Abstract
The glycoprotein (G protein) of vesicular stomatitis virus (VSV) is primarily organized in plasma membranes of infected cells into membrane microdomains with diameters of 100 to 150 nm, with smaller amounts organized into microdomains of larger sizes. This organization has been observed in areas of the infected-cell plasma membrane that are outside of virus budding sites as well as in the envelopes of budding virions. These observations raise the question of whether the intracellular virion components play a role in organizing the G protein into membrane microdomains. Immunogold-labeling electron microscopy was used to analyze the distribution of the G protein in arbitrarily chosen areas of plasma membranes of transfected cells that expressed the G protein in the absence of other viral components. Similar to the results with virus-infected cells, the G protein was organized predominantly into membrane microdomains with diameters of approximately 100 to 150 nm. These results indicate that internal virion components are not required to concentrate the G protein into membrane microdomains with a density similar to that of virus envelopes. To determine if interactions between the G protein cytoplasmic domain and internal virion components were required to create a virus budding site, cells infected with recombinant VSVs encoding truncation mutations of the G protein cytoplasmic domain were analyzed by immunogold-labeling electron microscopy. Deletion of the cytoplasmic domain of the G protein did not alter its partitioning into the 100- to 150-nm microdomains, nor did it affect the incorporation of the G protein into virus envelopes. These data support a model for virus assembly in which the G protein has the inherent property of partitioning into membrane microdomains that then serve as the sites of assembly of internal virion components.
Understanding the mechanism by which viral glycoproteins are incorporated into a virus envelope is a central problem in virology. Two types of mechanisms have been proposed (3). One idea is that internal virion components, such as viral matrix or capsid proteins, act to concentrate the viral transmembrane glycoproteins from the surrounding membrane into the area of membrane that will become the virus envelope. A second possibility is that viral glycoproteins are concentrated into areas of the membrane that will give rise to the virus envelope independently of interactions with other virion components. Our work addresses this problem by using the prototypic rhabdovirus, vesicular stomatitis virus (VSV).
VSV encodes a single type of envelope glycoprotein, the G protein. The internal virion components consist of a helical nucleocapsid (NC) associated with approximately 2,000 copies of a matrix (M) protein (15), which condenses the NC into a tightly coiled NC-M protein complex (8, 15). We have shown that much of the G protein is organized into membrane microdomains with diameters of 100 to 150 nm in areas of the infected-cell plasma membrane outside of virus budding sites as well as in the envelopes of budding virions (E. L. Brown and D. S. Lyles, submitted for publication). Furthermore, the higher levels of G protein expression at late times postinfection (p.i.) lead to G protein incorporation into microdomains with a wider variety of sizes rather than to a higher concentration in 100- to 150-nm microdomains. The role that the intracellular virion components play in organizing the G protein into these membrane microdomains is addressed in the present paper. The VSV M protein has been shown to interact with the G protein both by chemical cross-linking (2) and fluorescence spectroscopy experiments (7). However, it has not been determined whether this M protein-G protein interaction is responsible for concentrating the G protein in the envelope or for recruiting the NC-M complex to areas of the membrane enriched in the G protein.
In the experiments presented here, the plasma membrane distribution of the G protein was analyzed by immunogold-labeling electron microscopy in cells transfected with plasmid DNA encoding the G protein in the absence of other viral components and in cells infected with recombinant VSVs encoding truncation mutations of the G protein cytoplasmic domain. In transfected cells, the G protein was organized predominantly into membrane microdomains with diameters of approximately 100 to 150 nm, similar to the microdomains that form the virus envelope. These results indicate that internal virion components are not required to concentrate the G protein into the membrane microdomains observed during virus infection. In cells infected with recombinant viruses encoding truncations of the G protein cytoplasmic domain, the mutant G proteins were predominantly organized into microdomains of 100 to 150 nm at early times p.i. At later times p.i., the higher level of G protein expression led to incorporation of mutant G proteins into microdomains with a wider variety of sizes, similar to the results with wild-type (wt) virus. Truncation of the cytoplasmic domain had little if any effect on incorporation of the G protein into the virus envelope. Collectively, these data support a model for virus assembly in which the G protein is organized into membrane microdomains that will form virus envelopes without interactions with internal virion components.
MATERIALS AND METHODS
Cells and viruses.
The wt VSV (Indiana serotype, San Juan strain) was grown at 37°C in BHK cells in Dulbecco's modified Eagle medium (DMEM) containing 2% fetal bovine serum (FBS) as described previously (7). The recombinant VSVs CT1 and CT9, encoding G proteins with cytoplasmic domain truncations from the 29 amino acids of the wt to 1 amino acid and 9 amino acids, respectively, were provided by John K. Rose from the Yale University School of Medicine (12). Recombinant virus stocks were prepared by growth in BHK cells in DMEM containing 2% FBS following two rounds of plaque purification. Cytoplasmic domain truncations of the G protein were confirmed by automated DNA sequencing of reverse transcription-PCR products.
Plasmid construction.
The pCAGGS vector (9) was engineered to express the VSV G protein under the transcriptional control of the chicken actin promoter (pCAGGS-G). The G protein cDNA described by Rose and Gallione (11) was amplified by PCR using the primers 5′-GCTCTAGACGATCTGTTTCCTTGACACT-3′ and 5′-CCCGCATGGTAACTCAAATCCTGCACAACA-3′ and then cloned into a plasmid vector between the XbaI and SphI sites. The plasmid was digested with XbaI; a blunt end was created with a Klenow DNA polymerase fragment, and an EcoRI linker was ligated. The G gene was excised by digestion with EcoRI and SphI and ligated into the pCAGGS vector to create pCAGGS-G.
Immunogold-labeling electron microscopy.
BHK cells were seeded in a six-well dish to approximately 70% confluence. Cells were infected with either VSV CT1 or CT9 at a multiplicity of infection (MOI) of 20 PFU/cell for 8 or 12 h. For analysis of transfected cells, BHK cells were seeded in a six-well dish to approximately 50% confluence. Cells were subsequently transfected with 1 μg of pCAGGS-G by using Lipofectin reagent (GIBCO-BRL) for 24 h. Cells were washed three times with phosphate-buffered saline (PBS), fixed with 4% formaldehyde for 10 min, washed three times with PBS containing 0.1 M glycine, and incubated in PBS containing 0.1 M glycine and 10% bovine serum albumin (BSA) at 4°C overnight. Samples were incubated with mouse anti-VSV G protein monoclonal antibody I1 (5) or an isotype-matched control monoclonal antibody diluted 1:100 for 1 h. Following three washes with PBS containing 10% BSA, cells were labeled with a secondary goat anti-mouse immunoglobulin G antibody conjugated to 5- or 6-nm gold beads (Jackson Immunoresearch Laboratories) for 1 h. Cells were then postfixed in 1% osmium tetroxide for 15 min, washed three times in PBS, scraped from the dish, and collected by centrifugation. Cells were embedded in Spurr resin by the standard embedding method. Thin sections (80 nm) of the embedded material were viewed with a Philips EM400 electron microscope operating at 80 keV. The gold-conjugated secondary antibody was adjusted to give low densities so that nonspecific labeling in the negative control was not detectable. Also, the low labeling density reduced the problem of interference between two beads separated by distances of <10 nm.
Analysis of virus budding sites.
A series of 25 electron micrographs of arbitrarily chosen VSV budding sites were collected from each of two separate experiments (total of 50 micrographs), and virus budding sites were analyzed as described elsewhere (Brown and Lyles, submitted). Since several micrographs contained more than one virus budding site, the actual numbers of budding sites analyzed in the 50 micrographs were 90 for CT1 at 8 h p.i., 77 for CT1 at 12 h p.i., 75 for CT9 at 8 h p.i., and 91 for CT9 at 12 h p.i. Briefly, virus budding sites were analyzed by measuring the distance of each gold particle from the center of the leading edge of the virus budding site along the trace of the plasma membrane. All gold particles were within 1 μm of the budding site. For budding sites separated by distances of <2 μm, the distance between them was divided at the midpoint. Data were presented as a histogram of the average density of gold particles on the y axis versus the distance from the center of the budding site in 20-nm increments on the x axis. The length of the virus envelope was determined by tracing the membrane from the tip of the budding virion to the end of the electron density of the densely stained NC-M complex. The left and right sides of the virus envelope were measured separately.
Analysis of areas of plasma membrane that exclude virus budding sites.
A series of 25 electron micrographs of arbitrarily chosen areas of plasma membrane that exclude virus budding sites were collected in two separate experiments (total of 50 micrographs) and analyzed as described elsewhere (Brown and Lyles, submitted). Both virus-infected and pCAGGS-G-transfected cells were analyzed. All virus-infected cells were identified by the presence of virus budding sites at the cell surface, while pCAGGS-G-transfected cells were identified by G protein cell surface expression. Briefly, micrographs were analyzed by tracing the plasma membrane from the left side of the image to the right while registering each gold particle that was encountered in the trace. The data were converted to pairwise distance measurements between all the gold particles in a micrograph and were then plotted as a histogram with the average density of gold particles on the y axis and the distance between gold particles on the x axis displayed as successive 20-nm windows. The number of changes in slope in each graph was determined by least-squares analysis by fitting the data to one, two, or three straight lines (zero, one, or two changes in slope, respectively) by using Mac Curve Fit, version 1.1.2 (Kevin Raner Software). A minimum of a twofold decrease in the sum of the squares was required to characterize a graph as having two changes in slope versus one.
Flow cytometry analysis.
For analysis of G protein cell surface expression in VSV-infected cells, BHK cells were seeded to approximately 70% confluence in a six-well dish. Cells were infected at an MOI of 10 PFU/cell for 4.5, 6, 8, 10, 12, or 14 h in DMEM containing 2% FBS. G protein cell surface expression in the absence of other viral components was determined by flow cytometry analysis of pCAGGS-G-transfected cells. BHK cells were seeded to approximately 50% confluence in a six-well dish. Cells were then transfected with 1 μg of pCAGGS-G DNA by using Lipofectin reagent for 12, 24, or 36 h. Following infection or transfection, cells were washed twice and then incubated for 1 h with 0.1 M glycine-10% BSA in PBS at 4°C. Surface-expressed G protein was labeled with anti-G protein antibody I1 at a 1:400 dilution for 1 h. Cells were washed three times and then incubated with a goat anti-mouse immunoglobulin G secondary antibody conjugated to fluorescein (ICN Biomedicals, Inc.) at a dilution of 1:200 for 1 h at 4°C. Cells were washed three times and then fixed for 10 min in 4% formaldehyde at 4°C and were collected in 1 ml of 10% BSA. G protein cell surface fluorescence was quantitated with a Becton Dickinson FACSCaliber flow cytometer.
Quantitation of virus budding.
BHK cells were grown to approximately 70% confluence in a six-well dish. Cells were infected with wt VSV, CT1, or CT9 at an MOI of 10 PFU/cell in the presence of 100 μCi of [35S]methionine/ml. At 8 or 12 h p.i., cells were washed three times with PBS to remove previously budded virus and the [35S]methionine label. Cells were then incubated in 0.5 ml of DMEM containing 2% FBS for 30 min at 37°C, and the culture supernatants and cell lysates were collected. Supernatants were centrifuged at 2,000 rpm in a Hereus Instruments 8155 rotor for 5 min at 4°C to remove cellular debris and then were centrifuged over a 15% sucrose cushion at 50,000 rpm for 40 min in a Beckman TLS-55 rotor. Virus pellets were resuspended in a total volume of 100 μl of 0.1% sodium dodecyl sulfate (SDS). Cell lysates were collected by adding 0.5 ml of radioimmunoprecipitation assay buffer (0.15 M NaCl, 1% deoxycholate, 1% Triton X-100, 0.1% SDS, 10 mM Tris [pH 7.4]) containing 20 nM aprotinin to each well on ice. Cellular debris was removed by centrifugation at 2,000 rpm for 5 min at 4°C. Equal volumes of virus and cell lysates were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) on 10% polyacrylamide gels. Gels were fixed, dried, and analyzed by phosphorescence imaging using Quantity One software (Bio-Rad).
RESULTS
The G protein partitions into membrane microdomains independently of intracellular viral components.
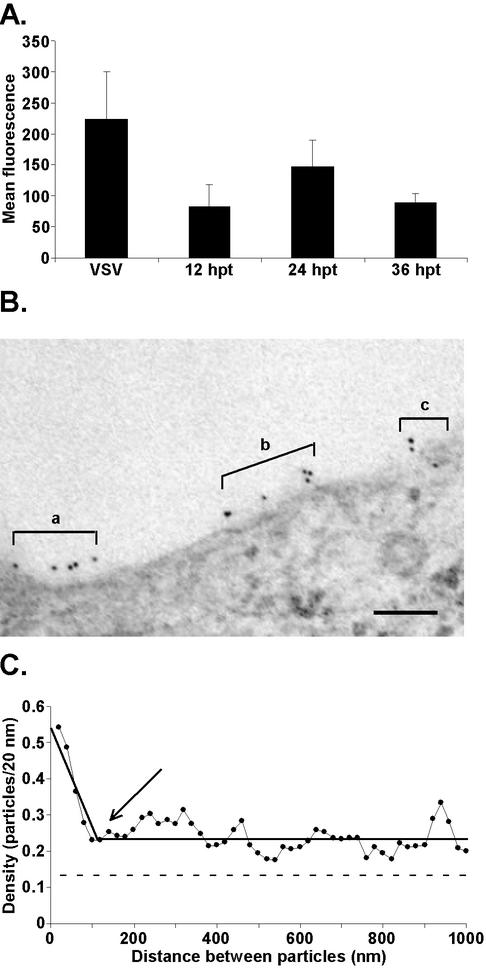
The VSV G protein is organized predominantly into membrane microdomains in virus-infected cells with diameters of 100 to 150 nm; smaller amounts of G protein are organized into microdomains of larger sizes (Brown and Lyles, submitted). Two hypotheses can explain the observed distribution of the G protein into membrane microdomains in the virus-infected cells. One explanation is that intracellular viral components alter the distribution of the G protein in the plasma membrane during the creation of a virus budding site. An alternative explanation is that the G protein partitions into these membrane microdomains independently of other viral components and the virus preferentially buds from these G protein-containing microdomains. To test these hypotheses, we analyzed cells transfected with a plasmid encoding the G protein to determine whether the G protein is organized into microdomains in the plasma membrane in the absence of other viral components. BHK cells were transiently transfected with a plasmid that expresses the G protein under the transcriptional control of the chicken actin promoter. This promoter has been shown previously to drive high-level gene expression in a variety of cell lines (9). Flow cytometry analysis was used to compare the G protein cell surface expression in VSV-infected cells at 4.5 h p.i. to that in transfected cells at 12, 24, and 36 h posttransfection (p.t.) (Fig. 1A). Transfected cells expressed the highest levels of G protein at the cell surface at 24 h p.t., which was 66% of the level in cells infected with wt VSV for 4.5 h.
FIG. 1.
The G protein partitions into membrane microdomains independently of other viral components. (A) Comparison of G protein cell surface expression in VSV-infected cells to that in cells transfected with pCAGGS-G DNA. BHK cells were infected with wt VSV for 4.5 h or were transfected with pCAGGS-G DNA for 12, 24, or 36 h. Following infection or transfection, cells were labeled for immunofluorescence with an anti-G protein antibody and a secondary antibody conjugated to fluorescein. G protein cell surface fluorescence was quantitated by flow cytometry. The data are presented as the mean levels of fluorescence of the G protein-positive population ± SD for three experiments. (B) Representative micrograph of immunogold labeling of the G protein in transfected cells. a to c, groups of gold particles that might represent G protein-containing microdomains. Bar = 100 nm. (C) Analysis of the G protein distribution in the plasma membranes of cells transfected with pCAGGS-G. A series of 50 electron micrographs of arbitrarily chosen areas of plasma membrane similar to those shown in panel B were digitized and analyzed by tracing the plasma membrane from the left side of the image to the right while registering each gold particle that was encountered in the trace. The data were converted to pairwise distance measurements between all the gold particles in a micrograph and were plotted as a histogram with the number of pairs of gold particles on the y axis and the distance between gold particles on the x axis displayed as successive 20-nm windows. The y values were normalized by dividing by the number of gold particles analyzed to give average densities. Horizontal dashed line, average concentration of the G protein in the plasma membrane; solid black line through the data points, best-fit line for the data; arrow, point at which slope changes, which gives the diameter of the microdomains.
To determine the distribution of the G protein in plasma membrane microdomains, BHK cells were transfected with plasmid DNA encoding the G protein for 24 h and then were labeled for immunoelectron microscopy. A representative micrograph is shown in Fig. 1B. There are three groups of gold particles in this micrograph (labeled a, b, and c), which give the impression that there are three different G protein-containing microdomains separated by areas of membrane lacking G protein labeling. However, in any one individual micrograph, it is not possible to tell whether this pattern reflects the organization of the G protein in the membrane or whether it occurred due to random placement of gold particles. By analysis of data from many such micrographs, the random variation in G protein labeling is averaged out and patterns of G protein labeling emerge.
A series of 25 micrographs of arbitrarily chosen areas of the plasma membrane were analyzed in two separate experiments using a new analytical method designed to determine the extent of organization of a protein into membrane microdomains. The method of data analysis is described in detail elsewhere (Brown and Lyles, submitted). Briefly, a pairwise measurement of the distance traced along the plasma membrane between each gold particle and every other gold particle in the micrograph was made. These data generated a data set of distances between all of the gold particles in all of the micrographs. The data were plotted as a histogram with the average number of gold particles separated by a given distance on the y axis versus the distance of separation on the x axis in 20-nm increments. In the resulting histogram, the y intercept is the average concentration of gold particles that are separated by a distance of 20 nm. The fact that nearly all of the gold particles within the microdomain have a high concentration of the G protein 20 nm away serves to raise this average much higher than the average concentration of the G protein in the plasma membrane as a whole. At a distance that is large compared to the size of the microdomains (e.g., 1 μm), the average concentration of gold particles separated by this distance should be the same as the average in the membrane as a whole. At intermediate distances of separation, the average density is between these two extremes. The diameters of membrane microdomains can be determined from the points at which the graph changes slope.
Figure 1C shows the pairwise distance distribution of the G protein in transfected cells at 24 h p.t. The results of the two experiments were in good agreement, so this graph includes combined data from both experiments. The average density of the G protein in the plasma membrane is 0.14 particles/20 nm. A single dramatic change in slope, which occurred at approximately 100 to 150 nm, was observed in this analysis (Fig. 1C). However, G protein densities higher than the average extend to at least 1,000 nm. The observation that G protein densities remain higher than the average for distances between 150 and 1,000 nm could be due to several causes. One possibility is that the average density of the G protein is artificially low due to the cell-to-cell variability of gene expression observed during transfection. It is also possible that the higher-than-average density for these distances reflects incorporation of smaller amounts of the G protein into microdomains with a wide variety of sizes. The important conclusion from the data in Fig. 1C is that most of the G protein partitions into discrete membrane microdomains of approximately 100 to 150 nm, with perhaps smaller amounts of the G protein distributed into microdomains of larger size. This distribution of the G protein occurs independently of other viral components.
Partitioning of the G protein into membrane microdomains occurs independently of the cytoplasmic domain in virus-infected cells.
It has been hypothesized that interactions between the G protein cytoplasmic domain and the internal virion components are an essential feature of the budding process. Therefore, to determine if the cytoplasmic domain of the G protein is important for its organization into membrane microdomains or formation of the virus envelope, we analyzed recombinant VSVs that encode G proteins with cytoplasmic domain truncations. The wt G protein contains a 29-amino-acid cytoplasmic domain. CT9 and CT1 are recombinant VSVs that encode G proteins with cytoplasmic domain truncations to either 9 amino acids or 1 amino acid, respectively. The CT1 G protein has previously been reported to promote virus production at a level lower than that promoted by the wt G protein (12). In contrast, the CT9 G protein sequence is the minimum required for efficient virus production and hence serves as the positive control for the truncation of the G protein in these experiments.
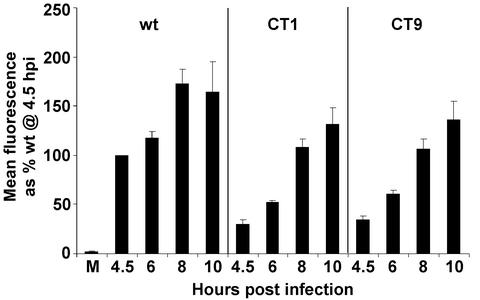
Previous data showed CT1 and CT9 G proteins are delayed in the transport to the cell surface (16). Therefore, flow cytometry analysis of G protein cell surface expression was performed on cells infected with wt, CT1, or CT9 viruses at various times p.i. to determine when CT1 and CT9 viruses express levels of G protein at the cell surface comparable to those expressed by cells infected with wt VSV (Fig. 2). Both CT1 and CT9 viruses expressed lower levels of G protein than wt VSV, consistent with their slower rate of transport to the cell surface. Cells infected with CT1 or CT9 viruses did not express levels of G protein comparable to those found at the surfaces of wild-type virus-infected cells at 4.5 h p.i. until nearly 4 h later, at 8 h p.i., indicating an approximately 4-h lag in G protein cell surface expression in the mutant-virus-infected cells. A similar flow cytometry analysis was performed in a separate series of experiments to determine that mutant-virus-infected cells did not express the G protein at the cell surface to the same extent as wt VSV at 8 h p.i. until 12 h p.i. (data not shown), consistent with the 4-h delay in G protein cell surface expression observed at earlier times p.i. These results with CT9 virus agree with similar flow cytometry data published by Schnell et al. (12). For CT1 virus, Schnell et al. detected a level of G protein cell surface expression greater than that for CT9 virus. This minor difference between our results and theirs may reflect the fact that we did not observe as much of a defect in the budding of CT1 virus compared to that of CT9 virus (see Fig. 5); thus more of the CT1 G protein was released into virions in our experiments.
FIG. 2.
Time course of G protein cell surface expression in BHK cells infected with wt, CT1, or CT9 virus. M, mock infection. At the indicated times, cells were labeled for immunofluorescence with the anti-G protein antibody and a secondary antibody conjugated to fluorescein. G protein cell surface fluorescence was quantitated by flow cytometry. The data were normalized to the mean fluorescence of wt VSV-infected cells at 4.5 h p.i. and are presented as the averages of three experiments with SD.
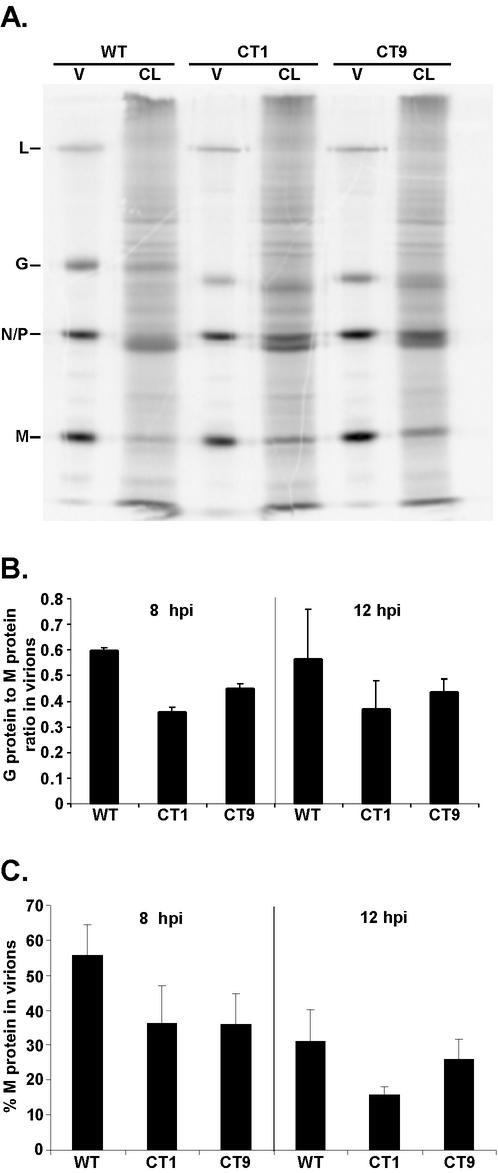
FIG. 5.
Analysis of the budding efficiency of CT1 and CT9. BHK cells were infected with wt VSV, CT1, or CT9 in the presence of [35S]methionine. At 8 or 12 h p.i., cells were washed to remove previously budded virus and the [35S]methionine label and then incubated at 37°C for 30 min in media lacking 35S. Virus-containing supernatants were collected, and virions were purified over a 15% sucrose cushion. Both purified virions (V) and cell lysates (CL) were analyzed by SDS-PAGE and phosphorescence imaging. Aliquots of virions were 30% of the total, and cell lysates were 6% of the total. (A) Representative image obtained at 8 h p.i. Viral protein bands are indicated. (B) Quantitation of G protein-to-M protein ratio at 8 and 12 h p.i. Data are presented as the averages of three experiments with SD. (C) Quantitation of budding efficiency by determining the amount of labeled M protein that was released into the extracellular virions as a percentage of the total M protein in cells plus virions. Data are presented as the averages of three experiments with SD.
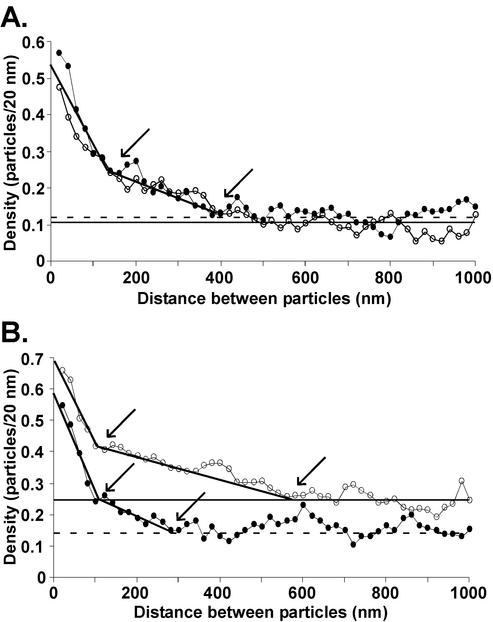
CT1 and CT9 G proteins were analyzed in areas of the membrane outside of virus budding sites by using the same pairwise distance distribution used for transfected cells. The purpose of this analysis was to determine if the cytoplasmic domain of the G protein alters its distribution into membrane microdomains outside of virus budding sites (Fig. 3). Cells were analyzed at both 8 (Fig. 3A) and 12 h (Fig. 3B) p.i. The distribution of gold particle distances in CT1 virus-infected cells at 8 h p.i. was nearly indistinguishable from that for CT9 virus-infected cells (Fig. 3A). Therefore, only one best-fit curve was drawn to represent the two data sets. Both curves exhibit dramatic changes in slope at about 100 to 150 nm, similar to that which was observed in the wt-virus-infected cells at 4.5 h p.i. (Brown and Lyles, submitted). A second, less distinct slope (Fig. 3A) can be discerned for densities between 150 and 400 nm. The striking feature of these curves is how similar they are to each other. This result indicates that the incorporation of CT1 and CT9 G proteins into membrane microdomains is independent of the cytoplasmic domain. These data are also similar to those from transfected cells expressing the wt G protein (Fig. 1C). In both cases, most of the G protein was present in 100- to 150-nm microdomains. The most noticeable difference was the additional G protein in microdomains of approximately 400 nm in cells infected with CT1 and CT9 viruses. This probably reflects the higher levels of G protein expression in virus-infected cells than in transfected cells, which leads to incorporation of some of the G protein into larger microdomains.
FIG. 3.
The cytoplasmic domain is not required for G protein partitioning into membrane microdomains. BHK cells were infected with either CT1 (solid circles) or CT9 (open circles) for 8 (A) or 12 h (B). Immunogold labeling and quantitation of the gold particle distribution in the plasma membranes of the virus-infected cells were performed as described for Fig. 1C. Horizontal dashed and solid lines, average concentrations of G protein in the plasma membrane for CT1 and CT9, respectively; solid black line through the data points, best-fit line for the data for both viruses; arrows, points at which slope changes, giving the diameters of the microdomains.
Figure 3B shows the pairwise distance analysis of the G protein distribution in CT1 and CT9 virus-infected cells at 12 h p.i. For CT9 virus-infected cells, the average density increased from 0.11 to 0.25 particles/20 nm between 8 and 12 h (Fig. 3B versus A), which resulted in a slightly higher y intercept of the graph at 12 h. The increased density of the G protein in the plasma membrane also resulted in the change in slope at around 150 nm becoming less distinct, indicating a broader distribution of the G protein into microdomains of different sizes. However, the 100- to 150-nm domain as well as larger microdomains (extending in this case to approximately 600 nm) can still be discerned (Fig. 3B). For CT1 virus-infected cells, the average density of G protein labeling at late times p.i. did not increase as much as for the CT9 virus-infected cells (Fig. 3A and B), indicating that there was less of an increase in G protein surface expression in cells infected with CT1 virus. A distinct change in slope can still be discerned at approximately 100 to 150 nm at the 12-h time point as well as a less-distinct change at approximately 300 nm, similar to the results at 8 h p.i. These data indicate that CT1 and CT9 G proteins are incorporated into both 100- to 150-nm and 300- to 400-nm microdomains at 8 h p.i. and that the higher levels of CT9 G protein expression at 12 h p.i. lead to incorporation into microdomains with a wider variety of sizes, similar to results with wt VSV (Brown and Lyles, submitted).
Budding from G protein microdomains occurs independently of the cytoplasmic domain of the G protein.
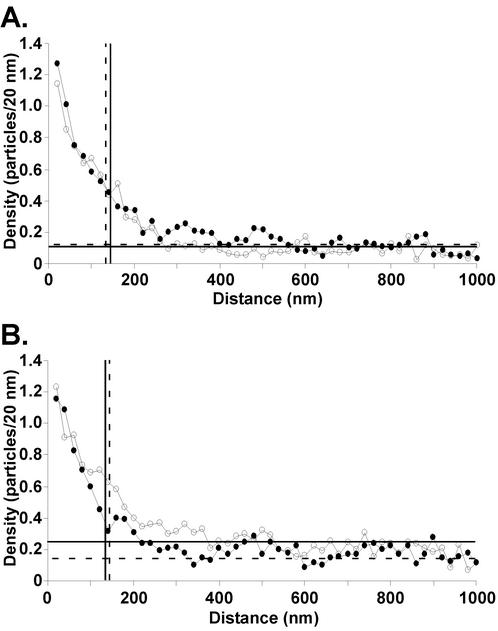
To determine if the cytoplasmic domain of the G protein is important for its distribution at VSV budding sites, we analyzed both CT1 and CT9 virus budding sites by immunogold-labeling electron microscopy at 8 and 12 h p.i. (Fig. 4). BHK cells were infected with CT1 or CT9 viruses, and, at 8 (Fig. 4A) or 12 h p.i. (Fig. 4B), cells were fixed and immunolabeled with an antibody against the G protein and a secondary antibody conjugated to 6-nm gold beads. Twenty-five electron micrographs of arbitrarily chosen VSV budding sites were collected in each of two separate experiments (50 micrographs total). The micrographs were analyzed by measuring the distance of each gold particle from the center of the leading edge of the budding virion to a distance of about 1 μm on each side. The results of the quantitation are shown in Fig. 4 as the density of gold particles versus the distance from the center of the budding site in 20-nm increments. The results of the two experiments were in good agreement, so this graph includes combined data from both experiments. The vertical lines represent the average lengths of the virus envelope in all budding sites analyzed as defined by the end of the densely stained NC-M complex. Therefore, all data points to the left of the vertical line represent data that were collected from within the virus envelope.
FIG. 4.
Budding from G protein microdomains occurs independently of the cytoplasmic domain of the G protein. BHK cells were infected with CT1 (solid circles) or CT9 (open circles). At 8 (A) or 12 h (B) p.i., cells were fixed and labeled with an anti-VSV G protein monoclonal antibody and a secondary antibody conjugated to 6-nm gold beads. A series of 25 electron micrographs of arbitrarily chosen VSV budding sites were collected from two separate experiments. Negatives were digitized and analyzed by measuring the distance of each gold particle from the center of the leading edge of the virus budding site to a distance of 1 μm on each side. The average data from the two experiments were plotted as a histogram of the density of gold particles (y axis) versus the distance from the center of the budding site in 20-nm increments (x axis). The average density of G protein in the plasma membrane was determined from the 50 micrographs of arbitrarily chosen areas of plasma membrane analyzed in Fig. 3 (CT1, horizontal dashed lines; CT9, horizontal solid lines). The average length of the virus envelope was determined by tracing the membrane from the tip of the budding virion to the end of the electron density of the densely stained NC-M complex (CT1, vertical dashed lines; CT9, vertical solid lines).
Both CT1 and CT9 virus budding sites contained the highest densities of G protein at the tip of the virus envelope, with lower densities found at the base (Fig. 4). Also, a higher-than-average density extended to about 300 nm. As shown for wt VSV (Brown and Lyles, submitted), the higher average density at the tip of the virus envelope than at the base is due, in part, to viruses that bud from microdomains that are smaller than the virus envelope and thus do not extend to the base. The extension of densities higher than the average to distances around 300 nm indicates that both CT1 and CT9 viruses bud from G protein-containing membrane microdomains with diameters up to 300 to 400 nm. This distribution of G protein is similar to that described for wt VSV (Brown and Lyles, submitted). The most striking result is that the curves for CT1 and CT9 viruses are nearly indistinguishable from each other. These data indicate that the cytoplasmic domain of the G protein is not important for incorporation of the G protein into the VSV envelope.
The distribution of the CT1 and CT9 G proteins at virus budding sites at 12 h p.i. (Fig. 4B) was similar to that observed at 8 h p.i. (Fig. 4A) with the exception that the average density of the CT9 G protein at 12 h p.i. was approximately twofold higher than that of the CT1 G protein, 0.25 versus 0.14 particles/20 nm, respectively (Fig. 4B). However, the increased density of G protein in the plasma membrane had no effect on the distribution of the G protein at the virus budding sites. There was a slightly higher density at the base of CT9 budding sites and in the immediately surrounding membrane, which probably reflects a higher percentage of virions that bud from microdomains that are larger than the virus envelope. These results confirm the observation made for wt-VSV-infected cells (Brown and Lyles, submitted) that the distribution of the G protein at virus budding sites is largely independent of the level of G protein in the plasma membrane. Also, these results further indicate that the cytoplasmic domain of the G protein does not alter its organization into virus budding sites.
Since there was no difference in the distribution of the G protein in budding sites of CT1 versus CT9 viruses, these results suggest that the CT1 virus is not defective in the incorporation of the G protein into the virus envelope. This prediction was tested by measuring the ratio of the G protein to internal viral proteins at both 8 and 12 h p.i. in radiolabeling experiments. Cells were infected with wt, CT1, or CT9 virus in the presence of [35S]methionine for either 8 or 12 h. Cells were labeled continuously so that all viral proteins were radioactively labeled to uniform specific activity. At 8 or 12 h, labeled media were removed, and cells were washed to remove all previously budded virus as well as the remaining 35S label and then incubated for 30 min in media lacking 35S to measure virus production. Following this 30-min period, virus-containing supernatants were collected and virions were purified by centrifugation over a sucrose cushion. Both cell lysates and purified virions were analyzed by SDS-PAGE and phosphorescence imaging.
Figure 5A shows a representative image obtained at 8 h p.i. The G proteins of wild-type, CT1, and CT9 viruses have different mobilities as a result of the truncation of the CT1 and CT9 G protein cytoplasmic domains. This image shows that there was little difference in the release of virion proteins between cells infected with CT1 and CT9 viruses and those infected with wt VSV. Data from three experiments similar to these were quantitated and are shown in Fig. 5B and C.
Figure 5B shows the ratio of G protein to M protein in virions. Similar results were obtained by analysis of the ratio of G protein to the NC proteins (N and P proteins) (data not shown). CT1 and CT9 viruses incorporated similar amounts of G protein into virions, although both incorporated slightly less G protein into virions than wt VSV at both times p.i. These results are in agreement with the previously published results (12). The important result is that there was little if any difference between CT1 and CT9 viruses at either time p.i. This result confirms the results of the electron microscopy analysis (Fig. 4).
In Fig. 5C, budding efficiency was quantitated by determining the labeled M protein that was released into the extracellular virions as a percentage of the total M protein in cells plus virions. At 8 h p.i., there was no detectable difference in budding efficiency between CT1 and CT9 viruses. However, a slight reduction in the budding efficiencies of both viruses compared to that of the wt virus was observed. This decreased budding efficiency observed for CT1 and CT9 viruses may reflect the delay in G protein cell surface expression observed in the flow cytometry analysis (Fig. 2). At 12 h p.i., there was little if any difference in the budding efficiencies of wt and CT9 viruses. However, CT1 virus was released at approximately 51% of the efficiency of wt virus (16 versus 31%; Fig. 5C), suggesting a minor defect in the budding of CT1 virus at 12 h p.i. These results differ somewhat from previously published work, which showed that CT9 and CT1 viruses produced total amounts of virus that were 51 and 11% of the level of wt virus after 24 h of infection, respectively (12). We analyzed total virus production in 24 h using the same experimental protocol as that described in the previously published experiments and observed levels of virus production for CT9 and CT1 viruses of 102% ± 5% and 52% ± 19% of the level of wt virus, respectively (means ± standard deviations [SD] for three separate experiments). These data are in good agreement with the data obtained at 12 h p.i. in Fig. 5C. Thus the difference in the magnitude of the defect in production of CT1 and CT9 viruses compared to previous data reflects laboratory-to-laboratory variation rather than differences in experimental design. We do not consider this to be a major discrepancy with the previously published data, since such differences in virus yield (within 1 log unit) are often observed between different laboratories.
DISCUSSION
Analysis of the distribution of the G protein in areas of the membrane that exclude virus budding sites indicated that the G protein partitions into membrane microdomains that are approximately 100 to 150 nm, with smaller amounts distributed into larger microdomains (Brown and Lyles, submitted). These microdomains are remarkably similar to those observed at the sites of budding, which occur in G protein-containing microdomains with a range of sizes, some of which are smaller than the virus envelope (<150 nm) while others extend to a maximum of 300 to 400 nm from the tip of the virus budding site. These results suggested a model for virus assembly in which the G protein assembles into microdomains independently of other viral components and these microdomains serve as sites for virus assembly. The data presented here showed that the partitioning of the G protein into these membrane microdomains occurs independently of other viral components (Fig. 1). This result indicates that internal virion components are not required to concentrate the G protein into membrane microdomains with sizes and densities similar to those of virus envelopes. This idea was further supported by the observation that deletion of the cytoplasmic domain of the G protein did not alter its partitioning into the 100- to 150-nm microdomains (Fig. 3). Interestingly, deletion of the cytoplasmic domain also did not affect the incorporation of the G protein into virus envelopes (Fig. 4 and 5).
The data presented in this paper support a model for virus assembly in which the G protein partitions into membrane microdomains that resemble virus envelopes independently of other viral components and these microdomains serve as the sites of assembly of internal virion components. This model differs from previous models of virus assembly, primarily by the absence of a requirement for internal virion components to organize the G protein into the membrane microdomains that will become the virus envelope. Most previous models for assembly of enveloped viruses have postulated a role for the interaction of the cytoplasmic domains of envelope glycoproteins with internal virion proteins, such as the matrix or capsid protein, in order to concentrate viral glycoproteins in the envelope. For alphaviruses, there is clear evidence for an interaction with capsid proteins in forming an icosahedral arrangement of viral glycoproteins in the envelope (3). For VSV, the evidence for an interaction between the G protein and M protein includes chemical cross-linking experiments (2) with purified virions and previous fluorescence experiments by Lyles et al. using purified G protein and M protein or NC-M complexes (7). However, these experiments do not address whether this interaction is important for concentrating the G protein in the envelope. Instead the interaction between the G protein and the M protein may play a role in recruiting NC-M complexes to preformed G protein-containing microdomains.
The data presented here do not rule out a role for G protein-M protein interaction in concentrating the G protein in the envelope. For example, interaction of the G protein with the M protein at the tip of the NC-M complex might contribute to the high concentration of G protein at the tip of the virus budding site. This would be consistent with data indicating that the M protein at the tip of the NC-M complex is more accessible to the membrane than the M protein in the tightly coiled shaft (1). However, our results with transfected cells indicate that G protein-containing microdomains can achieve a concentration of G protein similar to that observed in virions without the participation of other viral components. Thus there may be little if any role for the G protein-M protein interaction in concentrating the G protein at the sites of virus budding. Instead, these data favor a model in which the G protein-M protein interaction serves to recruit NC-M complexes to G protein-containing microdomains.
A model in which the G protein serves to recruit NC-M complexes to the sites of virus assembly would also be consistent with data showing that G gene deletions or mutations affecting temperature sensitivity reduce virus budding by 10-fold or more (4, 13, 14). Previously published data indicated that CT1 virus produces virions at only 11% of the level of wt virus after 24 h (12), similar to the results obtained in the absence of the G protein, suggesting that the cytoplasmic domain is necessary for enhancement of virus budding by the G protein. However, we observed only a minor defect in the budding of CT1 virus: 51% that of the wt (Fig. 5C). This result was obtained regardless of whether budding was analyzed at 8 or 12 h (Fig. 5C) or after 24 h, as in the published data. However, both our work (Fig. 5B) and the previous work on CT1 virus (12) found similar ratios of G protein/M protein in both wt and mutant viruses.
These results are consistent with our electron microscopy data, which show that the CT1 and CT9 mutant viruses are not defective in their ability to incorporate the G protein into the virus envelope (Fig. 4). These results show that the cytoplasmic domain of the G protein is not required for VSV budding from membrane microdomains that are enriched in the G protein. Other data have shown that sequences in the ectodomain of the G protein are important for enhancing virus budding (10, 12). Since the M protein does not span the membrane lipid bilayer (6), these results have led to the idea that the G protein might interact with the M protein or NC-M complexes indirectly through alterations in the structure of the membrane lipid bilayer (10). Alternatively, sequences in the ectodomain might induce the transmembrane domain of the G protein to form a binding site recognized by the M protein. Future experiments will test these hypotheses.
Acknowledgments
We acknowledge the Microscopy Core Laboratory, especially Ken Grant for his expert technical assistance with the electron microscope and Paula Moore for assistance with the preparation of the immunogold-labeled electron microscopy samples. We also thank Jay Jerome, Mark Willingham, Griffith Parks, and David Ornelles for critical advice.
This research was supported by Public Health Service Grant AI 15892 from the National Institute for Allergy and Infectious Diseases. E.L.B. was supported by National Institutes of Health Training Grant T32 AI07401. The Microscopy Core Laboratory was supported in part by the core grant for the Comprehensive Cancer Center of Wake Forest University CA12197 from the National Cancer Institute.
REFERENCES
- 1.Barge, A., Y. Gaudin, P. Coulon, and R. W. Ruigrok. 1993. Vesicular stomatitis virus M protein may be inside the ribonucleocapsid coil. J. Virol. 67:7246-7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dubovi, E. J., and R. R. Wagner. 1977. Spatial relationships of the proteins of vesicular stomatitis virus: induction of reversible oligomers by cleavable protein cross-linkers and oxidation. J. Virol. 22:500-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garoff, H., R. Hewson, and D. J. E. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62:1171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knipe, D. M., D. Baltimore, and H. F. Lodish. 1977. Maturation of viral proteins in cells infected with temperature-sensitive mutants of vesicular stomatitis virus. J. Virol. 21:1149-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lefrancois, L., and D. S. Lyles. 1982. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. II. Monoclonal antibodies of nonneutralizing and cross-reactive epitopes of Indiana and New Jersey serotypes. Virology 121:168-174. [DOI] [PubMed] [Google Scholar]
- 6.Lenard, J. 1996. Negative-strand virus M and retrovirus MA proteins: all in a family? Virology 216:289-298. [DOI] [PubMed] [Google Scholar]
- 7.Lyles, D. S., M. McKenzie, and J. W. Parce. 1992. Subunit interactions of vesicular stomatitis virus envelope glycoprotein stabilized by binding to viral matrix protein. J. Virol. 66:349-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newcomb, W. W., G. J. Tobin, J. J. McGowan, and J. C. Brown. 1982. In vitro reassembly of vesicular stomatitis virus skeletons. J. Virol. 41:1055-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 10.Robison, C. S., and M. A. Whitt. 2000. The membrane-proximal stem region of vesicular stomatitis virus G protein confers efficient virus assembly. J. Virol. 74:2239-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rose, J. K., and C. J. Gallione. 1981. Nucleotide sequences of the mRNA's encoding the vesicular stomatitis virus G and M proteins determined from cDNA clones containing the complete coding regions. J. Virol. 39:519-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schnell, M. J., L. Buonocore, E. Boritz, H. P. Ghosh, R. Chernish, and J. K. Rose. 1998. Requirement for a non-specific glycoprotein cytoplasmic domain sequence to drive efficient budding of vesicular stomatitis virus. EMBO J. 17:1289-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schnell, M. J., J. E. Johnson, L. Buonocore, and J. K. Rose. 1997. Construction of a novel virus that targets HIV-1-infected cells and controls HIV-1 infection. Cell 90:849-857. [DOI] [PubMed] [Google Scholar]
- 14.Takada, A., C. Robison, H. Goto, A. Sanchez, K. G. Murti, M. A. Whitt, and Y. Kawaoka. 1997. A system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. USA 94:14764-14769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas, D., W. W. Newcomb, J. C. Brown, J. S. Wall, J. F. Hainfeld, B. L. Trus, and A. C. Steven. 1985. Mass and molecular composition of vesicular stomatitis virus: a scanning transmission electron microscopy analysis. J. Virol. 54:598-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitt, M. A., L. Chong, and J. K. Rose. 1989. Glycoprotein cytoplasmic domain sequences required for rescue of a vesicular stomatitis virus glycoprotein mutant. J. Virol. 63:3569-3578. [DOI] [PMC free article] [PubMed] [Google Scholar]