Abstract
Single-envelope human immunodeficiency virus (HIV) vaccines have been studied for more than a decade, with some successes in homologous challenge experiments in nonhuman primates but with no clear successes in clinical trials. To gain insight into the breadth of the immunity elicited by such vaccines, we have dissected the T-helper cell response of C57BL/6 mice to an individual, molecularly cloned envelope protein. Here, we report that T-helper cells responsive to HIV type 1 1035 envelope are very highly restricted in C57BL/6 animals: seven different hybridomas recovered from five separate mice recognized the same peptide, PKVSFEPIPIHYCAP, located in the C2 region of gp120. Three of these hybridomas were tested on a natural variant of the peptide but failed to respond. A more extensive analysis of whole splenic populations from other C57BL/6 mice immunized with the 1035 envelope reproducibly confirmed that the gp120-specific T-helper response was almost exclusively focused on a single epitope. We conclude that single-envelope vaccines may frequently fail to provoke an immune response sufficiently diverse to recognize variant sequences among circulating HIV. The results encourage the inclusion of more than one envelope in future vaccines to enhance the potential diversity and respective surveillance capacities of responding T-helper cell populations.
Envelope molecules are the only virus-encoded proteins on the surface of human immunodeficiency virus (HIV) and therefore serve as critical targets for HIV vaccines. For many years, attempts have been made to elicit protective immunity in humans with vaccines composed of single-envelope proteins (2, 12, 13, 18, 30). Though similar strategies have protected nonhuman primates against homologous challenges with HIV or simian immunodeficiency virus (4, 15), the single-envelope vaccines have not demonstrated full protection from natural HIV infection in clinical trials (3, 13).
To mimic these human trials, we have vaccinated C57BL/6 mice with molecularly cloned envelope proteins and analyzed the resultant T-helper cell responses. We chose to study virus-specific T-helper lymphocytes because these cells can directly target HIV (11) and additionally support both B- and cytotoxic-T-lymphocyte activities (5, 34). Previous studies with two different envelope proteins (one from clade B and one from clade D) showed that epitopes recognized by T-helper cells were limited to peptides in four distinct regions. Three of these regions appeared together on one face of the folded gp120 protein, while the fourth consisted of a 20-mer peptide in the gp41 stalk (33).
Here, we describe the T-helper cell response to an envelope from another clade B virus, HIV type 1 (HIV-1) 1035, which we found to be even more strikingly skewed. In this case, the gp120-specific response was essentially focused on a single peptide. Our illustration of such narrow reactivity may explain, at least in part, the lack of full protection against HIV in previous clinical trials (3, 13). Clearly, HIV isolates vary in sequence, and an effective vaccine must elicit T cells responsive to each of these variants (14, 22). If very few peptides are recognized by vaccine-induced T-helper cells, the likelihood that a matched peptide will be presented by every challenge virus is low. As vaccine epitopes increase in number, the likelihood that challenge viruses will share at least one epitope with the vaccine increases similarly. To enhance the total number of determinants to which activated T-helper cells respond, we suggest that future HIV vaccines encompass a variety of distinct envelope proteins.
MATERIALS AND METHODS
Animals.
Adult female C57BL/6J mice were purchased from Jackson Laboratories (Bar Harbor, Maine) and housed in the St. Jude Children's Research Hospital animal facilities under conditions specified by Association for Assessment and Accreditation of Laboratory Animal Care guidelines.
Immunizations for hybridoma production.
Each vaccine component expressed gp140 (encompassing gp120 and the extracellular region of gp41) derived from a clade B primary isolate (1035, from an HIV-infected individual in Memphis, Tenn.). Two distinct immunization regimens were used for hybridoma production. In the first case, mice were immunized with HIV gp140 envelope using a previously described vaccine strategy (25). Briefly, injections were done with recombinant DNA (100 μg by the intramuscular route 24 h after a bupivacaine injection), followed 3 to 4 weeks later with recombinant vaccinia virus (VV; 107 PFU by the intraperitoneal route) (33). The recombinant VV was prepared by substituting the 1035 envelope sequence for the BH10 envelope sequence in a pSC11-based VV recombination vector. The plasmid was transfected into VV (Western Reserve)-infected TK-143B cells, and recombinant VVs were selected in bromodeoxyuridine and then plaqued and further selected by Western blot analyses using anti-HIV antibodies as developing reagents (26). Three weeks after the VV inoculation, the spleens were removed for fusion. We selected this time point for fusion because the response to VV peaks relatively late after immunization (24). In the second case, mice were immunized as described above but were boosted 3 to 4 weeks after the VV inoculation with purified CHO-derived envelope protein (1 to 10 μg of protein with complete Freund's adjuvant injected in the base of the tail [6, 33]). Ten days later, draining lymph nodes were removed for fusion.
Hybridoma production.
Effector cells from lymph nodes or spleens were stimulated in vitro with homologous recombinant CHO cell lysates (prepared from a cell pellet which was freeze-thawed and resuspended to yield a final concentration of 105 original cells/ml) for 2 to 4 days in complete medium (17), fused with BW5147 α− β− (35) or BWZ.36 (a lacZ-inducible fusion partner) (27), and grown in selection medium. Hybridoma lines were harvested from plates in which fewer than one-third of wells scored positive for growth. Hybridomas that responded specifically to the HIV envelope recombinant VV were expanded for further screening against overlapping peptides representing the full 1035 gp140 protein. Cells were first tested against peptide pools and then against individual peptides within the positive pools. In total, cells from five immunized mice were prepared individually for hybridoma preparation and analyses.
Peptides.
Peptides (8- to 15-mers) were made by Chiron (Emeryville, Calif.) or the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children's Research Hospital. Initial hybridoma screening was performed with peptides at concentrations of 1 to 10 μg/ml. Subsequent assays were done with serially diluted peptides, starting at concentrations of 25 μg/ml.
Peptide specificity assays.
Hybridomas were assayed for peptide-specific responses using either C57BL/6 spleen cells or I-Ab-transfected L cells (AF7-1C6 [20]) as antigen-presenting cells (APC). Hybridomas (105 cells/well) were plated in 96-well plates with peptides and spleen cells (5 × 105/well) or L cells (1 × 105/well). The assays were developed after 18 to 24 h, using the interleukin 2 (IL-2) assay (for BW5147 α− β− fusions) or the blue-spot assay (for BWZ.36 fusions [see below]). Assays for IL-2 used an IL-2-dependent HT-2 cell line and an oxidation-reduction indicator (Alamar Blue; Alamar Biosciences Inc., Sacramento, Calif.) read at 570 nm with a reference wavelength of 595 nm (36). Recombinant human IL-2 (R&D Systems, Minneapolis, Minn.) served as the positive control. Blue-spot assays were performed as described previously (27, 32). Briefly, stimulated cells were washed with phosphate-buffered saline (PBS) at room temperature and fixed at 4°C with 1% formaldehyde-0.2% gluteraldehyde for 5 min. The cells were washed again, and β-galactosidase activity was detected by adding 50 μl of PBS containing 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 2 mM MgCl2, and 1 mg of the substrate X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)/ml to each well. The plates were incubated at 37°C for at least 8 h or overnight, and the blue cells were counted using a tissue culture inverted microscope.
Flow cytometry for TCR typing.
A panel of unconjugated, biotinylated, and phycoerythrin-labeled monoclonal antibodies was used to characterize the spectrum of T-cell receptors (TCR) on hybridomas. Individual monoclonal antibodies were specific for the constant region of the TCR β-chain and for Vβs 2 to 14. The secondary staining reagents included fluorochrome-labeled avidin, goat anti-rat immunoglobulin M (IgM), goat anti-mouse IgG, and goat anti-rat IgG. These reagents were prepared from hybridoma supernatants or were purchased from Pharmingen (San Diego, Calif.), Caltag (Burlingame, Calif.), Jackson ImmunoResearch (West Grove, Pa.), or Biosource International (Camarillo, Calif.). Stained cells were analyzed with a FACSCan (Becton Dickinson, Mountain View, Calif.).
Immunizations and ELISPOT assay.
Mice were injected two to four times at 1-month intervals with 100 μg of recombinant DNA given intramuscularly. Approximately 2 weeks after the last immunization, the spleens were taken and CD4+ T-cells were enriched. Briefly, the cells were treated with rat anti-mouse major histocompatibility complex (MHC) class II (TIB 120 cell supernatants) and rat anti-mouse CD8 (53-6.72 cell supernatants) antibodies. The cells were then mixed with sheep anti-mouse and sheep anti-rat IgG-coated dynabeads (Dynal ASA, Oslo, Norway). Cell samples were exposed to a magnet to remove MHC class II-positive, CD8-positive, and Ig-positive populations. APC were prepared from naïve mouse spleens by depleting T cells with an anti-mouse Thy1.2 antibody (AT83) and complement (one part rabbit and five parts guinea pig complement [Cedarlane, Ontario, Canada] in Hanks balanced salt solution plus 0.1% bovine serum albumin) and irradiating them with 2,500 rads. Enzyme-linked immunospot (ELISPOT) plates (96-well filtration plates; Millipore, Bedford, Mass.) were prepared by overnight incubation with 10 μg of anti-mouse gamma interferon (IFN-γ) (clone R4-6A2; BD Biosciences, San Diego, Calif.)/ml in PBS (100 μl/well) at 4°C. The plates were washed three times with PBS and blocked for at least 1 h at 37°C with complete medium; 1 × 106 CD4+ T cells and 5 × 105 APC per well were then added with peptides (2 μg/peptide/ml). Concanavalin A (4 μg/ml; Sigma, St. Louis, Mo.) was used as a positive control. After a 24- to 40-h incubation at 37°C and 10% CO2, the wells were washed five times with PBS and five times with wash buffer (PBS with 0.05% Tween 20). The plates were then incubated for 2 h with 100 μl of 5-μg/ml biotinylated rat anti-mouse IFN-γ (clone XMG1.2; BD Biosciences) in PBS with 0.05% Tween 20 and 1% fetal calf serum at room temperature. The wells were washed five times with wash buffer and incubated with streptavidin-conjugated alkaline phosphatase (1:500 in wash buffer) for 1 h. After five rinses with wash buffer, the spots were developed with 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium alkaline phosphatase substrate (Sigma). The plates were rinsed with water and air dried, and the spots were counted with an ELISPOT reader (Axioplan 2 imaging; Zeiss, München-Halbergmoos, Germany). These experiments were performed in triplicate with up to eight mice per experiment.
Nucleotide sequence accession number.
The HIV-1 1035 envelope sequence was submitted to GenBank (National Center for Biotechnology Information, Bethesda, Md.; accession no. AF532615).
RESULTS AND DISCUSSION
C57BL/6 mice were primed with a clade B gp140 envelope (HIV-1 1035) using a DNA-poxvirus prime-boost regimen (25), followed in some cases by a boost with purified 1035 envelope protein (6). Responding T-helper cell populations were restimulated in vitro with the HIV 1035 envelope and fused for the production of hybridomas. Five independent mouse immunizations and T-cell fusions were performed, from which seven hybridomas were derived. The hybridomas were cloned, expanded, characterized for TCR Vβ usage, and tested for reactivity against overlapping peptides spanning the entire gp140 sequence. Although each of the seven hybridomas was distinct (defined by mouse origin or unique TCR), each showed responsiveness to PKVSFEPIPIHYCAP, a peptide in the C2 region of HIV envelope (Table 1).
TABLE 1.
T-helper cell hybridomas respond to a single peptide in 1035 gp120
| Hybridoma name | Immunization regimena | Mouse no. | Fusion parent lineb | Positive peptidec | TCR Vβ usaged |
|---|---|---|---|---|---|
| H1035L-46 | D-V | 1 | BW5147 α− β− | PKVSFEPIPIHYCAP | Vβ 11 |
| H1035L-53 | D-V | 1 | BW5147 α− β− | PKVSFEPIPIHYCAP | Vβ 13 |
| H1035L-133 | D-V | 2 | BW5147 α− β− | PKVSFEPIPIHYCAP | Vβ 12 |
| H1035L-213 | D-V | 2 | BW5147 α− β− | PKVSFEPIPIHYCAP | Undefined |
| H1035P1-70 | D-V-P | 3 | BWZ.36 | PKVSFEPIPIHYCAP | ND |
| H1035P2-52 | D-V-P | 4 | BWZ.36 | PKVSFEPIPIHYCAP | Vβ 12 |
| H1035P3-34 | D-V-P | 5 | BWZ.36 | PKVSFEPIPIHYCAP | Vβ 12 |
Mice received two different immunization regimens. In one case, the animals were immunized with recombinant DNA and boosted with recombinant VV (D-V), after which the spleens were removed for restimulation in vitro and fusion. In the second case, the animals received D-V injections, followed by a boost with purified protein (D-V-P). In the latter instance, draining lymph nodes were taken for restimulation and fusion.
Parent lines were BW5147 α− β− and BWZ.36.
Peptide specificity was measured with IL-2 assays for BW5147 α− β− fusions and blue-spot assays for BWZ.36 fusions. TCR were stained with fluorochrome-labeled antibodies and analyzed by flow cytometry.
Undefined, not positive with any of the tested antibodies; ND, not determined.
We next tested one of the hybridomas (H1035P1-70) on a variety of 15-mer and truncated peptides to define the minimal T-cell epitope. As shown by representative results in Table 2, the minimal determinant recognized by this hybridoma was FEPIPIHYC.
TABLE 2.
Fine definition of a gp120 envelope T-helper determinant
| Peptidea | Blue-spot count at peptide dilutionb:
|
|||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
| HYCAPAGFAILKCND | 0 | 0 | 0 | 0 | 0 | 0 |
| EPIPIHYCAPAGFAI | 0 | 0 | 0 | 0 | 0 | 0 |
| FEPIPIHYCAPAGFA | >300 | >300 | 123 | 16 | 0 | 0 |
| SFEPIPIHYCAPAGF | >300 | >300 | >300 | 33 | 0 | 0 |
| VSFEPIPIHYCAPAG | >300 | >300 | >300 | >300 | 139 | 16 |
| KVSFEPIPIHYCAPA | >300 | >300 | >300 | >300 | >300 | 65 |
| CPKVSFEPIPIHYCA | >300 | >300 | >300 | 95 | 7 | 0 |
| ACPKVSFEPIPIHYC | >300 | 259 | 30 | 0 | 0 | 0 |
| QACPKVSFEPIPIHY | 0 | 0 | 0 | 0 | 0 | 0 |
| TQACPKVSFEPIPIH | 0 | 0 | 0 | 0 | 0 | 0 |
| FEPIPIHYCAP | >300 | 67 | 0 | 0 | 0 | 0 |
| EPIPIHYCAP | 0 | 0 | 0 | 0 | 0 | 0 |
| PKVSFEPIPIHYCAP | >300 | >300 | >300 | >300 | >300 | 243 |
| PKITFEPIPIHYCAP | >300 | >300 | >300 | >300 | >300 | 117 |
Boldface letters represent the minimal determinant recognized by hybridoma H1035P1-70.
Experiments were performed with hybridoma H1035P1-70. Blue-spot counts per well are shown with the background subtracted. There were ∼10 background counts/well. The peptides were serially diluted (1:3) starting at 25 μg/ml (dilution 1).
To determine how natural variation in envelope sequences affected recognition of the target epitope, we prepared synthetic peptides from sequences of two distinct isolates of HIV (1007 [33] and D760 [22]). The first peptide variant (PKITFEPIPIHYCAP from HIV 1007) contained amino acid substitutions in a region flanking the minimal epitope. As shown in Table 2 (last two lines), this alteration did not affect target peptide recognition by hybridoma H1035P1-70. The second peptide, from isolate D760, had amino acid substitutions in both the flanking region and the epitope core. This natural sequence variation abrogated recognition by hybridoma H1035P1-70 (Table 3). Two additional hybrids, H1035P2-52 and H1035P3-34, were also tested on this peptide but failed to respond (Table 3). Thus, natural variations in the target sequence were sufficient to abrogate T-helper cell activity.
TABLE 3.
T-helper hybridomas fail to recognize a natural gp120 peptide variant
| Hybridoma | Peptide | Blue-spot count at peptide dilutiona:
|
|||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | ||
| H1035P1-70 | KVSFEPIPIHYCAPA | >300 | >300 | 177 | − | − | − |
| KVTFDPIPIHYCAPA | 0 | 0 | 0 | − | − | − | |
| H1035P2-52 | KVSFEPIPIHYCAPA | >300 | >300 | >300 | >300 | >300 | >300 |
| KVTFDPIPIHYCAPA | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1035P3-34 | KVSFEPIPIHYCAPA | >300 | >300 | >300 | >300 | >300 | >300 |
| KVTFDPIPIHYCAPA | 0 | 0 | 0 | 0 | 0 | 0 | |
Blue-spot counts per well are shown with the background subtracted. The peptides were serially diluted (1:3) starting at 25 μg/ml (dilution 1). −, not tested.
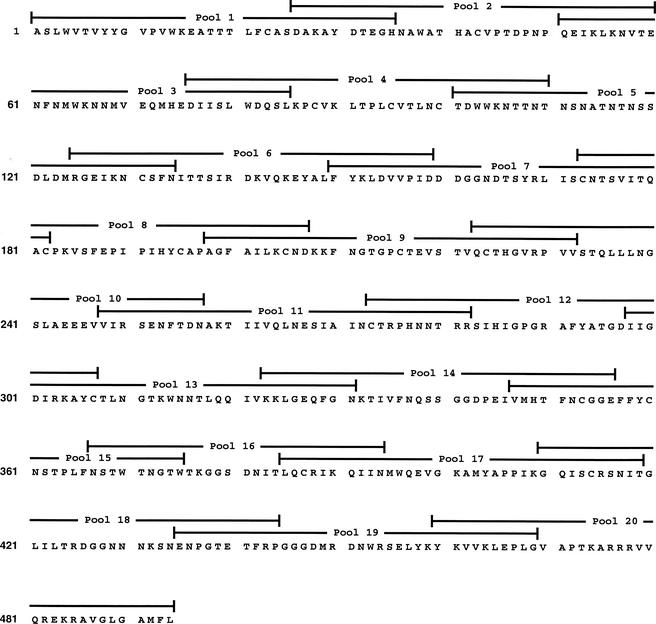
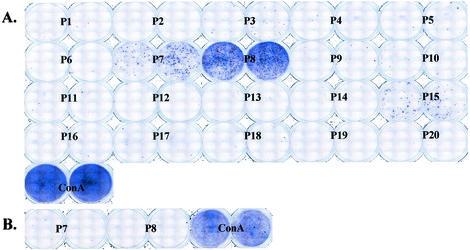
To confirm that hybridomas accurately represented whole T-cell populations, we immunized new sets of C57BL/6 animals with the 1035 envelope and examined fresh lymphocytes with an ELISPOT assay. In this case, mice received two to four injections with recombinant 1035 DNA at 1-month intervals, and the spleens were collected ∼2 weeks after the last injection. T-helper cells were plated in duplicate for testing against 20 pools of overlapping peptides representing the entire gp120 sequence of envelope 1035 (Fig. 1). Each pool contained five 8- to 15-mer peptides. As shown by representative results from one of three experiments (Fig. 2), the responses to gp120 pools were extremely restricted. The chief response was directed toward pool 8. Dissection of responses within pool 8 showed that there was only one stimulatory peptide in this pool, PKVSFEPIPIHYCAP.
FIG. 1.
HIV 1035 gp120 envelope sequence. The 1035 gp120 envelope sequence is shown, and sequences represented by each of 20 peptide pools are indicated; 8- to 15-mer peptides spanned the envelope sequence with overlaps of ∼10 amino acids.
FIG. 2.
T-helper cells from C56BL/6 mice respond predominantly to a single peptide in the HIV 1035 gp120 sequence. (A) Spleen cells from eight 1035 envelope-primed mice were pooled and measured by ELISPOT assay against 20 peptide pools spanning the gp120 sequence. Each pool contained five sequential peptides (8- to15-mers). The blue spots represent IFN-γ-producing, peptide-specific cells from immunized spleens. A concanavalin A control is also shown. (B) Results with negative control cells from a naive spleen are shown.
ELISPOT assays were also performed with overlapping peptides representing the gp41 component of the gp140 immunogen. These assays (not shown) revealed only one response, which was to TNVPWNASWSNKSLE, a peptide that was identified as a C57BL/6 T-helper cell epitope in previous hybridoma studies (33).
Why is the gp120-specific T-helper cell response so narrow in 1035 envelope-primed C57BL/6 mice? T-helper responses to the PKVSFEPIPIHYCAP region have been previously identified in independent analyses of other HIV envelopes (28, 31, 33), but responses to a number of different gp120 peptides were also recognized. In previous C57BL/6 experiments with two other gp140 vaccines (from HIV-1 1007 and UG92005 isolates [33]), the additional T-helper responses were against peptides not matched in the 1035 protein sequence (due to natural sequence variation). All of the gp120 epitopes (including PKVSFEPIPIHYCAP) were found to be located within one exposed face of the folded protein, suggesting that peptides in this location were preferentially released from the protein and thus rendered accessible to downstream antigen-processing mechanisms. In the context of the 1035 protein, and in the C57BL/6 mouse, the PKVSFEPIPIHYCAP region may have been most readily released for processing (and/or most readily bound to the class II I-Ab molecule), which would explain its preferred recognition by T cells. Our finding that the structural contexts of peptides may contribute to epitope immunodominance has been confirmed by others, with regard to both HIV and non-HIV antigens (9, 10, 19).
The limited C57BL/6 T-helper response described in this report is not solely dependent on a particular envelope sequence, nor is it solely dependent on a particular mouse strain (and the respective MHC class II peptide binding groove [23]). Rather, a combination of immunogen and mouse strain dictates the T-helper cell epitope profile; a change in either component will alter the pattern. This phenomenon is recognized not only for HIV antigens but in other antigenic systems, and it affects both mice and humans (1, 29). That CD8 T cells can be highly focused is well established (dependent in this case on a combination of an immunogen sequence and a particular MHC class I peptide binding groove [7, 8, 16]). Our studies (and some others [29]) simply emphasize that CD4 T-cell responses can be similarly skewed.
The restricted T-cell response identified for this antigen-MHC pair has implications for HIV vaccine design. Of the three envelopes that we have evaluated in the C57BL/6 mouse, the 1035 envelope elicited the most focused T-helper response. This extremely narrow response could not have been predicted by a survey of envelope sequences or by previous studies of the C57BL/6 mouse (which developed broader responses to two different envelope proteins). As an extrapolation of these results, we propose that at least some human T-cell responses to single-envelope (or envelope/gag/pol) vaccines are similarly narrow and non-cross-reactive with natural HIV variants, perhaps contributing to the incidence of breakthrough infections in clinical trials (3, 13).
The inclusion of determinants from multiple distinct HIV isolates in a vaccine (e.g., our polyenvelope vaccine, currently in clinical trials [21, 30]) may help to avoid the generation of such narrowly restricted responses among vaccinees. Diverse antigens within a cocktail may be processed by different APC (precluding intermolecular competition between envelopes) and trigger an array of T-helper effectors responsive to numerous distinct epitopes. As the diversity of targeted epitopes increases, so does the likelihood that naturally encountered viruses will contain at least a subset of matching epitopes and that broad protective immunity can be achieved.
Acknowledgments
We thank Amy Zirkel, Brita Brown, and Bart Jones for excellent technical assistance.
This work was supported in part by NIH NIAID P01-AI45142, NCI Cancer Center Support Core Grant P30-CA21765, The James B. Pendleton Charitable Trust, and the American Lebanese Syrian Associated Charities (ALSAC).
REFERENCES
- 1.Allen, P. M., B. P. Babbitt, and E. R. Unanue. 1987. T-cell recognition of lysozyme: the biochemical basis of presentation. Immunol. Rev. 98:171-187. [DOI] [PubMed] [Google Scholar]
- 2.Belshe, R. B., B. S. Graham, M. C. Keefer, G. J. Gorse, P. Wright, R. Dolin, T. Matthews, K. Weinhold, D. P. Bolognesi, R. Sposto, D. M. Stablein, T. Twaddell, P. W. Berman, T. Gregory, A. E. Izu, M. C. Walker, and P. Fast. 1994. Neutralizing antibodies to HIV-1 in seronegative volunteers immunized with recombinant gp120 from the MN strain of HIV-1. JAMA 272:475-480. [DOI] [PubMed] [Google Scholar]
- 3.Berman, P. W., A. M. Gray, T. Wrin, J. C. Vennari, D. J. Eastman, G. R. Nakamura, D. P. Francis, G. Gorse, and D. H. Schwartz. 1997. Genetic and immunologic characterization of viruses infecting MN-rgp120-vaccinated volunteers. J. Infect. Dis. 176:384-397. [DOI] [PubMed] [Google Scholar]
- 4.Berman, P. W., T. J. Gregory, L. Riddle, G. R. Nakamura, M. A. Champe, J. P. Porter, F. M. Wurm, R. D. Hershberg, E. K. Cobb, and J. W. Eichberg. 1990. Protection of chimpanzees from infection by HIV-1 after vaccination with recombinant glycoprotein gp120 but not gp160. Nature 345:622-625. [DOI] [PubMed] [Google Scholar]
- 5.Brander, C., and B. D. Walker. 1999. T lymphocyte responses in HIV-1 infection: implications for vaccine development. Curr. Opin. Immunol. 11:451-459. [DOI] [PubMed] [Google Scholar]
- 6.Caver, T. E., T. D. Lockey, R. V. Srinivas, R. G. Webster, and J. L. Hurwitz. 1999. A novel vaccine regimen utilizing DNA, vaccinia virus and protein immunizations for HIV-1 envelope presentation. Vaccine 17:1567-1572. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Z. W., L. Shen, M. D. Miller, S. H. Ghim, A. L. Hughes, and N. L. Letvin. 1992. Cytotoxic T lymphocytes do not appear to select for mutations in an immunodominant epitope of simian immunodeficiency virus gag. J. Immunol. 149:4060-4066. [PubMed] [Google Scholar]
- 8.Currier, J. R., M. deSouza, P. Chanbancherd, W. Bernstein, D. L. Birx, and J. H. Cox. 2002. Comprehensive screening for human immunodeficiency virus type 1 subtype-specific CD8 cytotoxic T lymphocytes and definition of degenerate epitopes restricted by HLA-A0207 and -C(W)0304 alleles. J. Virol. 76:4971-4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai, G., S. Carmicle, N. K. Steede, and S. J. Landry. 2002. Structural basis for helper T-cell and antibody epitope immunodominance in bacteriophage T4 Hsp10. Role of disordered loops. J. Biol. Chem. 277:161-168. [DOI] [PubMed] [Google Scholar]
- 10.Dai, G., N. K. Steede, and S. J. Landry. 2001. Allocation of helper T-cell epitope immunodominance according to three-dimensional structure in the human immunodeficiency virus type I envelope glycoprotein gp120. J. Biol. Chem. 276:41913-41920. [DOI] [PubMed] [Google Scholar]
- 11.Ehret, R., M. Heinkelein, R. F. Siliciano, and C. Jassoy. 1997. Human immunodeficiency virus glycoprotein-specific CD4+ cytotoxic T lymphocytes are involved in two types of cytotoxicity: antigen-specific and cell-cell fusion-related cell lysis. AIDS Res. Hum. Retrovir. 13:1017-1021. [DOI] [PubMed] [Google Scholar]
- 12.Gorse, G. J. 1994. Phase I/II trials of preventive HIV vaccine candidates. Dose and schedule: summary. Aids Res. Hum. Retrovir. 10(Suppl. 2):S141-S143. [PubMed] [Google Scholar]
- 13.Graham, B. S., M. J. McElrath, R. I. Connor, D. H. Schwartz, G. J. Gorse, M. C. Keefer, M. J. Mulligan, T. J. Matthews, S. M. Wolinsky, D. C. Montefiori, S. H. Vermund, J. S. Lambert, L. Corey, R. B. Belshe, R. Dolin, P. F. Wright, B. T. Korber, M. C. Wolff, and P. E. Fast. 1998. Analysis of intercurrent human immunodeficiency virus type 1 infections in phase I and II trials of candidate AIDS vaccines. J. Infect. Dis. 177:310-319. [DOI] [PubMed] [Google Scholar]
- 14.Harcourt, G. C., S. Garrard, M. P. Davenport, A. Edwards, and R. E. Phillips. 1998. HIV-1 variation diminishes CD4 T lymphocyte recognition. J. Exp. Med. 188:1785-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu, S.-L., K. Abrams, G. N. Barber, P. Moran, J. M. Zarling, A. J. Langlois, L. Kuller, W. R. Morton, and R. E. Benveniste. 1992. Protection of macaques against SIV infection by subunit vaccines of SIV envelope glycoprotein gp160. Science 255:456-459. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, R. P., S. A. Hammond, A. Trocha, R. F. Siliciano, and B. D. Walker. 1994. Induction of a major histocompatibility complex class I-restricted cytotoxic T-lymphocyte response to a highly conserved region of human immunodeficiency virus type 1 (HIV-1) gp120 in seronegative humans immunized with a candidate HIV-1 vaccine. J. Virol. 68:3145-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kappler, J. W., B. Skidmore, J. White, and P. Marrack. 1981. Antigen-inducible, H-2-restricted interleukin-2-producing T cell hybridomas: lack of independent antigen and H-2 recognition. J. Exp. Med. 153:1198-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keefer, M. C., R. Belshe, B. Graham, J. McElrath, M. L. Clements, R. Sposto, P. Fast, and the NIAID AIDS Vaccine Clinical Trials Network. 1994. Safety profile of HIV vaccination: first 1000 volunteers of AIDS vaccine evaluation group. Aids Res. Hum. Retrovir. 10(Suppl. 2):S139-S140. [PubMed] [Google Scholar]
- 19.Landry, S. J. 1997. Local protein instability predictive of helper T-cell epitopes. Immunol. Today 18:527-532. [DOI] [PubMed] [Google Scholar]
- 20.Lechler, R. I., M. A. Norcoss, and R. N. Germain. 1985. Qualitative and quantitative studies of antigen-presenting cell function by using I-A-expressing L cells. J. Immunol. 135:2914-2922. [PubMed] [Google Scholar]
- 21.Lockey, T. D., K. S. Slobod, T. E. Caver, S. D'Costa, R. J. Owens, H. M. McClure, R. W. Compans, and J. L. Hurwitz. 2000. Multi-envelope HIV vaccine safety and immunogenicity in small animals and chimpanzees. Immunol. Res. 21:7-21. [DOI] [PubMed] [Google Scholar]
- 22.Myers, G., B. Korber, S. Wain-Hobson, R. F. Smith, and G. N. Pavlakis. 1993. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 23.Rammensee, H. G., T. Friede, and S. Stevanoviic. 1995. MHC ligands and peptide motifs: first listing. Immunogenetics 41:178-228. [DOI] [PubMed] [Google Scholar]
- 24.Rencher, S. D., T. D. Lockey, R. V. Srinivas, R. J. Owens, and J. L. Hurwitz. 1997. Eliciting HIV-1 envelope-specific antibodies with mixed vaccinia virus recombinants. Vaccine 15:265-272. [DOI] [PubMed] [Google Scholar]
- 25.Richmond, J. F. L., F. Mustafa, S. Lu, J. C. Santoro, J. Weng, M. O'Connell, E. M. Fenyo, J. L. Hurwitz, D. C. Montefiori, and H. L. Robinson. 1997. Screening of HIV-1 Env glycoproteins for the ability to raise neutralizing antibody using DNA immunization and recombinant vaccinia virus boosting. Virology 230:265-274. [DOI] [PubMed] [Google Scholar]
- 26.Ryan, K. W., R. J. Owens, and J. L. Hurwitz. 1997. Preparation and use of vaccinia virus vectors for HIV protein expression and immunization, p. 1995-2015. In I. Lefkovits (ed.), Immunology methods manual. Academic Press, London, England.
- 27.Sanderson, S., and N. Shastri. 1994. LacZ inducible, antigen/MHC-specific T cell hybrids. Int. Immunol. 6:369-376. [DOI] [PubMed] [Google Scholar]
- 28.Sastry, K. J., and R. B. Arlinghaus. 1991. Identification of T-cell epitopes without B-cell activity in the first and second conserved regions of the HIV Env protein. AIDS 5:699-707. [DOI] [PubMed] [Google Scholar]
- 29.Schrier, R. D., J. W. Gnann, Jr., A. J. Langlois, K. Shriver, J. A. Nelson, and M. B. Oldstone. 1988. B- and T-lymphocyte responses to an immunodominant epitope of human immunodeficiency virus. J. Virol. 62:2531-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schultz, A. M., and J. A. Bradac. 2001. The HIV vaccine pipeline, from preclinical to phase III. AIDS 15(Suppl. 5):S147-S158. [DOI] [PubMed] [Google Scholar]
- 31.Sjolander, S., A. Bolmstedt, L. Akerblom, P. Horal, S. Olofsson, B. Morein, and A. Sjolander. 1996. N-linked glycans in the CD4-binding domain of human immunodeficiency virus type 1 envelope glycoprotein gp160 are essential for the in vivo priming of T cells recognizing an epitope located in their vicinity. Virology 215:124-133. [DOI] [PubMed] [Google Scholar]
- 32.Surman, S., and J. L. Hurwitz. 2002. A highly sensitive single-cell assay detects T-helper cell responses missed by conventional methods. J. Immunol. Methods 260:279-283. [DOI] [PubMed] [Google Scholar]
- 33.Surman, S., T. D. Lockey, K. S. Slobod, B. Jones, J. M. Riberdy, S. W. White, P. C. Doherty, and J. L. Hurwitz. 2001. Localization of CD4+ T cell epitope hotspots to exposed strands of HIV envelope glycoprotein suggests structural influences on antigen processing. Proc. Natl. Acad. Sci. USA 98:4587-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torres, B. A., G. Q. Perrin, M. G. Mujtaba, P. S. Subramaniam, A. K. Anderson, and H. M. Johnson. 2002. Superantigen enhancement of specific immunity: antibody production and signaling pathways. J. Immunol. 169:2907-2914. [DOI] [PubMed] [Google Scholar]
- 35.White, J., M. Blackman, J. Bill, J. Kappler, P. Marrack, D. P. Gold, and W. Born. 1989. Two better cell lines for making hybridomas expressing specific T cell receptors. J. Immunol. 143:1822-1825. [PubMed] [Google Scholar]
- 36.Woodland, D. L., M. P. Happ, J. Bill, and E. Palmer. 1990. Requirement for cotolerogenic gene products in the clonal deletion of I-E reactive cells. Science 247:964-967. [DOI] [PubMed] [Google Scholar]