Abstract
Chronic infection by hepatitis B virus results from an inability to clear the virus, which is associated with liver disease and liver cancer. Tumor necrosis factor alpha (TNF-α) is associated with noncytopathic clearance of hepatitis B virus in animal models. Here we demonstrate that the nuclear factor κB (NF-κB) signaling pathway is a central mediator of inhibition of hepatitis B virus by TNF-α and we describe the molecular mechanism. TNF-α is shown to suppress hepatitis B virus DNA replication without cell killing by disrupting the formation or stability of cytoplasmic viral capsids through a pathway requiring the NF-κB-activating inhibitor of κB kinase complex IKK-α/β and active transcription factor NF-κB. Hepatitis B virus replication could also be inhibited and viral capsid formation could be disrupted in the absence of TNF-α solely by overexpression of IKK-α/β or strong activation of NF-κB. In contrast, inhibition of NF-κB signaling stimulated viral replication, demonstrating that HBV replication is both positively and negatively regulated by the level of activity of the NF-κB pathway. Studies are presented that exclude the possibility that HBV inhibition by NF-κB is carried out by secondary production of gamma interferon or alpha/beta interferon. These results identify a novel mechanism for noncytopathic suppression of hepatitis B virus replication that is mediated by the NF-κB signaling pathway and activated by TNF-α.
Tumor necrosis factor alpha (TNF-α) is a proinflammatory cytokine best known for inducing immune-mediated apoptotic cell death. However, TNF-α also inhibits the replication of certain viruses in a noncytopathic manner (in the absence of cell killing), as shown for herpes simplex virus type 1, mouse cytomegalovirus, and hepatitis B virus (HBV) (reviewed in references 15 and 23). There is little molecular understanding of the noncytopathic antiviral activity of TNF-α. TNF-α can strongly activate nuclear factor κB (NF-κB), a transcription factor (reviewed in reference 28), and NF-κB, in turn, can impair cell death mediated by TNF-α (28). Activation of NF-κB by TNF-α involves the formation of a complex between TNF receptor-associated protein 2 and receptor-interacting protein, which activate the inhibitor of κB kinase (IKK) complex, containing kinases IKK-α and IKK-β (28). IKK-β and IKK-α phosphorylate the inhibitor of κB repressor, resulting in its degradation; release of NF-κB family members, such as p65 RelA/p50 complexes; and transcriptional activation (28).
Several proinflammatory cytokines have been shown to suppress HBV replication in a noncytopathic manner in experimental systems (reviewed in reference 15). In the livers of transgenic mice expressing human HBV, inflammatory cytokines such as alpha/beta interferon (IFN-α/β) can be experimentally induced by double-stranded RNA, which transiently and noncytopathically blocks HBV replication (15, 18, 37, 52). In this HBV transgenic mouse model, HBV replication is also downregulated by adoptive transfer of immune cells expressing IFN-γ or TNF-α (14, 16, 37). In a chimpanzee model of human HBV infection and in HBV transgenic mice, noncytopathic inhibition of HBV is associated with activated CD4+ and CD8+ T cells, as well as natural killer cells, which produce IFN-γ and TNF-α (7, 8, 17, 20). These studies demonstrate that HBV can be suppressed by production of specific cytokines without cell killing.
The extent to which HBV infection is cleared by noncytopathic means or by destruction of infected hepatocytes during natural infection is unresolved. In the woodchuck HBV model, viral inhibition and clearance from the liver were found to be associated with increased hepatocyte death in addition to increased expression of IFNs and TNF-α (21, 26, 66). Thus, both cytopathic and noncytopathic mechanisms likely contribute to the control and clearance of HBV during natural infection. A common feature of HBV inhibition by both mechanisms is production of IFNs and TNF-α. Suppression of HBV replication in transgenic mice by IFN-γ was linked to stimulation of inducible nitric oxide (NO) synthase (iNOS) and production of NO (19). However, IFNs and TNF-α act independently to inhibit HBV replication because neutralizing antibodies to both TNF-α and IFNs are required to fully block cytokine inhibition of viral replication in mice (reviewed in reference 15). It has been shown that IFN-α/β inhibits the replication of human HBV in the livers of transgenic mice and that of duck and human HBVs in infected hepatocyte cultures. IFN-α/β is reported to downregulate HBV replication by suppression of transcription (42, 43, 45) and likely other mechanisms as well (48, 49, 61).
HBV infects the mammalian and avian host liver as the primary target, often leading to chronic infection. Chronic infection by HBV is associated with development of liver disease and liver cancer in mammals (reviewed in reference 13). HBVs are pararetroviruses that replicate by reverse transcription of the viral pregenomic RNA (pgRNA), an RNA that is complementary to the entire viral DNA genome plus a terminal redundancy. HBVs differ from retroviruses in that they replicate in the cytoplasm within viral capsids and package the DNA replication intermediate, a circular partially double-stranded DNA (dsDNA) (13). The pgRNA encodes the viral reverse transcriptase (Pol protein) and the core protein (hepatitis B core antigen [HBcAg]) and serves as the template for reverse transcription. The cytoplasmic viral capsid, which is composed of the HBV core protein, encapsidates the pgRNA. Reverse transcription of the pgRNA and DNA replication of the viral genome occur within cytoplasmic capsids (core particles) (13). The HBV regulatory protein HBx is also required for viral replication (9, 67). HBx has a number of activities, including moderate activation of transcription factors, including NF-κB (34-36), and stimulation of cytoplasmic signal transduction pathways (2, 3, 10, 39, 54, 55), which promotes reverse transcription of the pgRNA and viral DNA replication (5, 31).
HBx has been shown to directly induce apoptosis in cells, probably when overexpressed and at lower levels to sensitize cells to proapoptotic stimuli such as TNF-α or etoposide (4, 11, 50, 53, 55, 56, 58). In the context of HBV replication in differentiated hepatic HepG2 cell lines, HBx was found to sensitize cells to killing by TNF-α only if the proto-oncogene myc was strongly expressed and the c-Jun N-terminal kinase was activated (55, 56). These data are consistent with the slightly enhanced ability of HBx to promote hepatocyte turnover in the livers of transgenic mice that overexpress the c-myc gene (57). However, other studies demonstrated that HBx stimulation of NF-κB prevents its ability to sensitize cells to TNF-α-mediated cell killing (56). Thus, the proapoptotic activity of HBx appears to require a particular set of conditions unless it is overexpressed, such as the absence of NF-κB stimulation in conjunction with elevated myc and/or c-Jun N-terminal kinase activity.
In this report, we demonstrate that activation of IKK-α/β and downstream NF-κB by TNF-α or by other means suppresses HBV replication without killing cultured hepatic cells. Activation of NF-κB signaling inhibits assembly or destabilizes the formation of cytoplasmic viral capsids, structures in which HBV replication occurs. We therefore identify a novel pathway for the noncytopathic inhibition of HBV replication by TNF-α that is mediated by NF-κB.
MATERIALS AND METHODS
Cell culture.
HepG2 cells, a differentiated hepatoblastoma cell line, were propagated in minimal essential medium with 10% fetal bovine serum, 1 mM sodium pyruvate, nonessential amino acids (Cellgro Mediatec Inc.), 2 mM glutamine, and 25 μg of gentamicin (Gibco-BRL) per ml. Human TNF-α (Sigma) was added to culture medium at the indicated doses and replenished every 24 h for 4 days. IFN-α (Schering-Plough) was added to culture medium at 500 or 1,000 U/ml immediately following transfection and replaced every 24 h for 3 days. Neutralizing antibodies to IFN-α/β (R&D Systems) were added to cell culture medium at 5 μg/ml every day following transfection of cells and maintained until cell harvest. For determination of transfection efficiency, green fluorescent protein (GFP) expression from transfected plasmid pGFP was monitored in living cells in culture with a Zeiss Axiovert fluorescence microscope at a magnification of ×100. Cells were photographed at 2 days after transfection by bright-field and UV light.
Transfection and plasmids.
HepG2 cells at 80% confluency were transfected with a 3:1 microgram ratio of liposomes to DNA (Fugene reagent; Roche). Cells were washed three times in serum-free medium prior to transfection. Freshly diluted recombinant human TNF-α (Calbiochem) was added to cells following transfection, and medium with recombinant human TNF-α was changed every 24 h. Plasmid pGEM7-HBVwt contains a 120% DNA copy of the HBV strain ayw genome. Plasmid pGEM7-HBV*7 is identical but contains a mutated HBx gene that does not express the HBx protein, herein referred to as HBV HBx− (38, 47). Plasmid pCMV-IκBα-SR (provided by W. Greene, University of North Carolina) expresses a superrepressor form of IκBα in which serines 32 and 36 were mutated to alanine (60). Plasmids pRc-β-actin-3HA-IKK-α (12) and -IKK-β (65) were described previously (provided by M. Karin, University of California San Diego). The NF-κB-Luc reporter vector pNF-κB-Luc and the GFP expression vector (pGFP) were purchased from Clontech Laboratories. Plasmid pRelA (provided by D. Baltimore, California Institute of Technology) expresses the p65 RelA subunit of NF-κB (1).
Analysis of viral replicative intermediates and proteins.
Viral core particles were isolated from cytoplasmic fractions of equal numbers of transiently transfected cells and subjected to Southern blot analysis as previously described (41), with modifications. Briefly, 107 HepG2 cells were lysed in 50 mM Tris HCl [pH 7.4]-100 mM NaCl-1 mM EDTA-10 mM magnesium acetate-1% NP-40, nuclei and debris were removed by centrifugation at 10,000 × g for 5 min at 4°C, protein concentrations and luciferase activities in protein-normalized samples were determined (Luciferase Assay Kit; Promega), and the supernatant was digested with 150 μg of DNase I per ml and 100 μg of RNase A per ml (Sigma) at 37°C for 60 min. Samples were centrifuged at 10,000 × g, and viral core particles were precipitated with 200 μl of 35% polyethylene glycol 8000 per ml in 1.75 M NaCl-10 mM EDTA at 4°C for 1 h. Pellets were resuspended in 10 mM Tris HCl (pH 7.4)-100 mM NaCl-1 mM EDTA-5 mM CaCl2-0.5% sodium dodecyl sulfate (SDS) and digested with 60 μg of proteinase K per ml at 55°C for 1 h. DNA was extracted with phenol-chloroform, followed by butanol-isopropanol (7:3), and precipitated with 2 volumes of ethanol; pellets were resuspended in 10 mM Tris HCl [pH 7.4]-1 mM EDTA, electrophoresed in 1.2% agarose gels, and then transferred to GeneScreen Plus nylon membranes (NEN Life Science Products, Inc.). For hybridization, the 3.2-kb genomic HBV DNA was used to produce 32P-labeled probes. Hybridization was performed at 68°C for 8 h with Perfecthyb plus (Sigma) and was followed by autoradiography. Blots representative of at least three independent experiments are shown. Data were quantified by densitometry of at least three independent experiment, and the results varied by less than 25% between studies.
Assay of encapsidated viral pgRNA and core particles.
Viral core particles were isolated from 107 cells as described above, encapsidated viral RNA was extracted from particles with Trizol (Gibco-BRL), and viral DNA was digested with 25 U of RNase-free RQ-1 DNase (Boehringer Mannheim) at 37°C for 30 min to fully degrade encapsidated viral DNA. RNase protection analysis was performed with one-fourth of each sample by using the RPA II RNase Kit (Ambion Inc., Austin, Tex.) and a 32P-labeled riboprobe of a 280-bp BamHI-EcoRI fragment HBV fragment that overlaps the 5′ end of the pgRNA (46). RNA fragments protected by RNase digestion were separated by electrophoresis in denaturing 5% polyacrylamide-8 M urea gels and visualized by autoradiography. Viral core particles were resolved and quantified by native agarose gel electrophoresis and immunoblot detection as previously described (44). Briefly, cells were lysed and treated with RNase and DNase as described for analysis of replicative intermediates above, and then 1% of the lysate was resolved by electrophoresis in a 1% native agarose gel in TBE buffer. Proteins were transferred to nitrocellulose filters, and core nucleocapsids were visualized by immunoblotting with rabbit polyclonal anti-core antibodies (a gift of M. Nassal) and a secondary chemiluminescent peroxidase-conjugated antibody (ECL; Amersham) as described above. Typical results of at least three independent experiments, which were quantified by densitometry, are shown.
Northern blot analysis.
RNAs were isolated by Trizol (Gibco) extraction in accordance with the manufacturer's instructions, poly(A)+ RNA was purified by oligo(dT)-cellulose chromatography, and RNAs were resolved by 1% agarose formaldehyde gel electrophoresis. After transfer to membrane, RNAs were hybridization to 32P-labeled HBV or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probes. Results were quantified by densitometry.
EMSA.
The electrophoretic mobility shift assay (EMSA) was carried out as described previously (55). Briefly, nuclear extracts (5 μg) were incubated for 30 min at 23°C in 15 μl of buffer [10 mM HEPES (pH 7.9), 50 mM NaCl, 0.5 mM EDTA, 5 mM MgCl2, 1 mM dithiothreitol, 10% glycerol, 3 μg of poly(dI-dC)] with 10 fmol of 5′-end-32P-labeled dsDNA oligonucleotide (106 cpm/reaction). The dsDNA oligonucleotide for the probe or competitor containing an NF-κB binding site corresponded to the sequence 5′-GATCCAGAGGGGCCACTTTCCGAGAGGA-3′. DNA-NF-κB complexes were separated from free labeled DNA by electrophoresis in 4.5% polyacrylamide gels containing 50 mM Tris-HCl (pH 8.5), 200 mM glycine, and 1 mM EDTA. Gels were dried, autoradiographed, and quantitated by PhosphorImager analysis. Unlabeled-competitor assays were carried out by adding a 100-fold molar excess of unlabeled dsDNA NF-κB oligonucleotide simultaneously with the labeled probe.
Cell death assay.
Cell killing was measured by using the lactate dehydrogenase (LDH) assay as recommended by the manufacturer (Promega). Briefly, an equal number of HepG2 cells were assayed and the amount of LDH released from dying or dead cells was detected with the Cytox 96 kit (Promega). Percent cell death was calculated as follows: (experimental LDH release − spontaneous LDH release)/(total LDH release − spontaneous LDH release) × 100.
RESULTS
Noncytopathic inhibition of HBV DNA replication by TNF-α in cultured hepatic cells.
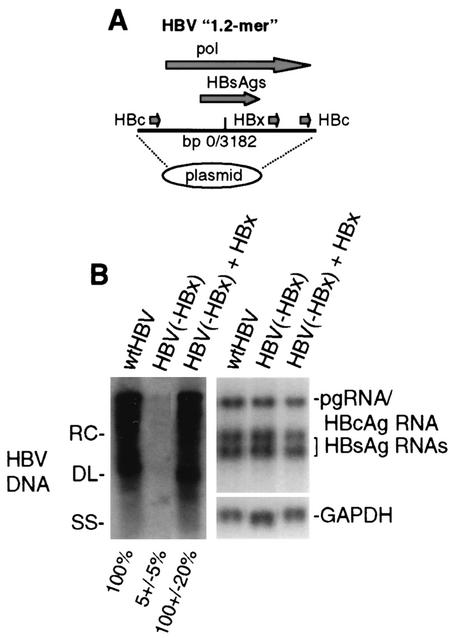
Although HBV cannot infect cultured hepatic cells, a somewhat greater than 120% head-to-tail DNA copy of the HBV genome (1.2-mer) replicates in an HBx-dependent manner when transfected into HepG2 cells (Fig. 1A) (6, 38). To examine the effect of TNF-α treatment on HBV replication, HepG2 cells were transfected with wild-type HBV genomic DNA or HBx− HBV genomic DNA, with or without trans complementation by an HBx expression plasmid. At 4 days posttransfection, cytoplasmic viral core particles were isolated and the level of viral replicative DNA intermediates was determined by Southern blot hybridization with 32P-labeled probes (Fig. 1B). All forms of viral replicative DNA intermediates were present in core particles: relaxed circular, double stranded linear, and single stranded. DNA replication of the HBV HBx− mutant was reduced to 5 to 10% of that of the wild type and could be trans complemented by cotransfection with an HBx expression plasmid, demonstrating that replication is responsive to stimulatory and inhibitory signals. As shown previously (6), HBV mRNA cytoplasmic abundance was relatively unaffected by the absence of HBx in this system (Fig. 1B), showing only a 50% reduction compared to a GAPDH mRNA control.
FIG. 1.
HBV replication and transcription in HepG2 cells. (A) Schematic representation of replication-competent HBV genomic DNA. Shown is a head-to-tail replicon of 1.2 copies (1.2-mer) of the HBV genome. Indicated are the duplicated viral core protein (HBc) coding regions, the single HBx coding region, the polymerase (pol) coding region, and the base pair junction of HBV strain ayw. (B) Southern blot analysis of cytoplasmic HBV core particle-associated viral genomic DNA from equal numbers of cells transfected with wild-type HBV, the HBV HBx− mutant, and the mutant trans complemented with an HBx expression vector for 4 days. Replicative DNA intermediates correspond to RC (relaxed circular), DL (double-stranded linear), and SS (single-stranded linear) DNAs. A Northern blot analysis of viral pregenomic (pg), core (HBc), and envelope (HBsAg) mRNAs and cellular GAPDH mRNA obtained from equal numbers of cells in duplicate plates is shown. Quantification of three independent experiments was obtained by densitometry and is shown at the bottom. Data were normalized to those obtained with the wild-type (wt) HBV sample.
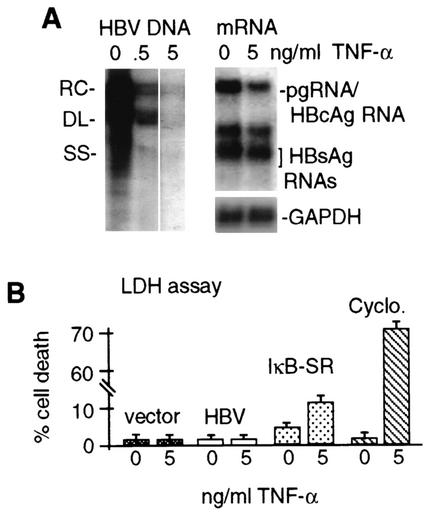
To determine whether there are TNF-α-sensitive steps during HBV replication in culture, HepG2 cells were transfected with wild-type HBV genomic DNA and treated with 0.5 or 5 ng of recombinant human TNF-α per ml for 4 days. The level of HBV DNA replication was then determined in purified cytoplasmic core particles and viral mRNA abundance was examined by Northern analysis (Fig. 2A). At 0.5 ng/ml, TNF-α reduced the level of HBV replicative DNA intermediates to ∼10% of that of untreated control samples, which was further reduced to ∼3% by a TNF-α concentration of 5 ng/ml. There was only a slight decrease in HBV mRNA levels relative to cellular GAPDH mRNA (Fig. 2A). By inclusion of a GFP expression vector with HBV genomic DNA, it was determined that 70 to 80% of HepG2 cells were transfected (data not shown). It should be noted that two prior studies detected only a slight downregulation of viral replication by TNF-α in culture (22, 40). We suspect that the hepatic cells used in those studies were only marginally sensitive to TNF-α, and no independent corroboration of sensitivity was provided. In one case, studies used HepG2.2.15 cells, a line stably transformed with replicating HBV genomic DNA (22). HepG2.2.15 cells produce largely inactive and nonsecreted TNF-α (32), and they are largely resistant to killing by high concentrations of TNF-α (22). Compared to uninfected HepG2 cells, HepG2.2.15 cells contain high levels of the protein GP73, which is typically downregulated by TNF-α (30). Thus, HepG2.2.15 cells appear to be only weakly sensitive to TNF-α. Another study was carried out with met oncogene-transformed hepatic cells containing a transgenomic replicating human HBV DNA (40). However, ectopic expression of the met oncogene has been shown to confer resistance to the biological effects of TNF-α (25). Moreover, accumulation of serum-derived α-fetoprotein on HepG2 cells has been reported to render them insensitive to TNF-α but washing in serum-free medium, as performed here, restores sensitivity (51). Consequently, we have restricted our studies to HepG2 cells transiently transfected with genomic HBV DNA.
FIG. 2.
Effect of TNF-α on HBV replication and cell viability. HepG2 cells were transfected with HBV 1.2-mer genomic DNA and treated with TNF-α for 4 days, with daily replenishment at the indicated doses. (A) Cytoplasmic core particles were purified from equal numbers of cells, and associated HBV DNA was extracted and detected by Southern blot analysis. Poly(A)+ mRNA was selected and detected by Northern blot analysis from duplicate plates of cells. RC, relaxed circular; DL, double stranded linear; SS, single stranded linear. (B) HepG2 cells were transfected with the vector alone or with wild-type HBV genomic DNA, with or without treatment with 5 ng of TNF-α per ml. At 4 days posttransfection, cells were assayed for percent death by the LDH assay. Untransfected cells were treated with 5 ng of TNF-α per ml and 10 μg of cycloheximide (Cyclo.) per ml for 10 h and then assayed for cell death. Results of both experiments were averaged from three independent assays and quantified by densitometry. Data were normalized to the untreated sample.
Inhibition of HBV replication by TNF-α was found to occur noncytopathically, as shown by assay of levels of LDH, a stable cytoplasmic enzyme that is released from dead or dying cells and quantitatively measures cell death, in medium (Fig. 2B). No significant cell death was detectable in vector-transfected or HBV-expressing cells, with or without treatment with 5 ng of TNF-α per ml for 4 days. Inhibition of NF-κB by transfection of a plasmid expressing the superrepressor form of IκB-α (IκB-SR), which blocks NF-κB activation (60), and treatment with 5 ng of TNF-α per ml resulted in approximately 15 to 20% cell killing. Addition of cycloheximide and TNF-α resulted in significant killing (80%), demonstrating that these cells are not resistant to TNF-α-mediated killing. Thus, when the ability to activate NF-κB is maintained, HBV DNA replication is inhibited by TNF-α without cell killing in this system. It should be noted that previous studies showed that HBx alone, in the absence of HBV replication, could sensitize HepG2 cells to TNF-α-mediated apoptotic killing (55, 56), whereas we found no evidence of increased cell death in the HBV replicon system. It is likely that the important difference is the lower level of HBx expression from viral genomic DNA in the context of HBV replication than that expressed from a transfected plasmid.
TNF-α inhibits HBV replication by activating NF-κB signaling.
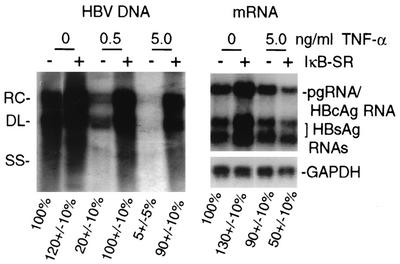
A primary effect of TNF-α is activation of the NF-κB signaling pathway, which is mediated by IKK-β and to some extent by IKK-α (28). We therefore investigated whether the NF-κB signaling pathway is involved in inhibition of HBV replication by TNF-α. HepG2 cells were transfected with a replication-competent wild-type HBV genomic DNA, with or without the IκB-SR plasmid. At 1 day posttransfection, cells were treated with TNF-α continuously for 4 days, cytoplasmic core particles were purified, and viral DNA replication and poly(A)+ RNAs were examined (Fig. 3). At 5 ng/ml, TNF-α inhibited HBV replication by 90 to 95% compared to untreated controls without significantly reducing viral mRNA levels. Coexpression of IκB-SR (which blocked NF-κB activation shown later in Fig. 5B) restored HBV replication to untreated-control levels despite treatment with 5 ng of TNF-α per ml while actually decreasing the abundance of viral mRNAs by 50%. These results indicate that TNF-α suppression of HBV DNA replication requires activation of the NF-κB signaling pathway.
FIG. 3.
Role of NF-κB inhibition in TNF-α suppression of HBV replication. HepG2 cells were transfected with HBV 1.2-mer genomic DNA for 1 day and then treated for 4 days with the indicated dose of TNF-α. An expression plasmid for the IκB-SR superrepressor of NF-κB was included in the initial transfection as indicated. Cytoplasmic viral core particles were purified from equal numbers of cells, and associated HBV DNA was detected by Southern blot analysis. Northern blot analysis was done with poly(A)+ RNA extracted from equal numbers of cells in duplicate plates. Autoradiograms were quantified by densitometry of at least three independent experiments and normalized to the untreated control without IκB-SR (first lane). RC, relaxed circular; DL, double stranded linear; SS, single stranded linear.
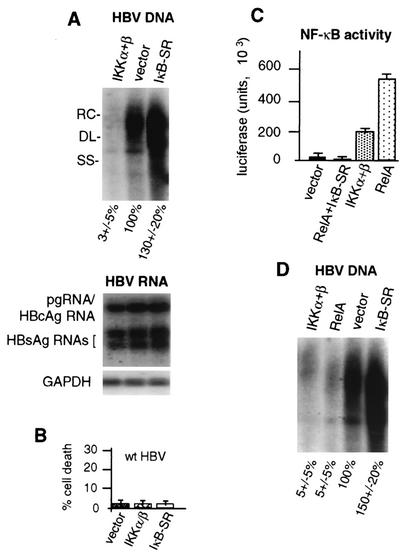
FIG. 5.
Role of NF-κB pathway in suppression of HBV replication. HepG2 cells were transfected with wild-type 1.2-mer HBV genomic DNA and the vector alone or the vector expressing NF-κB-activating kinase IKK-α/β or the NF-κB superrepressor IκB-SR. (A) Southern blot analysis on HBV core particle-associated viral DNA, and Northern blot analysis was performed on poly(A)+ RNA obtained from equal numbers of cells 4 days posttransfection in duplicate plates. RC, relaxed circular; DL, double stranded linear; SS, single stranded linear. (B) HepG2 cells were cotransfected with wild-type (wt) HBV genomic DNA and the vector expressing IKK-α/β or IκB-SR or the vector alone and analyzed for the level of cell death by the LDH assay at 3 days posttransfection. The data shown are averages of three independent experiments. (C) HepG2 cells were transfected with an NF-κB-dependent luciferase transcriptional reporter and the vector alone, the RelA NF-κB subunit expression vector, the vector expressing IKK-α/β, or the vectors expressing RelA and IκB-SR. NF-κB activity was determined by three independent luciferase assays using equal amounts of cellular extracts at 3 days posttransfection. (D) HepG2 cells were transfected with wild-type HBV genomic DNA and the expression vector for IKK-α/β or RelA, the vector alone, or IκB-SR. Viral core particles were isolated from equal numbers of cells at 4 days posttransfection, and viral DNA was assayed by Southern blot analysis. Autoradiograms were quantified by densitometry of at least three independent experiments and normalized to vector-plus-HBV samples.
Strong activation of the NF-κB signaling pathway inhibits HBV replication in the absence of TNF-α.
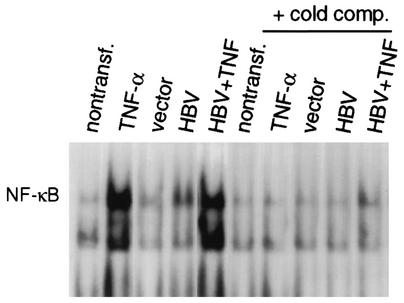
We investigated whether activation of the NF-κB signaling pathway is sufficient to suppress HBV replication in the absence of TNF-α treatment. One potentially confounding issue is that the HBV HBx protein is reported to stimulate transcription factor NF-κB in a variety of cell types, although HBx activity has not been widely studied when expressed in the context of an HBV replicon. We therefore compared the level of NF-κB activation during HBV replication to that of TNF-α-treated cells (Fig. 4). Formation of NF-κB DNA binding complexes was examined by EMSA with equal amounts of nuclear extracts and a 32P-labeled dsDNA oligonucleotide containing an NF-κB binding site. Nontransfected and vector- and HBV HBx−-transfected HepG2 cells contained low levels of active NF-κB. NF-κB levels were stimulated 2- to 3-fold by transfection of wild-type HBV and approximately 20-fold by TNF-α treatment of uninfected or HBV-transfected cells. Inclusion of excess unlabeled NF-κB DNA oligonucleotide (cold comp.) abolished DNA binding complexes, demonstrating NF-κB binding specificity. Thus, HepG2 cells in the absence of HBV do not contain constitutively high levels of NF-κB, transfection itself does not induce NF-κB, and NF-κB is only very modestly activated (2-fold) during HBV replication in this system but strongly induced (20-fold) by TNF-α.
FIG. 4.
Characterization of NF-κB activation by TNF-α or during HBV replication. HepG2 cells were either nontransfected (nontransf.) or transfected with wild-type or HBx− mutant HBV genomic DNA for 3 days. Cells were treated with 5 ng of TNF-α per ml for 30 min as indicated. Nuclear extracts were prepared, and equal amounts (10 μg) were used to assay NF-κB DNA binding activity by EMSA with a 32P-labeled double-stranded oligonucleotide probe containing a single NF-κB binding site. Unlabeled-competitor (cold comp.) ablation of NF-κB binding was performed by using a 100-fold molar excess of unlabeled NF-κB oligonucleotide. Complexes were resolved by electrophoresis and autoradiography as described previously (56). Complexes were quantified by densitometry.
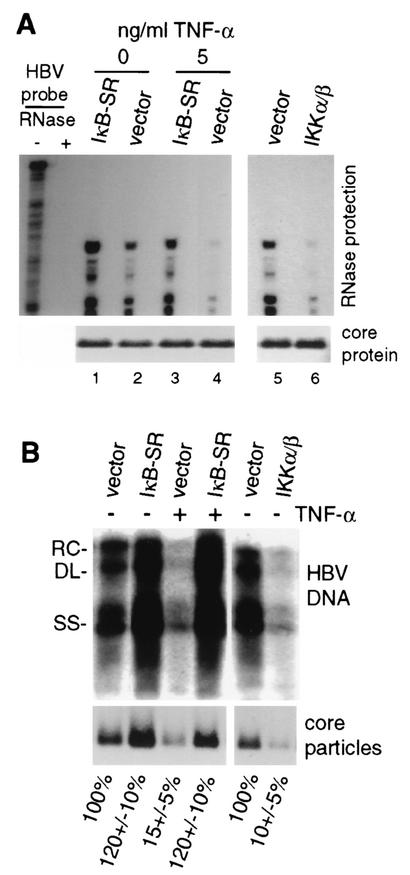
Since NF-κB is only marginally activated during HBV replication, we examined the effect of strong NF-κB activation or repression on HBV replication. Cells were cotransfected with HBV genomic DNA and an expression vector for NF-κB-activating IKK-α/β or IκB-SR, and Southern blot DNA analysis was performed on purified cytoplasmic HBV capsids (core particles) 4 days after transfection (Fig. 5A). Overexpression of IKK-α/β reduced viral replication to <5% of that of the HBV (vector) control. Overexpression of IκB-SR promoted higher levels of wild-type HBV DNA replication. These data indicate that strong activation of NF-κB suppresses HBV DNA replication with little change in HBV mRNA levels during the time course of these studies. Studies were carried out to examine the possibility that the effect of NF-κB on HBV DNA replication is related to increased cell death. HepG2 cells were transfected with wild-type HBV and either a vector-only, IKK-α/β, or IκB-SR expression construct, and the extent of cell death was determined by LDH assay (Fig. 5B). At 3 days posttransfection, little, if any, cell death was detectable in cultures transfected with the vector control, IKK-α/β, or IκB-SR and HBV genomic DNA. Thus, suppression of HBV replication associated with elevated NF-κB levels occurs noncytopathically.
Since a major effect of coexpression of IKK-α/β is strong activation of NF-κB, and given the ability of IκB-SR overexpression to promote HBV replication, we investigated whether inhibition of HBV replication is mediated by activated NF-κB transcription factor RelA. To quantitate the effect of RelA overexpression on NF-κB activity, HepG2 cells were transfected with an NF-κB-dependent luciferase reporter and expression vectors for RelA, IKK-α/β, and/or IκB-SR (Fig. 5C). Coexpression of RelA increased NF-κB-dependent transcription by >100-fold compared to the vector control, more than 3-fold higher than that of IKK-α/β. NF-κB activation by RelA or IKK-α/β was inhibited by overexpression of IκB-SR. We therefore determined the effect of RelA overexpression on HBV DNA replication by using Southern blot hybridization of isolated DNA from core particles (Fig. 5D). Cells were transfected with wild-type HBV genomic DNA and either the empty vector or an expression plasmid for RelA, IKK-α/β, or IκB-SR. RelA overexpression reduced HBV replication to 5 to 10% of that of wild-type HBV, similar in magnitude to overexpression of IKK-α/β. Thus, strong expression of active NF-κB inhibits HBV replication, whether by TNF-α treatment, overexpression of activating kinases IKK-α/β, or the NF-κB transcription factor.
TNF-α and NF-κB block HBV replication by inhibiting accumulation of viral core particles.
Since HBV mRNA levels were not significantly reduced within the time frame of these studies, despite TNF-α or NF-κB inhibition of viral replication, these results implicate a posttranscriptional event in inhibition of HBV replication. It is thought that the HBV pgRNA must be encapsidated within cytoplasmic viral core particles (capsids) to undergo reverse transcription and DNA replication (reviewed in reference 13). We first investigated the abundance of encapsidated pgRNA within cytoplasmic viral core particles. Denaturing SDS-polyacrylamide gel electrophoresis and immunoblot analysis demonstrated equal amounts of total core protein in HBV-transfected cells regardless of treatment with TNF-α or coexpression with IκB-SR (Fig. 6A). Core particles were isolated from equal numbers of the same cells used for detection of total core protein, representing untreated, TNF-α-treated, or IKK-α/β-transfected cells, and pgRNA was extracted and its level was determined by quantitative RNase protection analysis. Treatment of cells with 5 ng of TNF-α per ml for 3 days reduced the encapsidated pgRNA to very low levels (∼20-fold) compared to those in untreated samples (Fig. 6A), which was blocked by cotransfection of IκB-SR (Fig. 6A), consistent with its ability to promote HBV replication. Similarly, expression of IKK-α/β also reduced the level of encapsidated pgRNA by ∼20-fold in the absence of TNF-α treatment (compare lanes 5 and 6). There was no change in total core protein levels. We determined whether the reduced level of encapsidated pgRNA mediated by TNF-α or IKK-α/β expression results from a decrease in the abundance of cytoplasmic core particles by using duplicate plates of cells. Core particles migrate much more slowly than free core protein in native agarose gels and were proven to be core particles by coelectrophoresis with bacterium-produced core particles (gift of D. Standring; data not shown). Core particles were isolated from equal numbers of cells by nondenaturing agarose gel electrophoresis and then detected by immunoblot analysis (Fig. 6B). Compared to untreated vector control samples, treatment of cells with 5 ng of TNF-α per ml, or expression of IKK-α/β, reduced the abundance of core particles to 10 to 15% (or less) of that of wild-type HBV. The level of replicated HBV DNA within core particles was similarly reduced by treatment with TNF-α or expression of IKK-α/β. Both the abundance of core particles and HBV DNA replication levels were restored by transfection of IκB-SR and inhibition of NF-κB. The reduced abundance of core particles can account for the decreased levels of HBV DNA replication and the low level of encapsidated pgRNA. These results therefore show that strong activation of NF-κB by TNF-α treatment, or by overexpression of IKK-α/β, inhibits HBV DNA replication by impairing the formation or stability of cytoplasmic HBV capsids.
FIG. 6.
Effects of TNF-α and NF-κB on HBV replication, core protein, and particles. (A) HepG2 cells were transfected with wild-type HBV genomic DNA and the vector alone or the expression vector for IKK-α/β or IκB-SR for 4 days. Cytoplasmic core particles were purified from equal numbers of cells, and RNase protection analysis was carried out on the 5′ end of the encapsidated pgRNA. Protected RNA fragments were resolved by denaturing urea-acrylamide gel electrophoresis and autoradiography. Total core protein was determined by SDS-polyacrylamide gel electrophoresis of equal amounts of cellular lysates and detected by immunoblot analysis with antibodies to HBcAg. TNF-α treatment was performed for 1 to 4 days with daily replenishment. (B) HepG2 cells were transfected and left untreated or treated with 5 ng of TNF-α per ml as described above. Southern blot analysis was conducted on HBV DNA extracted from core particles obtained from equal numbers of cells. Core particles were resolved by native agarose gel electrophoresis using the same cell lysates examined for HBV replication and subjected to immunoblot analysis with antibodies to HBcAg. See Materials and Methods for details. Results were quantified by densitometry of at least three independent autoradiograms and normalized to the untreated vector control. RC, relaxed circular; DL, double stranded linear; SS, single stranded linear.
NF-κB inhibition of HBV replication does not act through IFN-γ or IFN-α/β.
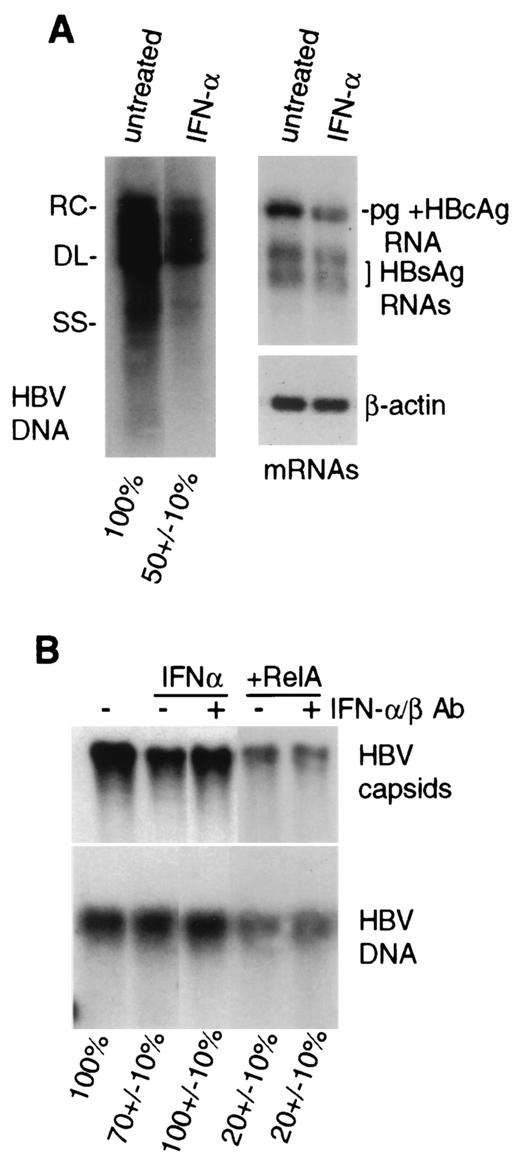
Induction of NF-κB is not generally sufficient for synthesis of IFN-α/β or IFN-γ (28). Nevertheless, we investigated whether strong activation of NF-κB inhibits HBV replication by inducing secretion of IFNs. IFN-γ has been shown to downregulate HBV replication by activating the NO synthase (iNOS) gene and inducing NO (19). Only very low and unchanged levels of iNOS mRNA were found in vector control- and HBV-transfected HepG2 cells, and there was no detectable activation of a cotransfected IFN-γ promoter (GAS)-dependent transcriptional reporter (data not shown). Thus, IFN-γ is not likely to be directly involved in the inhibition of HBV replication in this system. IFN-α/β also impairs HBV replication, possibly through several cell type-specific mechanisms (42, 43, 45, 48, 49, 61). To determine whether IFN-α/β is responsible for the suppression of HBV replication mediated by NF-κB, cells were continuously treated with 1,000 U of IFN-α per ml for 3 days and the effects on HBV replication and transcription were determined. Control studies demonstrated activation of the IFN-α/β-inducible protein kinase PKR only in HBV-replicating cells treated with IFN-α (data not shown), indicating that untreated cells are highly unlikely to produce sufficient levels of IFN to be effective. IFN-α at these substantial doses only reduced viral replication by about half in HepG2 cells, with a similar effect on viral, but not cellular, β-actin mRNA levels (Fig. 7A). Moreover, addition of high levels of neutralizing antibodies to IFN-α/β to the culture medium of cells transfected with wild-type HBV genomic DNA and a RelA expression vector failed to prevent downregulation of HBV cytoplasmic core particle abundance, whereas it did block the modest downregulation mediated by direct addition of IFN-α to the culture medium (Fig. 7B). Analysis of HBV replication was conducted on the transferred core particles by Southern blot analysis of disrupted capsids. The modest downregulation of HBV core particles by IFN-α/β and its reversal by anti-IFN-α/β antibodies were reflected in the levels of viral DNA replication as well. Additional studies (data not shown) that exclude the possibility that NF-κB activation acts via production of IF-α/β include the inability to observe activation of PKR in HBV-replicating cells cotransfected with RelA. TNF-α has been reported to transiently activate PKR, so this parameter was not measured in those cells (64). These results therefore indicate that NF-κB inhibition of HBV replication does not directly involve secretion of IFNs in this system. Moreover, it is clear that transfection itself with the lipophilic agent Fugene, used here, does not induce IFN production.
FIG. 7.
IFNs are not the mediators of NF-κB inhibition of HBV replication. (A) HepG2 cells were transfected with HBV genomic DNA and 1 day later mock treated or treated with 1,000 U of recombinant IFN-α per ml for 3 days, with daily replenishment. Core-associated viral DNA and poly(A)+ mRNA Northern analyses were done with equal numbers of cells from duplicate plates. RC, relaxed circular; DL, double stranded linear; SS, single stranded linear. (B) HepG2 cells were transfected with HBV genomic DNA and the vector alone or the expression vector for RelA. IFN treatment was carried out as described above. When added, neutralizing antibodies (Ab) to IFN-α/β were included at 5 μg/ml throughout the period of infection. Cells were harvested after 4 days and assayed for core particles by native agarose gel electrophoresis and immunoblot analysis. Core particle-associated viral DNA replication was measured by Southern blot analysis of disrupted core particles that had been transferred to nitrocellulose. Autoradiograms from three independent experiments were quantified by densitometry and normalized to the untreated control.
DISCUSSION
There is only a very poor understanding of the molecular mechanisms by which cytokines suppress HBV replication within infected hepatic cells. An HBV transgenic mouse model has provided important insights, demonstrating noncytopathic inhibition of HBV replication by type I and II IFNs and by TNF-α (reviewed in reference 15). Here we demonstrated the inhibition of HBV replication by TNF-α in a tissue culture system, which has enabled dissection of the innate antiviral mechanism.
We provided several lines of evidence that showed that TNF-α suppresses HBV replication without cell killing by activating the NF-κB signaling pathway, which predominantly blocks the formation or destabilizes the integrity of HBV capsids, thereby blocking viral replication. First, the antiviral effects of TNF-α occurred at cytokine concentrations that did not induce apoptosis (Fig. 2) and was sustainable over the course of 5 days (the longest time tested). Second, the noncytopathic antiviral effect of TNF-α could be blocked by inhibition of NF-κB (Fig. 3). Third, strong intracellular upregulation of the NF-κB signaling pathway in the absence of TNF-α treatment fully mimicked the antiviral effect and mechanism of action of TNF-α and could be blocked by inhibition of NF-κB (Fig. 5). These results indicate that the NF-κB signaling pathway functions as a central mediator of an innate antiviral immune response to HBV. After the completion of our study, Chisari and coworkers (40) demonstrated downregulation of HBV replication and viral capsid abundance by IFN-γ treatment of met oncogene-transformed murine hepatic cells harboring transgenic human HBV. Thus, there may be a common innate antiviral response to HBV that is mediated independently by TNF-α and IFN-γ. The molecular mechanism of IFN-γ inhibition of HBV is not known (40). Importantly, we found no evidence of activation of iNOS or an IFN-γ-responsive transcriptional reporter (data not shown), suggesting that our observations are not related to those previously reported for IFN-γ (15, 40). Inclusion of neutralizing antibodies to IFN-α/β also did not block HBV downregulation by strong activation of NF-κB but did block viral downregulation mediated by addition of IFN-α to the medium (Fig. 7). Although the possibility cannot be excluded that low levels of autocrine IFNs act in conjunction with TNF-α to synergistically block HBV replication in this system, since addition of antibodies cannot neutralize very low levels of cytokines, the levels of IFNs would have to be too low to be detectable by most biological criteria and unlikely to act in the absence of TNF-α. Thus, the most direct interpretation of these results is that HBV replication is inhibited by activation of NF-κB, whether by overexpression of RelA, IKK-α/β overexpression, or treatment of cells with TNF-α, and that this represents a novel innate antiviral immune response. One caveat to the tissue culture studies concerns whether the complex life cycle and requirements for HBV replication in animals are recapitulated here. Although the viral transcription template, covalently closed circular DNA, is not produced in this system, viral transcription is derived from the transfected plasmid and appears to correctly represent the normal viral pattern. More importantly, hepatic cell lines in culture can only partially reproduce the complex response of hepatocytes to cytokines in the liver. Nevertheless, the noncytopathic inhibition of HBV replication observed in our studies is consistent with the effect of TNF-α in HBV transgenic mice that is associated with transient suppression of the virus (15).
TNF-α stimulates a variety of cellular genes, many through activation of NF-κB (23). NF-κB-inducible genes number well over 100 and play a fundamental role in inflammatory responses, as many NF-κB-responsive genes encode proinflammatory proteins such as Cox-2 or cytokines such as IL-1β, IL-6, IL-8, and TNF-α (reviewed in reference 28). NF-κB is an important inhibitor of apoptosis, which is mediated through target genes activated by NF-κB, including c-Flip, c-IAPs, Bcl-2 family members, and GADD45β, among others. The NF-κB pathway is central to innate immunity as well, whether in insect cells or in mammalian cells (29). To some extent, the antiapoptotic activity of NF-κB is intertwined with poorly understood roles in innate immunity. For instance, by inhibiting apoptosis and consequent anti-inflammatory collapse of the cell, NF-κB preserves antigen presentation capabilities and activation of Toll-like receptors that initiate immune responses (29). In addition, conditional inhibition of NF-κB in the mouse liver impairs expression of chemokines such as IL-8 that recruit inflammatory cells and aid in clearing microbial infections (33). However, these are all NF-κB-dependent responses that aid in the development of systemic antibacterial and antiviral immunity. There is very little understanding of innate NF-κB-dependent antiviral mechanisms that act within the infected cell, apart from IFN- and double-stranded RNA-mediated effects, which have been excluded in our study.
Studies now need to investigate the mechanism by which TNF-α and NF-κB activity impair HBV capsid integrity. It is clear that core protein is not degraded more rapidly and virus particles are not exported more efficiently because there was no decrease in cytoplasmic core protein levels (Fig. 6). Studies have shown that protein kinase C phosphorylation of the C terminus of the core protein in vitro prevents capsid formation and promotes nuclear uptake of core proteins (27). It is not known whether a similar mechanism functions during HBV replication, but it is unlikely to account for our results, as there was no observable nuclear increase in HBV core protein following TNF-α treatment (data not shown). While it remains possible that phosphorylation of capsids is involved in their loss of integrity and inhibition of HBV replication, the TNF-α-NF-κB response more likely occurs by induction of gene products that inhibit HBV particle assembly. Studies are ongoing to identify potential NF-κB-stimulated cellular genes that act to inhibit HBV particle integrity.
An important aspect of this study that needs to be investigated in future work concerns the multiple roles played by the HBV HBx protein in viral replication. When expressed from HBV genomic DNA in this system, HBx was not a strong activator of viral transcription or of NF-κB. However, in a recent HBV transgenic mouse model, HBx was found to strongly stimulate the viral core gene promoter, which controls pregenomic and core mRNA transcription (62). There are a number of fundamental differences between these two model systems for HBV replication that may account for the different effects observed. However, it is possible that the much higher levels of HBV genomic DNA present in transfected HepG2 cells eliminate the pronounced transcriptional effect of HBx observed in transgenic mouse hepatocytes, which likely contain only several copies of the HBV genome. It is not known whether HBx activates the NF-κB signaling pathway in the HBV transgenic mouse model. If it does, it will be important to determine whether HBx both activates HBV replication and governs the level of viral replication in this system. It is interesting that a number of cases of fulminant HBV infection, in which the virus replicates to very high levels that become pathological, may correspond to mutations in the HBx coding region (24, 59, 63) in addition to established mutations in the precore gene. It needs to be determined whether HBx gene mutations found in fulminant HBVs fail to activate NF-κB but continue to activate HBV replication and therefore may be involved in the fulminant viral phenotype.
Acknowledgments
We thank W. Greene and M. Karin for plasmids used in this study. Special thanks go to members of the laboratory for critical reading of the manuscript. Anti-core antibodies were kindly provided by M. Nassal (University of Freiburg), and core particles were kindly provided by D. Standring (Novirio).
This study was supported by grants from the National Institutes of Health (CA56533 to R.J.S.) and the Deutsche Forschungsgemeinschaft (to M.B). R.P was supported by National Institutes of Health training grant T32AI07180.
REFERENCES
- 1.Beg, A. A., W. C. Sha, R. T. Bronson, S. Ghosh, and D. Baltimore. 1995. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature 376:167-170. [DOI] [PubMed] [Google Scholar]
- 2.Benn, J., and R. J. Schneider. 1994. Hepatitis B virus HBx protein activates Ras-GTP complex formation and establishes a Ras, Raf, MAP kinase signalling cascade. Proc. Natl. Acad. Sci. USA 91:10350-10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benn, J., F. Su, M. Doria, and R. J. Schneider. 1996. Hepatitis B virus HBx protein induces transcription factor AP-1 by activation of extracellular signal-regulated and c-Jun N-terminal mitogen-activated protein kinases. J. Virol. 70:4978-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergametti, F., S. Prigent, B. Luber, A. Benoit, P. Tiollais, A. Sarasin, and C. Transy. 1999. The proapoptotic effect of hepatitis B virus HBx protein correlates with its transactivation activity in stably transfected cell lines. Oncogene 18:2860-2871. [DOI] [PubMed] [Google Scholar]
- 5.Bouchard, M., S. Giannakopoulos, E. H. Wang, N. Tanese, and R. J. Schneider. 2001. Hepatitis B virus HBx protein activation of cyclin A-cyclin-dependent kinase 2 complexes and G1 transit via a Src kinase pathway. J. Virol. 75:4247-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouchard, M. J., L. H. Wang, and R. J. Schneider. 2001. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 294:2376-2378. [DOI] [PubMed] [Google Scholar]
- 7.Cavanaugh, V. J., L. G. Guidotti, and F. V. Chisari. 1998. Inhibition of hepatitis B virus replication during adenovirus and cytomegalovirus infections in transgenic mice. J. Virol. 72:2630-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavanaugh, V. J., L. G. Guidotti, and F. V. Chisari. 1997. Interleukin-12 inhibits hepatitis B virus replication in transgenic mice. J. Virol. 71:3236-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, H.-S., S. Kaneko, R. Girones, R. W. Anderson, W. E. Hornbuckle, B. C. Tennant, P. J. Cote, J. L. Gerin, R. H. Purcell, and R. H. Miller. 1993. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J. Virol. 67:1218-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chirillo, P., M. Falco, P. L. Puri, M. Artini, C. Balsano, M. Levrero, and G. Natoli. 1996. Hepatitis B virus pX activates NF-κB-dependent transcription through a Raf-independent pathway. J. Virol. 70:641-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chirillo, P., S. Pagano, G. Natoli, P. L. Puri, V. L. Burgio, C. Balsano, and M. Levrero. 1997. The hepatitis B virus X gene induces p53-mediated programmed cell death. Proc. Natl. Acad. Sci. USA 94:8162-8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiDonato, J. A., M. Hayakawa, D. M. Rothwarf, E. Zandi, and M. Karin. 1997. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature 388:548-554. [DOI] [PubMed] [Google Scholar]
- 13.Ganem, D., and R. J. Schneider. 2001. The molecular biology of the hepatitis B viruses, p. 2923-2970. In D. Knipe and P. Howley (ed.), Field's virology, 4th ed., vol. 2. Lippincott, New York, N.Y. [Google Scholar]
- 14.Gilles, P. N., G. Fey, and F. V. Chisari. 1992. Tumor necrosis factor alpha negatively regulates hepatitis B virus gene expression in transgenic mice. J. Virol. 66:3955-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guidotti, L. G., and F. V. Chisari. 2001. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu. Rev. Immunol. 19:65-91. [DOI] [PubMed] [Google Scholar]
- 16.Guidotti, L. G., S. Guilhot, and F. V. Chisari. 1994. Interleukin-2 and alpha/beta interferon down-regulate hepatitis B virus gene expression in vivo by tumor necrosis factor-dependent and -independent pathways. J. Virol. 68:1265-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guidotti, L. G., T. Ishikawa, M. V. Hobbs., B. Matzke, R. Schreiber, and F. V. Chisari. 1996. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity 4:25-36. [DOI] [PubMed] [Google Scholar]
- 18.Guidotti, L. G., B. Matzke, H. Schaller, and F. V. Chisari. 1995. High-level hepatitis B virus replication in transgenic mice. J. Virol. 69:6158-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guidotti, L. G., H. McClary, J. M. Loudis, and F. V. Chisari. 2000. Nitric oxide inhibits hepatitis B virus replication in the livers of transgenic mice. J. Exp. Med. 191:1247-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guidotti, L. G., R. Rochford, J. Chung, M. Shapiro, R. Purcell, and F. V. Chisari. 1999. Viral clearance without destruction of infected cells during acute HBV infection. Science 284:825-829. [DOI] [PubMed] [Google Scholar]
- 21.Guo, J. T., H. Zhou, C. Liu, C. Aldrich, J. Saputelli, T. Whitaker, M. I. Barrasa, W. S. Mason, and C. Seeger. 2000. Apoptosis and regeneration of hepatocytes during recovery from transient hepadnavirus infections. J. Virol. 74:1495-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagelstein, J., A. Kist, W. Stremmel, and P. R. Galle. 1998. Antiviral potential of interferon-omega on hepatitis B virus replication in human hepatoma cells. Arzneim.-Forsch. 48:343-347. [PubMed] [Google Scholar]
- 23.Herbein, G., and W. A. O'Brien. 2000. Tumor necrosis factor (TNF)-alpha and TNF receptors in viral pathogenesis. Proc. Soc. Exp. Biol. Med. 223:241-257. [DOI] [PubMed] [Google Scholar]
- 24.Honda, A., O. Yokosuka, K. Suzuki, and H. Saisho. 2000. Detection of mutations in hepatitis B virus enhancer 2/core promoter and X protein regions in patients with fatal hepatitis B virus infection. J. Med. Virol. 62:167-176. [DOI] [PubMed] [Google Scholar]
- 25.Hudziak, R. M., G. D. Lewis, W. E. Holmes, A. Ullrich, and H. M. Shepard. 1990. Selection for transformation and met protooncogene amplification in NIH 3T3 fibroblasts using tumor necrosis factor alpha. Cell Growth Differ. 1:129-134. [PubMed] [Google Scholar]
- 26.Kajino, K., A. R. Jilbert, J. Saputelli, C. E. Aldrich, J. Cullen, and W. Mason. 1994. Woodchuck hepatitis virus infections: very rapid recovery after a prolonged viremia and infection of virtually every hepatocyte. J. Virol. 68:5792-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kann, M., B. Sodeik, A. Vlachou, W. H. Gerlich, and A. Helenius. 1999. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J. Cell Biol. 145:45-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karin, M., and M. Delhase. 2000. The IκB kinase (IKK) and NF-κB: key elements of proinflammatory signalling. Semin. Immunol. 12:85-98. [DOI] [PubMed] [Google Scholar]
- 29.Karin, M., and A. Lin. 2002. NF-κB at the crossroads of life and death. Nat. Immunol. 3:221-227. [DOI] [PubMed] [Google Scholar]
- 30.Kladney, R. D., X. Cui, G. A. Bulla, E. M. Brunt, and C. J. Fimmel. 2002. Expression of GP73, a resident Golgi membrane protein, in viral and nonviral liver disease. Hepatology 35:1431-1440. [DOI] [PubMed] [Google Scholar]
- 31.Klein, N., M. Bouchard, L.-H. Wang, C. Kobarg, and R. J. Schneider. 1999. Src kinases involved in hepatitis B virus replication. EMBO J. 18:5019-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lara-Pezzi, E., P. L. Majano, M. Gomez-Gonzalo, C. Garcia-Monzon, R. Moreno-Otero, M. Leverero, and M. Lopez-Cabrera. 1998. The hepatitis B virus X protein up-regulates tumor necrosis factor-α gene expression in hepatocytes. Hepatology 28:1013-1021. [DOI] [PubMed] [Google Scholar]
- 33.Lavon, I., I. Goldberg, S. Amit, L. Landsman, S. Jung, B. Z. Tsuberi, I. Barshack, J. Kopolovic, E. Galun, H. Bujard, and Y. Ben-Neriah. 2000. High susceptibility to bacterial infection, but no liver dysfunction, in mice compromised for hepatocyte NF-κB activation. Nat. Med. 6:573-577. [DOI] [PubMed] [Google Scholar]
- 34.Levrero, M., C. Balsano, G. Natoli, M. L. Avantaggiati, and E. Elfassi. 1990. Hepatitis B virus X protein transactivates the long terminal repeats of human immunodeficiency virus types 1 and 2. J. Virol. 64:3082-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lucito, R., and R. J. Schneider. 1992. Hepatitis B virus X protein activates transcription factor NF-κB without a requirement for protein kinase C. J.Virol. 66:983-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahe, Y., N. Mukaida, K. Kuno, M. Akiyama, N. Ikeda, K. Matshushima, and S. Murakami. 1991. Hepatitis B virus X protein transactivates human interleukin-8 gene through acting on nuclear factor κB and CCAAT/enhancer-binding protein-like cis elements. J. Biol. Chem. 266:13759-13763. [PubMed] [Google Scholar]
- 37.McClary, H., R. Koch, F. V. Chisari, and L. G. Guidotti. 2000. Relative sensitivity of hepatitis B virus and other hepatotropic viruses to the antiviral effects of cytokines. J. Virol. 74:2255-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melegari, M., P. P. Scaglioni, and J. R. Wands. 1998. Cloning and characterization of a novel hepatitis B virus x binding protein that inhibits viral replication. J. Virol. 72:1737-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Natoli, G., M. L. Avantaggiati, P. Chirillo, P. L. Puri, A. Ianni, C. Balsano, and M. Levrero. 1994. Ras- and raf-dependent activation of c-jun transcriptional activity by the hepatitis B virus transactivator pX. Oncogene 9:2837-2843. [PubMed] [Google Scholar]
- 40.Pasquetto, V., S. F. Wieland, S. L. Uprichard, M. Tripodi, and F. V. Chisari. 2002. Cytokine-sensitive replication of hepatitis B virus in immortalized mouse hepatocyte cultures. J. Virol. 76:5646-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pugh, J. C., K. Yaginuma, K. Koike, and J. Summers. 1988. Duck hepatitis B virus (DHBV) particles produced by transient expression of DHBV DNA in a human hepatoma cell line are infectious in vitro. J. Virol. 62:3513-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rang, A., S. Gunther, and H. Will. 1999. Effect of interferon alpha on hepatitis B virus replication and gene expression in transiently transfected human hepatoma cells. J. Hepatol. 31:791-799. [DOI] [PubMed] [Google Scholar]
- 43.Rang, A., T. Heise, and H. Will. 2001. Lack of a role of the interferon-stimulated response element-like region in interferon alpha-induced suppression of hepatitis B virus in vitro. J. Biol. Chem. 276:3531-3535. [DOI] [PubMed] [Google Scholar]
- 44.Ren, S., and M. Nassal. 2001. Hepatitis B virus (HBV) virion and covalently closed circular DNA formation in primary tupaia hepatocytes and human hepatoma cell lines upon HBV genome transduction with replication-defective adenovirus vectors. J. Virol. 75:1104-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romero, R., and J. E. Lavine. 1996. Cytokine inhibition of the hepatitis B virus core promoter. Hepatology 23:17-23. [DOI] [PubMed] [Google Scholar]
- 46.Scaglioni, P. P., M. Melegari, and J. R. Wands. 1997. Biologic properties of hepatitis B viral genomes with mutations in the precore promoter and precore open reading frame. Virology 233:374-381. [DOI] [PubMed] [Google Scholar]
- 47.Scaglioni, P. P., M. Melegari, and J. R. Wands. 1997. Posttranscriptional regulation of hepatitis B virus replication by the precore protein. J. Virol. 71:345-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schultz, U., and F. V. Chisari. 1999. Recombinant duck interferon gamma inhibits duck hepatitis B virus replication in primary hepatocytes. J. Virol. 73:3162-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schultz, U., J. Summers, P. Staeheli, and F. V. Chisari. 1999. Elimination of duck hepatitis B virus RNA-containing capsids in duck alpha interferon-treated hepatocytes. J. Virol. 73:5459-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schuster, R., W. H. Gerlich, and S. Schaefer. 2000. Induction of apoptosis by the transactivating domains of the hepatitis B virus X gene leads to suppression of oncogenic transformation of primary rat embryo fibroblasts. Oncogene 19:1173-1180. [DOI] [PubMed] [Google Scholar]
- 51.Semenkova, L. N., E. I. Dudich, I. V. Dudich, L. N. Shingarova, and V. G. Korobko. 1997. Alpha-fetoprotein as a TNF resistance factor for the human hepatocarcinoma cell line HepG2. Tumour Biol. 18:30-40. [DOI] [PubMed] [Google Scholar]
- 52.Shimizu, Y., L. G. Guidotti, P. Fowler, and F. V. Chisari. 1998. Dendritic cell immunization breaks cytotoxic T lymphocyte tolerance in hepatitis B virus transgenic mice. J. Immunol. 161:4520-4529. [PubMed] [Google Scholar]
- 53.Sirma, H., C. Giannini, K. Poussin, P. Paterlini, D. Kremsdorf, and C. Brechot. 1999. Hepatitis B virus X mutants present in hepatocellular carcinoma tissue abrogate both the antiproliferative and transactivation effects of HBx. Oncogene 18:4848-4859. [DOI] [PubMed] [Google Scholar]
- 54.Su, F., and R. J. Schneider. 1996. Hepatitis B virus HBx protein activates transcription factor NF-κB by acting on multiple cytoplasmic inhibitors of rel-related proteins. J. Virol. 70:4558-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Su, F., and R. J. Schneider. 1997. Hepatitis B virus HBx protein sensitizes cells to apoptotic killing by TNFα. Proc. Natl. Acad. Sci. USA 94:8744-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Su, F., C. N. Theodosis, and R. J. Schneider. 2001. Role of NF-κB and myc proteins in apoptosis induced by hepatitis B virus HBx protein. J. Virol. 75:215-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Terradillos, O., O. Billet, C.-A. Rnard, R. Levy, T. Molina, P. Briand, and M. A. Buendia. 1997. The hepatitis B virus X gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene 14:395-404. [DOI] [PubMed] [Google Scholar]
- 58.Terradillos, O., T. Pollicino, H. Lecoeur, M. Tripodi, M. L. Gougeon, P. Tiollais, and M. A. Buendia. 1998. p53-independent apoptotic effects of the hepatitis B virus HBx protein in vivo and in vitro. Oncogene 17:2115-2123. [DOI] [PubMed] [Google Scholar]
- 59.Uchida, T., T. Saitoh, and H. Shinzawa. 1997. Mutations in the X region of hepatitis B virus and their clinical implications. Pathol. Int. 47:183-193. [DOI] [PubMed] [Google Scholar]
- 60.Wang, C.-Y., M. W. Mayo, and A. S. Baldwin, Jr. 1996. TNF-α and cancer therapy-induced apoptosis: potentiation by inhibition of NF-κB. Science 274:784-787. [DOI] [PubMed] [Google Scholar]
- 61.Wieland, S. F., L. G. Guidotti, and F. V. Chisari. 2000. Intrahepatic induction of alpha/beta interferon eliminates viral RNA-containing capsids in hepatitis B virus transgenic mice. J. Virol. 74:4165-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu, Z., T. S. Yen, L. Wu, C. R. Madden, W. Tan, B. L. Slagle, and J. H. Ou. 2002. Enhancement of hepatitis B virus replication by its X protein in transgenic mice. J. Virol. 76:2579-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yuasa, R., K. Takahashi, B. V. Dien, N. H. Binh, T. Morishita, K. Sato, N. Yamamoto, S. Isomura, K. Yoshioka, T. Ishikawa, S. Mishiro, and S. Kakumu. 2000. Properties of hepatitis B virus genome recovered from Vietnamese patients with fulminant hepatitis in comparison with those of acute hepatitis. J. Med. Virol. 61:23-28. [PubMed] [Google Scholar]
- 64.Zamanian-Daryoush, M., T. H. Mogensen, J. A. DiDonato, B. R. Williams. 2000. NF-κB activation by double-stranded-RNA-activated protein kinase (PKR) is mediated through NF-κB-inducing kinase and IκB kinase. Mol. Cell. Biol. 20:1278-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zandi, E., D. M. Rothwarf, M. Delhase, M. Hayakawa, and M. Karin. 1997. The IκB kinase complex (IKK) contains two kinase subunits, IKKα and IKKβ, necessary for IκB phosphorylation and NF-κB activation. Cell 91:243-252. [DOI] [PubMed] [Google Scholar]
- 66.Zhou, T., J. T. Guo, F. A. Nunes, K. L. Molnar-Kimber, J. M. Wilson, C. E. Aldrich, J. Saputelli, S. Litwin, L. D. Condreay, C. Seeger, and W. S. Mason. 2000. Combination therapy with lamivudine and adenovirus causes transient suppression of chronic woodchuck hepatitis virus infections. J. Virol. 74:11754-11763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zoulim, F., J. Saputelli, and C. Seeger. 1994. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J. Virol. 68:2026-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]