Abstract
The ordered assembly of the herpes simplex virus (HSV) type 1 replication apparatus leading to replication compartments likely involves the initial assembly of five viral replication proteins, ICP8, UL9, and the heterotrimeric helicase-primase complex (UL5-UL8-UL52), into replication foci. The polymerase and polymerase accessory protein are subsequently recruited to these foci. Four stages of viral infection (stages I to IV) have been described previously (J. Burkham, D. M. Coen, and S. K. Weller, J. Virol. 72:10100-10107, 1998). Of these, stage III foci are equivalent to the previously described promyelocytic leukemia protein (PML)-associated prereplicative sites and contain all seven replication proteins. We constructed a series of mutations in the putative primase subunit, UL52, of the helicase-primase and have analyzed the mutant proteins for their abilities to form intermediates leading to the formation of replication compartments. The results shown in this paper are consistent with the model that the five proteins, ICP8, UL5, UL8, UL9, and UL52, form a scaffold and that formation of this scaffold does not rely on enzymatic functions of the helicase and primase. Furthermore, we demonstrate that recruitment of polymerase to this scaffold requires the presence of an active primase subunit. These results suggest that polymerase recruitment to replication foci requires primer synthesis. Furthermore, they support the existence of two types of stage III intermediates in the formation of replication compartments: stage IIIa foci, which form the scaffold, and stage IIIb foci, which contain, in addition, HSV polymerase, the polymerase accessory subunit, and cellular factors such as PML.
Herpes simplex virus type 1 (HSV-1) encodes seven gene products that are essential for viral DNA synthesis: the origin binding protein (UL9), a heterotrimeric helicase-primase complex (UL5, UL8, and UL52), the HSV-1 single-stranded DNA binding protein (ICP8, or UL29), and a two-subunit polymerase (Pol) complex consisting of the Pol (UL30) and the Pol accessory protein (UL42) (36, 46). HSV-1 DNA replication and gene expression occur in large globular domains within the nucleus called replication compartments, originally defined by the staining patterns of ICP8 (37). Replication compartments can also form when cells are transfected with plasmids bearing the seven essential replication genes (30, 44, 47).
It has been previously shown that replication compartments form in proximity to cellular domains called ND10 (29, 32, 44), which are defined by the accumulation of promyelocytic leukemia protein (PML) and other cellular proteins involved in growth control, gene expression, and possibly DNA recombination (reviewed in reference 6). HSV-1 infection results in disruption of ND10 and degradation of some isoforms of PML; however, colocalization of other isoforms of PML with ICP8 in replication compartments has been observed, as well as the intermediates leading to their formation (9, 29). Based on the staining patterns of ICP8 and PML, four stages of wild-type HSV-1 infection have been defined previously (9), as follows. In stage I, PML (ND10) foci are intact and no ICP8 staining is visible. In stage II, the ND10 foci have been disrupted, and diffuse ICP8 staining is observed in the nucleus, indicating that immediate-early and early protein synthesis has occurred. Stage III is defined by the formation of a limited number of ICP8-containing foci. Formation of stage III foci has been shown to be dependent on the presence of UL5, UL8, UL9, and UL52, since cells infected with mutants lacking these proteins never progressed past stage II (9). It is also assumed that ICP8 itself is required for stage III focus formation, since ICP8 mutants do not form prereplicative sites or replication compartments (12, 38). On the other hand, stage III foci could form in cells infected with mutants lacking Pol or UL42. In cells infected with the wild-type virus, Pol and UL42 are recruited to stage III foci, and recruitment of PML to these foci requires the presence of Pol (8, 9). These observations suggest that stage III can actually be divided into two stages, IIIa and IIIb. Stage IIIa foci contain the five viral proteins ICP8, UL5, UL8, UL9, and UL52, whereas stage IIIb foci contain these five proteins along with HSV Pol, UL42, and PML. We suggest that ICP8, UL5, UL8, UL9, and UL52 form a scaffold which is capable of recruiting HSV Pol, UL42, and some isoforms of PML. Stage IIIb foci are equivalent to the PML-associated, less-abundant prereplicative sites described by Lukonis et al. (29) and Uprichard and Knipe (44). These foci likely represent relevant intermediates in replication compartment formation. Another class of prereplicative sites which are more numerous than the PML-associated prereplicative sites do not localize with PML; however, they can be labeled with bromodeoxyuridine (BrdU), indicating that they represent sites of cellular DNA synthesis and not actual intermediates in replication compartment formation (29, 44). If replication is allowed to proceed, replication compartments (37), which stain with both the ICP8 and PML antibodies, are observed (stage IV).
Initiation of replication and the recruitment of replication proteins to replication forks is a highly ordered process in all biological systems studied to date. In Esherichia coli, replisome assembly is initiated by the unwinding of oriC by the initiator protein DnaA (7) (19). Upon origin unwinding, the replicative helicase DnaB is assembled as a hexamer at the origin by the helicase loader DnaC (3, 7, 17). The DNA is unwound ∼100 nucleotides, and the primase (DnaG) is recruited. Once DnaG lays down an RNA primer, the Pol III holoenzyme can be recruited to the replication fork (16). In eukaryotic cells the recruitment of the replicative Pol to the replication fork is also dependent upon the activity of primase; once the Pol α-primase lays down an RNA-DNA primer, Pol δ and ɛ can be recruited as elongating Pols (34). For the HSV system, little is known about how the Pol and its accessory subunit UL42 are recruited to replication foci. By analogy to the E. coli and eukaryotic systems described above, we hypothesize that the HSV-1 helicase-primase complex plays a role in the formation of various subassemblies of replication proteins and in the recruitment of the HSV Pol to this subassembly. We are particularly interested in the role played by the primase subunit UL52.
The HSV-1 primase is part of a heterotrimer composed of the products of the UL5, UL8, and UL52 genes (11). The known functions of the helicase-primase complex include DNA-dependent ATPase, DNA binding, and helicase and primase activities. UL5 and UL52 together exhibit all the enzymatic activities of the complex (13). UL5 was originally believed to be the helicase, based on the presence of seven conserved helicase motifs characteristic of helicases belonging to superfamily 1 (21, 22, 31). UL52 has a conserved motif believed to correspond to the catalytic site of the primase (14, 26) and a highly conserved zinc finger domain (CX4HX28CX4C) which is essential for DNA binding, helicase, and primase activities of the UL5-UL52 complex (4). Thus, the UL5 and UL52 subunits are both needed for the helicase activity of the complex. The UL8 gene product appears to stimulate the activities of UL5-UL52 and to facilitate the localization of UL5 and UL52 to the nucleus, and it may promote the interaction of the heterotrimer with other components of the replication machinery (15, 18, 33, 41, 43). In this article we report the construction of a series of mutations in the zinc finger motif of UL52 and describe immunofluorescence assays performed to investigate whether these mutants are replication competent and capable of forming the various subassemblies of replication proteins described above.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney fibroblasts (Vero) and HEp2 cells (both from the American Type Culture Collection) were propagated and maintained as described previously (8). G418-resistant BL-1 cells, containing the UL52 gene under the control of the ICP6 promoter, and the hr114 UL52 null virus have been described previously (20). The hr99 UL5 null virus has been described previously (48). The UL52-D628Q recombinant baculovirus was a kind gift from Mark Challberg (National Institutes of Health, Bethesda, Md.) (26).
Reagents.
Dulbecco's modified Eagle medium was purchased from GIBCO-Life Technologies, Inc. Fetal calf serum was obtained from Atlanta Biologicals. Penicillin-streptomycin solution was purchased from Sigma. Lipofectamine PLUS reagent was obtained from GIBCO-BRL. Glycerol gelatin and 1,4-diazobicyclo-[2,2,2]octane were purchased from Sigma.
Alignments.
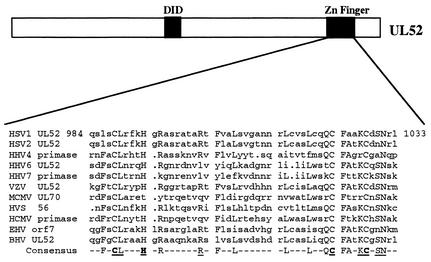
Sequence alignment of the UL52 zinc finger domain was performed by using the PILEUP program, a component of the GCG program (Wisconsin Package, version 9.1; Genetics Computer Group, Madison, Wis.). Consensus regions were generated with the PRETTY program, and the HSV-1 UL52 zinc finger region was aligned with those of 10 other alphaherpesviruses.
Plasmids.
The amplicon vector bearing a cytomegalovirus (CMV) promoter, pF1′CMV, was generously provided by Ann D. Kwong. pF1′CMV-UL52, expressing wild-type UL52 protein, has been described previously (4). pF1′CMV-UL5, expressing wild-type UL5 protein, was constructed by subcloning the BamHI fragment containing the UL5 gene from p6UL5 into the BamHI site of the amplicon vector pF1′CMV. p6UL5 has been described previously (48). Constructs which express UL8 and UL29 under the control of the CMV promoter, pCM-UL8 and pCM-DBP, respectively, have been described previously (25).
Antibodies.
The monospecific polyclonal anti-UL52 antibody 1248 was a kind gift from Mark Challberg (National Institutes of Health) (26). The polyclonal anti-ICP8 (αICP8) antibody was generously provided by William Ruyechan (State University of New York at Buffalo) (40). Monoclonal antibody PG-M3, recognizing PML, was obtained from Santa Cruz Biotechnology (Santa Cruz, Calif.). The monoclonal anti-UL30 antibody Mab1051c was generously provided by Charles W. Knopf (Deutsches Krebsforschungszentrum, Genomforschung und Bioinformatik, Heidelberg, Germany) (42). Secondary goat antibodies conjugated to either fluorescein isothiocyanate (FITC) or Texas red and directed against rabbit or mouse immunoglobulin were obtained from Cappel, Organon Teknika Corp. (Durham, N.C.).
Construction of point mutations.
The following UL52 mutants were constructed by a two-step PCR method (2): C1, in which cysteine 988 was changed to alanine; H2, in which histidine 993 was changed to alanine; L989A; R1002A; K1027A; and a double mutant, SNAA, in which serine 1030 and asparagine 1031 were changed to alanines. Table 1 shows the mutations constructed and the oligonucleotides used in the PCR for each mutant. In addition, the two outside primers were as follows: UL52C1-U (5′ GACGGGCCTGCGTGCGGAAGGCTG 3′) and OznM4 (previously described [4]). PCR products were digested with either the KpnI and HindIII or the BlpI and HindIII restriction enzymes and were inserted into the pF1′CMV-UL52 vector cut with the corresponding restriction enzymes. All the mutations introduced were sequenced to verify that no other mutations were introduced.
TABLE 1.
Oligonucleotides used in the construction of zinc finger mutants
| Amino acid change | Primera (5′ to 3′)
|
Restriction site introduced | |
|---|---|---|---|
| Upper | Lower | ||
| C988A | AAGCCTGTCGGCACTGCGCTTCAAGCACGG | GGCCGTGCTTGAAGCGCAGTGCCGACAGGC TTTGC | MseI site deleted |
| L989A | CAAAGCCTGTCGTGTGCACGCTTCAAGCAC | GTGCTTGAAGCGTGCACACGACAGGCTTTG | MseI site deleted |
| H993A | TGTCTGCGCTTCAAGGCAGGCCGGGCGAGT | ACTCGCCCGGCCTGCCTTGAAGCGCAGACA | MseI site deleted |
| R1002A | GCCACGGCGGCAACATTCGTCGCGCTGAGC GTCGGGGCCAACAATCGATTGTCCG | CGGACAATCGATTGTTGGCCCGACGCTCAG CGCGACGAATGTTGCCGCCGTGGC | ClaI site introduced |
| K1027A | TGTGTCAACAGTGCTTTGCCGCCGCATGCG ACAGCAACCG | CGGTTGCTGTCGCATGCGCGGCAAAGCACT GTTGACACA | HincII site introduced |
| S1030A N1031A | GTGTCAACAGTGCTTTGCCGCCAAATGCGA CGCAGCACGCCTGCACAC | GTGTGCAGGCGTGCTGCGTCGCATTTGGCG GCAAAGCACTGTTGACAC | HincII site introduced |
Silent mutations introduced for restriction digest analysis are indicated by underlined letters, and amino acid changes are shown in bold.
Transfection of mammalian cells.
Vero cells and HEp2 cells were transfected with 1 μg of plasmid DNA by using the Lipofectamine-PLUS reagent as directed by the manufacturer (GIBCO-BRL).
Transient complementation assay.
Vero cells were plated in a 60-mm-diameter tissue culture dish at a confluency of 60%. The cells were transfected with amplicon plasmids bearing wild-type or mutant versions of UL52 as previously described (4). Cells were superinfected 16 to 20 h later with the UL52 null virus hr114 at a multiplicity of infection (MOI) of 3 PFU/cell. Cells and media were harvested 20 to 24 h postinfection (p.i.) and sonicated as described previously (4). Cell debris was pelleted by centrifugation in a Beckman TJ-6 centrifuge for 10 min. Virus was titered on the UL52-complementing cell line BL-1.
Indirect immunofluorescence.
Mammalian cells were grown on glass coverslips, transfected, and superinfected as described above for the transient complementation assay. Cells were fixed in 3.7% formaldehyde in phosphate-buffered saline (PBS) for 30 min and then washed three times with PBS before being permeabilized with 1% Triton X-100 in PBS for 10 min. Cells were washed three times and then pretreated with 3% normal goat serum in PBS for 1 h. Cells were single stained with the primary antibody αICP8 for the four-protein transfection assay. For the transfection-infection assays, cells were double stained with the primary antibody αICP8 and either Mab1051c, PG-M3, or BrdU. A dilution of 1:200 in 3% normal goat serum for 30 min was used for all antibodies. Coverslips were washed several times with PBS before secondary antibodies were applied. Cells were stained with secondary antibodies for 30 min and washed six times in PBS before being mounted on glass coverslips with glycerol gelatin containing 2.5% 1,4-diazobicyclo-[2,2,2]octane to retard photobleaching.
Imaging.
Imaging was performed on a Zeiss Axiovert 135 laser scanning confocal microscope equipped with an argon-krypton laser. Texas red was excited at 568 nm; FITC was excited at 488 nm. Emissions were collected separately, and channels were overlaid by computer for dual images. Images were collected by using a 100× Zeiss Apochromat lens and were arranged and labeled by using Adobe Photoshop 5.0 and Adobe Illustrator 7.0.
RESULTS
Zinc finger motifs are believed to coordinate zinc atom binding, and they represent the most frequently observed DNA-binding motif of those found to date (39). These motifs are found in eukaryotic, prokaryotic, and viral primases (27), and residues in the zinc binding domain of the T7 primase have been shown to be involved in the specificity of primase recognition (27). A double mutation in the putative zinc finger of the primase subunit (UL52), in which cysteines 1023 and 1028 were replaced with alanines, was previously constructed (4). This mutant, CC3,4AA, was unable to complement the growth of the UL52 null mutant. Mutant UL52 and wild-type UL5 were coexpressed and purified from insect cells infected with recombinant baculoviruses, and the subcomplex was shown to be defective in primase, helicase, and single-stranded DNA binding activities (4). In the present study, in order to determine the role of the putative primase subunit in the formation of subassemblies of replication proteins, we asked whether this and other zinc finger mutants are capable of forming these subassemblies.
Zinc finger motif mutants vary in their abilities to complement a UL52 null virus.
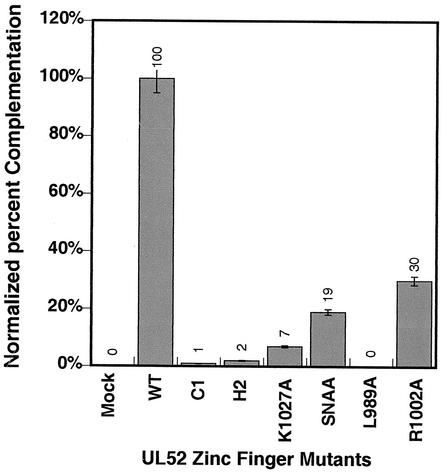
The putative zinc finger region of UL52 is highly conserved among the UL52 homologues within the herpesvirus family. An alignment of homologues from several alphaherpesviruses is shown in Fig. 1. Mutations were introduced into the most highly conserved residues within this region, and mutant genes were expressed in Vero cells from the CMV promoter-based amplicon plasmid (pF1′CMV). The engineered mutations were tested for their abilities to complement the UL52 null virus hr114 for growth. Vero cells were transfected with the expression plasmids and superinfected with hr114. Progeny virus was harvested as described in Materials and Methods and titered on the UL52-complementing cell line BL-1. It was previously reported that CC3,4AA was unable to complement the UL52 null virus (4). Figure 2 shows that mutants C1, H2, and L989A also failed to complement hr114. Thus, the invariant cysteine and histidine residues and L989 appear to be absolutely essential for viral replication. On the other hand, the R1002A, K1027A, and SNAA mutants showed partial complementation. On the basis of the low level of complementation exhibited by the K1027 mutant and the results shown below, the K1027 mutant will be classified as a noncomplementing mutant. In order to determine whether the noncomplementing mutations affected overall protein stability, the amplicon plasmids bearing wild-type or mutant versions of UL52 were assayed for protein production in transfected Vero cells. Western blot analysis of transfected cell lysates indicated that all the zinc finger mutants were able to make stable protein at levels similar to that of the wild type (data not shown). Thus, these mutations do not alter the global stability of the UL52 protein.
FIG. 1.
Sequence alignment of the herpesvirus primase zinc finger region homologues. Residues conserved in herpesvirus family primases (capitalized) were aligned with the GCG PILEUP program. Accession numbers for each virus, as obtained from the National Center for Biotechnology Information database, are as follows: HSV-1, NP044655; HSV-2, NP044523; human herpesvirus 4 (HHV-4), or Epstein-Barr virus (EBV), P03193; HHV-6, NP042936; HHV-7, AAC40757; varicella-zoster virus (VZV), P09270; murine cytomegalovirus (MCMV), Q69153; herpesvirus saimiri (HVS), P14346; human cytomegalovirus (HCMV), P17149; equine herpesvirus 1 (EHV-1), NP041016; and bovine herpesvirus (BHV), NP045306. Putative zinc binding residues are highlighted, and mutated residues are underlined.
FIG. 2.
Transient complementation of the UL52 null virus hr114. Complementation abilities of zinc finger mutants were determined as described in Materials and Methods. The complementation index of wild-type UL52 was set at 100%, which represents an approximate 100-fold increase in viral yield with the wild type over that with noncomplementing mutants. The complementation index was obtained by dividing the titer of virus produced in cells transfected with wild-type or mutant versions of UL52 plasmids and superinfected with hr114 by the titer in supernatants from mock-transfected cells. Data presented are averages from three or more assays performed with each mutant. The mean percent complementation index is shown. Error bars reflect the standard deviations calculated from the independent complementation assays performed.
All mutations can support the formation of a four-protein complex in transfected cells.
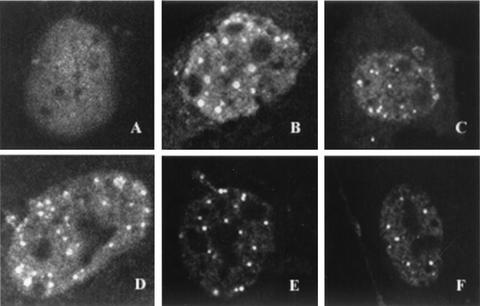
It was previously shown that cells transfected with plasmids expressing UL5, UL8, UL52, and ICP8 were able to form foci, as detected by staining with ICP8 antisera (28, 29); these foci were found to contain all four proteins. In cells lacking UL5, UL8, or UL52, ICP8 is generally found to be diffusely localized within the nucleus, suggesting that the formation of this four-protein complex is dependent on the presence of all four proteins (28, 29). We reasoned that the ability to form this complex can be used to determine whether the mutations introduced into the zinc finger motif of UL52 exhibit alterations in the protein-protein interactions needed to form this structure. We tested the abilities of the zinc finger mutants to form a four-protein complex in Vero cells transfected with plasmids bearing wild-type ICP8, UL5, and UL8 and wild-type or mutant versions of UL52. Sixteen hours posttransfection, cells were fixed and stained with the αICP8 antibody as described in Materials and Methods. In agreement with previously published results, a diffuse ICP8 staining pattern was observed (Fig. 3A) when cells were cotransfected with ICP8, UL5, UL8, and an empty pF1′CMV plasmid (no UL52). Cells cotransfected with ICP8, UL5, UL8, and wild-type UL52 exhibit punctate ICP8 foci as seen previously (Fig. 3B). Punctate ICP8 foci were also observed in cells transfected with the L989A, CC3,4AA, H2, and SNAA mutants (Fig. 3C, D, E, and F, respectively). Similar results were obtained with the C1 and K1027 mutants (data not shown). Thus, all mutant versions of UL52 were able to form punctate ICP8 foci in this assay. This result implies that no gross alterations in the structure of the helicase-primase complex occurred due to the mutations introduced. Furthermore, it appears that the zinc finger region is not involved in the protein-protein interactions leading to formation of the complex. In addition, the data also indicate that the helicase, primase, and DNA binding activities are not required for the formation of the four-protein complex, since ICP8 foci are present in cells transfected with the double mutant CC3,4AA, which lacks these activities (Fig. 3D).
FIG. 3.
Four-protein complex formation visualized by immunofluorescence. Vero cells were cotransfected with wild-type plasmids containing ICP8, UL5, and UL8 and wild-type or mutant versions of UL52. Cells were fixed and stained 16 h posttransfection with the polyclonal αICP8 antibody. The secondary antibody was conjugated to FITC, and staining patterns were visualized by fluorescence microscopy. All images depict cells containing wild-type ICP8, UL5, and UL8 and either pF1′CMV (A), wild-type UL52 (B), or the L989A (C), CC3,4AA (D), H2 (E), or SNAA (F) mutant.
All of the zinc finger mutants are able to progress to stage III of infection.
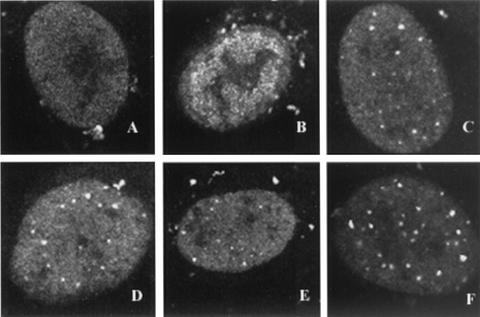
As mentioned in the introduction, it has previously been shown that in cells infected with the UL52 null virus (hr114), ICP8 localizes diffusely, indicating that the infection does not progress past stage II (9). We next asked whether the formation of stage IIIa foci depended on the enzymatic activities of the components of the helicase-primase complex or whether the mere presence of these five proteins in the context of viral infection is adequate. To answer this question, Vero cells were transfected with wild-type or mutant versions of UL52 and infected at 16 h posttransfection with the UL52 null mutant, hr114. At 5.5 to 6 h p.i., cells were fixed and stained with αICP8. In infected cells transfected with wild-type UL52, only replication compartments were observed (Fig. 4B); however, stage III foci could be observed if the cells were fixed and stained at earlier times points (data not shown). ICP8 foci were observed in infected cells transfected with any of the zinc finger mutant plasmids; the C1, CC3,4AA, K1027A, and SNAA mutants are shown as representative examples (Fig. 4C, D, E, and F, respectively). This staining pattern is reminiscent of that of the stage III foci reported previously (9), indicating that all zinc finger motif mutants are capable of forming at least stage IIIa foci. We propose that stage IIIa foci form a scaffold for the recruitment of other proteins and that enzymatic activity of the helicase-primase complex is not required for this scaffold to form.
FIG. 4.
Stage III foci in Vero cells transfected with plasmids bearing wild-type or mutant versions of UL52. Vero cells were transfected as described in Materials and Methods and superinfected with hr114 for 5.5 h p.i. Cells were stained with the polyclonal αICP8 antibody. The secondary antibody was a goat anti-rabbit immunoglobulin conjugated to FITC. FITC staining was visualized by fluorescence microsocopy. Cells were transfected with either pF1′CMV (A), wild-type UL52 (B), or the C1 (C), CC3,4AA (D), K1027A (E), or SNAA (F) mutant.
Figure 4 shows that cells transfected with mutant or wild-type versions of UL52 and infected with hr114 contained either stage III or stage IV ICP8 foci, as opposed to the diffuse ICP8 staining seen in cells infected with hr114 alone. Cells transfected and not infected did not stain for ICP8 and resembled uninfected cells. Thus, the transfection efficiency can be calculated by the number of cells which contain stage III or stage IV ICP8 foci. In these experiments, the transfection efficiency was approximately 30%. Of the 30% of cells that were transfected and superinfected, the majority (approximately 60%) exhibited the ICP8 foci shown in Fig. 4C through F; however, another staining pattern was seen for a sizable portion of cells which contained foci (around 40%). The second staining pattern resembled that of the numerous prereplicative sites previously shown to represent sites of cellular DNA synthesis in cells that are in S phase (29, 44). Figure 5 shows the “numerous” staining pattern in cells transfected with the H2 mutant (Fig. 5A to C) and superinfected with hr114. Interestingly, these cells stained for BrdU, indicating that they were in S phase; however, the BrdU staining did not colocalize with the ICP8 sites. This staining pattern was seen in cells transfected with any of the noncomplementing or partially complementing mutant versions of UL52 but was not seen in cells transfected with wild-type UL52. Cells infected with wild-type virus contained replication compartments which colabeled with ICP8 and BrdU (Fig. 5D to F). These results suggest that noncomplementing and partially complementing mutant UL52 proteins are able to form a pattern which is reminiscent of the numerous prepreplicative sites; surprisingly, however, they do not colocalize with BrdU, indicating that they are not localized at sites of cellular DNA synthesis. Further studies will be required to fully understand the significance of the pattern of numerous foci in this subset of cells transfected with UL52 mutants in the context of a viral infection.
FIG. 5.
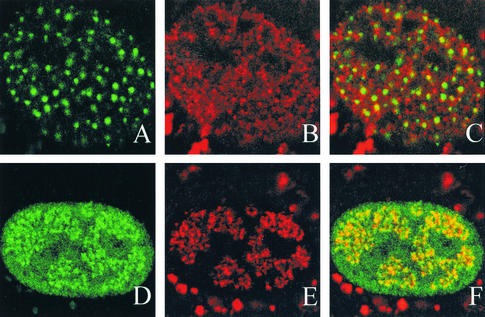
Vero cells transfected with the H2 mutant and superinfected with hr114 stained with ICP8 and BrdU. Vero cells were transfected with a plasmid bearing either the H2 mutant (A through C) or the wild-type (D through F) version of UL52 and were then superinfected with hr114 for 5.5 h p.i. BrdU was added during the last 30 min of the infection. Cells were stained with the polyclonal αICP8 antibody (A and D) or the monoclonal BrdU antibody (B and E), and images of the two stains were merged (C and F). The secondary antibodies were goat anti-rabbit immunoglobulin conjugated to FITC for ICP8, and goat anti-mouse immunoglobulin conjugated to Texas red for BrdU. The bright staining observed in the cytoplasm is likely due to nonspecific staining by the Texas red secondary antibody; however, the nuclear staining pattern of BrdU appears to be specific for DNA synthesis, since cells infected with wild-type virus contain replication compartments which colabel with ICP8 and BrdU.
Recruitment of PML to ICP8 foci.
We were interested in whether the ICP8 foci shown in Fig. 4 represent stage IIIa foci, which contain ICP8, helicase-primase, and UL9, or stage IIIb foci, which contain these five proteins along with HSV Pol, UL42, and PML. It was previously shown that PML recruitment to stage IIIb foci depends on the presence of HSV Pol (9). In this experiment we asked whether PML was recruited to the stage IIIa foci observed in cells transfected with various UL52 mutants and superinfected with hr114. Since the anti-PML antibody PG-M3 recognizes only the PML found in human cell lines, HEp2 cells were transfected and superinfected as described above and costained with αICP8 and PG-M3. In HEp2 cells transfected with the CC3,4AA mutant, ICP8 was observed in punctate foci; however, PML staining did not colocalize to these foci (Fig. 6A to C). In these cells, PML is either diffusely localized or localized in punctate foci, which do not colocalize with ICP8. A similar staining pattern was observed for cells transfected with the C1, H2, K1027A, or L989A mutant (data not shown). Interestingly, mutants which failed to recruit PML to ICP8 foci also exhibited little (K1027) or no (C1, H2, and L989) complementation of hr114 in the transient complementation assay. On the other hand, cells transfected with the partially complementing R1002A or SNAA mutant exhibited ICP8 staining patterns which colocalized with PML; in Fig. 6D to F, the pattern for the R1002A mutant is shown as a representative example. Since DNA synthesis was not inhibited in these cells, the foci seen in Fig. 6D to F actually represent small replication compartments. These small replication compartments appear larger than the stage IIIb foci seen in Fig. 4 for the SNAA mutant, another partially complementing mutant. This apparent difference reflects the fact that infection seems to progress somewhat faster in HEp2 cells than in Vero cells. The SNAA and R1002A mutants gave rise to similar foci in HEp2 cells (data not shown). Taken together, these results indicate that cells transfected with the noncomplementing mutants did not recruit PML to ICP8 foci, whereas, in cells transfected with the partially complementing mutants as in cells transfected with the wild type, ICP8 and PML colocalized in replication compartments.
FIG. 6.
PML recruitment to stage III foci. HEp2 cells were transfected with plasmids bearing wild-type or mutant versions of UL52 and then superinfected with hr114 for 5.5 h p.i. Cells were stained with the polyclonal αICP8 antibody and the monoclonal anti-PML antibody PG-M3. The secondary antibodies were goat anti-rabbit immunoglobulin conjugated to FITC for ICP8 and goat anti-mouse immunoglobulin conjugated to Texas red for PML. Staining was visualized by fluorescence microscopy. (A through C) Cells were transfected with the CC3,4AA mutant and stained with either ICP8 (A) or PML (B); the images were merged (C). (D through F) Cells were transfected with the R1002A mutant and stained with either ICP8 (D) or PML; the images were merged (F).
Recruitment of Pol to ICP8 foci.
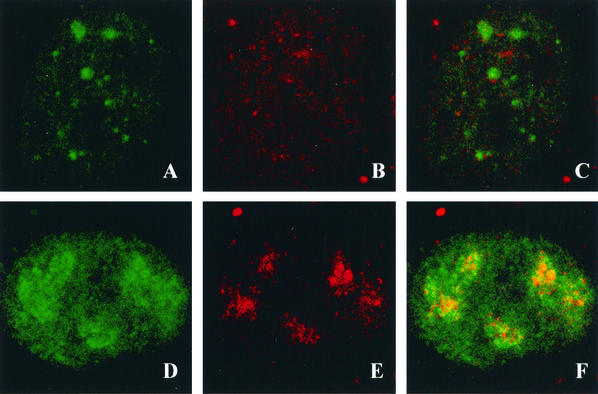
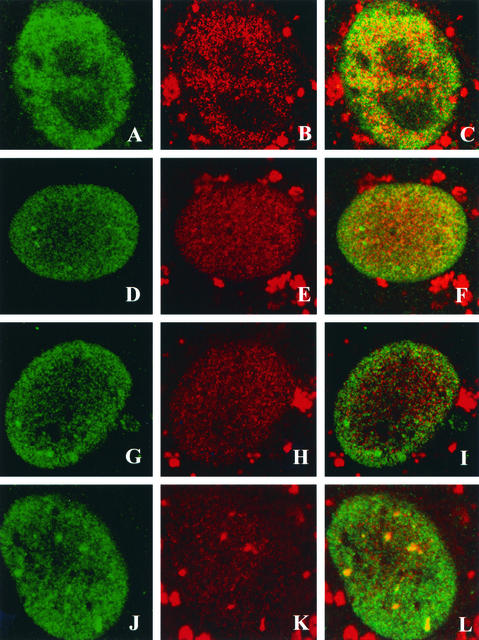
The lack of PML recruitment in the cells transfected with the noncomplementing mutants seen above is similar to the PML staining patterns observed in cells infected with the Pol null mutant HP66 (29). We were interested in learning whether HSV Pol could be recruited to the stage IIIa foci formed in cells transfected with the UL52 mutants. Vero cells were transfected with wild-type or mutant UL52 plasmids, superinfected with hr114, and costained with αICP8 and the anti-HSV Pol antibody Mab1051c. In cells transfected with wild-type UL52, ICP8 and HSV Pol were found colocalized in replication compartments (Fig. 7A to C). Cells transfected with the noncomplementing CC3,4AA or K1027A mutant formed ICP8 foci as expected; however, in these cells, Pol exhibited a diffuse nuclear staining pattern (Fig. 7D to I). In contrast, cells transfected with the partially complementing SNAA mutant exhibited ICP8 foci which also stained with the anti-HSV Pol antisera (Fig. 7J to L). The ability to recruit Pol and PML to stage IIIa foci (forming stage IIIb foci) correlated with the complementation ability of the individual UL52 mutant: in cells transfected with noncomplementing UL52 mutants, only stage IIIa foci were formed; however, in cells transfected with the partially complementing mutant, Pol and PML were colocalized in replication structures (stages IIIb and IV). All the zinc finger mutants can thus direct the formation of stage IIIa foci; however, recruitment of viral Pol appears to require an activity or condition found only in cells transfected with wild-type UL52 or the partially complementing mutant. Thus, some event(s) other than the formation of the five-protein scaffold may be required for recruitment of Pol to replication foci.
FIG. 7.
Pol recruitment to stage III foci. Vero cells were transfected with plasmids bearing wild-type or mutant versions of UL52 and then superinfected with hr114 for 5.5 h p.i. Cells were stained with the polyclonal αICP8 antibody or the monoclonal anti-Pol antibody Mab1051c. The secondary antibodies were goat anti-rabbit immunoglobulin conjugated to FITC for ICP8 and goat anti-mouse immunoglobulin conjugated to Texas red for Pol. All cells were infected with hr114; they were transfected with either wild-type UL52 (A through C) or the CC3,4AA (D through F), K1027A (G through I), or SNAA (J through L) mutant and stained for ICP8 (A, D, G, and J) or Pol (B, E, H, and K). Images were merged (C, F, I, and L).
Primase activity is required for Pol recruitment.
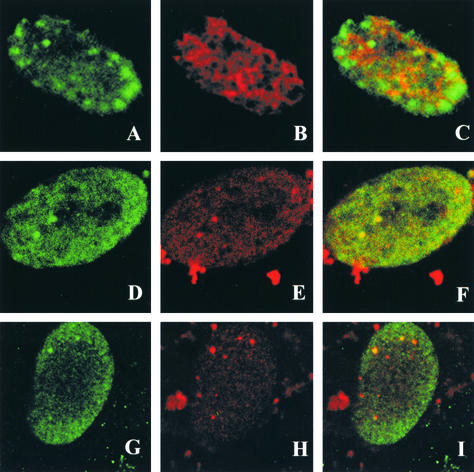
As mentioned above, in E. coli as well as in eukaryotic systems, an active primase appears to be required for recruitment of the replicative Pol to the replication fork. We were therefore interested in the role of UL52 in the recruitment of the HSV Pol to replication foci. Although no direct interaction between UL52 and the HSV Pol has ever been reported, it remains possible that such an interaction occurs but has escaped detection. Alternatively, it is possible that the UL52 zinc finger region undergoes conformational changes in the scaffold itself which are required for Pol recruitment. Another possibility is that the catalytic activity of the helicase and/or primase is needed in order to recruit Pol. To further understand the role of UL52 and other components of the helicase-primase complex, a mutation in another region of the UL52 gene, as well as a mutation in the UL5 subunit, was tested for its ability to recruit Pol. Klinedinst and Challberg (26) previously reported the isolation and characterization of a mutation in the DID motif of UL52, which is believed to represent the catalytic domain for primase activity. The D628Q mutant protein was coexpressed in insect cells with wild-type UL5, and the subcomplex was shown to exhibit helicase but not primase activity (26). The subcomplex likely retains DNA binding based on the presence of a functional helicase.
The D628Q mutation was subcloned into the amplicon vector. Vero cells transfected with this mutant and infected with hr114 contained ICP8 foci; however, Pol was not recruited to these foci (Fig. 8A to C). Thus, only stage IIIa foci form in cells transfected with this mutant. We also tested a UL5 mutation in the Walker A box (motif I) which resulted in a UL5-UL52 subcomplex that is defective in helicase activity but exhibits wild-type primase activity (23). Figure 8D to I show two cells transfected with the motif I mutant and superinfected with the UL5 null mutant hr99. In both cells a small number of ICP8 foci which colabeled with the Pol antibody (stage IIIb foci) were formed. These results suggest that the helicase activity of UL5 is dispensable, while catalytically active UL52 is essential, for UL30 recruitment to stage IIIa foci. This may indicate that Pol recruitment requires the presence of a primer template at the replication fork.
FIG. 8.
Pol recruitment with the UL52 D628Q and UL5 motif I mutants. Vero cells were transfected with plasmids bearing the UL52 D628Q catalytic mutant or the UL5 motif I mutant and then superinfected with hr114 or hr99, respectively, for 5.5 h p.i. Cells were stained with the polyclonal αICP8 antibody or the monoclonal anti-Pol antibody Mab1051c. The secondary antibodies were goat anti-rabbit immunoglobulin conjugated to FITC for ICP8 and goat anti-mouse immunoglobulin conjugated to Texas red for Pol. (A through C) Cells transfected with the D628Q mutant. (D through I) Two different cells (D through F and G through I) transfected with the UL5 motif I mutant. Cells were stained with ICP8 (A, D, and G) or Pol (B, E, and H), and images were merged (C, F, and I).
DISCUSSION
In this study we have engineered and characterized a series of mutations in the gene encoding the putative primase subunit (UL52) of the HSV-1 helicase-primase heterotrimeric complex. Table 2 summarizes the results of experiments performed with expression plasmids bearing wild-type and mutant versions of UL52. The following major observations were made. (i) All primase mutants were able to form a four-protein complex in cells transfected with ICP8, UL5, UL8, and mutant versions of UL52. (ii) All mutants were able to form stage IIIa foci in cells transfected with wild-type or mutant versions of UL52 and superinfected with the null mutant hr114. (iii) Partially complementing mutants were able to form stage IIIb and stage IV foci; in other words, they were able to recruit HSV-1 Pol and PML to stage IIIa foci and to progress to stage IV (replication compartments). (iv) Noncomplementing mutants were unable to recruit Pol and PML to stage IIIa foci. (v) A UL52 mutant with a mutation in the catalytic domain of the primase (D628Q) which is helicase positive and primase negative was able to form stage IIIa foci but was unable to recruit Pol and thus unable to form stage IIIb foci. (vi) A UL5 mutant with a mutation in motif I which is helicase negative and primase positive was able to recruit Pol, forming stage IIIb foci. Taken together, these results indicate that the ability to recruit Pol correlates with the primase activity of the UL52 subunit.
TABLE 2.
Activities of UL52 zinc finger motif mutants
| Zn finger proteina | Ability to form:
|
||
|---|---|---|---|
| Four-protein complex | Stage IIIa foci | Stage IIIb foci | |
| Wild type | + | + | + |
| C1 | + | + | − |
| H2 | + | + | − |
| CC3.4AA | + | + | − |
| L989A | + | + | − |
| R1002A | + | + | + |
| K1027A | + | + | − |
| SNAA | + | + | + |
The wild-type protein and the partially complementing mutants are shown in bold. The CC3,4AA mutant was previously characterized (4).
The requirement for an active primase in order for HSV Pol to be recruited to replication foci could be explained in several ways. As mentioned above, it may indicate that Pol can be recruited only to a replication fork which already contains a primer template. We cannot, however, rule out the possibility that Pol recognizes a conformational change in the viral scaffold containing an active primase subunit capable of binding DNA or other protein-protein interactions not detected in the experiments performed in this study. Further experiments will be required to distinguish between these possibilities.
The results presented in this paper have implications in terms of the formation of relevant subassemblies at the replication fork. The data for the four-viral-protein complex which forms in cells cotransfected with ICP8, UL5, UL8, and UL52 suggest that these four proteins are able to interact with one another in vivo. A possible interaction between these four proteins is supported by the observation that the UL8 component of the UL5-UL8-UL52 complex is needed to obtain optimal activity of the helicase-primase on ICP8-coated single-stranded DNA (15). Another indication that the four-protein complex represents a relevant subassembly of replication proteins is the recent report that ICP8, UL5, UL8, and UL52 can carry out a limited type of strand exchange reaction in vitro (35). Furthermore, these four proteins have also been found to be sufficient to provide helper virus activity for adeno-associated virus (AAV) DNA replication (45). In fact, the AAV Rep protein and replicating AAV DNA localize to viral foci formed by these four HSV proteins (M. D. Weitzmann, personal communication). Several of the zinc finger mutants described in this paper are still able to induce productive AAV infection, including the CC3,4AA mutant, which lacks enzymatic activities (M. D. Weitzmann, personal communication). These results support the notion that the four-protein complex represents a meaningful subassembly which can recruit viral and cellular proteins important for both AAV and HSV replication. Formation of this subassembly does not require the enzymatic activities of the helicase or primase.
It has been reported previously that stage III foci, also known as PML-associated prereplicative sites, represent intermediates in the formation of replication compartments (9, 29, 44). The immunofluorescence assays presented in this report indicate that stage III foci can actually be subdivided into two types of foci: stage IIIa foci contain ICP8, UL9, and the three components of the helicase-primase complex; whereas stage IIIb foci contain these five proteins and the viral Pol, the Pol accessory protein, and the cellular protein PML. Several protein-protein interactions among these five proteins have been reported, including interactions between ICP8 and UL9 (5), ICP8 and UL8 (15, 18, 24), and UL8 and UL9 (33), in addition to the reported interactions within the UL5-UL8-UL52 heterotrimer (10, 11). These interactions are likely to contribute to the formation of stage IIIa foci; however, the precise order of addition is not known. Based on the results of the four-protein transfection assays, it is possible that ICP8, UL5, UL8, and UL52 assemble into a discrete subassembly which is then recruited to UL9 at the origin of replication. Alternatively, all five proteins may assemble together on the origin of replication, resulting in the formation of stage IIIa foci. In the present report we have demonstrated that all the zinc finger mutants can form stage IIIa foci regardless of biological function or enzymatic activity. These results provide further support for the notion that the five-protein subassembly may act as a scaffold for the recruitment of other viral and cellular proteins.
The most striking observation made in this paper is that the ability to form stage IIIb foci, intermediates which contain ICP8, helicase-primase, UL9, HSV Pol, and UL42 as well as the cellular protein PML, appears to depend on the activity of the primase. Lending support to the notion that Pol recruitment requires active primase are experiments performed by Dracheva et al. In those experiments two different primase-negative mutants failed to support Pol activity in an in vitro coupled primase-Pol assay (14). The requirement of an active primase for recruitment of the HSV Pol is reminiscent of the finding that during E. coli replication initiation, an active primase is required in order for Pol III to assemble at the replication fork (1, 16). In that system it is not clear whether it is the primase itself or the primed DNA which is recognized by Pol III; however, the fact that Pol III can initiate DNA synthesis when oligonucleotide primers are supplied may suggest that Pol III recruitment needs primed DNA rather than the primase. In eukaryotic initiation, a crucial event in DNA replication is the switch from the Pol α-primase to the elongating Pols δ and ɛ. Mossi et al. have suggested that this switch depends on the length of the primer synthesized by the Pol α-primase complex (34). The fact that in both prokaryotic and eukaryotic systems there is a requirement for a primer in order for the Pol to be recruited to the replisome indicates that this may be a general mechanism for primary replicative Pol recruitment.
Based on the previous results from our laboratory and others, and on the data presented in this paper, a model can be envisioned for the assembly of HSV replication proteins into a functional replication complex. The assembly of a five-protein complex consisting of ICP8, UL9, and the helicase-primase has been reported previously (9, 28). We presume that this complex can form at an HSV origin of replication and stimulate limited unwinding. The presence of two helicases in this complex, UL9 and UL5, may indicate that a switch occurs between initial unwinding events and unwinding by the presumed replicative helicase, UL5. By analogy with the E. coli system, we suggest that limited unwinding may reveal primase recognition sites and provide a template for the primase. According to the results presented here, we suggest that once an RNA primer is synthesized, the Pol and the Pol accessory protein, UL42, can be recruited to the replication fork, followed by the recruitment of PML and perhaps other cellular proteins. To our knowledge, this is the first report defining the functional requirements for HSV Pol recruitment to replication foci.
Acknowledgments
We thank members of our laboratory, as well as Don Coen and Rob Kuchta, for helpful comments on the manuscript. We also thank Jennifer Burkham for helpful suggestions in designing experiments for this study and Matthew Weitzmann for communicating unpublished results.
This investigation was supported by Public Health Service grant AI21747.
REFERENCES
- 1.Ason, B., J. G. Bertram, M. M. Hingorani, J. M. Beechem, M. O'Donnell, M. F. Goodman, and L. B. Bloom. 2000. A model for Escherichia coli DNA polymerase III holoenzyme assembly at primer/template ends. DNA triggers a change in binding specificity of the gamma complex clamp loader. J. Biol. Chem. 275:3006-3015. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A., Smith, and K. Struhl. 1990. Introduction of a point mutation by sequential PCR steps, p. 8.5.7-8.5.9. In K. Janssen (ed.), Current protocols in molecular biology, vol. 2. John Wiley & Sons Inc., New York, N.Y. [Google Scholar]
- 3.Baker, T. A., B. E. Funnell, and A. Kornberg. 1987. Helicase action of dnaB protein during replication from the Escherichia coli chromosomal origin in vitro. J. Biol. Chem. 262:6877-6885. [PubMed] [Google Scholar]
- 4.Biswas, N., and S. K. Weller. 1999. A mutation in the C-terminal putative Zn2+ finger motif of UL52 severely affects the biochemical activities of the HSV-1 helicase-primase subcomplex. J. Biol. Chem. 274:8068-8076. [DOI] [PubMed] [Google Scholar]
- 5.Boehmer, P. E., and I. R. Lehman. 1993. Herpes simplex virus type 1 ICP8: helix-destabilizing properties. J. Virol. 67:711-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borden, K. L. 2002. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 22:5259-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bramhill, D., and A. Kornberg. 1988. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell 52:743-755. [DOI] [PubMed] [Google Scholar]
- 8.Burkham, J., D. M. Coen, C. B. Hwang, and S. K. Weller. 2001. Interactions of herpes simplex virus type 1 with ND10 and recruitment of PML to replication compartments. J. Virol. 75:2353-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burkham, J., D. M. Coen, and S. K. Weller. 1998. ND10 protein PML is recruited to herpes simplex virus type 1 prereplicative sites and replication compartments in the presence of viral DNA polymerase. J. Virol. 72:10100-10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crute, J. J., and I. R. Lehman. 1991. Herpes simplex virus-1 helicase-primase. Physical and catalytic properties. J. Biol. Chem. 266:4484-4488. [PubMed] [Google Scholar]
- 11.Crute, J. J., T. Tsurumi, L. A. Zhu, S. K. Weller, P. D. Olivo, M. D. Challberg, E. S. Mocarski, and I. R. Lehman. 1989. Herpes simplex virus 1 helicase-primase: a complex of three herpes-encoded gene products. Proc. Natl. Acad. Sci. USA 86:2186-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Bruyn Kops, A., and D. M. Knipe. 1988. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell 55:857-868. [DOI] [PubMed] [Google Scholar]
- 13.Dodson, M. S., and I. R. Lehman. 1991. Association of DNA helicase and primase activities with a subassembly of the herpes simplex virus 1 helicase-primase composed of the UL5 and UL52 gene products. Proc. Natl. Acad. Sci. USA 88:1105-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dracheva, S., E. V. Koonin, and J. J. Crute. 1995. Identification of the primase active site of the herpes simplex virus type 1 helicase-primase. J. Biol. Chem. 270:14148-14153. [DOI] [PubMed] [Google Scholar]
- 15.Falkenberg, M., D. A. Bushnell, P. Elias, and I. R. Lehman. 1997. The UL8 subunit of the heterotrimeric herpes simplex virus type 1 helicase-primase is required for the unwinding of single strand DNA-binding protein (ICP8)-coated DNA substrates. J. Biol. Chem. 272:22766-22770. [DOI] [PubMed] [Google Scholar]
- 16.Fang, L., M. J. Davey, and M. O'Donnell. 1999. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol. Cell 4:541-553. [DOI] [PubMed] [Google Scholar]
- 17.Funnell, B. E., T. A. Baker, and A. Kornberg. 1987. In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J. Biol. Chem. 262:10327-10334. [PubMed] [Google Scholar]
- 18.Gac, N. T. L., G. Villani, J. S. Hoffmann, and P. E. Boehmer. 1996. The UL8 subunit of the herpes simplex virus type-1 DNA helicase-primase optimizes utilization of DNA templates covered by the homologous single-strand DNA-binding protein ICP8. J. Biol. Chem. 271:21645-21651. [DOI] [PubMed] [Google Scholar]
- 19.Gille, H., and W. Messer. 1991. Localized DNA melting and structural perturbations in the origin of replication, oriC, of Escherichia coli in vitro and in vivo. EMBO J. 10:1579-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldstein, D. J., and S. K. Weller. 1988. An ICP6::lacZ insertional mutagen is used to demonstrate that the UL52 gene of herpes simplex virus type 1 is required for virus growth and DNA synthesis. J. Virol. 62:2970-2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorbalenya, A. E., and E. V. Koonin. 1988. One more conserved sequence motif in helicases. Nucleic Acids Res. 16:7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1988. A conserved NTP-motif in putative helicases. Nature 333:22. [DOI] [PubMed] [Google Scholar]
- 23.Graves-Woodward, K. L., J. Gottlieb, M. D. Challberg, and S. K. Weller. 1997. Biochemical analyses of mutations in the HSV-1 helicase-primase that alter ATP hydrolysis, DNA unwinding, and coupling between hydrolysis and unwinding. J. Biol. Chem. 272:4623-4630. [DOI] [PubMed] [Google Scholar]
- 24.Hamatake, R. K., M. Bifano, W. W. Hurlburt, and D. J. Tenney. 1997. A functional interaction of ICP8, the herpes simplex virus single-stranded DNA-binding protein, and the helicase-primase complex that is dependent on the presence of the UL8 subunit. J. Gen. Virol. 78:857-865. [DOI] [PubMed] [Google Scholar]
- 25.Heilbronn, R., A. Burkle, S. Stephan, and H. zur Hausen. 1990. The adeno-associated virus rep gene suppresses herpes simplex virus-induced DNA amplification. J. Virol. 64:3012-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klinedinst, D. K., and M. D. Challberg. 1994. Helicase-primase complex of herpes simplex virus type 1: a mutation in the UL52 subunit abolishes primase activity. J. Virol. 68:3693-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kusakabe, T., A. V. Hine, S. G. Hyberts, and C. C. Richardson. 1999. The Cys4 zinc finger of bacteriophage T7 primase in sequence-specific single-stranded DNA recognition. Proc. Natl. Acad. Sci. USA 96:4295-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liptak, L. M., S. L. Uprichard, and D. M. Knipe. 1996. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J. Virol. 70:1759-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukonis, C. J., J. Burkham, and S. K. Weller. 1997. Herpes simplex virus type 1 prereplicative sites are a heterogeneous population: only a subset are likely to be precursors to replication compartments. J. Virol. 71:4771-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukonis, C. J., and S. K. Weller. 1997. Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J. Virol. 71:2390-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marintcheva, B., and S. K. Weller. 2001. A tale of two HSV-1 helicases: roles of phage and animal virus helicases in DNA replication and recombination. Prog. Nucleic Acid Res. Mol. Biol. 70:77-118. [DOI] [PubMed] [Google Scholar]
- 32.Maul, G. G., A. M. Ishov, and R. D. Everett. 1996. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology 217:67-75. [DOI] [PubMed] [Google Scholar]
- 33.McLean, G. W., A. P. Abbotts, M. E. Parry, H. S. Marsden, and N. D. Stow. 1994. The herpes simplex virus type 1 origin-binding protein interacts specifically with the viral UL8 protein. J. Gen. Virol. 75:2699-2706. [DOI] [PubMed] [Google Scholar]
- 34.Mossi, R., R. C. Keller, E. Ferrari, and U. Hubscher. 2000. DNA polymerase switching. II. Replication factor C abrogates primer synthesis by DNA polymerase alpha at a critical length. J. Mol. Biol. 295:803-814. [DOI] [PubMed] [Google Scholar]
- 35.Nimonkar, A. V., and P. E. Boehmer. 2002. In vitro strand exchange promoted by the herpes simplex virus type-1 single strand DNA-binding protein (ICP8) and DNA helicase-primase. J. Biol. Chem. 277:15182-15189. [DOI] [PubMed] [Google Scholar]
- 36.Olivo, P. D., N. J. Nelson, and M. D. Challberg. 1989. Herpes simplex virus type 1 gene products required for DNA replication: identification and overexpression. J. Virol. 63:196-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quinlan, M. P., L. B. Chen, and D. M. Knipe. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36:857-868. [DOI] [PubMed] [Google Scholar]
- 38.Quinlan, M. P., and D. M. Knipe. 1983. Nuclear localization of herpesvirus proteins: potential role for the cellular framework. Mol. Cell. Biol. 3:315-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenfeld, R., and H. Margalit. 1993. Zinc fingers: conserved properties that can distinguish between spurious and actual DNA-binding motifs. J. Biomol. Struct. Dyn. 11:557-570. [DOI] [PubMed] [Google Scholar]
- 40.Shelton, L. S., A. G. Albright, W. T. Ruyechan, and F. J. Jenkins. 1994. Retention of the herpes simplex virus type 1 (HSV-1) UL37 protein on single-stranded DNA columns requires the HSV-1 ICP8 protein. J. Virol. 68:521-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherman, G., J. Gottlieb, and M. D. Challberg. 1992. The UL8 subunit of the herpes simplex virus helicase-primase complex is required for efficient primer utilization. J. Virol. 66:4884-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strick, R., J. Hansen, R. Bracht, D. Komitowski, and C. W. Knopf. 1997. Epitope mapping and functional characterization of monoclonal antibodies specific for herpes simplex virus type I DNA polymerase. Intervirology 40:41-49. [DOI] [PubMed] [Google Scholar]
- 43.Tenney, D. J., W. W. Hurlburt, P. A. Micheletti, M. Bifano, and R. K. Hamatake. 1994. The UL8 component of the herpes simplex virus helicase-primase complex stimulates primer synthesis by a subassembly of the UL5 and UL52 components. J. Biol. Chem. 269:5030-5035. [PubMed] [Google Scholar]
- 44.Uprichard, S. L., and D. M. Knipe. 1997. Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology 229:113-125. [DOI] [PubMed] [Google Scholar]
- 45.Weindler, F. W., and R. Heilbronn. 1991. A subset of herpes simplex virus replication genes provides helper functions for productive adeno-associated virus replication. J. Virol. 65:2476-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weller, S. K. 1995. Herpes simplex virus DNA replication and genome maturation, p. 189-213. In G. M. Cooper, R. G. Temin, and B. Sugden (ed.), Implications of the DNA provirus: Howard Temin's scientific legacy. ASM Press, Washington, D.C.
- 47.Zhong, L., and G. S. Hayward. 1997. Assembly of complete, functionally active herpes simplex virus DNA replication compartments and recruitment of associated viral and cellular proteins in transient cotransfection assays. J. Virol. 71:3146-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu, L. A., and S. K. Weller. 1992. The UL5 gene of herpes simplex virus type 1: isolation of a lacZ insertion mutant and association of the UL5 gene product with other members of the helicase-primase complex. J. Virol. 66:458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]