Abstract
During human immunodeficiency virus type 1 (HIV-1) assembly in HIV-1-transfected COS7 cells, almost all steady-state Gag/Gag and Gag/GagPol complexes are membrane bound. However, exposure to 1% Triton X-100 gives results indicating that while all Gag/GagPol complexes remain associated with the detergent-resistant membrane (DRM), only 30% of Gag/Gag complexes are associated with the DRM. Analysis of the localization of newly synthesized Gag/Gag and Gag/GagPol to the membrane indicates that after a 10-min pulse with radioactive [35S]Cys-[35S]Met, all newly synthesized Gag/GagPol is found at the DRM. Only 30% of newly synthesized Gag/Gag moves to the membrane, and at 0 min of chase, only 38% of this membrane-bound Gag/Gag is associated with the DRM. During the first 30 min of chase, most membrane-bound Gag/Gag moves to the DRM, while between 30 and 60 min of chase, there is a significant decrease in membrane-bound Gag/Gag and Gag/GagPol. Since the localization of newly synthesized Gag/Gag to the DRM and the interaction of GagPol with Gag both depend upon Gag multimerization, the rapid localization of GagPol to the DRM probably reflects the interaction of all newly synthesized GagPol with the first newly synthesized polymeric Gag to associate with the DRM.
Virus-like particles can be produced by human immunodeficiency virus type 1 (HIV-1) Gag (14, 16, 21, 49), and putative regions of interactions between Gag molecules have been identified as occurring in the C-terminal half of Gag and including the C-terminal half of the capsid (CA) (3, 12, 24, 30), p2 (1, 23, 32), nucleocapsid (7, 8, 11, 45), and p6 (13) proteins. It is generally assumed that in order to obtain interactions required for assembly, the Gag molecules must first be concentrated at a cellular site. The membrane has been proposed to play a role in the concentration and alignment of Gag molecules, and in HIV-1-transfected COS1 cells, almost all steady-state Gag is membrane bound (18, 52). In many retroviruses, the Gag precursor protein attaches to the lipid membrane via a myristic acid moiety added posttranslationally to the amino terminus of the matrix sequence (6, 15, 17, 39, 42) and through ionic interactions between the phospholipids in the membrane and a sequence of basic amino acid residues within the matrix sequence (40, 41, 54, 56). However, a minimal 16-kDa Gag construct lacking matrix sequences has been shown to be capable of forming virus-like particles in vivo and is composed of an N-terminal myristylation signal, the C-terminal half of CA, p2, a leucine zipper domain of yeast GCN4 replacing nucleocapsid, and a PPPPY motif replacing p6 (1).
Gag localization in the cell membrane seems to be punctate rather than uniform (18, 20, 35, 45), suggesting its localization in discrete regions of the membrane. In support of this thesis, it has also been found that the HIV-1 membrane has a lipid and protein composition discreet from that of the general plasma membrane and more characteristic of the composition of specialized areas in the plasma membrane known as lipid rafts. Lipid raft membrane subdomains are highly enriched in cholesterol, sphingolipids, and glycosylphosphatidylinositol-linked proteins (4, 5, 38, 48). Hydrogen bonding plays an important role in the association of protein with lipid rafts and is facilitated by attachment of saturated fatty acids, such as myristate and palmitate, to the proteins (43, 44, 46, 53). Proteins associated with lipid rafts can be distinguished from nonraft membrane proteins by their resistance to extraction with cold, nonionic detergents, such as NP-40 and Triton X-100 (5, 27). After nonionic-detergent extraction, lipid raft-associated proteins can be distinguished from soluble proteins by their lower buoyant densities in density gradients.
There are a number of reports indicating that HIV-1 buds from lipid raft domains. Compared to the host cell plasma membrane, the membrane surrounding HIV-1 is enriched in cholesterol and sphingolipids (2). Glycosylphosphatidylinositol-anchored proteins, associated with lipid rafts, are found in HIV-1 (36), while CD45, a protein excluded from lipid rafts, is not (33). Recent work (26, 34) has shown that 30 to 50% of membrane-bound Gag in HIV-1-transfected cells is associated with the detergent-resistant membrane (DRM). In COS1 cells, DRM-associated Gag bands in OptiPrep gradients with a greater buoyant density than that found for the lipid raft-associated caveolin-1, and this has been thought to be due to the greater molecular weights of multimeric Gag complexes (26). Removal of cholesterol from the cell membrane also inhibits HIV-1 budding and reduces viral infectivity (29, 34).
During HIV-1 assembly, the formation of Gag complexes is accompanied by the incorporation of another important precursor protein, GagPol, which contains the sequences for the important viral enzymes protease, reverse transcriptase (RT), and integrase (IN). Gag appears to be responsible for bringing GagPol to the membrane, since unmyristylated Gag or GagPol molecules can also be rescued into assembly complexes by myristylated Gag (31, 37, 49). The factors facilitating this interaction between Gag and GagPol have been less studied than those involved in Gag multimerization. It has recently been shown that the interaction of GagPol with Gag requires the RNA-facilitated polymerization of Gag but that the interaction of GagPol with RNA is not required (22). It has been assumed that Gag and GagPol interact with each other through sequences similar to those involved in Gag-Gag interactions, but recent work has indicated that coding sequences upstream of the RT domain in GagPol are not essential for GagPol incorporation into Gag virus-like particles (10).
In this work, we will show that, unlike bulk Gag/Gag complex, steady-state Gag/GagPol complex is almost completely associated with the DRM, and all newly synthesized GagPol is rapidly taken up as a DRM-associated Gag/GagPol complex.
(This work was performed by R.H. in partial fulfillment of the requirements for the Ph.D. degree at McGill University, Montreal, Canada.)
MATERIALS AND METHODS
Plasmid construction.
Table 1 provides an overview of the expression constructs used in the study, including appropriate references. pSVGAG-RRE-R, pSVGAGPOLprotD25G, and pSVGAGPOLFS5TprotD25G were kind gifts of David Rekosh (49-51) and were renamed pSVGAG, pSVGAG/GAGPOL.P−, and pSVGAGPOL.P−, respectively. The IN deletion mutant, pSVGAGPOL.P−(IN−), was constructed by deleting DNA sequence within the BspMI sites in the GagPol coding sequence. pSVGAGPOL.P− was first partially digested with BspMI, and the 12-kbp fragment containing all sequences but the 746-bp BspMI fragment was separated from the remaining digestion products in a 1% agarose gel. The 12-kbp fragment was electroeluted and purified. An oligonucleotide adapter containing a new translational termination site (5′TGTATAGGGTCA3′) was mixed with the 12-kbp fragment for ligation (28). pSVGAGPOL.P−(IN−) expresses a truncated GagPol sequence up to amino acid 30 of the IN sequence, which is immediately followed by a termination codon, UAG. Protein expression from these constructs requires cotransfection with a plasmid coding for Rev protein, pCMV-rev (49).
TABLE 1.
HIV-1 proviral DNA constructs
| Construct | Viral sequencea | Mutation in coding sequencea | Major Viral Proteins | Reference(s) |
|---|---|---|---|---|
| pSVC21 BH10 | A | None | All | 15 |
| pSVC21 BH10.P− | B | None | All | 15 |
| pSVGAG | C | None | Pr55gag | 49 |
| pSVGAG/GAGPOL.P− | D | None | Pr55gag and Pr160gag-pol | 49 |
| PSVGAGPOL P− | E | None | Pr160gag-pol | 49, 50 |
| pSVGAGPOL P-(IN-) | E | 258 amino acid (31-288) deletion of IN | Pr128gag-pol (truncated Gag-Pol) | |
| pCMV-rev | F | None | Rev protein | 49 |
Some constructs contain mutations that render them protease deficient or frameshift positive. A, full-length HIV-1 proviral DNA; B, full-length HIV-1 proviral DNA, but protease negative (D25R substitution); C, proviral DNA starting at HXB2 nucleotide sequence 679 (SacI site) and terminating immediately after the Gag open reading frame (ORF), HXB2 nucleotide sequence 2428 (BclI site) (proviral DNA is thus missing all the 5′ long terminal repeat [LTR] and leader sequences, as well as all coding regions downstream of Gag, but includes the Rev response element sequence; this construct requires cotransfection with pCMV-rev for protein expression); D, proviral DNA starting at HXB2 nucleotide sequence 679 (SacI site) and terminating immediately after the VifORF, HXB2 nucleotide sequence 5785 (SalI site) (proviral DNA is thus missing all the 5′ LTR and leader sequences, as well as all coding regions downstream of Vif, but includes the Rev response element sequence; contains an inactive protease [D25G substitution] which results in the synthesis of unprocessed Pr55gag and Pr160gag-pol only; this construct requires cotransfection with pCMV-rev for protein expression); E, proviral DNA starting at HXB2 nucleotide sequence 679 (SacI site) and terminating immediately after the Vif ORF, HXB2 nucleotide sequence 5785 (SalI site) (proviral DNA is thus missing all the 5′ LTR and leader sequences, as well as all coding regions downstream of Vif, but includes the RRE sequence; contains an inactive protease [D25G] and a deletion of five Ts [nucleotides 2082 to 2086], which results in only Pr160gag-pol, and not Pr55gag, being synthesized; this construct requires cotransfection with pCMV-rev for protein expression); F, proviral DNA starting at HXB2 nucleotide sequence 5954 and terminating immediately after the Rev ORF.
pSVC21 BH10 contains wild-type HIV-1 proviral DNA sequence. pSVC21 BH10.P− differs from pSVC21 BH10 by a single point mutation at position 25 of the protease region, converting Asp25 to Arg25. Transfection of pSVC21 BH10.P− produces noninfectious viral particles containing wild-type genomic RNA and the unprocessed precursor proteins Gag and GagPol (15). pSVC21 BH10 and pSVC21 BH10.P− were gifts from E. Cohen, University of Montreal.
Cell culture, transfection, and subcellular fractionation.
Culture and transfection of COS7 cells by the calcium-phosphate method were performed as previously described (9, 19). COS7 cells were lysed 48 h posttransfection at 4°C in two ways: (i) in hypotonic medium, where lysis was done by Dounce homogenization in 1.0 ml of hypotonic TE buffer (20 mM Tris-HCl, pH 7.4, 1 mM EDTA, 0.01% β-mercaptoethanol) supplemented with protease inhibitor cocktail (Complete; Boehringer Mannheim), and (ii) in nonionic detergent, where cells were lysed in 1.0 ml of TNT buffer (20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1% Triton X-100) supplemented with protease inhibitor cocktail (“Complete”). For either method, the cell homogenate was then centrifuged at 1,500 × g for 30 min to remove nuclei and unbroken cells. The supernatant (S1) was then centrifuged for 1 h at 100,000 × g in an SW55Ti rotor (Beckman, Columbia, Md.) at 4°C, resulting in the supernatant (S100) and the pellet (P100).
Further fractionation of P100 into membrane-free and membrane-bound proteins was done by a membrane floatation assay (47). The P100 was resuspended in 1 ml of 73% sucrose. Two milliliters of 65% sucrose in TNE (20 mM Tris, pH 7.8, 100 mM NaCl, 1 mM EDTA) was layered on top of the 73% sucrose, and 2 ml of 10% sucrose was layered on top of the 65% sucrose. The gradients were then centrifuged at 100,000 × g in a Beckman SW55Ti rotor overnight at 4°C. Fractions (0.8 ml) were collected and diluted with an equal volume of 2× TNT, and each fraction was immunoprecipitated at 4°C, first with a polyclonal antibody to IN (anti-IN) and then with monoclonal antibodies to CA (anti-CA) (Intracell). Immunoprecipitates from each fraction were dissolved in sodium dodecyl sulfate (SDS) sample buffer and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting.
Resolution of the P100 into detergent-resistant and detergent-susceptible proteins was performed using OptiPrep gradient centrifugation as described by Lindwasser and Resh (26). The P100 fraction was adjusted to 0.75 ml of 50% OptiPrep, and this volume was overlaid with 1.25 ml each of 40, 30, and 20% OptiPrep in TNT and, finally, 0.5 ml of 10% OptiPrep. After centrifugation at 170,000 × g for 4 h at 4°C, eight fractions were collected and diluted with an equal volume of 2× TNT, and each fraction was immunoprecipitated at 4°C, first with anti-IN and then with anti-CA. The immunoprecipitate from each fraction was dissolved in SDS sample buffer and analyzed by SDS-PAGE and Western blotting.
Immunoprecipitation of Gag/Gag and Gag/GagPol complexes.
Immunoprecipitation from the S100 and P100 fractions, or from sucrose or OptiPrep gradient fractions, was done by first using anti-IN to immunoprecipitate the Gag/GagPol complexes. Anti-IN was directed against either the first 16 amino acids of IN (National Institutes of Health AIDS Research and Reference Reagent Program) or amino acids 276 to 288 of IN (a gift from Mark Muesing, Aaron Diamond AIDS Research Center). After treatment with anti-IN, these same fractions were then exposed to anti-CA to immunoprecipitate Gag/Gag complexes. Before use, anti-IN or anti-CA was first cross-linked to Sepharose beads. Forty microliters of antibody and 400 μl of 50% (wt/vol) protein A-Sepharose (Pharmacia) were incubated together in 10 ml of 0.2 M triethanolamine (pH 9). Fifty-two milligrams of dimethyl pimelimidate cross-linker (Pierce) was then added, and the mixture was incubated for 1 h at room temperature. The beads were then washed with 5 ml of 0.2 M triethanolamine (pH 9) and incubated in 10 ml of 0.2 M triethanolamine for another 2 h at room temperature. Equal amounts of protein, 200 to 500 μg (Bio-Rad assay), were incubated with 30 μl of antibody cross-linked to protein A-Sepharose for 1 h at 4°C. The immunoprecipitate was then washed three times with TNT buffer and twice with phosphate-buffered saline (PBS). After the final supernatant was removed, 30 μl of 2× sample buffer (120 mM Tris HCl, pH 6.8, 20% glycerol, 4% SDS, and 0.02% bromphenol blue) was added, and the precipitate was then boiled for 5 min to release the precipitated proteins. After microcentrifugation, the resulting supernatant was analyzed using Western blots.
Western blot analysis.
Equal amounts of cell lysates or immunoprecipitated proteins were separated by SDS-PAGE (10% acrylamide) followed by blotting onto nitrocellulose membranes (Gelmann Science). Western blots were probed with the following antibodies: (i) mouse anti-RT monoclonal antibodies (25), (ii) mouse anti-CA monoclonal antibodies (Cellular Products, Inc., Buffalo, N.Y.), (iii) rabbit anti-caveolin-1 polyclonal antibodies and mouse anti-CD45 monoclonal antibodies (Santa Cruz Biotechnology, Santa Cruz, Calif.), (iv) goat anti-gp120 (National Institutes of Health AIDS Research and Reference Reagent Program), (v) mouse anti-Fyn (BD Transduction Labs, Lexington, Ky.), and (vi) anti-CD59 (BD PharMingen, San Diego, Calif.). These were used as the primary antibodies, and horseradish peroxidase-linked goat anti-mouse, donkey anti-rabbit (Amersham Pharmacia Biotech, Quebec, Canada), and rabbit anti-goat (Sigma, St. Louis, Mo.) were used as secondary antibodies. Antibody binding was detected by enhanced chemiluminescence (ECL kit; Pharmacia Amersham Biotech). The sizes of the detected protein bands were estimated using prestained high-molecular-weight protein markers (New England Biolabs). Western blots were quantitated by phosphorimage analysis, and because in most cases standard curves were not done, the values obtained were approximate.
Metabolic labeling of Gag/Gag and Gag/GagPol complexes.
Metabolic labeling was performed using 60 μCi of Tran 35S-label (obtained from ICN or NEN)/ml of culture medium. Equal numbers of HIV-1-transfected COS7 cells were labeled 64 h posttransfection with [35S]methionine-[35S]cysteine for 10 min and then chased for various lengths of time in Dulbecco modified Eagle medium containing 10% fetal bovine serum and 100 μM cysteine and methionine. After being washed, the cells were lysed hypotonically by Dounce homogenization in 1 ml of hypotonic TE buffer at 4°C, and the cell lysates were centrifuged at 1,500 × g for 30 min to remove nuclei and unbroken cells. The resulting S1 supernatant (1 ml) was fractionated into S100 and P100 fractions by centrifuging 0.5 ml for 1 h at 100,000 × g in an SW55Ti rotor at 4°C. The remaining 0.5 ml was split into two halves: one half was resolved by discontinuous sucrose centrifugation (floatation analysis), while the other half was brought to 1% Triton X-100 and resolved with OptiPrep gradients. Gradient fractions were immunoprecipitated, first with anti-IN and then with anti-CA, for 1 h at 4°C. The immunoprecipitated proteins were then subjected to SDS-10% PAGE and autoradiography.
RESULTS
Detection of Gag/GagPol complexes by coimmunoprecipitation with anti-IN.
In this work, we have examined the distribution of Gag/Gag and Gag/GagPol complexes in cells lysed either hypotonically or in the nonionic detergent Triton X-100. Gag/GagPol complexes were immunoprecipitated with anti-IN and detected on Western blots with both anti-CA and anti-IN, while Gag complexes were immunoprecipitated with anti-CA and detected on Western blots with anti-CA (additional probing with anti-RT yields no additional bands). Figure 1A and B demonstrate the specificity of immunoprecipitation of Gag/GagPol complexes from cells lysed in Triton X-100. Similar results are obtained when cells are lysed hypotonically. The use of anti-IN to immunoprecipitate allows us to detect Gag/GagPol complexes by monitoring Gag as well as GagPol, since Gag will not be immunoprecipitated by anti-IN in the absence of GagPol. This is shown in Fig. 1A and B, which show Western blots of viral proteins present in transfected COS7 cell lysates (A) and viral proteins immunoprecipitated from these cell lysates with anti-IN (B). Lanes 1 represent cells transfected with pSVGAG, a plasmid containing the HIV-1 DNA sequences that code only for Gag, which can be seen in Fig. 1A. Immunoprecipitation of this lysate with anti-IN, as well as several other cell lysates represented in Fig. 1A and B, results in a small amount of precipitation of a species migrating similarly to Gag (Fig. 1B). This species, which may be the heavy chain of anti-IN detected by the secondary antibody, is considered background in these experiments, since it also appears in immunoprecipitates of lysates from cells in which no Gag is synthesized (Fig. 1B, lanes 3, 4, and 5), including untransfected COS7 cell lysates (lane 5).
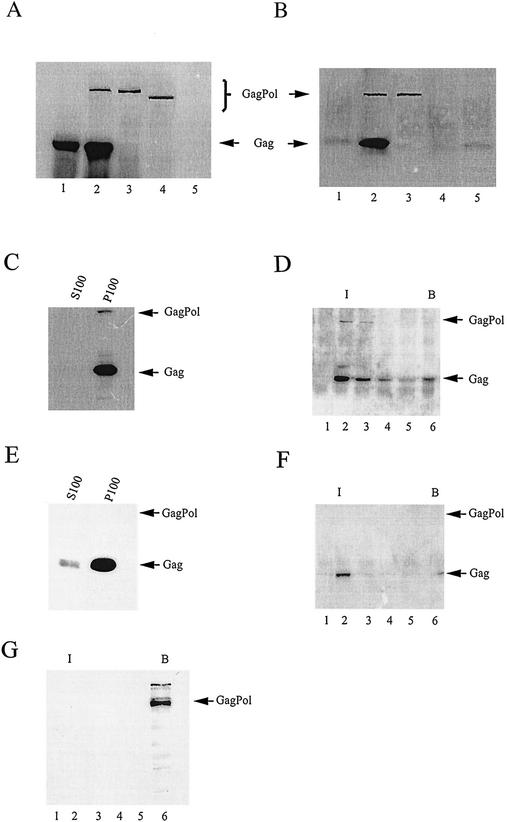
FIG. 1.
Distribution of Gag/Gag and Gag/GagPol complexes in COS7 cells. (A and B) Detection of Gag/GagPol complex by immunoprecipitation with anti-IN. (A) Cell lysate. COS7 cells were either untransfected (lanes 5) or transfected with a plasmid expressing Gag alone (pSVGAG) (lanes 1), a protease-deficient virus (pSVC21 BH10.P−) (lanes 2), GagPol only (pSVGAGPOL.P−) (lanes 3), or a truncated GagPol lacking the C-terminal 258 amino acids of IN [pSVGAGPOL.P−(IN−)] (lanes 4). Sixty-three hours posttransfection, the cells were washed with ice-cold PBS, collected, and lysed on ice in TNT buffer. The lysates were clarified by centrifugation at 1,500 × g, resolved on SDS-10% PAGE, and transferred to nitrocellulose membranes. Proteins were visualized by immunoblotting, using a mixture of anti-RT and anti-CA. (B) Immunoprecipitation. Cell lysates were immunoprecipitated using anti-IN cross-linked to protein A-Sepharose. The immunoprecipitates were washed with TNT lysis buffer and PBS and heated to release the immunoprecipitated proteins, and after centrifugation, the supernatants were resolved using SDS-10% PAGE and then transferred to nitrocellulose membranes. Viral proteins were visualized by immunoblotting, using a mixture of anti-RT and anti-CA. (C to G) Distribution of Gag/Gag and Gag/GagPol complexes in COS7 cells lysed in the absence of detergent. The cells were lysed at 4°C by Dounce homogenization in hypotonic TE buffer, and after clarification by centrifugation at 1,500 × g, were centrifuged at 100,000 × g for 1 h at 4°C. The pellet (P100) was suspended in 1.0 ml of TE buffer, equal to the S100 volume. (C and E) Western blots of immunoprecipitates of S100 and P100, first treated with anti-IN (C) and then with anti-CA (E). (D and F) Western blots of immunoprecipitates of sucrose gradient fractions. The P100 fraction was resolved by discontinuous sucrose gradient centrifugation into membrane-bound and membrane-free proteins (floatation analysis). Each gradient fraction was immunoprecipitated, first with anti-IN (D) and then with anti-CA (F). (G) COS7 cells were transfected with a plasmid, pSVGAGPOL.P−, which codes only for GagPol. After lysis in hypotonic TE buffer and clarification by centrifugation at 1,500 × g, the S1 supernatant was resolved by discontinuous sucrose gradient centrifugation into membrane-bound and membrane-free proteins (floatation analysis). Aliquots from each fraction were then resolved using SDS-PAGE and transferred to nitrocellulose membranes. Proteins were visualized by immunoblotting, using a mixture of anti-RT and anti-CA. For all floatation analyses, I is the interface between 10 and 65% sucrose layers, where membrane-bound protein localizes, and B is the bottom fractions of the gradient, where membrane-free protein remains during centrifugation. The fraction numbers increase from top to bottom of the gradient.
Figure 1A and B, lanes 2, represent an experiment in which COS7 cells are transfected with the plasmid pSVC21 BH10.P−, which contains full-length HIV-1 proviral DNA containing an inactive protease and so codes for both unprocessed Gag and GagPol. Both proteins are detected in the cell lysates in Fig. 1A, lane 2, and both proteins are coimmunoprecipitated with anti-IN (Fig. 1B, lane 2), indicating an interaction between Gag and GagPol. Lanes 3 represent cells transfected only with the plasmid pSVGAGPOL.P−, which codes for GagPol containing a defective viral protease and, because of a mutation causing a permanent frame shift, does not code for Gag. Therefore, only the presence of wild-type GagPol is seen in either the cell lysate (Fig. 1A, lane 3) or the anti-IN immunoprecipitate (Fig. 1B, lane 3). Lanes 4 represent an experiment in which cells are transfected with a plasmid, pSVGAGPOL.P−(IN−), which codes only for a mutant GagPol precursor in which the IN sequence has been deleted and the protease inactivated. The truncated GagPol is present in the cell lysates (Fig. 1A, lane 4) but is not immunoprecipitated with anti-IN (Fig. 1B, lane 4). In lanes 5, neither precursor can be detected in untransfected COS7 cell lysate (Fig. 1A, lane 5) or immunoprecipitated lysate (Fig. 1B, lane 5).
The data in Fig. 1A and B, therefore, indicate that immunoprecipitation with anti-IN will immunoprecipitate GagPol in the absence of Gag but will not immunoprecipitate Gag unless it is bound to GagPol. As will be clear below, the signal for Gag in the immunoprecipitated complex is generally much stronger than that for GagPol. This is probably because GagPol interacts with larger multimeric Gag complexes. The result is that the measurement of Gag in an anti-IN immunoprecipitate increases the sensitivity of detection of the Gag/GagPol complex.
In HIV-1-infected COS7 cells, steady-state Gag and Gag/GagPol are primarily pelletable, membrane-bound complexes.
In Fig. 1C to G, we show the distribution of Gag/Gag and Gag/GagPol complexes in three cellular fractions of COS7 cells transfected with pSVC21 BH10.P−: nonpelletable and membrane free, pelletable and membrane-free, and pelletable and membrane bound. Hermida-Matsumoto and Resh and Tritel and Resh reported that nearly all steady-state Gag in HIV-1-transfected COS1 cells is pelletable and membrane bound (18, 52). The data in Fig. 1C to G support a similar conclusion for the distribution of Gag in HIV-1-transfected COS7 cells and also show a similar distribution for Gag/GagPol complexes in the cell. For the experiments represented in Fig. 1C to G, cells were swollen in hypotonic buffer without detergent and lysed by Dounce homogenization for 25 to 30 strokes. The lysate was first centrifuged at low speed (1,500 × g) for 30 min, and the resulting supernatant (S1) was then centrifuged at 100,000 × g for 1 h, resulting in a pellet (P100) and supernatant (S100). Nonpelletable, membrane-free Gag and GagPol were defined as the molecules remaining in the S100 supernatant, while the pelletable material (P100) was further resolved by discontinuous sucrose-gradient centrifugation into membrane-free components remaining at the bottom of the gradient and membrane-bound components located at the interface between the 65 and 10% sucrose layers (floatation assay). For immunoprecipitation experiments, the P100 and S100 fractions were in the same volumes of TNT buffer.
In Fig. 1C, anti-IN was used first to immunoprecipitate Gag/GagPol from either the S100 or P100 fraction, and then the remaining Gag in these fractions was further immunoprecipitated with anti-CA (Fig. 1E). Western blot analysis of the immunoprecipitates, using as the primary antibody either anti-CA- anti-RT (Fig. 1C) or anti-CA (Fig. 1E), shows that almost all Gag/GagPol (Fig. 1C) and Gag/Gag (Fig. 1E) in transfected COS7 cells is found in the P100 fraction. In Fig. 1D and F, the P100 fraction was further resolved into membrane-free and membrane-bound fractions by discontinuous sucrose gradient centrifugation. Gradient fractions were first immunoprecipitated with anti-IN (Fig. 1D), and these gradient fractions were then immunoprecipitated with anti-CA (Fig. 1F). Western blot analysis of each immunoprecipitated fraction, using anti-CA- anti-RT (Fig. 1D) or anti-CA (Fig. 1F) as the primary antibody, shows that almost all Gag/GagPol (Fig. 1D) or Gag/Gag (Fig. 1F) complexes are membrane bound. It is not known if the small amount of membrane-free Gag/GagPol detected in the P100 fraction (Fig. 1D) dissociated from the membrane or was never associated with it. We thus conclude that in transfected COS7 cells, the majority of Gag/Gag and Gag/GagPol complexes are pelletable, membrane-bound complexes.
The appearance of Gag in the anti-IN immunoprecipitates indicates that Gag is involved in the complex with GagPol. To show directly that the appearance of GagPol at the membrane requires Gag, COS7 cells were transfected with a plasmid coding only for GagPol (pSVGagPol.P−). Lysates of cells lysed hypotonically were centrifuged at low speed, and the S1 was resolved by floatation analysis. Western blots of each gradient fraction were probed with anti-RT- anti-CA, and as shown in Fig. 1G, GagPol does not associate with the membrane in the absence of Gag. Since GagPol has the same membrane-binding sequences at its N terminus as Gag, it is not clear why it does not bind readily to the membrane. This could be due to a steric blockage of the N terminus by the much larger Pol region. This possibility is supported by data indicating (i) that GagPol does not bind to genomic RNA when it associates with Gag (22) and (ii) that the incorporation of GagPol into Gag virus-like particles can occur in the absence of the Gag sequences within the GagPol molecule (10).
Membrane domains associated with Gag and Gag/GagPol complexes.
The resistance of membrane-bound Gag/GagPol and Gag/Gag complexes to nonionic detergent was examined. When cells are lysed at 4°C in TNT buffer, the distribution of Gag/GagPol and Gag/Gag complexes within the three lysate fractions (nonpelletable and membrane free, pelletable and membrane free, and pelletable and membrane bound) are different, and these data are shown in Fig. 2.
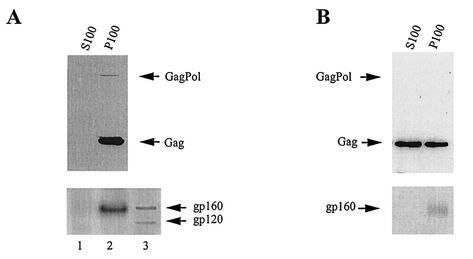
FIG. 2.
Distribution of Gag/Gag and Gag/GagPol complexes in COS7 cells lysed in the presence of 1% Triton X-100. COS7 cells transfected with BH10.P− DNA were lysed on ice in TNT buffer (20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1% Triton X-100), and after clarification by centrifugation at 1,500 × g, the S1 supernatant was further centrifuged at 100,000 × g for 1 h at 4°C. The pellet (P100) was suspended in 1.0 ml of TNT buffer, equal to the volume of the supernatant (S100). Gag/GagPol (A) and Gag/Gag (B) were immunoprecipitated with anti-IN and then anti-CA, respectively, from the S100 and P100 fractions. The immunoprecipitates were analyzed by Western blotting as described in the legend to Fig. 1 and were probed with anti-RT, anti-CA, and anti-gp120 (A) or anti-CA and anti-gp120 (B). Lane 3 in panel A contains HIV-1-infected cell lysate.
Although Gag/GagPol and Gag/Gag complexes are found in the pelletable and membrane-bound fraction when COS7 cells are lysed in hypotonic medium by Dounce homogenization, lysing cells in 1% Triton X-100 specifically alters the distribution of Gag/Gag complexes. Gag/GagPol complexes are immunoprecipitated from the S100 and P100 fractions using anti-IN, and Western blots are probed with anti-CA- anti-RT (Fig. 2A). Gag/Gag complexes are then immunoprecipitated from these same fractions with anti-CA, and the Western blots are probed with anti-CA (Fig. 2B). Figure 2A shows that the Gag/GagPol complexes are detected only in the P100 fraction. In contrast, Fig. 2B shows that exposure of cells to 1% Triton X-100 releases 56% of Gag complexes into the S100 fraction. We have also probed the Western blots shown in Fig. 2A and B with anti-gp120. The precursor to gp120, gp160, is not detectable in the immunoprecipitates produced from the S100 fraction, but both anti-IN and anti-CA immunoprecipitates from the P100 fraction contain gp160. Figure 2A, bottom, shows a Western blot probed with anti-gp120 and compares the electrophoretic mobility of gp120 and gp160 found in infected cell lysate with the mobility of the protein band detected by anti-gp120 in the P100 anti-IN immunoprecipitate fraction. The presence of gp160 in the anti-IN (Fig. 2A) or anti-CA (Fig. 2B) immunoprecipitate could indicate that some membrane may still be present. However, some Gag cores that do not contain membrane have been shown to retain Env protein even after extraction with Triton X-100 (55), so the presence of gp160 does not in itself indicate the presence of membrane, but work with known membrane markers discussed below supports this conclusion.
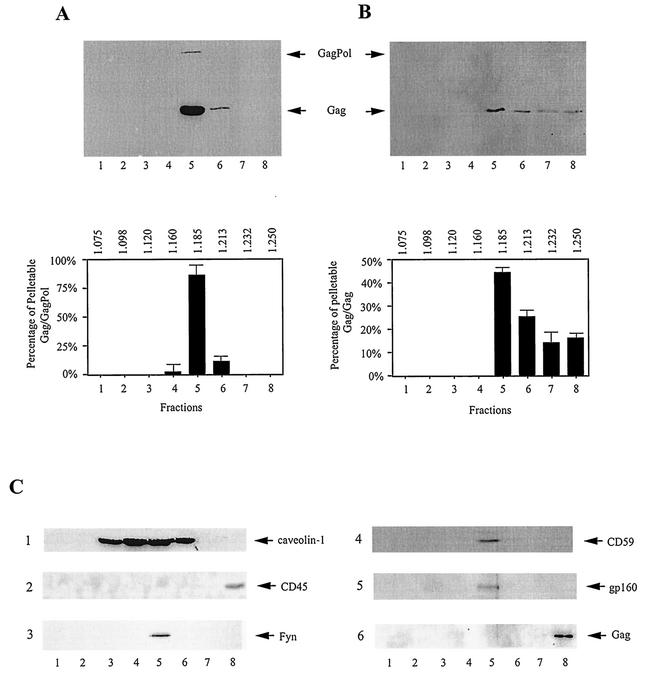
Evidence for the association of pelletable Gag and GagPol with lipid raft domains is shown in Fig. 3, in which their buoyant densities on OptiPrep gradients were used to monitor their associations with the DRM. OptiPrep gradients were set up according to the method of Lindwasser and Resh (26), who showed that these DRM-Gag complexes migrate in this medium at a lighter buoyant density than membrane-free protein, which remains at the bottom of the gradient. In Fig. 3, the P100 fraction was resolved by centrifugation in OptiPrep gradients, and collected fractions were immunoprecipitated, first with anti-IN (Fig. 3A) and then with anti-CA (Fig. 3B). Western blots of these immunoprecipitates were probed with either anti-CA- anti-RT (Fig. 3A) or anti-CA (Fig. 3B). Figure 3A shows that ∼97% of Gag/GagPol in the P100 fraction has a density associated with the DRM (∼86 and 11% in fractions 5 and 6), while Fig. 3B shows that ∼69% of the Gag complexes in the P100 fraction have a density associated with the DRM (∼44 and 25% in fractions 5 and 6).
FIG. 3.
Buoyant densities of DRM-associated Gag/GagPol and Gag/Gag complexes. COS7 cells transfected with BH10.P− DNA were lysed on ice in TNT buffer, and after clarification by centrifugation at 1,500 × g, were centrifuged at 100,000 × g for 1 h at 4°C to produce the S100 and P100 fractions. The pellet (P100) was resuspended and resolved by centrifugation in OptiPrep gradients, prepared as described in Materials and Methods. Each gradient fraction was immunoprecipitated at 4°C, first with anti-IN (A) and then with anti-CA (B). The immunoprecipitates from each fraction were resolved by SDS-PAGE, and Western blots were probed with either anti-CA- anti-RT (A) or anti-CA (B). The results are presented graphically at the bottom of each panel, with buoyant densities listed at the top of each graph. The graphs represent the means of experiments performed three times or more, and the error bars represent standard deviations. (C) Distribution of other proteins on OptiPrep gradients. Western blots: 1, P100 fraction probed with anti-caveolin-1; 2, P100 fraction probed with anti-CD45; 3, anti-IN immunoprecipitate from the P100 fraction probed with anti-Fyn; 4, anti-IN immunoprecipitate from the P100 fraction probed with anti-CD 59; 5, anti-IN immunoprecipitate from the P100 fraction probed with anti-gp120; 6, anti-CA immunoprecipitate from the S100 fraction probed with anti-CA. The fraction numbers increase from top to bottom, with DRM-associated protein localizing near the middle of the gradient (fraction 5) and non-raft-associated proteins localizing at the bottom of the gradient.
The DRM with which Gag has been found to be associated also has been found to have the high cholesterol, sphingomyelin, and glycosphingolipids associated with specialized membrane areas known as lipid rafts (4, 5). The correlations between lipid raft and non-lipid raft proteins and the densities on OptiPrep gradients are shown in Fig. 3C. Blots 2 and 6 show that CD45 in the P100 fraction (gradient 2), a membrane protein not associated with lipid rafts (33), and Gag present in the S100 fraction (blot 6) both band at the bottom of the gradient (fraction 8). Blots 3 and 4 show that the lipid raft-associated proteins Fyn and CD59 (26, 33), along with gp160 (blot 5), remain associated with the Gag/GagPol complex immunoprecipitated from the P100 fraction with anti-IN, i.e., they band at the same bouyant densities. Blot 1 shows that the distribution in the P100 fraction of another lipid raft marker, caveolin-1, is rather broad and peaks in fraction 4 at a lighter buoyant density than that found for the immunoprecipitated Gag/GagPol complex and associated lipid raft markers (fraction 5). This has previously been reported for Gag/Gag complexes, although caveolin-1 was found to band at fraction 3 in similar Optiprep gradients rather than at fraction 4 (26), which could be due to the authors' use of 0.5% Triton X-100 instead of 1% Triton X-100.
Thus, the distributions of steady-state Gag and GagPol at the membrane differ. All detectable membrane-bound Gag/GagPol complexes appear to be associated with the DRM, which probably represents lipid raft domains. On the other hand, only ∼44% of membrane-bound Gag is resistant to dissociation from the membrane with 1% Triton X-100, and ∼70% of this appears to be associated with lipid rafts. Thus, ∼30% of membrane-bound Gag in HIV-transfected COS7 cells is associated with the DRM.
Association of newly synthesized Gag/GagPol and Gag/Gag with membrane domains.
HIV-1-transfected COS7 cells were labeled 64 h posttransfection with [35S]methionine-[35S]cysteine for 10 min, followed by a chase period with cold methionine-cysteine. Aliquots of cells were taken during the chase up to 60 min. After being washed, the cells were lysed hypotonically at 4°C, and the cell lysates were centrifuged at 1,500 × g to remove nuclei and broken cells; 0.5 ml of S1 supernatant was further fractionated into S100 and P100 fractions. The remaining 0.5 ml was split into two halves: one half was resolved by discontinuous sucrose centrifugation (floatation analysis) to determine membrane-free and membrane-bound proteins; the other half was brought to 1% Triton X-100, and detergent-resistant and detergent susceptible-proteins were separated by centrifugation through OptiPrep gradients. The gradient fractions were immunoprecipitated, first with anti-IN and then with anti-CA, for analysis of the distribution of radioactive Gag/GagPol and Gag/Gag complexes, respectively, using one-dimensional (1-D) PAGE and autoradiography.
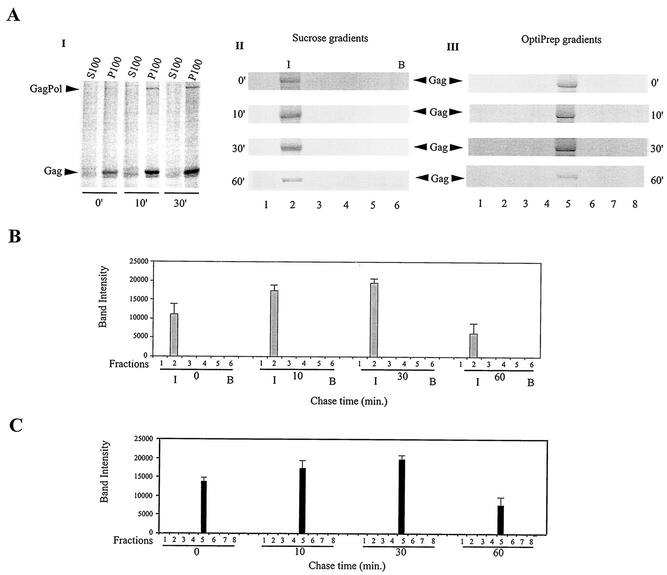
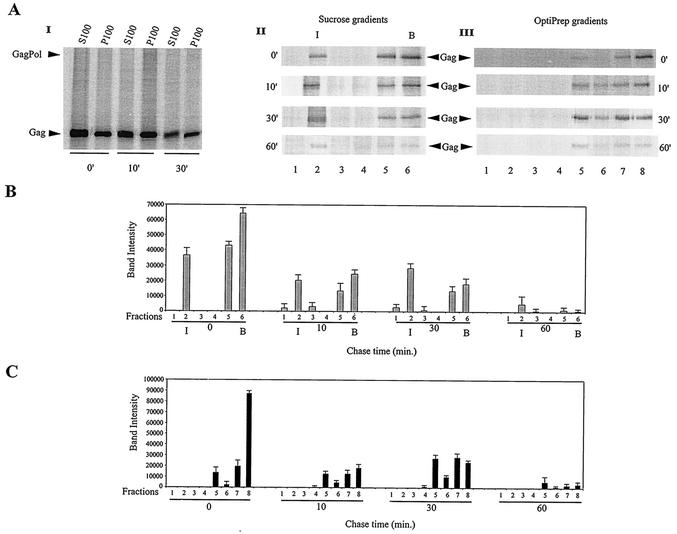
Figure 4A, gel I, shows the distribution of newly synthesized Gag/GagPol in the S100 and P100 fractions over a 30-min chase. Cells were lysed hypotonically. Gag/GagPol was immunoprecipitated from each fraction with anti-IN and resolved by 1-D PAGE. There was a steady increase in pelletable Gag/GagPol over the first 30 min of chase, but the absence of newly synthesized Gag/GagPol in the S100 suggests that the formation of this complex from newly synthesized Gag and GagPol is rapid and that it moves rapidly into the pelletable fraction.
FIG. 4.
Association of newly synthesized Gag/GagPol with bulk membrane and DRM. (A) HIV-1-transfected COS7 cells were metabolically labeled 64 h posttransfection with [35S]methionine-[35S]cysteine for 10 min and then chased for various lengths of time in medium containing 100 μM cysteine and methionine, as described in Materials and Methods. Aliquots of cells obtained at various times were lysed hypotonically in TE buffer and centrifuged at 1,500 × g to produce the S1 supernatant, and 0.5 ml of S1 was further fractionated by centrifugation to produce the S100 and P100 fractions. These fractions were exposed to anti-IN to immunoprecipitate Gag/GagPol and were analyzed by 1-D PAGE and autoradiography (gel I). The remaining S1 was split into two halves: one half was resolved by discontinuous sucrose centrifugation (floatation analysis), while the other half was brought to 1% Triton X-100 and resolved by centrifugation on OptiPrep gradients. The gradient fractions were exposed to anti-IN to immunoprecipitate Gag/GagPol and analyzed by 1-D PAGE and autoradiography. Gel II, 1-D PAGE and autoradiography of immunoprecipitates from sucrose gradient fractions; gel III, 1-D PAGE and autoradiography of immunoprecipitates from OptiPrep gradients. (B) Graphic representation of panel A, gel II. (C) Graphic representation of panel A, gel III. The bar graphs represent the means of experiments performed three times or more, and the error bars represent standard deviations.
Figure 4A, gel II, shows the distribution of newly synthesized Gag/GagPol between the cytoplasm and membrane over time, using floatation analysis to analyze the S1 supernatant not containing detergent, and the results are presented graphically in Fig. 4B. Gag/GagPol in each fraction was immunoprecipitated with anti-IN and was resolved by 1-D PAGE. Floatation analysis (using the radioactive Gag band as the measure of Gag/GagPol present) indicates that all newly synthesized Gag/GagPol is present at the membrane at 0 min of chase, i.e., after a 10-min pulse. The amount of Gag/GagPol at the membrane increases during the first 30 min and then undergoes a significant decrease between 30 and 60 min. The increase in newly synthesized Gag/GagPol over the first 30 min of the chase period probably reflects continued formation of this complex during this period, and the absence of newly synthesized Gag/GagPol in the S100 (Fig. 4A, gradient I) indicates that formation of the complex is rapid.
Figure 4A, gel III, follows the appearance of radioactive Gag/GagPol at the DRM over time by centrifugation of the S1 supernatant containing 1% Triton X-100 in an OptiPrep gradient. Gag/GagPol was immunoprecipitated from each fraction with anti-IN and was resolved by 1-D PAGE. These results are presented graphically in Fig. 4C. The results closely mirror the results shown in Fig. 4B, i.e., the Gag/GagPol appearing at the membrane in Fig. 4B is all associated with the DRM (Fig. 4C).
The results in Fig. 5 indicate a more complex behavior for newly synthesized Gag. Figure 5A, gel I, shows the distribution of newly synthesized Gag/Gag in the S100 and P100 fractions over a 30-min chase. Cells were lysed hypotonically. Gag/Gag was immunoprecipitated from each fraction with anti-CA and resolved by 1-D PAGE. At 0 min of chase, newly synthesized Gag/Gag is found in both S100 and P100, with a larger fraction in S100, and by 30 min of chase, newly synthesized Gag in both S100 and P100 have diminished.
FIG. 5.
Association of newly synthesized Gag/Gag with bulk membrane and DRM. After immunoprecipitation with anti-IN of the cell lysate fractions and gradient fractions described in the legend to Fig. 4, the same fractions were treated further with anti-CA to immunoprecipitate Gag/Gag complexes, which were analyzed by 1-D PAGE and autoradiography. (A) Gel I, 1-D PAGE and autoradiography of anti-CA immunoprecipitates from S100 and P100; gel II, 1-D PAGE and autoradiography of anti-CA immunoprecipitates from sucrose gradient fractions; gel III, 1-D PAGE and autoradiography of anti-CA immunoprecipitates from OptiPrep gradient fractions. (B) Graphic representation of the results shown in panel A, gel II. (C) Graphic representation of the results shown in panel A, gel III. The bar graphs represent the means of experiments performed three times or more, and the error bars represent standard deviations.
Figure 5A, gel II, shows the distribution of newly synthesized Gag protein between the cytoplasm and membrane over time, using floatation analysis to analyze the S1 supernatant not containing detergent. The results are plotted in Fig. 5B. The amount of radioactive Gag appearing at the membrane does not change greatly over the first 30 min but rapidly diminishes between 30 and 60 min of chase. The amount of radioactive Gag in the cytoplasm, which at 0 min of chase represents 76% of the labeled Gag, diminishes over time. At 60 min of chase, there is very little radioactive Gag left either at the membrane or in the cytoplasm.
Figure 5A, gel III, follows the appearance of radioactive Gag/Gag at the DRM over time by centrifugation of the S1 supernatant containing 1% Triton X-100 in an OptiPrep gradient. Radioactive Gag/Gag was immunoprecipitated from each fraction with anti-CA and was resolved by 1-D PAGE. These results are presented graphically in Fig. 5C. Taking fractions 5 and 6 as containing Gag associated with the DRM, there is an increase in labeled Gag over the first 30 min of chase, followed by a decrease between 30 and 60 min of chase. At 0 min of chase, the majority of Gag is not associated with the DRM, and over time, this material decreases without a concomitant increase in detergent-resistant Gag.
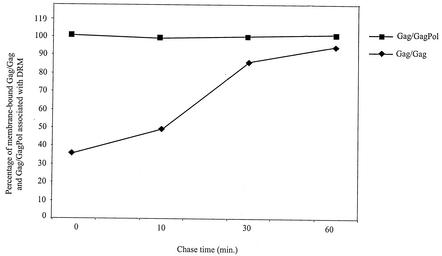
The association of newly synthesized, membrane-bound Gag/Gag complex with the DRM occurs more slowly than for Gag/GagPol, first passing through a detergent-susceptible membrane, or “non-DRM,” phase. This is shown in Fig. 6, which presents graphically the percentages of total membrane-bound Gag/GagPol (obtained from Fig. 4B) and membrane-bound Gag/Gag (obtained from Fig. 5B) that become associated with the DRM (obtained from Fig. 4C and 5C) over time. After a 10-min pulse, all newly synthesized Gag/GagPol (Fig. 6) is associated with the DRM, while only 38% of membrane-bound Gag/Gag is associated with the DRM, although this value reaches close to 100% after a 30-min chase.
FIG. 6.
Rates of association of membrane-bound Gag/GagPol and Gag/Gag with DRM. The percentage of total membrane-associated protein that is DRM-associated protein was calculated from Fig. 4 and 5 by dividing the amount of DRM-associated protein (Gag/GagPol [Fig. 4C]; Gag/Gag [Fig. 5C]) by the total amount of membrane-associated protein (Gag/GagPol [Fig. 4B]; Gag/Gag [Fig. 5B]).
DISCUSSION
The results presented in this paper indicate that most, and perhaps all, steady-state Gag/GagPol complex is associated with the DRM. This contrasts very much with the membrane association of Gag in the cell. In HIV-1-transfected COS1 cells, nearly all steady-state Gag appears to be associated with membrane in the absence of detergent (18, 52), a result similar to what we report here for COS7 cells but somewhat higher than that reported for HIV-1-transfected HeLa and Jurkat cells, in which only 50% of Gag was found associated with membrane (34). In COS7 cells lysed in 1% Triton X-100 (TNT buffer), ∼30% of this membrane-bound Gag is associated with the DRM, similar to previous findings for COS1 cells (26) but somewhat lower than the 50% value reported for HeLa cells (34). Since others have shown that HIV-1 and Gag virus-like particles bud from lipid raft domains (2, 26, 29, 34, 36), it is likely that the DRM we are studying may in fact be lipid raft membrane domains. This is further supported by the comigration of known lipid-raft proteins, such as Fyn, CD59, and caveolin-1, with Gag/Gag and Gag/GagPol immunoprecipitates on the Optiprep gradient (Fig. 3C).
Newly synthesized Gag/Gag also behaves differently than most newly synthesized Gag/GagPol. Much newly synthesized Gag does not appear to become membrane associated and may be degraded in the cytoplasm, as has been reported in COS1 cells (52). Of the newly synthesized Gag associated with the membrane, only 38% is associated with the DRM after a 10-min pulse, although this increases to nearly 100% over a 30-min chase. On the other hand, newly synthesized GagPol interacts rapidly with a small fraction of newly synthesized Gag, and this Gag/GagPol complex is located entirely in the DRM after a 10-min pulse. The kinetics of the association of Gag/GagPol and Gag/Gag with the DRM is shown in Fig. 6. Ono and Freed (34) have shown very similar data in HIV-1-transfected HeLa cells for the passage of newly synthesized Gag/Gag complexes from the non-DRM to the DRM over a similar time period. We find that at steady state, only 30% of membrane-associated Gag is DRM-associated. At least some of the membrane-bound Gag that is detergent susceptible may represent newly synthesized Gag that will eventually localize in the DRM.
A likely explanation for the rapid location of newly synthesized GagPol in the DRM is that it is all bound to the early Gag associated with the DRM. Ono and Freed (34) showed that the stable association of newly synthesized Gag/Gag with the DRM required regions of Gag associated with Gag multimerization, i.e., p2 and the N terminus of the nucleocapsid. It was suggested that Gag multimerization might facilitate its binding to the DRM. The gradual increase in membrane-bound Gag found at the DRM might reflect the speed of Gag multimerization. Since GagPol interaction with Gag has been shown to require an RNA-facilitated multimerization of Gag (22), and Gag is in great excess over GagPol, GagPol might interact with the earliest multimeric Gag complex formed, which would also be the first to associate with the DRM. However, other parameters could be responsible for the rapid localization of GagPol to the DRM. These could include a GagPol-facilitated multimerization of Gag or a conformational change of the polymeric Gag/Gag, which would allow Gag bound to GagPol to more rapidly localize in the DRM. What is not known is whether polymerization of Gag precedes its localization in the DRM or whether the DRM helps facilitate, or stabilize, this polymerization.
The non-DRM, with which much steady-state Gag associates, is not identified. While Gag polymerization could facilitate the movement of Gag from the non-DRM to the DRM, an alternative explanation has been suggested (34), in which the apparent movement from the non-DRM to the DRM could represent two stages of bonding to the DRM: (i) a weak association with the DRM of monomeric or slightly multimeric Gag, which will be susceptible to detergent, and (ii) a stronger, detergent-resistant association with the DRM as Gag forms larger polymeric structures.
After synthesis, a 30-min chase is required for all membrane-associated Gag/Gag to associate with the DRM (Fig. 6). There is then a significant loss of DRM-associated Gag/Gag and Gag/GagPol between 30 and 60 min of chase (Fig. 4C and 5C). These observations are consistent with an early localization of Gag/GagPol at the DRM but with budding commencing only after more newly synthesized Gag has moved into this site. The significant decrease in DRM-associated Gag/GagPol (Fig. 4C) or DRM-associated Gag/Gag (Fig. 5C) between 30 and 60 min of chase may reflect their loss from the DRM due to the budding of virions. However, preliminary experiments in our laboratory indicate that a major fraction of the DRM radioactivity lost during this time and later in the chase period is not accounted for by the amount of radioactivity appearing in virions, and it may be due to the release of Gag and GagPol back into the cytoplasm.
Acknowledgments
This work was supported by grants from the Canadian Institutes for Health Research and the Canadian Foundation for AIDS Research.
We thank Sandy Fraiberg for assistance in preparation of the manuscript.
REFERENCES
- 1.Accola, M. A., B. Strack, and H. G. Gottlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aloia, R. C., H. Tian, and F. C. Jensen. 1993. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc. Natl. Acad. Sci. USA 90:5181-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berthet-Colominas, C., S. Monaco, A. Novelli, G. Sibai, F. Mallet, and S. Cusack. 1999. Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with a monoclonal antibody Fab. EMBO J. 18:1124-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, D. A., and E. London. 1998. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 14:111-136. [DOI] [PubMed] [Google Scholar]
- 5.Brown, D. A., and E. London. 2000. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 275:17221-17224. [DOI] [PubMed] [Google Scholar]
- 6.Bryant, M., and L. Ratner. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 87:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, S., and V. M. Vogt. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cen, S., Y. Huang, A. Khorchid, J. L. Darlix, M. A. Wainberg, and L. Kleiman. 1999. The role of Pr55gag in the annealing of tRNALys3 to human immunodeficiency virus type 1 genomic RNA. J. Virol. 73:4485-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiu, H., S. Yao, and C. Wang. 2002. Coding sequences upstream of the HIV-1 reverse transcriptase domain in GagPol are not essential for incorporation of the Pr160gag-pol into virus particles. J. Virol. 76:3221-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craven, R. C., and L. J. Parent. 1996. Dynamic interactions of the Gag polyprotein. Curr. Top. Microbiol. Immuol. 214:65-94. [DOI] [PubMed] [Google Scholar]
- 12.Gamble, T. R., S. Yoo, F. Vajdos, U. K. Von Schwedler, J. McCutcheon, and W. I. Sundquist. 1997. Structure of the carboxy-terminal dimerization domain of HIV-1 capsid protein. Science 278:849-853. [DOI] [PubMed] [Google Scholar]
- 13.Garnier, L., L. Ratner, B. Rovinski, S. X. Cao, and J. W. Wills. 1998. Particle size determinants in the human immunodeficiency virus type 1 Gag protein. J. Virol. 72:4667-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gheysen, D., E. Jacobs, F. de Foresta, C. Thiriart, M. Francotte, D. Thines, and M. De Wilde. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103-112. [DOI] [PubMed] [Google Scholar]
- 15.Göttlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:5781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haffar, O., J. Garrigues, B. Travis, P. Moran, J. Zarling, and S. L. Hu. 1990. Human immunodeficiency virus-like, nonreplicating, gag-env particles assemble in a recombinant vaccinia virus expression system. J. Virol 64:2653-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson, L. E., H. C. Krutzsch, and S. Oroszlan. 1983. Myristyl amino-terminal acylation of murine retrovirus proteins: an unusual post-translational protein modification. Proc. Natl. Acad. Sci. USA 80:339-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hermida-Matsumoto, L., and M. D. Resh. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 74:8670-8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, Y., J. Mak, Q. Cao, Z. Li, M. A. Wainberg, and L. Kleiman. 1994. Incorporation of excess wild type and mutant tRNALys3 into HIV-1. J. Virol. 68:7676-7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones, T. A., G. Blaug, M. Hansen, and E. Barklis. 1990. Assembly of Gag-beta-galactosidase proteins into retrovirus particles. J. Virol. 64:2265-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karacostas, V., K. Nagashima, M. A. Gonda, and B. Moss. 1989. Human immunodeficiency virus-like particles produced by a vaccinia virus expression vector. Proc. Natl. Acad. Sci. USA 86:8964-8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khorchid, A., R. Halwani, M. A. Wainberg, and L. Kleiman. 2002. Role of RNA in facilitating Gag/Gag-Pol interaction. J. Virol. 76:4131-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kräusslich, H.-G., M. Fäcke, A.-M. Heuser, J. Konvalinka, and H. Zentgraf. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 69:3407-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, S., C. P. Hill, W. I. Sundquist, and J. T. Finch. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409-413. [DOI] [PubMed] [Google Scholar]
- 25.Li, X., E. Amandoron, M. A. Wainberg, and M. A. Parniak. 1993. Generation and characterization of murine monoclonal antibodies reactive against N-terminal and other regions of HIV-1 reverse transcriptase. J. Med. Virol. 39:251-259. [DOI] [PubMed] [Google Scholar]
- 26.Lindwasser, O. W., and M. D. Resh. 2001. Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains. J. Virol. 75:7913-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.London, E., and D. A. Brown. 2000. Insolubility of lipids in triton X-100: physical origin and relationship to sphingolipid/cholesterol membrane domains (rafts). Biochim. Biophys. Acta 1508:182-195. [DOI] [PubMed] [Google Scholar]
- 28.Mak, J., A. Khorchid, Q. Cao, Y. Huang, I. Lowy, M. A. Parniak, V. R. Prasad, M. A. Wainberg, and L. Kleiman. 1997. Effects of mutations in Pr160gag-pol upon tRNALys3 and Pr160gag-pol incorporation into HIV-1. J. Mol. Biol. 265:419-431. [DOI] [PubMed] [Google Scholar]
- 29.Maziere, J. C., J. C. Landureau, P. Giral, M. F. Auclair, L. Fall, A. Lachgar, A. Achour, and D. Zagury. 1994. Lovastatin inhibits HIV-1 expression in H9 human T lymphocytes cultured in cholesterol-poor medium. Biomed. Pharmacother. 48:63-67. [DOI] [PubMed] [Google Scholar]
- 30.Momany, C., L. Kovari, A. Prongay, W. Keller, R. Gitti, B. Lee, A. Gorbalenya, L. Tong, J. McClure, L. Ehrlich, M. Summers, C. Carter, and M. Rossmann. 1996. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol. 3:763-770. [DOI] [PubMed] [Google Scholar]
- 31.Morikawa, Y., S. Hinata, H. Tomoda, T. Goto, M. Nakai, C. Aizawa, H. Tanaka, and S. Omura. 1996. Complete inhibition of human immunodeficiency virus Gag myristoylation is necessary for inhibition of particle budding. J. Biol. Chem. 271:2868-2873. [DOI] [PubMed] [Google Scholar]
- 32.Morikawa, Y., D. J. Hockley, M. V. Nermut, and I. M. Jones. 2000. Roles of matrix, p2, and N-terminal myristoylation in human immunodeficiency virus type 1 Gag assembly. J. Virol. 74:16-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen, D. H., and J. E. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ono, A., and E. O. Freed. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA 98:13925-13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ono, A., J. M. Orenstein, and E. O. Freed. 2000. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J. Virol. 74:2855-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ott, D. E. 1997. Cellular proteins in HIV virions. Rev. Med. Virol. 7:167-180. [DOI] [PubMed] [Google Scholar]
- 37.Park, J., and C. D. Morrow. 1992. The nonmyristylated Pr160gag-pol polyprotein of human immunodeficiency virus type 1 interacts with Pr55gag and is incorporated into virus-like particles. J. Virol. 66:6304-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pralle, A., P. Keller, E. L. Florin, K. Simons, and J. K. Horber. 2000. Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J. Cell Biol. 148:997-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rein, A., M. R. McClure, N. R. Rice, R. B. Luftig, and A. M. Schultz. 1986. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc. Natl. Acad. Sci. USA 83:7246-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Resh, M. D. 1994. Myristylation and palmitylation of Src family members. The fats of the matter. Cell 76:411-413. [DOI] [PubMed] [Google Scholar]
- 41.Rhee, S. S., and E. Hunter. 1987. Structural role of the matrix protein of type D retroviruses in Gag polyprotein stability and capsid assembly. J. Virol. 64:4383-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhee, S. S., and E. Hunter. 1987. Myristylation is required for intracellular transport but not for assembly of D-type retrovirus capsids. J. Virol. 61:1045-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robbins, S. M., N. A. Quintrell, and J. M. Bishop. 1995. Myristoylation and differential palmitoylation of the HCK protein-tyrosine kinases govern their attachment to membranes and association with caveolae. Mol. Cell. Biol. 15:3507-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodgers, W., B. Crise, and J. K. Rose. 1994. Signals determining protein tyrosine kinase and glycosyl-phosphatidylinositol-anchored protein targeting to a glycolipid-enriched membrane fraction. Mol. Cell. Biol. 14:5384-5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandefur, S., V. Varthakavi, and P. Spearman. 1998. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55gag. J. Virol. 72:2723-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaul, P. W., E. J. Smart, L. J. Robinson, Z. German, I. S. Yuhanna, Y. Ying, R. G. Anderson, and T. Michel. 1996. Acylation targets endothelial nitric oxide synthase to plasmalemmal cavolae. J. Biol. Chem. 271:6518-6522. [DOI] [PubMed] [Google Scholar]
- 47.Simon, J. H. M., E. A. Carpenter, R. A. M. Fouchier, and M. H. Malim. 1999. Vif and the p55gag polyprotein of human immunodeficiency virus type 1 are present in colocalizing membrane-free cytoplasmic complexes. J. Virol. 73:2667-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature 387:569-572. [DOI] [PubMed] [Google Scholar]
- 49.Smith, A. J., M. I. Cho, M. L. Hammarskjöld, and D. Rekosh. 1990. Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into virus-like particles. J. Virol. 64:2743-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith, A. J., N. Srivivasakumar, M.-L. Hammarskjöld, and D. Rekosh. 1993. Requirements for incorporation of Pr160gag-pol from human immunodeficiency virus type 1 into virus-like particles. J. Virol. 67:2266-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Srinivasakumar, N., M.-L. Hammarskjöld, and D. Rekosh. 1995. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and Gag-Pol precursor incorporation. J. Virol. 69:6106-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tritel, M., and M. D. Resh. 2000. Kinetic analysis of human immunodeficiency virus type 1 assembly reveals the presence of sequential intermediates. J. Virol. 74:5845-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van't Hof, W., and M. D. Resh. 1999. Dual fatty acylation of p59(Fyn) is required for association with the T cell receptor zeta chain through phosphotyrosine-Src homology domain-2 interactions. J. Cell Biol. 145:377-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wills, J. W., R. C. Craven, R. A. Weldon, Jr., T. D. Nelle, and C. R. Erdie. 1991. Supression of retroviral MA deletion by the amino-terminal membrane-binding domain of p60src. J. Virol. 65:3804-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wyma, D. J., A. Kotov, and C. Aiken. 2000. Evidence for a stable interaction of gp41 with Pr55(Gag) in immature human immunodeficiency virus type 1 particles. J. Virol. 74:9381-9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou, W., L. J. Parent, J. W. Wills, and M. D. Resh. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 68:2556-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]