A (1 mg ml-1) and stained with propidium iodide (100 µg ml-1) for 30 minutes at 37°C. Cell cycle analysis was performed using a Becton-Dickinson (San Jose, CA) fluorescence-activated cell analyzer. Data were analyzed using the ModFitLT model provided by the manufacturer. Results represent a minimum of 10,000 cells assayed for each sample.
Caspase Assay
H1299 cells were transfected with pCMV vector, Chk1 full-length antisense cDNA or ribo910. Twenty-four hours after transfection, the cells were split onto 96-well plates at 7500 cells/well for additional 24-hour incubation. The cells were lysed in 120 µl of lysis buffer [16 mM Hepes-KOH, pH 7.5, 67 mM KCL, 8 mM MgCl2, 0.22 mM PMSF, 1.11 mM DTT, 1.11 µg ml-1 pepstatin A, 1.1 µg ml-1 leupeptin, 5.56 µg ml-1 aprotonin, 0.11 mM EGTA, 0.11 mM EDTA, 0.56% CHAPS (wt/vol)] for 20 minutes. ICE buffer (80 µl) was mixed with the cell lysates and the fluorescence was measured with a Cytofluor fluorometer (PerSeptive Biosystems (Foster City, CA) excitation=360/40, emission=460/40, gain=38, cycle=1) at 0 and 3 hours after incubation at 37°C. Caspase activity was calculated as units per hour by substracting the 0-hour reading from the 3-hour reading and dividing by 3.
Mitotic Index
Twenty-four hours after transfection, H1299 cells were treated for 8 hours with 300 ng ml-1 adriamycin or 20 µg ml-1 etoposide followed by an additional 16-hour incubation together with 40 ng ml-1 nocodazole (Calbiochem, San Diego, CA; Cat#: 487928). Cells were harvested by trypsinization, washed with PBS, centrifuged (1200 rpm, 3 minutes), and then resuspended in 0.5 ml of hypotonic buffer containing 75 mM KCl. Cells were centrifuged immediately (1200 rpm, 3 minutes) and again resuspended in 100 µl of hypotonic buffer. Samples were dropped onto glass slides, air-dried, and stained with the Diff-Qwik Stain Set (Dade, Deerfield, IL; Cat#: B4132-1) according to the vendor's instructions. For each sample, at least 600 cells were randomly counted by light microscopy and mitotic cells were scored based on evidence of mitotic chromosomal condensation and the lack of a nuclear membrane.
Results
Chk1 is Required for the Checkpoint Mechanism During the Normal Cell Cycle
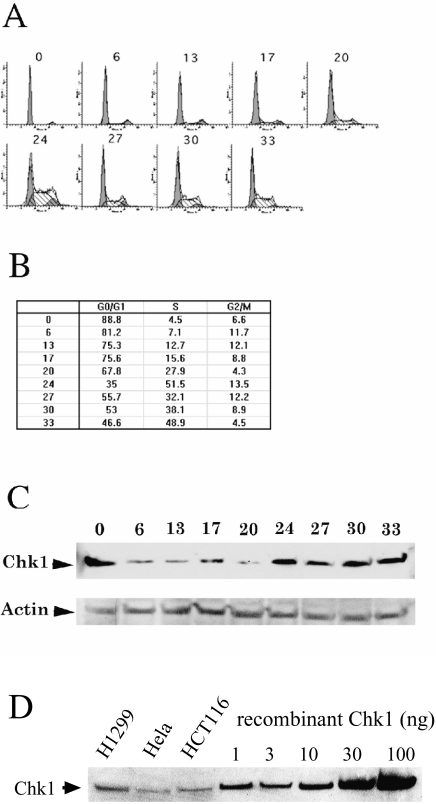
We examined the Chk1 expression profile during the cell cycle progression. H1299 cells were treated with mevastatin for 24 hours to achieve G1 synchronization. Cells were released from the mevastatin arrest and harvested at different time points. The Chk1 protein level was analyzed using Western blotting techniques and the cell cycle distribution was examined using fluorescence-activated cell sorting analysis (FACS). The Chk1 antibody recognizes the recombinant Chk1 expressed in baculovirus, which comigrates with the recognized antigen in the total cell lysates (Figure 1D). The majority of cells progressed into the S phase within 20 to 24 hours of mevastatin release. Chk1 protein levels were low during G1 and elevated during S and later (Figure 1A–C). The same cell cycle-dependent Chk1 expression pattern was seen in H1299 cells synchronized by contact inhibition (data not shown). The Chk1 level is high when the cells are arrested in G1 by either mevastatin (Figure 1) or contact inhibition (data not shown), suggesting that Chk1 could be upregulated in response to G1 arrest induced by these treatments and downregulated at G1 during normal cell cycling. In contrast to our findings, Kaneko et al. [29] showed that Chk1 was downregulated when the cells were in quiescence after serum starvation. This difference could be due to different treatments or differences in cell lines unique to each experimental condition. These results suggest a function for Chk1 in the S or the G2/M phases of the cell cycle. Indeed, it has been reported that Chk1 kinase activity is upregulated during the S and the G2/M phases [29]. To study Chk1 function, we used several approaches to block Chk1 expression in mammalian cells. We transfected vectors that expressed full-length sense or antisense Chk1 cDNA, antisense oligonucleotides, or ribozymes (Figure 2A) in H1299 cells using LipofectAMINE PLUS. Approximately 80% of the cells were transfected as judged by cotransfection with a green florescent protein expression vector (data not shown). Total RNA was extracted and real-time quantitative RT-PCR was used to measure the Chk1 mRNA level in the adherent cells. Full-length Chk1 sense and antisense cDNAs were expressed at 700- to 900-fold over the endogenous Chk1 mRNA level (data not shown).
Figure 1.
Chk1 expression during the cell cycle. Cells were harvested 0, 6, 13, 17, 20, 24, 27, 30, and 33 hours post mevastatin release and subjected to flow cytometry and western blot analysis as described. (A) FACS analysis. (B) The quantification of the FACS analysis. (C) Western blot of total cell lysates with Chk1 antibody or actin antibody. (D) Western blot of baculovirus-expressed recombinant Chk1 and total cell lysate from NCI-H1299, Hela and HCT116 cells.
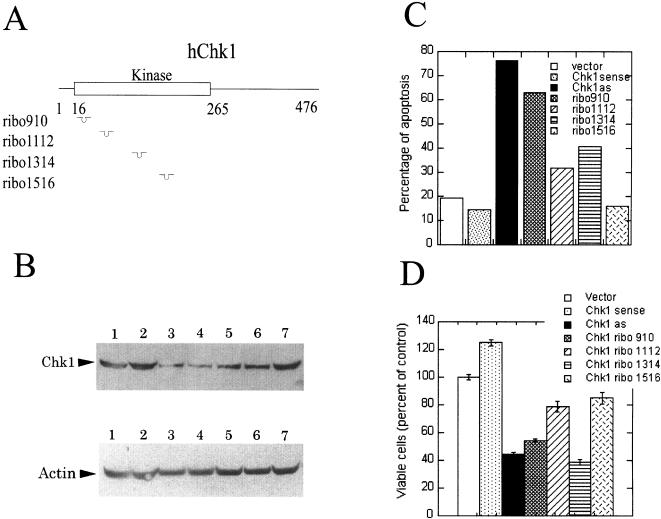
Figure 2.
Blocking Chk1 expression induces cell death. (A) Cartoon of human Chk1 gene and the hybridization positions for different ribozymes. (B) Chk1 protein levels in the transfected cells. H1299 cells were transfected with different plasmids as indicated below using LipofectAMINE PLUS. Western analysis of the total cell lysates was performed as described. The filter was blotted with anti-Chk1 antibody (top panel followed by blotting with anti-actin antibody (lower panel. Lane 1: vector control; lane 2: full-length Chk1 sense cDNA; lane 3: full-length Chk1 antisense cDNA; lane 4: ribozyme 910; lane 5: ribozyme 1112; lane 6; ribozyme 1314; lane 7: ribozyme 1516. (C) Blocking Chk1 expression induces cell death. Cells were harvested 24 hours after transfection. The apoptosis assay was carried out as described. (D) Blocking Chk1 expression decreases cell survival. H1299 cells were transfected with different plasmids as indicated. An Alamar Blue assay was carried out to determine the cell survival.
In order to be able to observe a decrease in Chk1 protein levels in transient transfection experiments, Chk1 must have a relatively short half-life. We measured the half-life of Chk1 to be 3 to 5 hours (data not shown). Western analysis revealed that the expression of Chk1 sense cDNA resulted in a two-fold increase in Chk1 protein levels (Figure 2B). The full-length Chk1 antisense cDNA reduced the Chk1 protein level to 30% to 50% of that of the empty vector control. Ribozyme 910 (ribo910) was as effective as full-length Chk1 antisense cDNA in reducing Chk1 expression. Ribozymes 1112 and 1314 had intermediate blocking effects (Figure 2B). We also transfected Chk1 antisense oligonucleotides into H1299 cells. We found that four of the 14 Chk1 antisense oligonucleotides were effective in reducing Chk1 expression (data not shown).
S. pombe cells with the Δchk1 allele are viable [9]. In contrast, we observed apoptosis in H1299 cells following antisense or ribo910 treatment. The extent of cell death correlates inversely with the Chk1 protein levels: full-length Chk1 antisense cDNA and ribo910 were the most effective in reducing Chk1 protein levels and they were also the most potent in inducing apoptosis. Ribozyme 1112 and 1314 had intermediate effects in reducing Chk1 expression and the extent of apoptosis was concomitantly intermediate. Ribozyme 1516 did not reduce the Chk1 protein level and had no effect on cell survival. Importantly, a two-fold overexpression of Chk1 protein in Chk1 sense cDNA transfectants appeared to moderately protect cells from apoptosis (Figure 2C). A direct correlation was observed between the Chk1 protein levels and the amount of viable cells (Figure 2D). Similar results were obtained in Hela cells (data not shown). We also observed similar effects at 48 hours posttransfection after removal of the dead cells at 24 hours posttransfection (data not shown).
The dramatic cell death induced in the LipofectAMINE PLUS-transfected cells masked the effect of DNA-damaging agents. Therefore, we used LipofectAMINE 2000 in the following experiments. Although the reduction of Chk1 protein levels and the extent of apoptosis were less pronounced using LipofectAMINE 2000 (compare Figures 3 and 6A to Figure 2), the milder condition allowed us to better examine the effect of DNA damage. We first characterized the apotosis induced by Chk1 antisense and ribo910 transfection. FACS analysis demonstrated that the antisense cDNA and ribo910-transfected cells had a significantly higher distribution in apoptosis than the vector-transfected cells. Additionally, there is a significant increase in caspase activity in the antisense cDNA and ribo910 transfected cells, indicating that cells die through apoptosis (Figure 3D). Furthermore, we consistently observed a moderate increase in the G2/M population of Chk1 sense-transfected cells (Figure 3B and C). This suggests that Chk1 is involved in the regulation of the timing of mitosis onset during the normal cell cycle. Active Chk1 keeps cells in check before they are ready for mitosis. As a consequence of decreasing Chk1 protein level, cells may die of premature mitosis.
Figure 3.
Blocking Chk1 expression induces cell death. H1299 cells were transfected with different plasmids as indicated using LipofectAMINE 2000. (A) Chk1 protein levels in the transfected cells. Lane 1: vector control; lane 2: full-length Chk1 sense cDNA; lane 3: full-length Chk1 antisense cDNA; lane 4: ribozyme 910. (B) FACS analysis of the transfected cells. H1299 cells were transfected with different plasmids as indicated. Twenty-four hours after transfection, both the floating and the attached cells were harvested and subjected to FACS analysis. (C) The quantification of the FACS analysis. (D) Caspase assay. H1299 cells were transfected with different plasmids as indicated. Fourty-eight hours after transfection, caspase activity was measure as described in the Materials and Methods section.
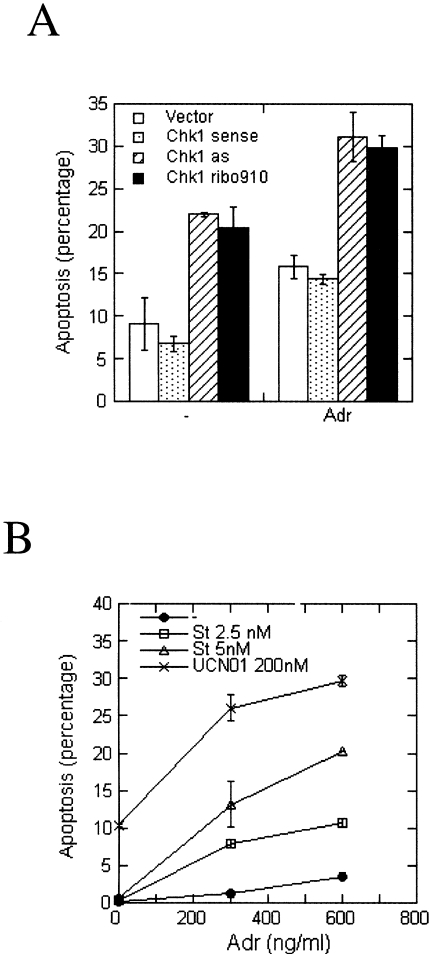
Figure 6.
(A) Blocking Chk1 expression and adriamycin have an additive effect in killing tumor cells. Twenty hours after transfection, the transfected cells were treated with or without adriamycin for 48 hours. The cells were harvested and subjected to the apoptosis assay. The percentage of apoptotic cells is shown. Each value is the average of values from two independent experiments. (B) UCN-01 or staurosporine sensitizes H1299 cells to adriamycin. H1299 cells were treated with different amounts of adriamycin in combination with UCN-01 or staurosporine as indicated in the figure. Forty-four hours after the treatment, the cells were harvested and subjected to the apoptosis assay. The percentage of apoptotic cells is plotted against the concentration of adriamycin. Each value is the average from two independent experiments.
We have shown that Chk1 expression is cell cycle-dependent and that Chk1 antisense or ribo910 can induce apoptosis. Taken together, these results indicate that Chk1 function is essential for normal cell cycle progression.
Chk1 is Required for the G2 Arrest after DNA Damage
We used adriamycin and etoposide to examine the DNA damage response in Chk1 antisense cDNA or ribo910-transfected cells. Adriamycin induces a G2 block in H1299 cells (Figure 5). We examined the effect of Chk1 antisense and ribo910 on the G2 checkpoint mechanism. The transfected cells were treated with 200 ng ml-1 adriamycin 24 hours after transfection to induce a DNA damage response. Eight hours after the addition of adriamycin, 40 ng ml-1 nocodazole was added to trap in mitosis those cells that had escaped the G2 arrest. The mitotic indices of cells that were transfected with Chk1 antisense cDNA or ribozyme 910 were much higher than those of cells transfected with either vector control or Chk1 sense cDNA (Figure 4). This result indicates that more antisense or ribozyme 910 transfectants progressed through the G2/M boundary without being arrested. Thus, the decreased Chk1 protein level results in a defect in G2 arrest after DNA damage. A similar result was obtained when the cells were treated with another DNA-damaging agent, etoposide (Figure 4).
Figure 5.
Staurosporine or UCN-01 overrides the G2 arrest induced by adriamycin. (A) FACS analysis. H1299 cells were treated with 300 ng ml-1 adriamycin for 24 hours with or without the following drugs: 1 nM staurosporine, 3 nM staurosporine, or 200 nM UCN-01. (B) The quantification of the FACS analysis. (C) H1299 cells were treated with 300 ng ml-1 adriamycin for 8 hours before the addition of 40 ng ml-1 nocodazole with or without 5 nM staurosporine or 200 nM UCN-01. Mitotic indices were measured as described. The percentage of mitotic cells is plotted. Each value is the average from two independent experiments.
Figure 4.
Blocking Chk1 expression allows cells to partially escape the G2 arrest induced by adriamycin or etoposide. The mitotic indices of the transfected cells were measured as described. (A) Morphology of the transfected cells treated with drugs. Cells with the typical mitotic chromosomal spread and the disappearance of nuclear membrane are marked with arrows. (B) The mitotic indices of the cells. The value of each data point is the average from two independent experiments.
UCN01 (7-hydroxy staurosporine) has been shown to specifically inhibit Chk1 and abrogate G2 checkpoint. UCN-01 also inhibits c-TAK1. However, the physiological function of c-TAK1 in G2/M checkpoint is not clear [30]. Both UCN-01 and staurosporine inhibit Chk1 with an IC50 of 4 to 10 nM [30–33]. We looked at the effect of UCN-01 or staurosporine on the G2 arrest induced by adriamycin. H1299 cells were treated with 300 ng ml-1 adriamycin for 8 hours before 40 ng ml-1 nocodazole was added to trap the mitotic cells. Cells treated with either adriamycin alone, or adriamycin plus nocodazole had very low mitotic indices, presumably because they were arrested in late G2 by adriamycin. UCN-01 (200 nM) or 5 nM staurosporine alone had no effect on mitotic index. However, H1299 cells treated first with adriamycin followed by treatment with staurosporine or UCN-01 plus nocodazole, showed a significant increase in the mitotic index. This result indicates that UCN-01 and staurosporine can override the G2 arrest induced by adriamycin (Figure 5C).
The cell cycle distribution was analyzed using FACS. H1299 cells were treated with 300 ng ml-1 adriamycin for 24 hours with or without the following drugs: 1 nM staurosporine, 3 nM staurosporine, or 200 nM UCN-01. Treatment with adriamycin alone induced a G2 arrest in 80% of the cells. However, we observed a significant decrease in the percentage of G2 cells upon addition of UCN-01 or staurosporine to adriamycin-treated cells. In addition, we observed that the S-phase fraction increased, which indicates that more cells were cycling as a result of UCN-01 or staurosporine addition. UCN-01 or staurosporine alone can induce G1 arrest [34,35]. In our experiment, the S-phase fraction increased suggesting that the decrease in G2 fraction was not due to G1 arrest caused by UCN-01 or staurosporine. In fact, UCN-01/staurosporine treatment was able to override the adriamycin-induced G2 arrest and pushed more cells into active cycling (Figure 5A and B). These results are consistent with Chk1 function indicated by the Chk1 antisense or ribo910 experiments.
Chk1 Is Required for Cell Survival after DNA Damage
In the event of DNA damage, cells are halted in either G1 or G2 to repair DNA. This is vital for maintaining genetic fidelity as well as for the survival of the cells. Thus, it is expected that abrogation of these checkpoint mechanisms would decrease cell survival. We examined the apoptotic rate of transfected cells upon treatment with adriamycin. We found that treatment with 600 ng ml-1 adriamycin killed H1299 cells in addition to the killing effect of Chk1 antisense cDNA or ribo910 (Figure 6A).
We also examined the cell survival rate of cells treated with adriamycin in the presence or absence of UCN-01 or staurosporine. H1299 cells were treated with different amounts of adriamycin in combination with UCN-01 or staurosporine. Forty-four hours after the treatment, the cells were harvested and subjected to the apoptosis assay. Both UCN-01 and staurosporine increased the efficacy of adriamycin against H1299 cells. The potentiation by staurosporine showed a dose-response effect (Figure 6B). Therefore, inhibition of Chk1 could sensitize tumor cells to chemotherapeutic agents.
Discussion
We have shown that blocking Chk1 expression induces apoptosis, abrogates G2 arrest after DNA damage, and sensitizes cells to DNA-damaging agents. This is in agreement with the recent reports showing that CHK1-/- mouse embryos or ES cells also exhibit a defect in the G2 checkpoint mechanism [18,27]. These data provide genetic evidence that Chk1 is essential for the G2 checkpoint mechanism during normal cell cycle as well as for the DNA damage response.
In our experiments, cells transfected with the full-length Chk1 antisense or ribo910 partially abrogated the G2 arrest induced by either adriamycin or etoposide. The incomplete G2 abrogation could be explained by two factors: (1) the presence of the untransfected cells; (2) G2 arrest is largely dependent on Cdc25C activity and Cdc25C can be phosphorylated and inhibited by both Chk1 and Chk2. Chk2 could compensate for the loss of Chk1 and induce G2 arrest after DNA damage [23–25].
More than 50% of tumors have mutations in p53. Furthermore, almost all have G1 checkpoint deficiencies [36]. Cells deficient in p53 cannot undergo p53-dependent apoptosis and are resistant to drugs that induce the p53-dependent apoptosis. In fact, such resistance has been reported in many p53 null or mutated human tumor cell lines as well as in clinical samples [37–41]. We have shown that Chk1 antisense or ribo910 induces apoptosis in H1299 cells that are deficient in p53. Thus, Chk1 inhibitors could provide a cytotoxic cancer therapeutic agent, especially for p53-deficient tumors.
It has been shown that G2 abrogation can sensitize p53-deficient cells to DNA-damaging cancer therapeutic agents [33,42–44]. In the present report, we show that decreasing the Chk1 protein level by using antisense or ribo910 sensitizes H1299 cells to adriamycin. Thus, we expect that an inhibitor of Chk1 should sensitize tumors to DNA-damaging cancer therapies. In fact, UCN01 or staurosporine sensitizes H1299 cells to adriamycin, presumably due to its G2 abrogation activity. Thus, Chk1 inhibition may provide a useful and specific approach for the treatment of cancer.
Acknowledgements
We thank Dr. Perry Nisen for helpful discussions. We also thank Jason Zizter and Kenneth B. Idler for sequencing the constructs.
Abbreviations
- Chk1
checkpoint kinase 1
- ribo910
ribozyme 910
- S. cerevisiae
Saccharomyces cerevisiae
- S. pombe
Schizosaccharomyces pombe
References
- 1.Walworth N, Davey S, Beach D. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature. 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- 2.Paulovich AG, Toczyski DP, Leland HH. When checkpoints fail. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 3.O'Connor PM, Ferris DK, Hoffmann I, Jackman J, Draetta G, Kohn KW. Role of the cdc25C phosphatase in G2 arrest induced by nitrogen mustard. Proc Natl Acad Sci USA. 1994;91:9480–9484. doi: 10.1073/pnas.91.20.9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watanabe N, Broome M, Hunter T. Regulation of the human WEE1Hu CDK tyrosine 15-kinase during the cell cycle. EMBO J. 1995;14:1878–1891. doi: 10.1002/j.1460-2075.1995.tb07180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGowan CH, Russell P. Human Wee1 kinase inhibits cell division by phosphorylating p34cdc2 exclusively on Tyr15. EMBO J. 1993;12:75–85. doi: 10.1002/j.1460-2075.1993.tb05633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunphy WG, Kumagai A. The cdc25 protein contains an intrinsic phosphatase activity. Cell. 1991;67:189–196. doi: 10.1016/0092-8674(91)90582-j. [DOI] [PubMed] [Google Scholar]
- 7.Strausfeld U, Labbe JC, Fesquet D, Cavadore JC, Picard A, Sadhu K, Russell P, Doree M. Dephosphorylation and activation of a p34cdc2/cyclin B complex in vitro by human CDC25 protein. Nature. 1991;351:242–245. doi: 10.1038/351242a0. [DOI] [PubMed] [Google Scholar]
- 8.Rhind N, Furnari B, Russell P. Cdc2 tyrosine phosphorylation is required for the DNA damage checkpoint in fission yeast. Genes Dev. 1997;11:504–511. doi: 10.1101/gad.11.4.504. [DOI] [PubMed] [Google Scholar]
- 9.Furnari B, Rhind N, Russell P. Cdc25 mitotic inducer targeted by chk1 DNA damage checkpoint kinase. Science. 1997;277:1495–1497. doi: 10.1126/science.277.5331.1495. [DOI] [PubMed] [Google Scholar]
- 10.Walworth NC, Bernards R. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- 11.Lopez-Girona A, Furnari B, Mondesert O, Russell P. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein [see comments] Nature. 1999;397:172–175. doi: 10.1038/16488. [DOI] [PubMed] [Google Scholar]
- 12.Furnari B, Blasina A, Boddy MN, McGowan CH, Russell P. Cdc25 inhibited in vivo and in vitro by checkpoint kinases Cds1 and Chk1. Mol Cell Biol. 1999;10:833–845. doi: 10.1091/mbc.10.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez-Girona A, Kanoh J, Russell P. Nuclear exclusion of cdc25 is not required for the DNA damge checkpoint in fission yeast. Curr Biol. 2001;11:50–54. doi: 10.1016/s0960-9822(00)00026-9. [DOI] [PubMed] [Google Scholar]
- 14.O'Connell MJ, Raleigh JM, Verkade HM, Nurse P. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 1997;16:545–554. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J, Kumagai A, Dunphy WG. Positive regulation of Wee1 by Chk1 and 14-3-3 proteins. Mol Cell Biol. 2001;12:551–563. doi: 10.1091/mbc.12.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, Elledge SJ. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 17.Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 18.Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, De Mayo F, Bradley A, Donehower LA, Elledge SJ. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev. 2000;14:2448–2459. [PMC free article] [PubMed] [Google Scholar]
- 19.Guo ZJ, Kumagai A, Wang SX, Dunphy WG. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen P, Gatei M, O'Connell MJ, Khanna KK, Bugg SJ, Hogg A, Scott SP, Hobson K, Lavin MF. Chk1 complements the G2/M checkpoint defect and radiosensitivity of ataxia-telangiectasia cells. Oncogene. 1999;18:249–256. doi: 10.1038/sj.onc.1202257. [DOI] [PubMed] [Google Scholar]
- 21.Mailand N, Falck J, Lukas C, Syljuasen RG, Welcker M, Bartek J, Lukas J. Rapid destruction of human Cdc25A in response to DNA damage. Science. 2000;288:1425–1429. doi: 10.1126/science.288.5470.1425. [DOI] [PubMed] [Google Scholar]
- 22.Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites [published erratum appears in Genes Dev 2000 Mar 15;14(6):750 ] Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:893–897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 24.Blasina A, de Weyer IV, Laus MC, Luyten WH, Parker AE, McGowan CH. A human homologue of the checkpoint kinase Cds1 directly inhibits Cdc25 phosphatase. Curr Biol. 1999;9:1–10. doi: 10.1016/s0960-9822(99)80041-4. [DOI] [PubMed] [Google Scholar]
- 25.Brown AL, Lee CH, Schwarz JK, Mitiku N, Piwnica-Worms H, Chung JH. A human Cds1-related kinase that functions downstream of ATM protein in the cellular response to DNA damage. Proc Natl Acad Sci USA. 1999;96:3745–3750. doi: 10.1073/pnas.96.7.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng CY, Graves PR, Ogg S, Thoma RS, Byrnes J, III, Wu Z, Stephenson MT, Piwnica-Worms H. C-TAK1 protein kinase phosphorylates human Cdc25C on serine 216 and promotes 14-3-3 protein binding. Cell Growth Differ. 1998;9:197–208. [PubMed] [Google Scholar]
- 27.Takai H, Tominaga K, Motoyama N, Minamishima YA, Nagahama H, Tsukiyama T, Ikeda K, Nakayama K, Nakanishi M, Nakayama K. Aberrant cell cycle checkpoint function and early embryonic death in Chk1 -/- mice. Genes Dev. 2000;14:1439–1447. [PMC free article] [PubMed] [Google Scholar]
- 28.Jarvis TC, Alby LJ, Beaudry AA, Wincott FE, Beigelman L, McSwiggen JA, Usman N, Stinchcomb DT. Inhibition of vascular smooth muscle cell proliferation by ribozymes that cleave c-myb mRNA. RNA. 1996;2:419–428. [PMC free article] [PubMed] [Google Scholar]
- 29.Kaneko YS, Watanabe N, Morisaki H, Akita H, Fujimoto A, Tominaga K, Terasawa M, Tachibana A, Ikeda K, Nakanishi M. Cell-cycle-dependent and ATM-independent expression of human Chk1 kinase [published erratum appears in Oncogene 1999 Sep 16; 18(37):5246] Oncogene. 1999;18:3673–3681. doi: 10.1038/sj.onc.1202706. [DOI] [PubMed] [Google Scholar]
- 30.Busby EC, Leistritz DF, Abraham RT, Karnitz LM, Sarkaria JN. The radiosensitizing agent 7-hydroxystaurosporine (UCN-01) inhibits the DNA damage checkpoint kinase hChk1. Cancer Res. 2000;60:2108–2112. [PubMed] [Google Scholar]
- 31.Jackson JR, Gilmartin A, Imburgia C, Winkler JD, Marshall LA, Roshak A. An indolocarbazole inhibitor of human checkpoint kinase (Chk1) abrogates cell cycle arrest caused by DNA damage. Cancer Res. 2000;60:566–572. [PubMed] [Google Scholar]
- 32.Graves PR, Yu L, Schwarz JK, Gales J, Sausville EA, O'Connor PM, Piwnica-Worms H. The Chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01. J Biol Chem. 2000;275:5600–5605. doi: 10.1074/jbc.275.8.5600. [DOI] [PubMed] [Google Scholar]
- 33.Luo Y, Rockow-Magnone SK, Joseph MK, Bradner J, Butler CC, Tahir SK, Han EK-H, Ng SC, Severin JM, Gubbins EJ, Reilly RM, Rueter A, Simmer RL, Holzman TF, Giranda VL. Abrogation of G2 checkpoint specifically sensitize p53 defective cells to cancer chemotherapeutic agents. AntiCancer Res. 2001;21:23–28. [PubMed] [Google Scholar]
- 34.Akinaga S, Nomura K, Gomi K, Okabe M. Effect of UCN-01, a selective inhibitor of protein kinase C, on the cell-cycle distribution of human epidermoid carcinoma, A431 cells. Cancer Chemother Pharmacol. 1994;33:273–280. doi: 10.1007/BF00685899. [DOI] [PubMed] [Google Scholar]
- 35.Gong J, Traganos F, Darzynkiewicz Z. Staurosporine blocks cell progression through G1 between the cyclin D and cyclin E restriction points. Cancer Res. 1994;54:3136–3139. [PubMed] [Google Scholar]
- 36.Kastan MB, Onyekwere O, Sidransky D, Vogelstein B, Craig RW. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 37.Weinstein JN, Myers TG, O'Connor PM, Friend SH, Fornace AJ, Jr, Kohn KW, Fojo T, Bates SE, Rubinstein LV, Anderson NL, Buolamwini JK, van Osdol WW, Monks AP, Scudiero DA, Sausville EA, Zaharevitz DW, Bunow B, Viswanadhan VN, Johnson GS, Wittes RE, Paull KD. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275:343–349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- 38.Lee JM, Bernstein A. p53 mutations increase resistance to ionizing radiation. Proc Natl Acad Sci USA. 1993;90:5742–5746. doi: 10.1073/pnas.90.12.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes [see comments] Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 40.Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 41.Rouby E, Thomas A, Costin D, Rosenberg CR, Potmesil M. p53 mutation in B-cell chronic lymphotic leukemia is associated with drug resistance and is independent of MDR1/MDR3 gene expression. Blood. 1993;82:3452–3459. [PubMed] [Google Scholar]
- 42.Russell KJ, Wiens LW, Demers GW, Galloway DA, Plon SE, Groudine M. Abrogation of the G2 checkpoint results in differential radiosensitization of G1 checkpoint-deficient and G1 checkpoint-competent cells. Cancer Res. 1995;55:1639–1642. [PubMed] [Google Scholar]
- 43.Yao SL, Akhtar AJ, McKenna KA, Bedi GC, Sidransky D, Mabry M, Ravi R, Collector MI, Jones RJ, Sharkis SJ, Fuchs EJ, Bedi A. Selective radiosensitization of p53-deficient cells by caffeine-mediated activation of p34cdc2 kinase. Nat Med. 1996;2:1140–1143. doi: 10.1038/nm1096-1140. [DOI] [PubMed] [Google Scholar]
- 44.Powell SN, DeFrank JS, Connell P, Eogan M, Preffer F, Dombkowski D, Tang W, Friend S. Differential sensitivity of p53(-) and p53(+) cells to caffeine-induced radiosensitization and override of G2 delay. Cancer Res. 1995;55:1643–1648. [PubMed] [Google Scholar]