Abstract
Human immunodeficiency virus type 1 (HIV-1) subtype C viruses with different coreceptor usage profiles were isolated from 29 South African patients with advanced AIDS. All 24 R5 isolates were inhibited by the CCR5-specific agents, PRO 140 and RANTES, while the two X4 viruses and the three R5X4 viruses were sensitive to the CXCR4-specific inhibitor, AMD3100. The five X4 or R5X4 viruses were all able to replicate in peripheral blood mononuclear cells that did not express CCR5. When tested using coreceptor-transfected cell lines, one R5 virus was also able to use CXCR6, and another R5X4 virus could use CCR3, BOB/GPR15, and CXCR6. The R5X4 and X4 viruses contained more-diverse V3 loop sequences, with a higher overall positive charge, than the R5 viruses. Hence, some HIV-1 subtype C viruses are able to use CCR5, CXCR4, or both CXCR4 and CCR5 for entry, and they are sensitive to specific inhibitors of entry via these coreceptors. These observations are relevant to understanding the rapid spread of HIV-1 subtype C in the developing world and to the design of intervention and treatment strategies.
The envelope glycoproteins of human immunodeficiency virus type 1 (HIV-1) mediate viral entry into cells. This process requires the sequential interaction of gp120 with two host surface proteins, namely CD4 and a chemokine receptor, most often CCR5 or CXCR4 (15). Transmission of HIV-1 is almost always associated with viruses that utilize CCR5 (R5 viruses), which predominate during the acute and asymptomatic phases of infection (31, 40). Disease progression is often associated with the emergence of viruses that have acquired the ability to use CXCR4 (X4 viruses) instead of, or as well as, CCR5 (R5X4 viruses) (10, 42). These patterns of coreceptor usage correspond to the phenotypes previously defined by the MT-2 assay; here, syncytium-inducing (SI) viruses use CXCR4, while non-syncytium-inducing (NSI) viruses use CCR5 (3). Some isolates from AIDS patients can also use other chemokine receptors as coreceptors in receptor-transfected cell lines, including CCR1, CCR2b, CCR3, BOB (GPR15), and CXCR6 (Bonzo/STRL33) (14, 18, 39, 45, 61, 63; G. Alkhatib, F. Liao, E. A. Berger, J. M. Farber, and K. W. Peden, Letter, Nature 388:238, 1997). Rarely, however, are such alternative coreceptors used in primary cells (45, 64).
Agents that target CCR5 or CXCR4 can block HIV-1 entry and prevent infection in vitro. These include RANTES, a natural chemokine ligand for CCR5; PRO 140, a mouse monoclonal antibody directed at CCR5; TAK-779, a small molecule that binds to a pocket within CCR5 transmembrane helices 1, 2, 3, and 7; and SCH-C, another small-molecule, CCR5 antagonist (2, 17, 36, 47, 53, 60; J. Reynes, Abstr. 2nd Collaborative Res. Semin. HIV Other Viral Entry Inhibitors, abstr., 2002). Among these, SCH-C is currently in clinical trials and has been found to have antiviral activity (J. Reynes, R. Rouzier, T. Kanouni, V. Baillat, B. Baroudy, A. Keung, C. Hogan, M. Markowitz, and M. Laughlin, Abstr. 9th Conf. Retrovir. Opportun. Infect., abstr. 1, 2002.). AMD3100 is a small-molecule CXCR4 antagonist with activity against X4 viruses in vitro and antiviral activity in vivo, although it is no longer being pursued clinically because of pharmacology and toxicology considerations (16, 25, 43; G. Bridger, Abstr. 2nd Collaborative Res. Semin. HIV Other Viral Entry Inhibitors, abstr., 2002; C. Hendrix, A. C. Collier, M. Lederman, R. Pollard, S. Brown, M. Glesby, C. Flexner, G. Bridger, K. Badel, R. MacFarland, G. Henson, G. Calandra, et al., Abstr. 9th Conf. Retrovir. Opportun. Infect., abstr. 391-T, 2002.). Collectively these molecules are members of a new class of antiretroviral drugs termed entry inhibitors, some of which are being tested for their abilities to prevent or treat HIV-1 infection (29, 33, 44).
HIV-1 subtype C now accounts for more than half of new infections worldwide and is now the most prevalent subtype (www.unaids.org). It is associated with the rapidly expanding epidemics of southern Africa, India, and China. Previous studies have reported that subtype C isolates almost exclusively use the CCR5 coreceptor, with CXCR4 usage being only very rarely observed. This includes studies of isolates from India, Ethiopia, Malawi, and South Africa (1, 4, 7, 34, 38). HIV-1 cellular tropism and coreceptor specificity are largely determined by the sequence of the third hypervariable loop (V3) of the viral gp120 glycoprotein, and distinct changes in this region have been associated with the NSI/SI phenotype among subtype B viruses (8, 12, 28, 46). For example, the presence of a neutral amino acid (S) at position 11 and a negatively charged amino acid (D or E) at position 25 correlates with the NSI phenotype. Conversely, a basic residue at one of these positions, which increases the overall positive charge of the V3 loop, can change the viral phenotype from NSI to SI (11, 26, 32). Compared to other subtypes, the V3 regions of the subtype C viruses studied to date tend to be highly conserved and have a low overall positive charge, which is consistent with the NSI phenotypes and restricted coreceptor usage of subtype C viruses (23).
The above observations have created the impression that subtype C viruses might have envelope glycoproteins that are atypical in structure or function compared to viruses from other HIV-1 subtypes. If this were truly the case, intervention strategies, including the design of vaccines based on neutralizing-antibody induction, as well as therapeutic approaches based on inhibiting viral entry, might need to be designed differently for use in areas where subtype C viruses predominate. Moreover, an atypical property of the subtype C envelope glycoproteins might, in principle, help to explain why subtype C viruses are now spreading so rapidly in southern Africa and elsewhere, as has been proposed (5, 20, 35). However, an important caveat to the early studies on the coreceptor usage of subtype C strains is that they focused predominantly on isolates from patients who were in the relatively early stages of disease or whose clinical status was poorly defined (1, 4, 7, 34, 38, 51). Yet in studies on North American or European cohorts, the acquisition of CXCR4 use by subtype B viruses increases with time after infection and as the disease becomes more severe. This raises the possibility that the predominant usage of CCR5 by subtype C isolates was due more to a sampling artifact than to any fundamental, biological property of these viruses. We have therefore isolated viruses from patients with subtype C infection and advanced AIDS to see whether SI viruses are identified more frequently under these conditions than hitherto. We have also explored whether subtype C isolates able to use either or both of CCR5 and CXCR4 are sensitive to coreceptor-specific entry inhibitors.
Isolation of NSI and SI HIV-1 subtype C viruses.
Patients admitted to the Sizwe Infectious Diseases Hospital in Johannesburg, South Africa, between August and December 1999 with HIV-1-related opportunistic infections and CD4-T-cell counts of less than 100 cells/μl were selected for this study. Ethical clearance was obtained from the University of the Witwatersrand Committee for Research on Human Subjects (Medical), and informed consent was obtained from all patients prior to drawing 5 ml of blood in EDTA. The study cohort comprised 13 men and 16 women with a median age of 34 years and a median CD4 count of 40 cells/μl (Table 1). Levels of virus in plasma were measured using the Versant HIV-1 RNA 3.0 assay (bDNA assay from Bayer Nucleic Acid Diagnostics), previously shown to amplify HIV-1 RNA from all HIV-1 group M genetic subtypes (19, 50). The median viral load in plasma for this cohort was high, at 321,070 HIV-1 RNA copies/ml, consistent with advanced HIV-1 disease (Table 1). Twenty-two patients had tuberculosis (TB), four had cryptococcal meningitis (CM), two had Pneumocystis carinii pneumonia (PCP), and one had mycobacteria other than tuberculosis. HIV-1 was isolated from 22 patients when their peripheral blood mononuclear cells (PBMC) were cocultured with phytohemagglutinin (PHA)-activated donor PBMC as previously described (34). Isolates from the remaining seven patients were obtained by culturing 100 μl of plasma overnight with PHA-activated PBMC. Culture supernatants were tested weekly using a commercial p24 antigen kit (Beckman Coulter Ltd., Buckinghamshire, United Kingdom) and expanded when high levels of p24 antigen were present. Cultures were replenished with fresh interleukin 2 (IL-2) medium every 3 to 4 days and with PHA-stimulated donor PBMC every 7 days for up to 3 weeks. Virus-containing supernatants were spun to remove cellular debris and frozen at −70°C until use. Some isolates were reexpanded using PBMC stimulated with both PHA and surface-immobilized anti-CD3 monoclonal antibody plus IL-2 as previously described (54) to generate high titer stocks for further experiments. Isolates therefore had limited expansion in PBMC in vitro and are likely to reflect the viral swarm present in vivo, although we cannot exclude that some selection, albeit limited, occurred during in vitro passage.
TABLE 1.
Characteristics of 29 South African AIDS patients with NSI or SI isolates
| Parameter | Value for groupa
|
||
|---|---|---|---|
| Total cohort | NSI isolates | SI isolates | |
| No. studied (% of total) | 29 | 24 (83) | 5 (17) |
| No. female | 16 | 18 | 3 |
| No. male | 13 | 11 | 2 |
| Median age | 34 | 35 | 29 |
| Median viral load | 321,070 | 776,090∗ | 68,410∗ |
| Median CD4 count | 40 | 48∗∗ | 10∗∗ |
| Diagnosis | |||
| Tuberculosis | 22 | 19 | 3 |
| Cryptococcal meningitis | 4 | 2 | 2 |
| PCP | 2 | 2 | 0 |
| MOTT/Mac | 1 | 1 | 0 |
∗, P < 0.05 by Mann-Whitney test. ∗∗, P < 0.05 by Mann-Whitney test.
Viruses were tested for their ability to cause syncytia in the MT-2 T-cell line (34). Among the 29 isolates, 5 (17%) were able to grow in MT-2 cells, leading to syncytium formation (SI isolates) (Table 1). The subset of patients with SI viruses had significantly reduced CD4-T-cell counts, which is similar to observations made with patients infected with subtype B strains (41). Viral loads among the patients with SI viruses were not particularly high compared to levels in the cohort as a whole, perhaps because too few memory CD4 T cells remained to support high-level viral replication (Table 1). However, viral load measurements were available from only three patients with SI viruses, so no conclusions can be drawn from the statistical perspective. SI viruses were isolated from 3 of 22 (14%) TB patients and 2 of 4 (50%) CM patients. However, overall there were no significant differences in the age, gender, or opportunistic infections of patients with SI isolates compared to those that harbored NSI isolates. A cohort containing larger numbers of patients with SI isolates will be needed to fully address such issues.
The HIV-1 isolates were subtyped in the C2-V5 region of the env gene by a heteroduplex mobility assay (HMA), and all were shown to belong to HIV-1 subtype C, the predominant subtype in South Africa (6, 58). Thirteen isolates were also subtyped in gag by HMA and shown to belong to subtype C (data not shown), suggesting that these isolates were unlikely to be intersubtype recombinants, although this cannot be excluded. Two viruses, CM9 and SW7, have been confirmed as being subtype C throughout its genome (37).
Coreceptor usage of HIV-1 subtype C isolates in transfected cell lines.
Viral isolates were tested for their abilities to replicate in U87.CD4 cells transfected with either CCR5 or CXCR4, as previously described (34). Cells were plated in 12-well plates at 105 cells/well in 2 ml of selection medium (Dulbecco's modified Eagle's medium containing 10% fetal calf serum, antibiotics plus 500 μg of G418 (Boehringer Mannheim GmbH)/ml, and 1 μg of puromycin (ICN Biomedicals Inc., Aurora, Ohio)/ml. The following day, 1,000 50% tissue culture infectious doses were added. After incubation overnight, the cultures were washed three times with Dulbecco's modified eagle's medium plus 10% fetal calf serum to remove unbound virus and then monitored for syncytium formation and p24 antigen production on days 4, 8, and 12. The in-house p24 antigen enzyme-linked immunosorbent assay has a detection limit of 250 pg/ml (52). Isolates that induced syncytium formation and generated increasing concentrations of p24 antigen were considered to be replication positive.
There was an absolute correlation between the NSI and SI phenotypes and the ability of an isolate to use CCR5 or CXCR4, respectively. Thus, all 24 NSI isolates were able to replicate in CCR5-expressing cells but not in the CXCR4-expressing cells (hence, they are conventionally designated R5 viruses) (Table 2). The five SI isolates all replicated in CXCR4-expressing cells. Among them, three could replicate in CCR5-expressing cells (designated R5X4 viruses) while two did not (designated X4 viruses). Although PCP1 was classified as an R5 virus, it did show low-level usage of CXCR4 that was below the cutoff for the p24 antigen assay (data not shown). The ability of this virus to use CXCR6 (see below) very efficiently may account for these low levels of replication, since U87.CD4 cells express this receptor, and also GPR1, endogenously (21, 56).
TABLE 2.
Coreceptor usage by HIV-1 subtype C isolates from AIDS patients
| Isolate | Source | CD4 count (cells/μl) | Viral load (RNA copies/ml) | Subtype of env gene | MT-2 assay result | p24 antigen levelb
|
Biotype | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| U87.CD4 cells expressing:
|
GHOST.3 cells expressing:
|
||||||||||
| CXCR4 | CCR5 | CXCR6 | CCR3 | BOB | |||||||
| PCP1 | PBMC | 2 | NAa | C | NSI | − | ++++ | +++ | − | − | R5 |
| CM1 | PBMC | 43 | 146,514 | C | NSI | − | ++ | − | − | − | R5 |
| CM7 | PBMC | 79 | 14,089 | C | NSI | − | ++++ | − | − | − | R5 |
| SW2 | Plasma | 84 | 157,150 | C | NSI | − | ++++ | − | − | − | R5 |
| SW3 | Plasma | 53 | 261,880 | C | NSI | − | ++++ | − | − | − | R5 |
| SW4 | Plasma | 76 | 1,496,620 | C | NSI | − | ++++ | − | − | − | R5 |
| SW5 | PBMC | 40 | 1,374,235 | C | NSI | − | ++++ | − | − | − | R5 |
| SW8 | Plasma | 67 | 1,198,880 | C | NSI | − | + | − | − | − | R5 |
| SW9 | Plasma | 65 | 301,605 | C | NSI | − | ++++ | − | − | − | R5 |
| SW14 | PBMC | 32 | 5,000,000 | C | NSI | − | +++ | − | − | − | R5 |
| SW16 | PBMC | 9 | 151,630 | C | NSI | − | + | − | − | − | R5 |
| SW22 | PBMC | 67 | 2,749,340 | C | NSI | − | ++ | − | − | − | R5 |
| SW23 | PBMC | 10 | 696,000 | C | NSI | − | ++++ | − | − | − | R5 |
| SW26 | PBMC | 25 | NA | C | NSI | − | ++++ | − | − | − | R5 |
| SW28 | Plasma | 43 | 3,814,130 | C | NSI | − | ++++ | − | − | − | R5 |
| SW29 | PBMC | 12 | 1,190,180 | C | NSI | − | ++++ | − | − | − | R5 |
| SW34 | PBMC | 40 | 127,000 | C | NSI | − | ++++ | − | − | − | R5 |
| SW35 | PBMC | 37 | 1,652,870 | C | NSI | − | ++++ | − | − | − | R5 |
| SW36 | PBMC | 55 | 201,340 | C | NSI | − | ++++ | − | − | − | R5 |
| SW37 | PBMC | 121 | 34,210 | C | NSI | − | + | − | − | − | R5 |
| SW38 | PBMC | 37 | 1,777,930 | C | NSI | − | ++++ | − | − | − | R5 |
| SW39 | PBMC | 94 | 321,070 | C | NSI | − | ++++ | − | − | − | R5 |
| SW40 | PBMC | 67 | 856,180 | C | NSI | − | ++++ | − | − | − | R5 |
| SW41 | PBMC | 60 | 3,162,240 | C | NSI | − | +++ | − | − | − | R5 |
| CM9 | PBMC | 24 | NA | C | SI | ++++ | ++++ | +++ | +++ | +++ | R5X4 |
| SW7 | PBMC | 10 | NA | C | SI | ++++ | − | − | − | − | X4 |
| SW12 | Plasma | 27 | 68,410 | C | SI | ++++ | − | − | − | − | X4 |
| SW20 | PBMC | 2 | 43,595 | C | SI | ++ | ++++ | − | − | − | R5X4 |
| SW30 | PBMC | 2 | 73,860 | C | SI | ++++ | ++++ | − | − | − | R5X4 |
NA, not available.
p24 antigen level on day 12 (endpoint p24 assay). −, <250 pg of p24 antigen/ml; +, 250 pg to 1 ng of p24 antigen/ml; ++, 1 to 2 ng of p24 antigen/ml; +++, 2 to 4 ng of p24 antigen/ml; ++++, >4 ng of p24 antigen/ml.
In order to explore whether viral isolates were also able to use alternate HIV-1 coreceptors, U87.CD4 cell lines expressing CCR1, CCR2b, or CCR3 and GHOST.3 cell lines expressing BOB/GPR15 or CXCR6 were used. The GHOST.3 cell assays were performed essentially as described for the U87.CD4 cell assays except that cells were grown in the presence of 100 μg of hygromycin (Sigma-Aldrich, Johannesburg, South Africa)/ml. Because GHOST.3 cell lines express endogenous CXCR4, it was necessary to add 1.3 μM AMD3100 to prevent entry via this coreceptor (62, 64). Viral replication in the presence of AMD3100 therefore indicated use of the transfected coreceptor (either BOB/GPR15, CCR3, or CXCR6). Two viruses were able to use alternate coreceptors efficiently. Thus, PCP1 could use CXCR6, while CM9 could use CXCR6, BOB/GPR15, and CCR3 (Table 2). In each case, entry via the alternate coreceptor was at a level of efficiency comparable to that mediated by CCR5 or CXCR4, in that the extent of virus production was similar in the different cell cultures (Table 2).
Sensitivity of subtype C isolates to CCR5 and CXCR4 inhibitors in PBMC.
To confirm the use of CCR5 or CXCR4 in PBMC, viral isolates were cultured in the presence of CCR5-specific (RANTES and PRO 140) or CXCR4-specific (AMD3100) entry inhibitors. A subset of 13 subtype C viruses able to use different coreceptors was selected for analysis, in comparison with a panel of 7 isolates from subtypes A, B, and E (from National Institutes of Health Reference and Reagent Repository). PBMC used in these experiments were depleted of CD8+ T cells using a RosetteSep CD8 antibody cocktail (StemCell Technologies, Vancouver, Canada). The reduction in p24 levels was calculated relative to cultures without inhibitors, and the percentage of inhibition at a high and low concentration of each inhibitor is reported. Results shown are from duplicate cultures, and each experiment was repeated twice as previously described (52).
The replication of seven of the eight R5 subtype C viruses was effectively inhibited (>90%) by the highest concentrations of RANTES and PRO 140 that were tested (Table 3). One such isolate (CM1) was marginally less sensitive to RANTES (87% inhibition) and PRO 140 (78% inhibition). In contrast, the CCR5 inhibitors were ineffective against R5X4 and X4 viruses, suggesting that these viruses can all use CXCR4 to enter PBMC efficiently. AMD3100 effectively inhibited all the R5X4 and X4 viruses, although two isolates (SW12 and SW30) were slightly less sensitive (86 and 85% inhibition, respectively) than the others. The ability of AMD3100 to restrict the replication of all three R5X4 viruses suggests that they use CXCR4 in preference to CCR5 to enter PBMC in vitro. As expected, AMD3100 was ineffective against all the R5 isolates, because it is known not to interfere with CCR5-mediated entry (16, 43). The ability of an isolate to use alternate coreceptors in transfected cell lines did not influence whether it was sensitive to any of the entry inhibitors in PBMC cultures. Thus, both PCP1 and CM9 were fully sensitive to RANTES, PRO 140, and AMD3100 despite being able to use alternate coreceptors (Table 2). Either expression levels of the alternate coreceptors on PBMC are lower than on the transfected cell lines or these coreceptors simply do not function as HIV-1 entry receptors on primary cells (45).
TABLE 3.
Inhibition of HIV-1 subtype C viruses by CCR5 and CXCR4 inhibitors
| Virus | Subtype | Phenotype | % Inhibition by CCR5 inhibitora
|
% Inhibition by CXCR4 inhibitor AMD3100 ata:
|
||||
|---|---|---|---|---|---|---|---|---|
| RANTES at:
|
PRO 140 at:
|
|||||||
| 19 nM | 4 nM | 167 nM | 33 nM | 500 nM | 100 nM | |||
| PCP1 | C | R5, CXCR6 | 97 | 68 | 98 | 88 | 34 | 18 |
| CM1 | C | R5 | 87 | 49 | 78 | 59 | 0 | 8 |
| CM7 | C | R5 | 94 | 89 | 97 | 100 | 3 | 19 |
| SW5 | C | R5 | 100 | 75 | 100 | 100 | 19 | 6 |
| SW23 | C | R5 | 100 | 89 | 98 | 94 | 0 | 0 |
| SW26 | C | R5 | 100 | 91 | 93 | 87 | 8 | 0 |
| SW29 | C | R5 | 98 | 74 | 98 | 91 | 13 | 0 |
| SW38 | C | R5 | 100 | 34 | 100 | 96 | 0 | 0 |
| CM9 | C | R5X4, CXCR6, BOB, R3 | 36 | 36 | 34 | 13 | 97 | 58 |
| SW7 | C | X4 | 0 | 24 | 0 | 5 | 100 | 87 |
| SW12 | C | X4 | 0 | 0 | 0 | 0 | 86 | 31 |
| SW20 | C | R5X4 | 0 | 0 | 79 | 0 | 100 | 0 |
| SW30 | C | R5X4 | 24 | 9 | 8 | 39 | 85 | 3 |
| DJ258 | A | R5 | 100 | 82 | 100 | 98 | 46 | 0 |
| 92RW026 | A | R5 | 98 | 69 | 100 | 100 | 63 | 23 |
| JR-FL | B | R5 | 99 | 71 | 100 | 98 | 0 | 36 |
| 92US714 | B | R5 | 95 | 60 | 100 | 100 | 0 | 0 |
| 92US077 | B | R5X4 | 17 | 48 | 48 | 41 | 99 | 76 |
| CM235 | E | R5 | 95 | 62 | 100 | 97 | 22 | 2 |
| 92TH001 | E | R5 | 100 | 88 | 99 | 98 | 60 | 21 |
Inhibition of >90% is bolded, and inhibition of <90% is in italics.
The CCR5 inhibitors were also effective against the R5 isolates from subtypes A, B, and E. One R5X4 subtype B isolate (92US077) was effectively inhibited by AMD3100 but not by RANTES and PRO 140, suggesting that it too uses CXCR4 preferentially to enter PBMC. Overall, there was a good, and subtype-independent, correlation between the classification of an isolate as R5 or X4 by use of the coreceptor-transfected cell lines and its sensitivity to CCR5- or CXCR4-specific entry inhibitors in PBMC (52, 55). However, the isolates classified as R5X4, because they could use both CCR5 and CXCR4 in coreceptor-transfected cells (Table 2), were sensitive only to the CXCR4 inhibitor AMD3100 in PBMC and were little affected by the CCR5 antagonists PRO 140 and RANTES (Table 3). Hence, CXCR4 is the coreceptor of choice for these R5X4 isolates in primary T cells, with only a minority of the viruses present entering the cells via CCR5. The quantitative limitations of coreceptor-transfected cells for viral phenotyping have been previously discussed (62-64).
Replication of NSI and SI viruses in wild-type and CCR5 Δ32/Δ32 PBMC.
The subtype C isolates used above were also tested for their ability to replicate in PBMC deficient in CCR5 (i.e., cells from an HIV-1-negative individual whom we confirmed by PCR to have the CCR5 Δ32/Δ32 genotype) (59). PBMC from the CCR5 Δ32/Δ32 donor and also from CCR5 +/+ control donors were isolated and stimulated using PHA and surface-immobilized anti-CD3 monoclonal antibody, as described above. On the following day, the cultures were washed three times and incubated in IL-2-containing medium. Supernatants were assayed for p24 antigen levels using a commercial assay with a kinetic readout (DuPont/NEN Life Sciences, Boston, Mass.) on days 4, 8, and 11.
None of the R5 isolates was able to replicate in the CCR5 Δ32/Δ32 PBMC, but all replicated well in the CCR5 +/+ PBMC, confirming their dependence on CCR5 to infect cells (Table 4). The R5X4 and X4 isolates were able to replicate in both the CCR5 Δ32/Δ32 and the CCR5 +/+ PBMC, although p24 production from the CCR5 Δ32/Δ32 cells was relatively low. In particular, the replication of isolate SW20 was sporadic in the CCR5 Δ32/Δ32 PBMC, with only some of the replicate wells producing p24 antigen. This contrasts with the consistent replication of the same virus in the CCR5 +/+ PBMC.
TABLE 4.
HIV-1 subtype C replication in PBMC from Δ32/Δ32 and wt/wt donors
| Isolate | p24 antigen level for genotypea
|
|
|---|---|---|
| Δ32/Δ32 | wt/wtb | |
| R5 | ||
| PCP1 | − | ++++ |
| CM1 | − | ++++ |
| CM7 | − | ++++ |
| SW5 | − | +++ |
| SW23 | − | ++++ |
| SW26 | − | ++++ |
| SW29 | − | ++++ |
| SW38 | − | ++++ |
| R5X4 | ||
| CM9 | + | ++ |
| SW20 | +/− | + |
| SW30 | + | ++++ |
| X4 | ||
| SW7 | + | +++ |
| SW12 | ++ | +++ |
p24 antigen level on day 8 (kinetic p24 assay). −, <250 pg of p24 antigen/ml; +, 250 pg to 10 ng of p24 antigen/ml; ++, 10 to 30 ng of p24 antigen/ml; +++, 30 to 60 ng of p24 antigen/ml; ++++, >60 ng/ml p24 antigen/ml.
wt, wild type.
CXCR4-using subtype C viruses show alterations in V3 loop.
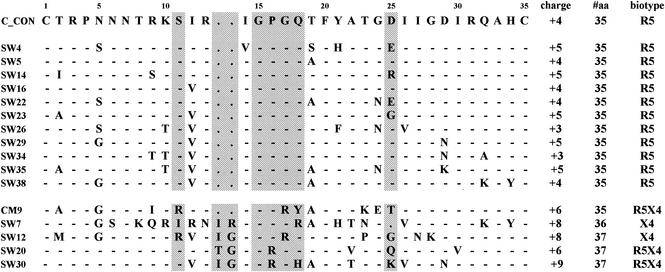
The V3 loop is involved in determining coreceptor usage (27). Certain genetic changes in this region are associated with CXCR4 use, in the context of viruses from subtypes B and D (13). To determine whether CXCR4-using subtype C isolates also have distinctive V3 loop sequences, we isolated RNA from cultured isolates and sequenced their V3 regions, as described previously (6). Phylogenetic analysis of the nucleotide sequences of all 16 V3 loop sequences showed that all clustered with HIV-1 subtype C (data not shown), confirming the HMA data. The V3 loops of all the R5 viruses were 35 amino acids in length and contained the GPGQ motif at the crown that is characteristic of HIV-1 subtype C viruses (Fig. 1). A neutral serine residue was always present at position 11 and, in most cases, an amino acid with a negatively charged side chain (either D or E) was present at position 25. The net charge of the V3 loop was +3 to +5. In contrast, the V3 loops of the R5X4 and X4 viruses varied markedly from the consensus sequence, containing substitutions, deletions, and insertions. A common, and unusual, feature was a two-residue insertion between positions 13 and 14, most frequently isoleucine and glycine, which increased the length of the loop to 37 amino acids. A tetrapeptide motif containing positively charged arginine residues was also frequently present in the V3 loops of the R5X4 and X4 isolates, resulting in a marked increase in the overall positive charge of the loops to +6 to +9. Furthermore, all the R5X4 and X4 isolates had either a neutral or positively charged amino acid substitution or a deletion at position 25. There were no sequence motifs that distinguished R5X4 from X4 viruses.
FIG. 1.
Predicted V3 amino acid sequence of 16 South African HIV-1 subtype C isolates (12 R5 isolates and 5 R5X4 or X4 isolates) compared to an HIV-1 subtype C consensus sequence. The overall positive charge, number of amino acids, and biotype of each isolate are shown on the right. Dashes indicate concurrence, and a period indicates a deletion or lack of an insertion. Changed amino acids are indicated. Positions 11 and 25 (associated with changes in biological phenotype), the tip of the V3 loop, and the region with insertions are highlighted in grey.
Discussion.
Here we describe HIV-1 subtype C isolates from patients with late-stage AIDS that grow in MT-2 cells (SI viruses) and use CXCR4 for entry into cells. These isolates were fully sensitive to AMD3100, a CXCR4-specific entry inhibitor, and were able to grow in PBMC devoid of CCR5. Most of these SI viruses had significant genetic changes in the V3 loop compared to NSI viruses isolated from the same cohort. This observation is similar to those reported for viruses from subtypes B and D (13, 26). Hence, the HIV-1 subtype C envelope glycoproteins can accommodate the amino acid changes needed to use CXCR4 as a coreceptor. Probably, the gp120 proteins from all subtypes interact with CXCR4 in a broadly similar manner.
Our observations stand in contrast to those of previous reports that subtype C infections rarely involved viruses with the SI phenotype (1, 4, 7, 34, 38, 51). These earlier descriptions of subtype C isolates mostly involved cohorts of patients in the relatively early stages of HIV-1 infection, identified relatively soon after subtype C viruses started to spread rapidly in the geographic area under study. Now, we have deliberately selected for study patients with advanced AIDS, since they were more likely to harbor SI viruses. The isolation of CXCR4-using viruses from some of our late-stage AIDS patients suggests that subtype C viruses may undergo a phenotypic switch during disease progression, as occurs during subtype B infection (10, 41, 42). However, because our cohort was observational, we cannot know whether any of the patients with SI viruses had NSI viruses at an earlier stage. Prolonged, longitudinal followup of individuals with acute subtype C infection will be needed to address this issue. The viral phenotypes identified here are unlikely to be the result of in vitro selection pressure. Although viral stocks were grown in PBMC, the V3 sequences of cultured isolates were identical to those from the corresponding uncultured plasma samples (data not shown).
Although only 5 of our 29 (17%) AIDS patients were infected with SI viruses, this represents a higher frequency than was observed in previous studies on subtype C infection (1, 4, 7, 34, 38, 51). In subtype B cohorts, from 50 to 90% of patients with late-stage AIDS have been reported to harbor SI isolates (41, 42). Whether the frequency of subtype C X4 viruses will increase over time or will remain relatively low compared to that of subtype B infections remains to be determined. An analogy can be made to the HIV-1 subtype E epidemic in Thailand; there, early isolates were mostly of the NSI phenotype, but as the epidemic progressed, SI viruses started to appear with ever-increasing frequency (13, 30, 48, 49, 57). Further studies on subtype C infection may reveal the same trend. In any case, the assumptions that subtype C viruses use CCR5 almost exclusively are unlikely to be valid.
Most of the patients in our study had TB, which is the most common opportunistic infection in AIDS patients in South Africa. CCR5 expression levels are affected by environmental factors, such as TB, among African patients, which could favor a relatively high frequency of R5 viruses throughout the course of disease (9, 22). The transmission of R5 viruses may be particularly efficient in areas such as South Africa where there is a high incidence of early HIV-1 infection (24), which is associated with high viral loads and R5 viruses. Furthermore, any individuals who harbor SI viruses early in infection may not survive long enough in an African health care setting to become enrolled in a clinical cohort. Hence, this factor might lead to underrepresentation of SI isolates in a cohort such as ours. Patients with cryptococcal meningitis were particularly likely to have CXCR4-using viruses, which is probably related to the association between this opportunistic infection and advanced HIV-1 disease.
We previously showed that subtype C viruses were as sensitive as subtype B viruses to the CCR5 inhibitors, PRO 140, RANTES, and TAK-779 (52). Here, we have extended our observations to a more diverse group of viruses and we have also tested the CXCR4 inhibitor, AMD3100. Together, these studies support the clinical evaluation of coreceptor-specific entry inhibitors in South Africa; the antiviral responses are likely to be similar to those that have been observed in studies on subtype B infections in North America and Europe (Hendrix et al., 9th Conf. Retrovir. Opportun. Infect., abstr. 391-T, 2002; Reynes et al., 9th Conf. Retrovir. Opportun. Infect., abstr. 1). Subtype C accounts for the vast majority of global HIV-1 infections that are spread by heterosexual and perinatal transmission in developing countries. The ability of coreceptor inhibitors to prevent infection makes them particularly attractive as interventions in high-incidence settings. Novel approaches could include the use of entry inhibitors as microbicides to prevent sexual transmission or as supplements to be added to breast milk to block perinatal infection. Such interventions, however, would need to be effective, cheap, and simple to administer. Our data suggest that therapeutic or preventative approaches based on coreceptor entry inhibitors could be useful in developing countries where subtype C circulates and where such interventions are most desperately needed.
Nucleotide sequence accession numbers.
The V3 sequences determined in this study were submitted to GenBank under the following accession numbers (isolate is identified in parenthesis): AY170657 (SW4), AY170658 (SW5), AY170659 (SW14), AY170660 (SW16), AY170661 (SW22), AY170662 (SW23), AY170663 (SW26), AY170664 (SW29), AY170665 (SW34), AY170666 (SW35), AY170667 (SW38), AF411967 (CM9), AF411966 (SW7), AY230878 (SW12), AY230879 (SW20), and AY230880 (SW30).
Acknowledgments
We thank Mary Phoswa and Janice Croft for technical assistance; Helba Bredell, Julie Nelson, and Maria Papathanasopoulos for help with the V3 sequence analysis; and Adrian Puren for viral loads assays. We are grateful to Dave Johnson and Rod Grant for supplying cells with the Δ32/Δ32 genotype and to Ron Swanstrom and Maria Papathanasopoulos for critical reading of the manuscript. Coreceptor-transfected cell lines were obtained from the NIH AIDS Research and Reference Reagent Repository and the United Kingdom MRC AIDS Reagent Program.
This work was funded by the Wellcome Trust, Poliomyelitis Research Foundation, Fogarty International Centre (TWO-0231), WHO-UNAIDS African AIDS Vaccine Program (AAVP), and National Institutes of Health (R01 AI41420 and P01 AI52048). L.M. is a Wellcome Trust Overseas Senior Research Fellow in Biomedical Science in South Africa, and J.P.M. is a Stavros S. Niarchos Scholar. The Department of Microbiology and Immunology at the Weill Medical College gratefully acknowledges the support of the William Randolph Hearst Foundation.
REFERENCES
- 1.Abebe, A., D. Demissie, J. Goudsmit, M. Brouwer, C. L. Kuiken, G. Pollakis, H. Schuitemaker, A. L. Fontanet, and T. F. Rinke de Wit. 1999. HIV-1 subtype C syncytium- and non-syncytium-inducing phenotypes and coreceptor usage among Ethiopian patients with AIDS. AIDS 13:1305-1311. [DOI] [PubMed] [Google Scholar]
- 2.Baba, M., O. Nishimura, N. Kanzaki, M. Okamoto, H. Sawada, Y. Iizawa, M. Shiraishi, Y. Aramaki, K. Okonogi, Y. Ogawa, K. Meguro, and M. Fujino. 1999. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc. Natl. Acad. Sci. USA 96:5698-5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjorndal, A., H. Deng, M. Jansson, J. R. Fiore, C. Colognesi, A. Karlsson, J. Albert, G. Scarlatti, D. R. Littman, and E. M. Fenyo. 1997. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 71:7478-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjorndal, A., A. Sonnerborg, C. Tscherning, J. Albert, and E. M. Fenyo. 1999. Phenotypic characteristics of human immunodeficiency virus type 1 subtype C isolates of Ethiopian AIDS patients. AIDS Res. Hum. Retrovir. 15:647-653. [DOI] [PubMed] [Google Scholar]
- 5.Blackard, J. T., B. Renjifo, W. Fawzi, E. Hertzmark, G. Msamanga, D. Mwakagile, D. Hunter, D. Spiegelman, N. Sharghi, C. Kagoma, and M. Essex. 2001. HIV-1 LTR subtype and perinatal transmission. Virology 287:261-265. [DOI] [PubMed] [Google Scholar]
- 6.Bredell, H., C. Williamson, P. Sonnenberg, D. J. Martin, and L. Morris. 1998. Genetic characterization of HIV type 1 from migrant workers in three South African gold mines. AIDS Res. Hum. Retrovir. 14:677-684. [DOI] [PubMed] [Google Scholar]
- 7.Cecilia, D., S. S. Kulkarni, S. P. Tripathy, R. R. Gangakhedkar, R. S. Paranjape, and D. A. Gadkari. 2000. Absence of coreceptor switch with disease progression in human immunodeficiency virus infections in India. Virology 271:253-258. [DOI] [PubMed] [Google Scholar]
- 8.Chan, S. Y., R. F. Speck, C. Power, S. L. Gaffen, B. Chesebro, and M. A. Goldsmith. 1999. V3 recombinants indicate a central role for CCR5 as a coreceptor in tissue infection by human immunodeficiency virus type 1. J. Virol. 73:2350-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clerici, M., S. Butto, M. Lukwiya, M. Saresella, S. Declich, D. Trabattoni, C. Pastori, S. Piconi, C. Fracasso, M. Fabiani, P. Ferrante, G. Rizzardini, and L. Lopalco, et al. 2000. Immune activation in Africa is environmentally-driven and is associated with upregulation of CCR5. AIDS 14:2083-2092. [DOI] [PubMed] [Google Scholar]
- 10.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Jong, J. J., A. De Ronde, W. Keulen, M. Tersmette, and J. Goudsmit. 1992. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J. Virol. 66:6777-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Jong, J. J., J. Goudsmit, W. Keulen, B. Klaver, W. Krone, M. Tersmette, and A. de Ronde. 1992. Human immunodeficiency virus type 1 clones chimeric for the envelope V3 domain differ in syncytium formation and replication capacity. J. Virol. 66:757-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Wolf, F., E. Hogervorst, J. Goudsmit, E. M. Fenyo, H. Rubsamen-Waigmann, H. Holmes, B. Galvao-Castro, E. Karita, C. Wasi, S. D. Sempala, et al. 1994. Syncytium-inducing and non-syncytium-inducing capacity of human immunodeficiency virus type 1 subtypes other than B: phenotypic and genotypic characteristics. AIDS Res. Hum. Retrovir. 10:1387-1400. [DOI] [PubMed] [Google Scholar]
- 14.Deng, H. K., D. Unutmaz, V. N. KewalRamani, and D. R. Littman. 1997. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature 388:296-300. [DOI] [PubMed] [Google Scholar]
- 15.Doms, R. W., and S. C. Peiper. 1997. Unwelcomed guests with master keys: how HIV uses chemokine receptors for cellular entry. Virology 235:179-190. [DOI] [PubMed] [Google Scholar]
- 16.Donzella, G. A., D. Schols, S. W. Lin, J. A. Este, K. A. Nagashima, P. J. Maddon, G. P. Allaway, T. P. Sakmar, G. Henson, E. De Clercq, and J. P. Moore. 1998. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat. Med. 4:72-77. [DOI] [PubMed] [Google Scholar]
- 17.Dragic, T., A. Trkola, D. A. Thompson, E. G. Cormier, F. A. Kajumo, E. Maxwell, S. W. Lin, W. Ying, S. O. Smith, T. P. Sakmar, and J. P. Moore. 2000. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc. Natl. Acad. Sci. USA 97:5639-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edinger, A. L., T. L. Hoffman, M. Sharron, B. Lee, B. O'Dowd, and R. W. Doms. 1998. Use of GPR1, GPR15, and STRL33 as coreceptors by diverse human immunodeficiency virus type 1 and simian immunodeficiency virus envelope proteins. Virology 249:367-378. [DOI] [PubMed] [Google Scholar]
- 19.Elbeik, T., W. G. Alvord, R. Trichavaroj, M. de Souza, R. Dewar, A. Brown, D. Chernoff, N. L. Michael, P. Nassos, K. Hadley, and V. L. Ng. 2002. Comparative analysis of HIV-1 viral load assays on subtype quantification: Bayer Versant HIV-1 RNA 3.0 versus Roche Amplicor HIV-1 Monitor version 1.5. J. Acquir. Immune Defic. Syndr. 29:330-339. [DOI] [PubMed] [Google Scholar]
- 20.Essex, M. 1999. Human immunodeficiency viruses in the developing world. Adv. Virus Res. 53:71-88. [DOI] [PubMed] [Google Scholar]
- 21.Farzan, M., H. Choe, K. Martin, L. Marcon, W. Hofmann, G. Karlsson, Y. Sun, P. Barrett, N. Marchand, N. Sullivan, N. Gerard, C. Gerard, and J. Sodroski. 1997. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J. Exp. Med. 186:405-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraziano, M., G. Cappelli, M. Santucci, F. Mariani, M. Amicosante, M. Casarini, S. Giosue, A. Bisetti, and V. Colizzi. 1999. Expression of CCR5 is increased in human monocyte-derived macrophages and alveolar macrophages in the course of in vivo and in vitro Mycobacterium tuberculosis infection. AIDS Res. Hum. Retrovir. 15:869-874. [DOI] [PubMed] [Google Scholar]
- 23.Gaschen, B., B. Korber, and B. Foley. 1999. Part VII. Global variation in the V3 region. Los Alamos National Laboratory, Los Alamos, N.M. http://hiv-web.lanl.gov/content/hiv-db/HTML/99compendium.html.
- 24.Gouws, E., B. G. Williams, H. W. Sheppard, B. Enge, and S. A. Karim. 2002. High incidence of HIV-1 in South Africa using a standardized algorithm for recent HIV seroconversion. J. Acquir. Immune Defic. Syndr. 29:531-535. [DOI] [PubMed] [Google Scholar]
- 25.Hendrix, C. W., C. Flexner, R. T. MacFarland, C. Giandomenico, E. J. Fuchs, E. Redpath, G. Bridger, and G. W. Henson. 2000. Pharmacokinetics and safety of AMD-3100, a novel antagonist of the CXCR-4 chemokine receptor, in human volunteers. Antimicrob. Agents Chemother. 44:1667-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffman, N. G., F. Seiller-Moiseiwitsch, J. Ahn, J. M. Walker, and R. Swanstrom. 2002. Variability in the human immunodeficiency virus type 1 gp120 Env protein linked to phenotype-associated changes in the V3 loop. J. Virol. 76:3852-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffman, T. L., and R. W. Doms. 1999. HIV-1 envelope determinants for cell tropism and chemokine receptor use. Mol. Membr. Biol. 16:57-65. [DOI] [PubMed] [Google Scholar]
- 28.Ivanoff, L. A., J. W. Dubay, J. F. Morris, S. J. Roberts, L. Gutshall, E. J. Sternberg, E. Hunter, T. J. Matthews, and S. R. Petteway, Jr. 1992. V3 loop region of the HIV-1 gp120 envelope protein is essential for virus infectivity. Virology 187:423-432. [DOI] [PubMed] [Google Scholar]
- 29.LaBranche, C. C., G. Galasso, J. P. Moore, D. P. Bolognesi, M. S. Hirsch, and S. M. Hammer. 2001. HIV fusion and its inhibition. Antivir. Res. 50:95-115. [DOI] [PubMed] [Google Scholar]
- 30.Menu, E., J. M. Reynes, M. C. Muller-Trutwin, L. Guillemot, P. Versmisse, M. Chiron, S. An, V. Trouplin, P. Charneau, H. Fleury, F. Barre-Sinoussi, and F. F. Sainte Marie. 1999. Predominance of CCR5-dependent HIV-1 subtype E isolates in Cambodia. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 20:481-487. [DOI] [PubMed] [Google Scholar]
- 31.Michael, N. L., G. Chang, L. G. Louie, J. R. Mascola, D. Dondero, D. L. Birx, and H. W. Sheppard. 1997. The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nat. Med. 3:338-340. [DOI] [PubMed] [Google Scholar]
- 32.Milich, L., B. H. Margolin, and R. Swanstrom. 1997. Patterns of amino acid variability in NSI-like and SI-like V3 sequences and a linked change in the CD4-binding domain of the HIV-1 Env protein. Virology 239:108-118. [DOI] [PubMed] [Google Scholar]
- 33.Moore, J. P., and M. Stevenson. 2000. New targets for inhibitors of HIV-1 replication. Nat. Rev. Mol. Cell. Biol. 1:40-49. [DOI] [PubMed] [Google Scholar]
- 34.Morris, L., T. Cilliers, H. Bredell, M. Phoswa, and D. Martin. 2001. CCR5 is the major coreceptor used by HIV-1 subtype C isolates from patients with active tuberculosis. AIDS Res. Human Retrovir. 17:697-701. [DOI] [PubMed] [Google Scholar]
- 35.Ndung'u, T., B. Renjifo, and M. Essex. 2001. Construction and analysis of an infectious human immunodeficiency virus type 1 subtype C molecular clone. J. Virol. 75:4964-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olson, W. C., G. E. Rabut, K. A. Nagashima, D. N. Tran, D. J. Anselma, S. P. Monard, J. P. Segal, D. A. Thompson, F. Kajumo, Y. Guo, J. P. Moore, P. J. Maddon, and T. Dragic. 1999. Differential inhibition of human immunodeficiency virus type 1 fusion, gp120 binding, and CC-chemokine activity by monoclonal antibodies to CCR5. J. Virol. 73:4145-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papathanasopoulos, M., T. Cilliers, L. Morris, J. Mokili, W. Dowling, D. Birx, and F. McCutchan. 2002. Full-length genome analysis of HIV-1 subtype C utilizing CXCR4 and intersubtype recombinants isolated in South Africa. AIDS Res. Hum. Retrovir. 18:879-886. [DOI] [PubMed] [Google Scholar]
- 38.Ping, L. H., J. A. Nelson, I. F. Hoffman, J. Schock, S. L. Lamers, M. Goodman, P. Vernazza, P. Kazembe, M. Maida, D. Zimba, M. M. Goodenow, J. J. Eron, Jr., S. A. Fiscus, M. S. Cohen, and R. Swanstrom. 1999. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: underrepresentation of X4 variants. J. Virol. 73:6271-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pohlmann, S., M. Krumbiegel, and F. Kirchhoff. 1999. Coreceptor usage of BOB/GPR15 and Bonzo/STRL33 by primary isolates of human immunodeficiency virus type 1. J. Gen. Virol. 80:1241-1251. [DOI] [PubMed] [Google Scholar]
- 40.Rana, S., G. Besson, D. G. Cook, J. Rucker, R. J. Smyth, Y. Yi, J. D. Turner, H. H. Guo, J. G. Du, S. C. Peiper, E. Lavi, M. Samson, F. Libert, C. Liesnard, G. Vassart, R. W. Doms, M. Parmentier, and R. G. Collman. 1997. Role of CCR5 in infection of primary macrophages and lymphocytes by macrophage-tropic strains of human immunodeficiency virus: resistance to patient-derived and prototype isolates resulting from the delta CCR5 mutation. J. Virol. 71:3219-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richman, D. D., and S. A. Bozzette. 1994. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J. Infect. Dis. 169:968-974. [DOI] [PubMed] [Google Scholar]
- 42.Scarlatti, G., E. Tresoldi, A. Bjorndal, R. Fredriksson, C. Colognesi, H. K. Deng, M. S. Malnati, A. Plebani, A. G. Siccardi, D. R. Littman, E. M. Fenyo, and P. Lusso. 1997. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat. Med. 3:1259-1265. [DOI] [PubMed] [Google Scholar]
- 43.Schols, D., S. Struyf, J. Van Damme, J. A. Este, G. Henson, and E. De Clercq. 1997. Inhibition of T-tropic HIV strains by selective antagonization of the chemokine receptor CXCR4. J. Exp. Med. 186:1383-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwarz, M., T. N. Wells, and A. E. Proudfoot. 2001. Chemokine receptors—the next therapeutic target for HIV? Recept. Channels 7:417-428. [PubMed] [Google Scholar]
- 45.Sharron, M., S. Pohlmann, K. Price, E. Lolis, M. Tsang, F. Kirchhoff, R. W. Doms, and B. Lee. 2000. Expression and coreceptor activity of STRL33/Bonzo on primary peripheral blood lymphocytes. Blood 96:41-49. [PubMed] [Google Scholar]
- 46.Shimizu, N., Y. Haraguchi, Y. Takeuchi, Y. Soda, K. Kanbe, and H. Hoshino. 1999. Changes in and discrepancies between cell tropisms and coreceptor uses of human immunodeficiency virus type 1 induced by single point mutations at the V3 tip of the env protein. Virology 259:324-333. [DOI] [PubMed] [Google Scholar]
- 47.Strizki, J. M., S. Xu, N. E. Wagner, L. Wojcik, J. Liu, Y. Hou, M. Endres, A. Palani, S. Shapiro, J. W. Clader, W. J. Greenlee, J. R. Tagat, S. McCombie, K. Cox, A. B. Fawzi, C. C. Chou, C. Pugliese-Sivo, L. Davies, M. E. Moreno, D. D. Ho, A. Trkola, C. A. Stoddart, J. P. Moore, G. R. Reyes, and B. M. Baroudy. 2001. SCH-C (SCH 351125), an orally bioavailable, small molecule antagonist of the chemokine receptor CCR5, is a potent inhibitor of HIV-1 infection in vitro and in vivo. Proc. Natl. Acad. Sci. USA 98:12718-12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sutthent, R., K. Chokephaibulkit, D. Piyasujabul, N. Vanprapa, A. Roogpisuthipong, and P. Chaisilwatana. 2002. Effect of perinatal short-course zidovudine on the clinical and virological manifestations of HIV-1 subtype E infection in infants. J. Clin. Virol. 25:47-56. [DOI] [PubMed] [Google Scholar]
- 49.Sutthent, R., K. Sumrangsurp, P. Wirachsilp, P. Chaisilwattana, A. Roongpisuthipong, P. Chaiyakul, P. Nooma, M. Honda, and P. Warachit. 2001. Diversity of HIV-1 subtype E in semen and cervicovaginal secretion. J. Hum. Virol. 4:260-268. [PubMed] [Google Scholar]
- 50.Swanson, P., V. Soriano, S. G. Devare, and J. Hackett, Jr. 2001. Comparative performance of three viral load assays on human immunodeficiency virus type 1 (HIV-1) isolates representing group M (subtypes A to G) and group O: LCx HIV RNA quantitative, AMPLICOR HIV-1 MONITOR version 1.5, and Quantiplex HIV-1 RNA version 3.0. J. Clin. Microbiol. 39:862-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tien, P. C., T. Chiu, A. Latif, S. Ray, M. Batra, C. H. Contag, L. Zejena, M. Mbizvo, E. L. Delwart, J. I. Mullins, and D. A. Katzenstein. 1999. Primary subtype C HIV-1 infection in Harare, Zimbabwe. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 20:147-153. [DOI] [PubMed] [Google Scholar]
- 52.Trkola, A., T. J. Ketas, K. A. Nagashima, L. Zhao, T. Cilliers, L. Morris, J. P. Moore, P. J. Maddon, and W. C. Olson. 2001. Potent, broad-spectrum inhibition of human immunodeficiency virus type 1 by the CCR5 monoclonal antibody PRO 140. J. Virol. 75:579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trkola, A., S. E. Kuhmann, J. M. Strizki, E. Maxwell, T. Ketas, T. Morgan, P. Pugach, S. Xu, L. Wojcik, J. Tagat, A. Palani, S. Shapiro, J. W. Clader, S. McCombie, G. R. Reyes, B. M. Baroudy, and J. P. Moore. 2002. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc. Natl. Acad. Sci. USA 99:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trkola, A., J. Matthews, C. Gordon, T. Ketas, and J. P. Moore. 1999. A cell line-based neutralization assay for primary human immunodeficiency virus type 1 isolates that use either the CCR5 or the CXCR4 coreceptor. J. Virol. 73:8966-8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trkola, A., W. A. Paxton, S. P. Monard, J. A. Hoxie, M. A. Siani, D. A. Thompson, L. Wu, C. R. Mackay, R. Horuk, and J. P. Moore. 1998. Genetic subtype-independent inhibition of human immunodeficiency virus type 1 replication by CC and CXC chemokines. J. Virol. 72:396-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Unutmaz, D., W. Xiang, M. J. Sunshine, J. Campbell, E. Butcher, and D. R. Littman. 2000. The primate lentiviral receptor Bonzo/STRL33 is coordinately regulated with CCR5 and its expression pattern is conserved between human and mouse. J. Immunol. 165:3284-3292. [DOI] [PubMed] [Google Scholar]
- 57.Utaipat, U., A. Duerr, D. L. Rudolph, C. Yang, S. T. Butera, D. Lupo, T. Pisell, A. Tangmunkongvorakul, N. Kamtorn, N. Nantachit, T. Nagachinta, V. Suriyanon, V. Robison, K. E. Nelson, N. Sittisombut, and R. B. Lal. 2002. Coreceptor utilization of HIV type 1 subtype E viral isolates from Thai men with HIV type 1-infected and uninfected wives. AIDS Res. Hum. Retrovir. 18:1-11. [DOI] [PubMed] [Google Scholar]
- 58.Van Harmelen, J. H., E. Van der Ryst, A. S. Loubser, D. York, S. Madurai, S. Lyons, R. Wood, and C. Williamson. 1999. A predominantly HIV type 1 subtype C-restricted epidemic in South African urban populations. AIDS Res. Hum. Retrovir. 15:395-398. [DOI] [PubMed] [Google Scholar]
- 59.Williamson, C., S. A. Loubser, B. Brice, G. Joubert, T. Smit, R. Thomas, M. Visagie, M. Cooper, and E. van der Ryst. 2000. Allelic frequencies of host genetic variants influencing susceptibility to HIV-1 infection and disease in South African populations. AIDS 14:449-451. [DOI] [PubMed] [Google Scholar]
- 60.Wu, L., G. LaRosa, N. Kassam, C. J. Gordon, H. Heath, N. Ruffing, H. Chen, J. Humblias, M. Samson, M. Parmentier, J. P. Moore, and C. R. Mackay. 1997. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J. Exp. Med. 186:1373-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao, L., D. L. Rudolph, S. M. Owen, T. J. Spira, and R. B. Lal. 1998. Adaptation to promiscuous usage of CC and CXC-chemokine coreceptors in vivo correlates with HIV-1 disease progression. AIDS 12:F137-F143. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, Y., B. Lou, R. B. Lal, A. Gettie, P. A. Marx, and J. P. Moore. 2000. Use of inhibitors to evaluate coreceptor usage by simian and simian/human immunodeficiency viruses and human immunodeficiency virus type 2 in primary cells. J. Virol. 74:6893-6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, Y. J., T. Dragic, Y. Cao, L. Kostrikis, D. S. Kwon, D. R. Littman, V. N. KewalRamani, and J. P. Moore. 1998. Use of coreceptors other than CCR5 by non-syncytium-inducing adult and pediatric isolates of human immunodeficiency virus type 1 is rare in vitro. J. Virol. 72:9337-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, Y. J., and J. P. Moore. 1999. Will multiple coreceptors need to be targeted by inhibitors of human immunodeficiency virus type 1 entry? J. Virol. 73:3443-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]