Abstract
Incorporation of the vaccinia virus A36R protein into the outer membrane of intracellular enveloped virions (IEV) is dependent on expression of the A33R protein. Possible interactions of the 200-amino-acid cytoplasmic domain of the A36R protein with itself or with the cytoplasmic domain of the A33R, A34R, B5R, or F12L IEV membrane protein was investigated by using the yeast two-hybrid system. A strong interaction was detected only between the cytoplasmic domains of the A36R and A33R proteins. Upon further analyses, the interaction site was mapped to residues 91 to 111 of the A36R protein. To investigate the role of the A36R:A33R interaction during viral infection, five recombinant vaccinia viruses containing B5R-GFP as a marker were constructed. Four had the full-length A36R gene replaced with various-length C-terminal truncations of A36R, of which two contained residues 91 to 111 and two were missing this region. The fifth recombinant virus had an A33R gene with most of the 40-amino-acid cytoplasmic tail deleted. Residues 91 to 111 of A36R and the cytoplasmic tail of A33R were required for a strong interaction between the two proteins during viral infection and for maximal amounts of A36R protein on IEV. Mutants lacking these regions of A33R or A36R formed IEV that exhibited only short sporadic intracellular movement, displayed no actin tails, and formed small plaques on cell monolayers equivalent to those of an A36R deletion mutant and smaller than those formed by point mutations that specifically abrogate actin tail formation. The A33R interaction site of the A36R protein is highly conserved among orthopoxviruses and may overlap binding sites for cellular proteins needed for microtubular movement and actin tail formation.
Vaccinia virus is one of the most complex viruses, as exemplified by the multiple infectious forms that arise during morphogenesis (23). The earliest infectious form, known as the intracellular mature virion (IMV), is assembled by poorly understood mechanisms within the cytoplasmic factory area (6, 13, 28, 37). Trans-Golgi or endosomal cisternae wrap a subset of IMV with a double membrane to form intracellular enveloped virions (IEV) (35, 39). IEV travel along microtubules to the cell periphery where the outer of the two IEV membranes fuses with the plasma membrane, thereby translocating the cell-associated enveloped virions (CEV) to the cell surface (12, 16, 27, 42, 43). The CEV spread to adjacent cells at the tips of motile actin-containing microvilli (3, 4, 14, 38). CEV that are released from the infected cell into the surrounding medium are called extracellular enveloped virions (EEV) and may mediate long-range spread (1, 25).
A large number of proteins has been localized to the IEV and extracellular viral membranes, and mutations of these proteins give complex phenotypes. The A33R (31), A34R (7), A56R (36), B5R (8, 18), and F13L (15) proteins are envelope components of the IEV and extracellular forms of vaccinia virus, whereas A36R (41) and F12L (40) proteins are restricted to IEV. The F13L (2) and B5R (9, 44) proteins are required for wrapping of the IMV, and deletion of either gene reduces IEV and extracellular virus production and plaque size on cell monolayers. Plaque size is also reduced when the A33R (30), A34R (7, 21, 45), A36R (24, 33, 48), or F12L (49) gene is deleted. Although a decreased number of actin tails is a feature of all mutants that produce small plaques (29, 30, 33, 45, 48), direct involvement in actin tail formation has been demonstrated only for A36R. Phosphorylation of A36R Tyr 112 and 132 results in the recruitment of the adaptor protein Nck, WASP-interacting protein, N-WASP, and Grb2, which leads to activation of the Arp2/Arp3 complex and nucleation of actin polymerization (11, 22, 34). Moreover, conservative mutations of Tyr 112 and 132 result in a specific block in actin tail formation by recombinant viruses (27, 42). While direct involvement has not been ruled out, the requirement for the A33R, A34R, and F12L proteins for efficient actin tail formation is likely to be indirect. Thus, the A33R protein is necessary for association of the A36R protein with the IEV envelope (47), deletion of the A34R protein enhances release of CEV (21), and deletion of the F12L protein (40) decreases movement of IEV to the periphery of the cell. IEV movement is also disrupted by deletion or mutation of the A36R protein (27, 42).
Previous coimmunoprecipitation experiments indicated the following protein:protein interactions: A33R:A33R, A33R:A36R, A34R:A36R, and A34R:B5R (32, 47). These physical associations complicate attempts to determine the individual roles of the various IEV/CEV proteins. During the present investigation we demonstrated an interaction between the cytoplasmic domains of the A33R and A36R proteins. The interaction site of the A36R protein was mapped to residues 91 to 111. Deletion of the interaction sites of either the A33R or A36R protein resulted in less A36R protein associated with IEV, an inability to detect rapid long-range movement of IEV to the periphery, absence of actin tails, and a small plaque phenotype similar to that of an A36R deletion mutant and more severe than that of point mutations that specifically abrogate actin tail formation.
MATERIALS AND METHODS
Yeast two-hybrid constructs and analyses.
The primer pairs GCATATGATGACACCAGAAAACGAC and CGAATTCAGATATTCTAATACATAGACCAAT, GCATATGAAATCGCTTAATAGACAAACT and CGAATTCAGCGGCCGGCACCGACAA, GCATATGATTTGTAGGAAAAAGATACGTACT and CGAATTCCACCAATGATACGACCGATGA, GCATATGGTTTGTTCCTGTGACAAAAAT and CGAATTCCGGTAGCAATTTATGGAAC, and GCATATGTTAAACAGGGTACAAATCTTG and CGAATTCTAATTTTACCATCTGACTCATGGA were used to amplify the coding sequences of the cytoplasmic domains of A33R, A34R, A36R, B5R, and F12L, respectively, and to add a 5′ NdeI and 3′ EcoRI (underlined) restriction endonuclease site. Amplified fragments were cloned into pGEM-T (Promega), sequenced, and subsequently excised from pGEM-T by digestion with NdeI-EcoRI and ligated into similarly cleaved pGBKT7 and pGADT7 (BD Biosciences Clontech). Primers CAGATCTACGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACCAATATTATCTACGTCATTGTT, CAGATCTACGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACCAGCGAAGCTATCATCGTC, CAGATCTACGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACCAATGTGTTCTGTGCTAGG, and CAGATCTACGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACCATTT ATTAGCAGCGTGCT were used in conjunction with the primer GCATATGATTTGTAGGAAAAAGATACGTACT to amplify the coding sequence of residues 24 to 80, 24 to 93, 24 to 111, and 24 to 123 of A36R and to add a 5′ NdeI (underlined) and 3′ V5 tag (italics) followed by a BglII (underlined) site. Primers GCATATGGAATCAGACTGGGAGGAT, GCATATGGAACAAAACAATGACGTA, GCATATGTCTAGGAATGAGATATTGGA, and GCATATGTTCGCTGGTAGTTTAATATG were used in conjunction with the primer CAGATCTACGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACCAATGTGTTCTGTGCTAGG to amplify the coding sequence of residues 61 to 111, 71 to 111, 81 to 111, and 91 to 111 of A36R and to add a 5′ NdeI (underlined) and 3′ V5 tag (italics) followed by a BglII (underlined) site. Amplified fragments were cloned into pGEM-T (Promega), sequenced, and subsequently excised from pGEM-T by digestion with NdeI-BglII and ligated into pGBKT7 that had been cleaved with NdeI-BamHI (BD Biosciences Clontech).
To test for interactions, yeast strain AH109 was cotransformed with purified plasmids by using the Yeastmaker yeast transformation system 2 (BD Biosciences Clontech) according to the manufacturer's instructions. Transformed yeast cells were plated onto standard dropout media minus leucine and tryptophan. Resulting yeast colonies were tested for interaction by being streaked on standard quadruple dropout medium minus leucine, tryptophan, histidine, and adenine. β-Galactosidase activity was measured by using the Gal-Screen assay system (Tropix).
Cells and viruses.
HeLa and BS-C-1 cell monolayers were grown in Dulbecco's modified Eagle's medium and Earle's minimum essential medium (Quality Biologicals), respectively, supplemented with 10% fetal bovine serum. All recombinant viruses were derived from the WR strain. Construction of vaccinia viruses vB5R-GFP, vB5R-GFP/ΔA36R, and vB5R-GFP/A36R-YdF and the plasmids pB5R-GFP, containing the B5R open reading frame (ORF) fused to green fluorescent protein (GFP) sequences and approximately 500 bp of flanking sequence on each side, and pBMW-32T, containing the A36R ORF and approximately 500 bp of flanking sequence on each side, have been previously described (42, 43).
The plasmid pBMW32-T and primers M13 forward (Promega) and CAGATCTTTAGTTTCCTTTTTATAAAATTGA were used to amplify approximately 500 bp of DNA downstream of A36R containing a 3′ BglII (underlined) site immediately after the A36R stop codon. Primers CAGATCTACGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACCAATATTATCTACGTCATTGTT, CAGATCTACGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACCAGCGAAGCTATCATCGTC, CAGATCTACGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACCAATGTGTTCTGTGCTAGG, and CAGATCTACGTAGAATCGAGACCGAGGAGAGGGTTAGGG ATAGGCTTACCATTTATTAGCAGCGTGCT were used in conjunction with the M13 reverse primer (Promega) to amplify approximately 500 bp of DNA upstream of and including codons 1 to 80, 1 to 93, 1 to 111, and 1 to 123 of A36R, respectively, and to add a 3′ V5 tag (italics) followed by a BglII (underlined) site. All amplified sequences were cloned into pGEM-T and sequenced. The left flank and the truncated coding sequence were excised from pGEM-T by digestion with SalI and BglII. The right flank was excised from pGEM-T by digestion of HindIII and BglII. Excised fragments were joined by a three-fragment ligation in the vector pBSgptgus (19) that had been cleaved with HindII and SalI. The truncated A36R coding sequences were inserted into the deleted A36R site of vB5R-GFP/ΔA36R by homologous recombination, and the recombinant virus was isolated by transient dominant selection, essentially as described previously (10). HeLa cells were infected at a multiplicity of infection of 0.05 with vB5R-GFP/ΔA36R and were transfected 2 h later with the appropriate pBSgptgus construct. After 2 days cells were harvested, frozen, and thawed three times and were used to infect BS-C-1 cells that were treated with mycophenolic acid (MPA), xanthine, and hypoxanthine at concentrations of 25, 250, and 15 μg per ml, respectively. Recombinant viruses that resulted from a single crossover would have the entire plasmid integrated into the viral genome and consequently would be resistant to MPA and positive for β-glucuronidase. MPA-resistant and β-glucuronidase-positive viruses were plaque purified twice in the presence of MPA before three more plaque purifications in the absence of MPA to obtain the desired double crossover mutant. X-Gluc staining was used to confirm the absence of the gus gene, and PCR amplification and sequencing were used to confirm the insertion of the mutated A36R ORF.
The primers CAAGCTTCGTTAACGACTTATTATTAATTCA and CGTCGACAGACGTGTTTAAATGCCTTCC were used to amplify the A33R ORF plus 500 bp of flanking sequence on each side and to add a HindIII site and SalI site (underlined). The amplified product was cloned into pGEM-T to yield pBMW-78T and was sequenced. Deletion of codons 5 to 40 of A33R was accomplished by using two-stage PCR with plasmid pBMW-78T as the template. The following primer pairs were used to amplify two overlapping fragments: GAAATAACCATTGGTGTCATCATGATAATAAA and M13 reverse, and TGACACCAATGGTTATTTCACTACTATCT and M13 forward. The resulting amplified products were purified and joined together by a third amplification, and the resulting product was cloned into pGEM-T and sequenced. Subsequently, the mutated A33R coding sequence plus 500 bp of flanking sequence was excised from pGEM-T by digestion with HindIII and SalI and was ligated into similarly cleaved pBSgptgus to yield pBMW79-BSgptgus. The mutated A33R coding sequence was inserted by recombination into the A33R site of vB5R-GFP, and the recombinant virus (vA33RΔ5-40) was isolated by transient dominant selection, as described above.
Virus plaque assay.
Plaque assays were carried out in BS-C-1 cells by using standard procedures. Images were obtained at two days after infection by using the Leica DMIRBE inverted fluorescent microscope with a cooled charged coupled device (Princeton Instruments) that was controlled with Image Pro software. After imaging, cells were stained with crystal violet, rinsed with water, and allowed to air dry. Stained plates were imaged with a Kodak Image Station 440cf with Kodak 1D Image Analysis software (Kodak Digital Science).
Electron microscopy.
For immunoelectron microscopy, RK13 cells were grown in 60-mm-diameter dishes and were infected with vaccinia virus at a multiplicity of infection of 10. After 24 h the cells were prepared for freezing as described previously (46), except that the final fixation step was in 8% paraformaldehyde. Ultrathin sections were cut, collected, immunostained, and viewed as described previously (5). The V5 monoclonal antibody (MAb) (Invitrogen) was used at a dilution of 1:1,000. Anti-A33R MAb, a kind gift of Alan Schmaljohn, was used at a dilution of 1:250. The anti-A36R serum (47) was used at a dilution of 1:400.
Western blots and coimmunoprecipitations.
Monolayers of HeLa cells were infected at a multiplicity of infection of 5. After 2 h the inoculum was removed and replaced with normal medium. The next day cells were harvested by scraping, collected by low-speed centrifugation, resuspended in half-strength phosphate-buffered saline containing 1% NP-40 and complete protease inhibitor tablets (PBS-N; Roche Molecular Biochemicals), and incubated on ice for 20 min. Lysates were clarified by centrifugation for 30 min at a relative centrifugal force of 20,000. For Western blots, clarified lysates were mixed with protein loading buffer, resolved by electrophoresis on a sodium dodecyl sulfate (SDS)-10 to 20% polyacrylamide gel (Invitrogen), and transferred to a nitrocellulose membrane. Membranes were incubated with either a polyclonal antibody to the A36R protein (47) followed by horseradish peroxidase-conjugated donkey anti-rabbit antibody (Amersham), horseradish peroxidase-conjugated anti-V5 MAb (Invitrogen), or anti-A33R MAb followed by horseradish peroxidase-conjugated sheep anti-mouse antibody (Amersham). Bound antibodies were detected with chemiluminescence reagents (Pierce) as directed by the manufacturer. For coimmunoprecipitation clarified lysates were pretreated with protein G-Sepharose for 2 h followed by low-speed centrifugation to collect bound proteins. Treated lysates were incubated with anti-A33R MAb, and antigen-antibody complexes were bound to protein G-Sepharose, washed three times in PBS-N, resuspended in protein loading buffer, boiled, resolved by SDS-polyacrylamide gel electrophoresis, and analyzed by Western blotting as described above.
Fluorescent microscopy.
HeLa cells were grown to confluence on coverslips and were infected at a multiplicity of infection of 1. For staining of permeabilized cells, infected cells were washed once in PBS and fixed with 4% paraformaldehyde and permeabilized with Triton X-100, both in PBS. Fixed cells were stained with either anti-A33R MAb or anti-A36R antiserum followed by either Texas Red-conjugated goat anti-mouse or Texas Red-conjugated goat anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories). Stained coverslips were mounted in mowoil containing 1 μg of 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Molecular Probes) per ml. For staining of unfixed cells, infected cells were washed once in fresh culture medium and were incubated for 1 h at 37°C in culture medium containing MAb 19C2 hybridoma culture supernatant diluted 1 to 100. Stained cells were washed three times in PBS, fixed with 4% paraformaldehyde, and quenched with 0.1 M glycine, both in PBS. Bound antibodies were detected with Alexa Fluor 568-conjugated goat anti-rat secondary antibody (Molecular Probes). Coverslips were mounted in PBS containing 0.1% azide and were sealed with rubber cement. Images were collected on a Leica TCS-NT/SP inverted confocal microscope system and were overlaid by using Adobe PhotoShop version 6.0.
Fluorescence microscopy of live cells.
Time-lapse confocal microscopy was carried out as described previously (43). Briefly, HeLa cells were plated at ∼80% confluence onto ΔTC3 dishes (Bioptechs, Inc.). On the next day, cells were infected with 0.2 PFU of virus per cell and were visualized on the following day by using a Bio-Rad MicroRadiance confocal scanning system attached to a Zeiss Axiovert 135 microscope. Throughout microscopy cells were maintained on a heated ΔTC3 stage (Bioptechs) with the temperature set at 35°C with fresh medium supplemented with 2.5% fetal calf serum and 25 mM HEPES perfused onto the dish at a rate of 0.1 ml/min. Maximum-intensity projections were created by using Imaris 3.0.2 (Bitplane AG) software. Virions were colorized by using Adobe PhotoShop version 6.0.
RESULTS
Cytoplasmic domains of the A36R and A33R proteins interact in the yeast two-hybrid system.
The A36R protein is a type Ib membrane protein with an approximately 200-amino-acid cytoplasmic domain that is anchored to the IEV membrane by the N-terminal 22 amino acids (32). We employed the yeast two-hybrid system to determine interactions between the cytoplasmic domain of the A36R protein with itself and four other IEV membrane proteins, namely, A33R, A34R, B5R, and F12L. DNA encoding amino acids 24 to 221 of A36R was fused to DNA of the GAL4 DNA binding domain. Likewise, DNA encoding the cytoplasmic domain of A33R, A34R, A36R, B5R, or F12L was fused to DNA of the GAL4 activation domain (Table 1). The A36R DNA binding domain plasmid was cotransfected into yeast with one of the activation domain plasmids, and a positive interaction was determined by scoring for growth on standard quadruple dropout medium lacking leucine, tryptophan, histidine, and adenine. As indicated in Table 1, an interaction was detected between the cytoplasmic tails of A36R and A33R but not between A36R and itself or the other three cytoplasmic tail constructs.
TABLE 1.
Interaction of the cytoplasmic domain of various IEV proteins with A36R in a yeast two-hybrid assay
| BDa-Fusion | ADb-Fusion | Growth on QDOc |
|---|---|---|
| A36R (24-221)d | A36R (24-221) | − |
| A36R (24-221) | A33R (1-40) | + |
| A36R (24-221) | A34R (1-20) | − |
| A36R (24-221) | B5R (300-317) | − |
| A36R (24-221) | F12L (1-635) | − |
BD, GAL4 DNA binding domain.
AD, GAL4 activation domain.
QDO, quadruple dropout medium.
Amino acids.
To determine the region of A36R responsible for the interaction with the 40-amino-acid cytoplasmic tail of A33R, we constructed four plasmids encoding truncated cytoplasmic domains of A36R fused to the GAL4 DNA binding domain. The truncated proteins carrying amino acids 24 to 123 and 24 to 111 interacted with the cytoplasmic tail of A33R, while the two carrying amino acids 24 to 93 and 24 to 80 did not interact, as determined by growth on quadruple dropout medium (Table 2). β-Galactosidase expression, another indicator of protein:protein interactions, was highest with the construct containing amino acids 24 to 111 (Table 2). Thus, the A33R interaction region was contained within the first 111 residues of A36R. Four more truncations of A36R were constructed in which the first 60, 70, 80, or 90 residues of the cytoplasmic domain were removed in conjunction with the deletion of the last 108 residues to give constructs expressing amino acids 61 to 111, 71 to 111, 81 to 111, or 91 to 111. Positive results were obtained with each of these truncated proteins (Table 2), indicating that residues 91 to 111 of A36R, comprising only 20 amino acids, were sufficient for interaction with the 40-amino-acid cytoplasmic domain of the A33R protein in the yeast two-hybrid system.
TABLE 2.
Interaction of ADa A33R (amino acids 1 to 40) with various BDb A36R constructs in a yeast two-hybrid assay
| BD-Fusion (amino acids) | Growth on QDOc | Relative β-galactosidase production |
|---|---|---|
| A36R (24-221) | + | + |
| A36R (24-123) | + | + |
| A36R (24-111) | + | ++ |
| A36R (24-93) | − | − |
| A36R (24-80) | − | − |
| A36R (61-111) | + | NDd |
| A36R (71-111) | + | ND |
| A36R (81-111) | + | ND |
| A36R (91-111) | + | + |
AD, GAL4 activation domain.
BD, GAL4 DNA binding domain.
QDO, quadruple dropout medium.
ND, Not determined.
Construction of recombinant vaccinia viruses with truncations of A36R or the cytoplasmic tail of A33R.
To confirm the data obtained with the yeast two-hybrid system and to delineate the role of the A33R:A36R interaction during infection, recombinant viruses were constructed. The starting virus for one set of recombinant viruses was an A36R deletion mutant (vB5R-GFP/ΔA36R) that expressed a functional B5R-GFP fusion protein (42). DNA encoding A36R amino acids 1 to 123, 1 to 111, 1 to 93, or 1 to 80, fused in each case to a V5 epitope tag, was inserted into the A36R deletion site by homologous recombination. Recombinant viruses were isolated by a transient dominant selection scheme (10) that left the truncated A36R ORF under the control of its normal promoter and did not add any selection or screening markers. These recombinant viruses, which all contained B5R fused to GFP and full-length A33R, were called vA36R1-123-V5, vA36R1-111-V5, vA36R1-93-V5, and vA36R1-80-V5. By using a similar method, recombinant virus vA33RΔ5-40 containing B5R-GFP, full-length A36R, and an A33R ORF with amino acids 5 to 40 deleted was constructed. The recombinant vB5R-GFP, which contained full-length A36R and A33R, served as a control virus.
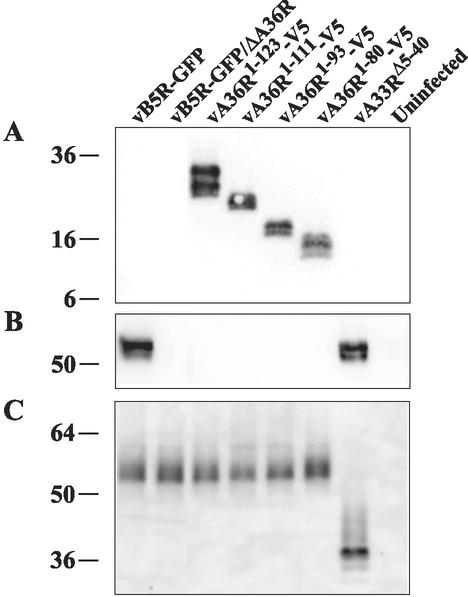
Expression of the truncated versions of the A36R proteins was demonstrated in the following manner. Lysates from uninfected or infected cells were subjected to SDS-polyacrylamide gel electrophoresis and then were blotted onto nitrocellulose and were probed with a MAb that specifically reacted with the V5 tag or antiserum to a peptide representing the C-terminal 13 residues of the A36R protein. The V5 MAb reacted with each truncated A36R protein, indicating that they were expressed and stable (Fig. 1A). The bands appeared as doublets, possibly due to heavily phosphorylated and unphosphorylated forms of the A36R proteins (47), with mobilities corresponding to the lengths of the ORFs. As expected, the A36R peptide antiserum failed to react with the truncated A36R proteins (Fig. 1B). The latter antiserum reacted with the full-length A36R protein expressed by the A33R cytoplasmic domain deletion mutant vA33RΔ5-40 as well as by vB5R-GFP, indicating that deletion of the cytoplasmic tail of the A33R protein did not noticeably affect the stability of the A36R protein (Fig. 1B).
FIG. 1.
Expression of truncated A36R and A33R proteins. HeLa cells mock infected or infected with the indicated recombinant viruses were incubated for 18 h, harvested, and analyzed by SDS-polyacrylamide gel electrophoresis under reducing (A and B) or nonreducing (C) conditions followed by Western blotting with anti-V5 MAb (A), anti-A36R antiserum (B), or anti-A33R MAb (C). The masses (in kilodaltons) and positions of migration of markers are indicated on the left.
Similar experiments were carried out to demonstrate expression of the truncated A33R protein. Because the A33R MAb (17) is conformation-specific and only reacts efficiently with disulfide-bonded dimers, the proteins were not reduced before SDS-polyacrylamide gel electrophoresis. The A33R MAb reacted with a diffuse band migrating at about 53 kDa from cells infected with vB5R-GFP expressing full-length A33R and an approximately 38-kDa band from cells infected with vA33RΔ5-40 (Fig. 1C). The slower than predicted mobility for both the full-length and truncated A33R protein dimers (41 and 34 kDa, respectively) relative to the standards is likely due to glycosylation and incomplete unfolding of the disulfide-bonded polypeptides. Immunoprecipitation of the truncated A33R protein with A33R MAb was also demonstrated (data not shown). Importantly, neither the size nor the intensity of the A33R bands was affected by the A36R truncations (Fig. 1C).
Coimmunoprecipitation of A33R and A36R.
Having determined that residues 91 to 111 of A36R were required for interaction with residues 1 to 40 of A33R in the yeast two-hybrid assay, we proceeded to confirm this interaction in cells infected with recombinant viruses by coimmunoprecipitation. Lysates, from uninfected cells or cells infected with one of the recombinant viruses, were incubated with the anti-A33R MAb followed by protein G-Sepharose. Bound proteins were separated by SDS-polyacrylamide gel electrophoreses, blotted onto nitrocellulose, and probed with the anti-V5 MAb. The V5-tagged A36R proteins of vA36R1-123-V5 and vA36R1-111-V5, containing amino acids 91 to 111, were coimmunoprecipitated with the full-length A33R protein (Fig. 2A). In contrast, barely detectable amounts of the A36R proteins lacking residues 91 to 111 coimmunoprecipitated (Fig. 2A). Because the A36R proteins of vA33RΔ5-40 and vB5R-GFP are not epitope tagged, a parallel blot was probed with the anti-A36R peptide antiserum following immunoprecipitation with anti-A33R MAb. Full-length A36R protein was not coimmunoprecipitated with the truncated A33R protein expressed by vA33RΔ5-40, though it was coimmunoprecipitated with the full-length A33R protein expressed by vB5R-GFP (Fig. 2B). Of course, the truncated A36R proteins were not detected because they lack the epitope of the anti-A36R peptide antiserum. Thus, the data from the yeast two-hybrid and coimmunoprecipitation experiments were in accord.
FIG. 2.
Coimmunoprecipitation. Lysates from HeLa cells that were either mock infected or infected with the indicated recombinant viruses were immunoprecipitated by anti-A33R MAb. Immune complexes were analyzed by SDS-polyacrylamide gel electrophoresis followed by Western blotting with either anti-V5 MAb (A) or anti-A36R antiserum (B). The masses (in kilodaltons) and positions of migration of markers are indicated on the left.
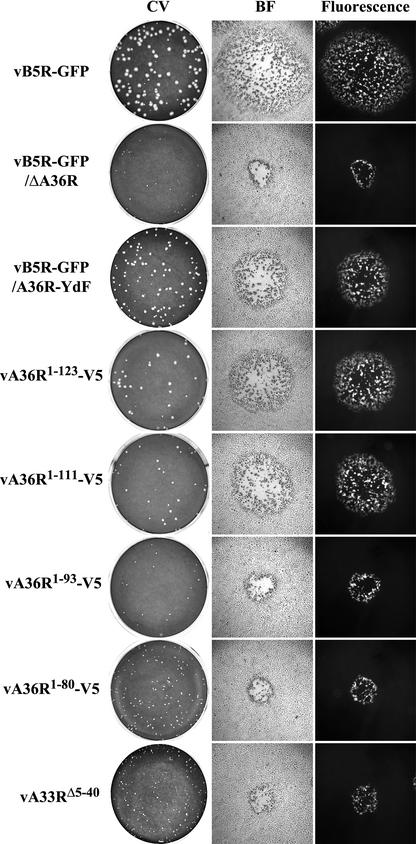
Plaque phenotypes of recombinant viruses.
Further experiments were designed to correlate the phenotypes of the recombinant viruses with the ability of their A33R and A36R proteins to interact. The plaques formed in BS-C-1 cells by the recombinant viruses containing truncations of the A36R or A33R ORF were compared to each other and to those formed by viruses containing unmutated A36R and A33R (vB5R-GFP), unmutated A33R and deleted A36R (vB5R-GFP/ΔA36R), and unmutated A33R and two conservative tyrosine-to-phenylalanine substitutions of A36R at residues 112 and 132 (vB5R-GFP/A36R-YdF). All of the recombinant viruses expressed B5R-GFP in place of the original B5R and were fluorescent (Fig. 3). As previously reported (42), vB5R-GFP and vB5R-GFP/ΔA36R formed large- and small-size plaques, respectively (Fig. 3), while vB5R-GFP/A36R-YdF formed intermediate-sized plaques due to its inability to form actin tails (Fig. 3). The plaques formed by both vA36R1-123-V5 and vA36R1-111-V5 were intermediate in size and similar to the plaques formed by vB5R-GFP/A36R-YdF (Fig. 3). It is noteworthy that vA36R1-123-V5 and vA36R1-111-V5 contain the A33R interaction site but are missing one (vA36R1-123-V5) or both (vA36R1-111-V5) tyrosines identified by Frischknecht et al. (11) as being important for actin nucleation and that both of these tyrosines were mutated to phenylalanine in vB5R-GFP/A36R-YdF, which also forms an intermediate-sized plaque. The plaques formed by vA36R1-93-V5 and vA36R1-80-V5, which are missing the defined A33R interaction site as well as the two tyrosines, were small in size and similar to the plaques formed by the A36R deletion mutant vB5R-GFP/ΔA36R (Fig. 3). Moreover, the plaques formed by vA33RΔ5-40, which lacks the A36R interaction site, were also small (Fig. 3). Because none of the viruses with mutated A36R or A33R ORFs were able to form actin tails (data not shown) and because the plaques formed by recombinant viruses lacking the A36R:A33R interaction site (vA36R1-93-V5, vA36R1-80-V5, and vA33RΔ5-40) are smaller than those formed by a recombinant virus with tyrosine point mutations (vB5R-GFP/A36R-YdF) that specifically abrogate actin tail formation, the A36R protein must have additional functions.
FIG. 3.
Plaque phenotypes. The indicated viruses were plated on monolayers of BS-C-1 cells. After 2 days, plaques were imaged by using light and fluorescence microscopy and then were stained with crystal violet (CV). BF, bright field microscopy.
Confocal microscopy of cells infected with mutant viruses expressing truncated A36R and A33R proteins.
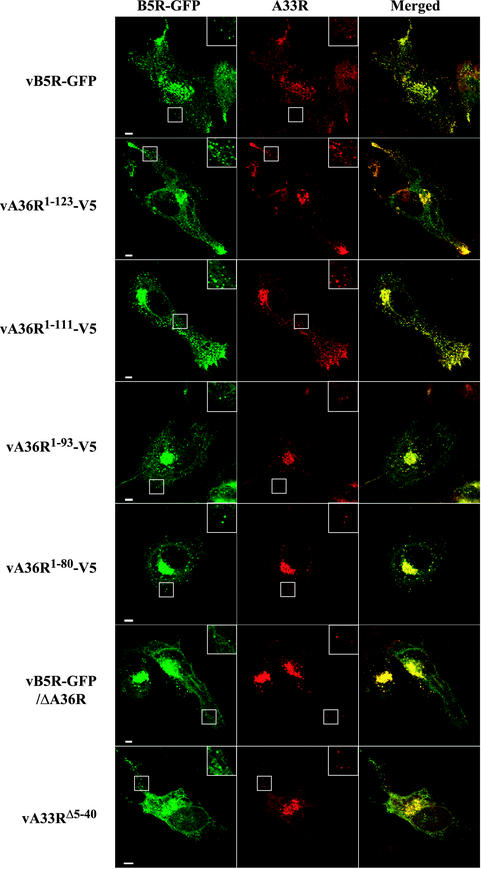
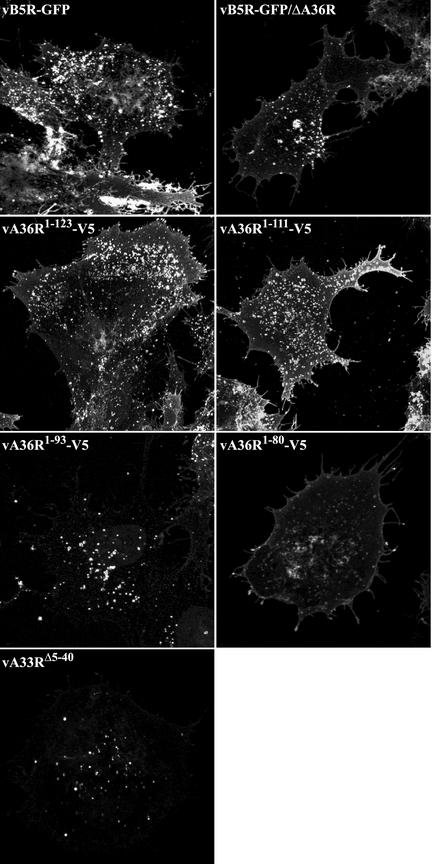
Previous confocal microscopy studies revealed a characteristic pattern of fluorescence in cells infected with vB5R-GFP (43). Individual IEV exhibited a discrete punctate fluorescence in the cytoplasm. After IEV were transported on microtubules to the cell periphery, they produced a very bright fluorescence in the vertices of the cells where they accumulated. Additional bright fluorescence occurred in the juxtanuclear Golgi region, representing either pre-IEV membrane structures or clusters of IEV that had not yet been transported through the cytoplasm.
All of the recombinant viruses in this study contained B5R-GFP, allowing us to compare their fluorescent patterns with those of vB5R-GFP. There was a similar distribution of green fluorescence in cells infected with vB5R-GFP, vA36R1-123-V5, or vA36R1-111-V5 (Fig. 4 and 5). Notably, there was punctate fluorescence in the cytoplasm, which colocalized with the DNA stain DAPI (data not shown), indicative of IEV as well as bright fluorescence in the vertices of cells, suggesting directed IEV movement to this location. In contrast, the bright fluorescence in the vertices was absent in cells infected with vB5R-GFP/ΔA36R, vA36R1-93-V5, vA36R1-80-V5, vB5R-GFP/ΔA36R, or vA33RΔ5-40, although there still was bright staining in the juxtanuclear region (Fig. 4 and 5).
FIG. 4.
Localization of the B5R and A33R proteins in infected cells by confocal microscopy. HeLa cells were infected with the indicated recombinant viruses and were stained with anti-A33R MAb followed by Texas Red-conjugated goat anti-mouse antibody (red). Green fluorescence represents B5R-GFP. Bar, 5 μm. Boxed regions are enlarged to show punctate fluorescence.
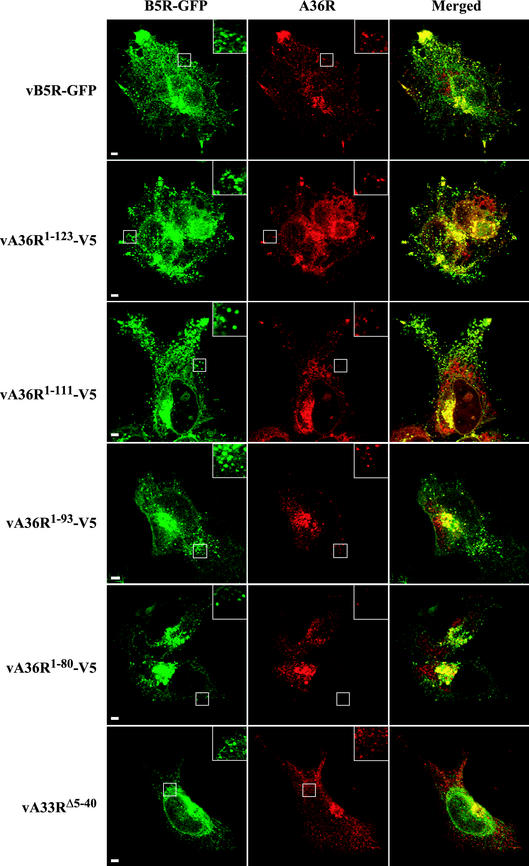
FIG. 5.
Localization of the B5R and A36R proteins in infected cells by confocal microscopy. HeLa cells were infected with the indicated recombinant viruses and were stained with either anti-A36R antiserum (vB5R-GFP, vA33RΔ5-40) or anti-V5 MAb (vA36R1-123-V5, vA36R1-111-V5, vA36R1-93-V5, vA36R1-80-V5) followed by either Texas Red-conjugated goat anti-rabbit or Texas Red-conjugated goat anti-mouse antibody, respectively (red). Green fluorescence represents B5R-GFP. Bar, 5 μm. Boxed regions are enlarged to show punctate fluorescence.
The distribution of the A33R and A36R proteins was also visualized. By using the A33R MAb, extensive colocalization of A33R and B5R-GFP occurred. Colocalized punctate fluorescence was discerned for all of the recombinants, as shown in the inset boxes and color merges in Fig. 4. These punctate structures also colocalized with the DNA stain DAPI, indicating that they were IEV (data not shown). This colocalization occurred even in cells infected with vA36R1-93-V5, vA36R1-80-V5, vB5R-GFP/ΔA36R, or vA33RΔ5-40, indicating that A36R:A33R interaction sites were not required for localization of the A33R protein on IEV.
The effects of the various truncations on the subcellular localization of the A36R protein was determined with anti-A36R peptide antiserum (for vB5R-GFP and vA33RΔ5-40) or anti-V5 MAb (for vA36R1-123-V5, vA36R1-111-V5, and vA36R1-93-V5 vA36R1-80-V5) followed by an appropriate Texas Red secondary antibody (Fig. 5). Complete colocalization with B5R-GFP was not expected, since the A36R protein is not a component of CEV. Nevertheless, there was extensive overlap of the A36R and B5R-GFP fluorescence in cells infected with vA36R1-123-V5 and vA36R1-111-V5, and the distribution of the signal appeared similar to that of cells infected with vB5R-GFP (Fig. 5, rows 1 to 3). Discrete punctate fluorescence, as well as bright fluorescence in the vertices and juxtanuclear regions, was discerned. In cells infected with vA36R1-93-V5 or vA36R1-80-V5, there was considerable B5R-GFP and A36R colocalization in the juxtanuclear region (Fig. 5, rows 4 and 5). However, the vertices did not exhibit the bright staining that was seen in cells infected with vB5R-GFP, vA36R1-123-V5, and vA36R1-111-V5. The altered pattern of fluorescence in cells infected with vA33RΔ5-40 was even more pronounced than that for vA36R1-93-V5 and vA36R1-80-V5. Although there was still bright A36R fluorescence in the juxtanuclear region, it was difficult to detect colocalizing punctate fluorescence in the cytoplasm and there was no accumulation of fluorescence in the vertices.
The absence of fluorescent signal in the vertices of cells infected with recombinant viruses that are missing the defined A33R-A36R interaction sights could indicate a reduced ability to produce enveloped virus on the surface of cells. To determine if the recombinant viruses produce extracellular virus, we stained infected unpermeabilized cells with a MAb to the luminal domain of the B5R envelope protein. Stained cells were then imaged as Z-series, and a representative infected cell from each recombinant is shown in Fig. 6 as a maximum-intensity projection. The absence of juxtanuclear staining indicates that only extracellular staining occurred. While all of the recombinant viruses produced punctate B5R signal on the cell surface, indicating that they were capable of producing extracellular virus, the level of surface staining was reduced in cells infected with vA33RΔ5-40, vA36R1-93-V5, and vA36R1-80-V5 as determined by the mean pixel fluorescence of each image (Fig. 6).
FIG. 6.
Surface staining of B5R on infected cells. HeLa cells were infected with the indicated recombinant viruses. Unpermeabilized cells were stained with MAb 19C2 to the B5R membrane protein followed by Alexa Fluor 568-conjugated goat anti-rat antibody. Cells were imaged by confocal microscopy as a series of optical sections and are displayed here as a maximum-intensity projection. The mean pixel fluorescence for each projection was calculated for vB5R-GFP, vB5R-GFP/ΔA36R, vA36R1-123-V5, vA36R1-111-V5, vA36R1-93-V5, vA36R1-80-V5, and vA33RΔ5-40 to be 42.8, 15.9, 39.8, 36.1, 11.4, 18.7, and 6.6, respectively.
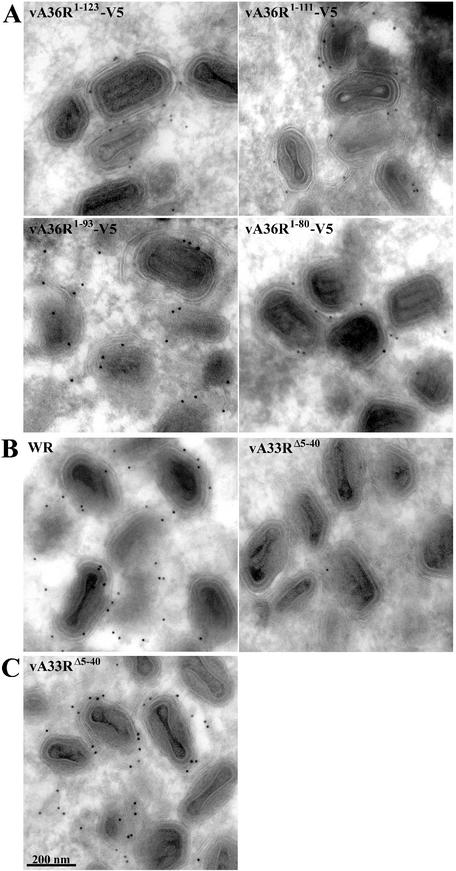
Electron microscopic localization of truncated proteins in infected cells.
To localize the truncated proteins at high resolution, immunoelectron microscopy with the V5 MAb followed by protein A-gold was performed. In cells infected with vA36R1-123-V5, vA36R1-111-V5, vA36R1-93-V5, or vA36R1-80-V5, gold grains were associated with IEV and adjacent membranes (Fig. 7A). The number of gold grains per 100 IEV was 0, 213, 202, 152, and 126 for cells infected by vB5R-GFP, vA36R1-123-V5, vA36R1-111-V5, vA36R1-93-V5, and vA36R1-80-V5, respectively. The difference between the group comprised of vA36R1-123-V5 and vA36R1-111-V5 and the group comprised of vA36R1-93-V5 and vA36R1-80-V5 was statistically significant (P = 0.035). Because vA33RΔ5-40 does not have a V5 epitope tag on A36R, we used the anti-A36R serum for immunostaining. Although abundant gold grains were found on IEV in cells infected with wild-type virus, only rare grains were associated with IEV in cells infected with vA33RΔ5-40 (Fig. 7B). Nevertheless, MAb A33R strongly and specifically stained IEV in cells infected with vA33RΔ5-40 (Fig. 7C), indicating that the cytoplasmic tail of A33R is not required for its incorporation into the viral envelope but is required for incorporation of A36R. The association of the truncated A33R with wrapped virions purified by CsCl gradient centrifugation provided further proof of its incorporation into enveloped virions (data not shown). Therefore, both the confocal and electron microscopy studies indicated that deletion of the A33R cytoplasmic tail has a more severe effect on the localization of A36R to IEV than does deletion of the defined A33R interaction region of A36R.
FIG. 7.
Immunoelectron microscopy of infected cells. Infected cells were cryosectioned and labeled with anti-V5 MAb (A), anti-A36R serum (B), or anti-A33R MAb (C) followed by protein A-gold (10 nm).
Intracellular movement of enveloped virions.
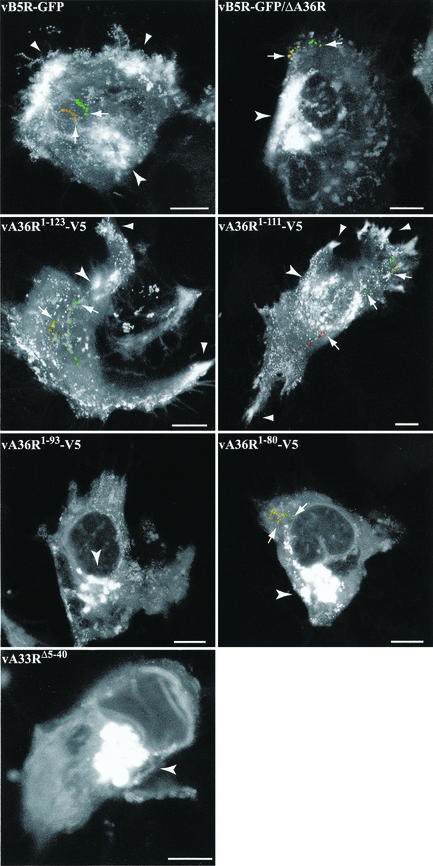
We previously reported difficulty in detecting typical IEV movement in cells infected with vB5R-GFP/ΔA36R (42) and anticipated a similar result with vA36R1-93-V5, vA36R1-80-V5, or vA33RΔ5-40 because of the absence of IEV clusters in the periphery of the cell. The intracellular movement of enveloped virions in living cells infected with the recombinant viruses was visualized by using time-lapse confocal microscopy. Images were collected at a rate of 1 image per 6 s for 10 min, and the full sequences are available at http://www.niaid.nih.gov/dir/labs/lvd/movies.htm. Individual time-lapse series were further analyzed by creating maximum-intensity projections comprising images of all time points (Fig. 8). In cells infected with vB5R-GFP, vA36R1-123-V5, and vA36R1-111-V5, movement of IEV was visualized as a trail moving from the juxtanuclear region to the cell periphery, where they collected (Fig. 8). In cells infected with vB5R-GFP/ΔA36R, vA36R1-93-V5, vA36R1-80-V5, and vA33RΔ5-40, however, the majority of the fluorescence remained in the juxtanuclear region with few particles elsewhere in the cytoplasm or in the periphery (Fig. 8). These few particles displayed abnormal, short sporadic movement that was difficult to trace over long distances in the cells.
FIG. 8.
Maximum-intensity projections of time-lapse microscopy. HeLa cells were infected with 0.2 PFU of the indicated recombinant virus per cell and were incubated overnight. The next day, time-lapse series of images were collected at 1 frame per 6 s for 10 min. Series of images were analyzed by maximum-intensity projections and are shown as a single image. As examples, the same virions from the original series of images are colored in the projection to facilitate the interpretation. Arrows point to the first colored virion from the original series. Arrowheads denote the bright areas of IEV accumulation at the vertices of the cells. Concave arrowheads denote the juxtanuclear region. Bar, 10 μm.
DISCUSSION
A previous vaccinia virus genome-wide yeast two-hybrid analysis did not detect interactions between IEV proteins, probably because full-length ORFs with hydrophobic transmembrane domains were used (20). By using the 200-amino-acid cytoplasmic domain of the A36R protein as bait in the yeast two-hybrid system, we screened the cytoplasmic domains of other IEV proteins. A strong interaction with A36R was found only with the 40-amino-acid cytoplasmic domain of the A33R protein. We were able to detect weak interactions between the cytoplasmic tails of A33R-A34R and A33R-A33R that permitted growth on triple dropout media but were not strong enough to support growth on quadruple dropout media (B. M. Ward, unpublished data). Neither of these weaker interactions was confirmed by other methods, however, so their relevance cannot be assessed.
In further studies with the two-hybrid system the interaction site of the A36R protein with the cytoplasmic domain of the A33R protein was mapped to amino acids 91 to 111. The yeast two-hybrid result was confirmed by preparing recombinant vaccinia viruses expressing truncated A36R or A33R proteins. No coimmunoprecipitation of the A36R protein with the A33R protein lacking the cytoplasmic tail could be detected. Good coimmunoprecipitation of A36R with A33R occurred when the cytoplasmic domain of A36R included residues 91 to 111. Very little coimmunoprecipitation was detected when the A36R protein had been truncated further. That weak interaction, however, raised the possibility of either a low-affinity site within the first 80 amino acids of the cytoplasmic domain that was not detected in the yeast two-hybrid system or a site in the transmembrane region which was not present in the yeast two-hybrid construct. Interaction between transmembrane segments is unlikely, however, as no interaction was detected when the cytoplasmic domain of the A33R protein was deleted, although the transmembrane domain was retained. The dimeric structure of the A33R protein, expressed in infected cells, may assist low-affinity interactions. There might also be indirect interaction between the cytoplasmic domains of the A36R and A33R proteins mediated by a third IEV protein. Notably, interactions between A34R and A36R and A34R and B5R have been suggested previously (32).
An earlier study had demonstrated that the A36R protein failed to associate with IEV when the A33R protein was not expressed (47). We determined in the present study that this association is dependent on the cytoplasmic tail of the A33R protein; when this 40-amino-acid segment was deleted, A36R could not be detected on IEV even though the truncated A33R protein was present on the IEV. The association of A36R with IEV was less strongly affected by deletion of the defined A33R interaction site, as the truncated A36R proteins comprised of amino acids 1 to 93 or 1 to 80 could still be detected on IEV, consistent with the weak interaction of these mutated A36R proteins with A33R detected by coimmunoprecipitation. Deletion of the A33R cytoplasmic domain or residues 90 to 111 of A36R caused a reduction in the amount of intracellular movement of IEV on microtubules from the juxtanuclear region to the cell periphery. The reduction of IEV movement correlated with changes in the distribution of B5R-GFP in cells infected with mutants exhibiting diminished A33R:A36R interactions, as they did not contain the intense fluorescence at the vertices and also had reduced levels of surface B5R. It was not surprising that formation of CEV was only partially inhibited by these mutations, as the A36R protein is not directly involved in exocytosis. Presumably, IEV that are formed near the plasma membrane or arrive there by mechanisms not involving rapid long-range movement on microtubules are externalized. Because it is difficult to enumerate IEV in the juxtanuclear region of the cell, it is difficult to rule out diminished IEV formation due to the disruption of the A33R:A36R interaction. We had previously noted a similar effect when the A36R gene was deleted (42).
By using a rescue protocol involving transfection of plasmids expressing mutated A36R-GFP ORFs into cells infected with an A36R inducible virus, Rietdorf et al. (27) recently reported that residues 71 to 100 of A36R were required for movement of IEV to the cell periphery. These investigators had found that the microtubule motor protein kinesin was associated with IEV and speculated that this region of A36R was directly involved in binding. Citing unpublished results, however, Rietdorf et al. (27) stated that they were unable to detect a direct interaction between the region of amino acids 71 to 100 of A36R and kinesin by using proteins produced in E. coli. The region of amino acids 71 to 100, defined by Rietdorf and coworkers (27), overlaps with the 90-to-111-amino-acid A33R interaction site that we found. Both sequences fall within a highly conserved segment of the A36R ORF; indeed, amino acids 89 to 101 are identical in all twelve orthopoxviruses examined (26). Moreover, the overlapping region, defined by residues 105 to 116, is responsible for binding the cellular Nck protein necessary for nucleation of actin tails. Thus, the region from residues 90 to 116 of A36R binds to both viral and cellular proteins. Perhaps overlapping binding sites regulate virion motility and actin tail nucleation so as to prevent them from occurring prematurely.
Another conclusion of our study was that the region of A36R comprising amino acids 112 to 222 was relatively unimportant. In the yeast two-hybrid system, fusion proteins containing residues 24 to 111 interacted with the cytoplasmic domain of the A33R protein more strongly than the fusion protein containing full-length A36R. Furthermore, the plaque sizes of recombinant viruses expressing A36R proteins of 1 to 123 or 1 to 111 residues were similar in size to those with full-length A36R containing two tyrosine point mutations. In addition, transfection studies had shown that residues 1 to 117 were sufficient to rescue actin tail formation, albeit at reduced levels (11). In a recent analysis of twelve A36R orthopoxvirus sequences, Pulford et al. (26) noted more variation in the C-terminal half than the N-terminal half of the ORF. More refined experiments will be needed to determine the role, if any, of this region of the A36R protein.
Acknowledgments
We thank members of the Laboratory of Viral Diseases for their interest and suggestions.
REFERENCES
- 1.Appleyard, G., A. J. Hapel, and E. A. Boulter. 1971. An antigenic difference between intracellular and extracellular rabbitpox virus. J. Gen. Virol. 13:9-17. [DOI] [PubMed] [Google Scholar]
- 2.Blasco, R., and B. Moss. 1991. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-Dalton outer envelope protein. J. Virol. 65:5910-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blasco, R., and B. Moss. 1992. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J. Virol. 66:4170-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cudmore, S., P. Cossart, G. Griffiths, and M. Way. 1995. Actin-based motility of vaccinia virus. Nature 378:636-638. [DOI] [PubMed] [Google Scholar]
- 5.Da Fonseca, F. G., A. Weisberg, E. J. Wolffe, and B. Moss. 2000. Characterization of the vaccinia virus H3L envelope protein: topology and post-translational membrane insertion via the C-terminal hydrophobic tail. J. Virol. 74:7508-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dales, S., and L. Siminovitch. 1961. The development of vaccinia virus in Earle's L strain cells as examined by electron microscopy. J. Biophys. Biochem. Cytol. 10:475-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duncan, S. A., and G. L. Smith. 1992. Identification and characterization of an extracellular envelope glycoprotein affecting vaccinia virus egress. J. Virol. 66:1610-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelstad, M., S. T. Howard, and G. L. Smith. 1992. A constitutively expressed vaccinia gene encodes a 42-kDa glycoprotein related to complement control factors that form part of the extracellular virus envelope. Virology 188:801-810. [DOI] [PubMed] [Google Scholar]
- 9.Engelstad, M., and G. L. Smith. 1993. The vaccinia virus 42-kDa envelope protein is required for the envelopment and egress of extracellular virus and for virus virulence. Virology 194:627-637. [DOI] [PubMed] [Google Scholar]
- 10.Falkner, F. G., and B. Moss. 1990. Transient dominant selection of recombinant vaccinia viruses. J. Virol. 64:3108-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frischknecht, F., V. Moreau, S. Rottger, S. Gonfloni, I. Reckmann, G. Superti-Furga, and M. Way. 1999. Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature 401:926-929. [DOI] [PubMed] [Google Scholar]
- 12.Geada, M. M., I. Galindo, M. M. Lorenzo, B. Perdiguero, and R. Blasco. 2001. Movements of vaccinia virus intracellular enveloped virions with GFP tagged to the F13L envelope protein. J. Gen. Virol. 82:2747-2760. [DOI] [PubMed] [Google Scholar]
- 13.Grimley, P. M., E. N. Rosenblum, S. J. Mims, and B. Moss. 1970. Interruption by rifampin of an early stage in vaccinia virus morphogenesis: accumulation of membranes which are precursors of virus envelopes. J. Virol. 6:519-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiller, G., K. Weber, L. Schneider, C. Parajsz, and C. Jungwirth. 1979. Interaction of assembled progeny pox viruses with the cellular cytoskeleton. Virology 98:142-153. [DOI] [PubMed] [Google Scholar]
- 15.Hirt, P., G. Hiller, and R. Wittek. 1986. Localization and fine structure of a vaccinia virus gene encoding an envelope antigen. J. Virol. 58:757-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollinshead, M., G. Rodger, H. Van Eijl, M. Law, R. Hollinshead, D. J. Vaux, and G. L. Smith. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 154:389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hooper, J. W., D. M. Custer, C. S. Schmaljohn, and A. L. Schmaljohn. 2000. DNA vaccination with vaccinia virus L1R and A33R genes protects mice against a lethal poxvirus challenge. Virology 266:329-339. [DOI] [PubMed] [Google Scholar]
- 18.Isaacs, S. N., E. J. Wolffe, L. G. Payne, and B. Moss. 1992. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J. Virol. 66:7217-7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishii, K., and B. Moss. 2001. Role of vaccinia virus A20R protein in DNA replication: construction and characterization of temperature-sensitive mutants. J. Virol. 75:1656-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCraith, S., T. Holtzman, B. Moss, and S. Fields. 2000. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc. Natl. Acad. Sci. USA 97:4879-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McIntosh, A. A., and G. L. Smith. 1996. Vaccinia virus glycoprotein A34R is required for infectivity of extracellular enveloped virus. J. Virol. 70:272-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreau, V., F. Frischknecht, I. Reckmann, R. Vincentelli, G. Rabut, D. Stewart, and M. Way. 2000. A complex of N-WASP and WIP integrates signalling cascades that lead to actin polymerization. Nat. Cell Biol. 2:441-448. [DOI] [PubMed] [Google Scholar]
- 23.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2883. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 24.Parkinson, J. E., and G. L. Smith. 1994. Vaccinia virus gene A36R encodes a Mr 43-50 K protein on the surface of extracellular enveloped virus. Virology 204:376-390. [DOI] [PubMed] [Google Scholar]
- 25.Payne, L. G. 1980. Significance of extracellular virus in the in vitro and in vivo dissemination of vaccinia virus. J. Gen. Virol. 50:89-100. [DOI] [PubMed] [Google Scholar]
- 26.Pulford, D. J., H. Meyer, and D. Ulaeto. 2002. Orthologs of the vaccinia A13L and A36R virion membrane protein genes display diversity in species of the genus Orthopoxvirus. Arch. Virol. 147:995-1015. [DOI] [PubMed] [Google Scholar]
- 27.Rietdorf, J., A. Ploubidou, I. Reckmann, A. Holmström, F. Frischknecht, M. Zettl, T. Zimmerman, and M. Way. 2001. Kinesin dependent movement on microtubules precedes actin based motility of vaccinia virus. Nat. Cell Biol. 3:992-1000. [DOI] [PubMed] [Google Scholar]
- 28.Risco, C., J. R. Rodriguez, C. Lopez-Iglesias, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 2002. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J. Virol. 76:1839-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodger, G., and G. L. Smith. 2002. Replacing the SCR domains of vaccinia virus protein B5R with EGFP causes a reduction in plaque size and actin tail formation but enveloped virions are still transported to the cell surface. J. Gen. Virol. 83:323-332. [DOI] [PubMed] [Google Scholar]
- 30.Roper, R., E. J. Wolffe, A. Weisberg, and B. Moss. 1998. The envelope protein encoded by the A33R gene is required for formation of actin-containing microvilli and efficient cell-to-cell spread of vaccinia virus. J. Virol. 72:4192-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roper, R. L., L. G. Payne, and B. Moss. 1996. Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene. J. Virol. 70:3753-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rottger, S., F. Frischknecht, I. Reckmann, G. L. Smith, and M. Way. 1999. Interactions between vaccinia virus IEV membrane proteins and their roles in IEV assembly and actin tail formation. J. Virol. 73:2863-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanderson, C. M., F. Frischknecht, M. Way, M. Hollinshead, and G. L. Smith. 1998. Roles of vaccinia virus EEV-specific proteins in intracellular actin tail formation and low pH-induced cell-cell fusion. J. Gen. Virol. 79:1415-1425. [DOI] [PubMed] [Google Scholar]
- 34.Scaplehorn, N., A. Holmstrom, V. Moreau, F. Frischknecht, I. Reckmann, and M. Way. 2002. Grb2 and Nck act cooperatively to promote actin-based motility of vaccinia virus. Curr. Biol. 12:740-745. [DOI] [PubMed] [Google Scholar]
- 35.Schmelz, M., B. Sodeik, M. Ericsson, E. J. Wolffe, H. Shida, G. Hiller, and G. Griffiths. 1994. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J. Virol. 68:130-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shida, H. 1986. Nucleotide sequence of the vaccinia virus hemagglutinin gene. Virology 150:451-462. [DOI] [PubMed] [Google Scholar]
- 37.Sodeik, B., and J. Krijnse-Locker. 2002. Assembly of vaccinia virus revisited: de novo membrane synthesis or acquisition from the host? Trends Microbiol. 10:15-24. [DOI] [PubMed] [Google Scholar]
- 38.Stokes, G. V. 1976. High-voltage electron microscope study of the release of vaccinia virus from whole cells. J. Virol. 18:636-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tooze, J., M. Hollinshead, B. Reis, K. Radsak, and H. Kern. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60:163-178. [PubMed] [Google Scholar]
- 40.van Eijl, H., M. Hollinshead, G. Rodger, W. H. Zhang, and G. L. Smith. 2002. The vaccinia virus F12L protein is associated with intracellular enveloped virus particles and is required for their egress to the cell surface. J. Gen. Virol. 83:195-207. [DOI] [PubMed] [Google Scholar]
- 41.van Eijl, H., M. Hollinshead, and G. L. Smith. 2000. The vaccinia virus A36R protein is a type Ib membrane protein present on intracellular but not extracellular enveloped virus particles. Virology 271:26-36. [DOI] [PubMed] [Google Scholar]
- 42.Ward, B. M., and B. Moss. 2001. Vaccinia virus intracellular movement is associated with microtubules and is independent of actin tails. J. Virol. 75:11651-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ward, B. M., and B. Moss. 2001. Visualization of intracellular movement of vaccinia virus virions containing a green fluorescent protein-B5R membrane protein chimera. J. Virol. 75:4802-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolffe, E. J., S. N. Isaacs, and B. Moss. 1993. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J. Virol. 67:4732-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolffe, E. J., E. Katz, A. Weisberg, and B. Moss. 1997. The A34R glycoprotein gene is required for induction of specialized actin-containing microvilli and efficient cell-to-cell transmission of vaccinia virus. J. Virol. 71:3904-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolffe, E. J., D. M. Moore, P. J. Peters, and B. Moss. 1996. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J. Virol. 70:2797-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolffe, E. J., A. Weisberg, and B. Moss. 2001. The vaccinia virus A33R protein provides a chaperone function for viral membrane localization and tyrosine phosphorylation of the A36R protein. J. Virol. 75:303-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolffe, E. J., A. S. Weisberg, and B. Moss. 1998. Role for the vaccinia virus A36R outer envelope protein in the formation of virus-tipped actin-containing microvilli and cell-to-cell virus spread. Virology 244:20-26. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, W. H., D. Wilcock, and G. L. Smith. 2000. Vaccinia virus F12L protein is required for actin tail formation, normal plaque size, and virulence. J. Virol. 74:11654-11662. [DOI] [PMC free article] [PubMed] [Google Scholar]