Abstract
We have found that coilin, the marker protein for Cajal bodies (coiled bodies, CBs), is a self-interacting protein, and we have mapped the domain responsible for this activity to the amino-terminus. Together with a nuclear localization signal, the self-interaction domain is necessary and sufficient for localization to CBs. Overexpression of various wild-type and mutant coilin constructs in HeLa cells results in disruption of both CBs and survival motor neurons (SMN) gems. Additionally, we have identified a cryptic nucleolar localization signal (NoLS), within the coilin protein, which may be exposed in specific coilin phospho-isoforms. The implications of these findings are discussed in light of the fact that other proteins known to localize within nuclear bodies (e.g., PML, SMN and Sam68) can also self-associate. Thus protein self-interaction appears to be a general feature of nuclear body marker proteins.
INTRODUCTION
The eukaryotic nucleus is highly organized. Individual chromosomes, for example, occupy discrete domains in the interphase nucleus called chromosome territories (Lamond and Earnshaw, 1998). The interchromatin space contains numerous morphologically distinct substructures and bodies, in addition to the readily observable nucleolus (Misteli and Spector, 1998; Matera, 1999). One such nuclear body is the Cajal body (CB), discovered in 1903 by Santiago Ramón y Cajal. Cajal noticed that these bodies often seemed to be in proximity to the nucleolus and termed them nucleolar ‘accessory’ bodies. In the ensuing 100 years, the precise functions of CBs have yet to be elucidated. Despite findings that CBs contain high concentrations of small nuclear ribonucleoproteins (snRNPs), splicing does not take place in the CB compartment (reviewed in Matera, 1999). Interestingly, recent work has shown that the survival motor neurons (SMN) protein is enriched in CBs (Liu and Dreyfuss, 1996; Matera and Frey, 1998; Carvalho et al., 1999; Young et al., 2000). SMN is also found to reside, in some cell types, in distinct nuclear foci called gems, for “Gemini of CBs.” Mutations in the SMN1 gene have been shown to be responsible for the autosomal recessive disease, spinal muscular atrophy (Lefebvre et al., 1995). Furthermore, SMN protein is part of a large complex that is involved in snRNP biogenesis (Fischer et al., 1997; Liu et al., 1997; Pellizzoni et al., 1999; Charroux et al., 1999; Charroux et al., 2000).
In addition to snRNPs and the SMN complex, CBs are enriched in a myriad of other factors including the nucleolar proteins Nopp140 and fibrillarin, various basal transcription factors, along with several kinases. Recently, we found that another kinase, CDK2-cyclin E, is enriched in CBs during the G1-to-S transition of the cell cycle (Liu et al., 2000). However, the only unambiguous marker for CBs is a protein called p80 coilin. Coilin was first identified by the use of autoimmune sera and shows a staining pattern consisting of several bright foci in the nucleus, as well as a diffuse nucleoplasmic pool (Andrade et al., 1991; Raska et al., 1991). Subsequent analysis revealed that coilin is a protein of 576 amino acids (aa), which contains two putative nuclear localization signals, but no other motifs from which function can be inferred (Chan et al., 1994; Wu et al., 1994; Bohmann et al., 1995). Coilin is a phosphoprotein, and this phosphorylation occurs only on serine residues (Carmo-Fonseca et al., 1993). During mitosis, the level of phosphorylation in coilin increases by at least two additional residues (Carmo-Fonseca et al., 1993). Cell cycle analysis reveals that CBs disassemble during mitosis and reform in the cell cycle at early- to mid-G1. Throughout the cell cycle, coilin levels remain relatively constant (Andrade et al., 1993), giving rise to the hypothesis that the phosphorylation status of coilin may play a role in CB formation. In support of this idea, treatment of HeLa cells with the specific serine/threonine protein phosphatase inhibitor, okadaic acid, results in the partial redistribution of coilin into the center of the nucleolus (Lyon et al., 1997). This same phenotype was reported after mutation of a single coilin serine residue at position 202 to aspartate (Lyon et al., 1997; Sleeman et al., 1998). These data imply that the phosphorylation state of coilin is important for proper subcellular localization and CB formation.
In this report, we provide additional evidence showing that proper coilin phosphorylation is required for the protein to correctly localize to the CB. Furthermore, several novel mutant coilin constructs suggest that coilin may transit through the nucleolus. We have identified a cryptic nucleolar localization signal (NoLS) in coilin. We also show that coilin is a self-interacting protein and that the self-interaction domain maps within the N-terminal 92 aa. Furthermore, hyperphosphorylation of the protein results in decreased self-interaction. Constructs lacking the coilin N-terminus fail to self-interact and do not accumulate in CBs. Taken together, these data point to a role for self-association in CB localization.
MATERIALS AND METHODS
In Vitro Transcription/Translation and In Vitro Coimmunoprecipitation
Human coilin was obtained from the plasmid Wu-1 (Wu et al., 1994) by digestion with NcoI + SacI. This fragment was then cloned into the in vitro transcription vector pTM1 (Moss et al., 1990), which was also digested with NcoI + SacI. Coilin was also cloned into an EE epitope-tagged version of pTM1, EE-TM1, (Horton and Templeton, 1997), using the same strategy. Cold EE-coilin was obtained by coupled in vitro transcription/translation (Promega, Madison, WI). 35S-labeled coilin was obtained by coupled in vitro transcription/translation (Promega), except that 35S-methionine was added to the reaction mix. Coimmunoprecipitation was conducted by mixing 5 μl of cold EE-tagged coilin or an EE-tagged heterologous sequence to 5 μl of 35S-labled coilin, followed by the addition of anti-EE antibodies. The mixture was allowed to incubate for 1 h at 4°C, followed by the addition of 30 μl of 50% protein G sepharose (Pharmacia, Piscataway, NJ), and another 1 h incubation at 4°C (with gentle inversion). The reaction was centrifuged, and the beads were washed four times with 500 μl RIPA buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1 mM EDTA). The beads were then resuspended in 30 μl of 2X SDS-loading buffer, boiled, and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, 8%), followed by drying and exposure to x-ray film. As a control, 1/15th input of 35S-labeled coilin was run on the same gel as the coimmunoprecipitation reactions.
Yeast Two-Hybrid Analysis
The NcoI + XhoI coilin fragment from pTM1-coilin was cloned into the prey vector pACT2, also digested with NcoI + XhoI. To clone coilin into the bait vector pAS2–1, a polymerase chain reaction fragment of coilin was generated, using primers containing NcoI and BamHI restriction sites and Wu-1 as the template. This fragment was cloned into pAS2–1 digested with NcoI and BamHI. Coilin bait truncations were generated using restriction sites within coilin. Coilin bait and prey constructs were transformed into the yeast strain PJ69–2A (Clontech, Palo Alto, CA) and plated onto medium lacking leucine and tryptophan to select for the plasmids. The resulting colonies were then picked to plates lacking leucine and tryptophan as well as adenine and histidine. Interacting proteins were assessed after 3 d incubation on this medium. Appropriate controls were used to ensure the validity of the interactions. To quantify the relative strength of the interactions, β-galactosidase assays were conducted using the yeast strain HF7c (Clontech), essentially as described (Isaac et al., 1998). The reported value represents the average number of Miller Units for three separate assays from three independent colonies, with individual values varying by no more than 20% from the average value.
Coimmunoprecipitation of Coilin from HeLa Cells
Coilin obtained by digestion of Wu-1 with BamHI + SacI was cloned into pEGFP-C3 (Clontech) digested with BglII + SacI. Green fluorescent protein (GFP)-coilin was transfected into HeLa cells using SuperFect (QIAGEN, Valencia, CA). After incubation for 24–36 h, cells were harvested, washed in PBS, and resuspended in 1 ml RIPA to lyse cells. The resuspended cells were incubated for 30 min at 4°C with gentle inversion, followed by centrifugation to pellet cellular debris. To 900 μl of lysate was added 50 μl of anti-GFP antibodies (Clontech). As a control, 900 μl of lysate was added to normal rabbit serum. Incubation was allowed to proceed with gentle shaking overnight at 4°C. Complexes were captured by the addition of 60 μl 50% Protein A Sepharose (Pharmacia) for a 2 h incubation with gentle shaking at 4°C. The beads were then washed 4 times with 1 ml RIPA buffer, resuspended in 30 μl 2X SDS loading buffer, boiled, and subjected to SDS-PAGE (8%). After transfer to nitrocellulose, the membrane was probed with anticoilin Ab R288 (1:500), followed by incubation with goat anti-rabbit horseradish peroxidase (Pierce Chemical, Rockford, IL ) (1:10,000) and chemiluminescence detection. Coimmunoprecipitation of coilin from HeLa cells transfected with myc-tagged coilin(94–576) was conducted in the same manner as described above, except that 5 μl of antic-myc antibodies (Santa Cruz, Biotechnology, Santa Cruz, CA) and Protein G Sepharose (Pharmacia) were used. Where indicated, HeLa cells were arrested in mitosis by the addition of nocodazole to the medium for 12–16 h at a final concentration of 0.4 μg/ml.
Immunochemical Methods and Antibodies
For indirect immunofluorescence, cells were grown on chambered slides (Nunc, Rochester, NY), fixed in paraformaldehyde, and permeabilized in Triton X-100 as described (Frey and Matera, 1995). The cells were then incubated with primary antibodies and detected using fluorochrome conjugated secondary antibodies. The following primary antibodies were used: anti-coilin rabbit serum (R288 at 1:100, Andrade et al., 1993; R508 at 1:200, Chan et al., 1994), anti-Nopp140 rabbit serum (RE10 at 1:200; Meier and Blobel, 1992), anti-myc (mAb 9E10 at 1:40; Santa Cruz), anti-Sm (mAb Y12 at 1:800; Lerner et al., 1981), antifibrillarin (mAb 72B9 at 1:800; gift from E. Chan), anti-GAR1 rabbit serum (1:500; gift from F. Dragon), anti-SMN (mAb 2B1 at 1:1000; Liu and Dreyfuss, 1996), anti-PML (mAB 5E10 at 1:10; Stuurman et al., 1992). Secondary antibodies anti-rabbit Alexa 594 (1:800) and anti-mouse Alexa 594 (1:400) were obtained from Molecular Probes (Eugene, OR). Anti-mouse fluorescein (1:200) was obtained from Vector Laboratories (Burlingame, CA). Images were obtained using a Zeiss (Thornwood, NY) Axioplan epifluorescence microscope equipped with a cooled charge-coupled device (CCD) camera (Photometrics, Tucson, AZ). Fluorescein isothiocyanate and Texas red filter sets (Chroma Tech, Brattleboro, VT) were used. Images were processed using Registration (Biological Detection Systems, Pittsburgh, PA) and Adobe Photoshop 5.5 (Adobe Systems, Mountain View, CA) programs. Prints were generated on a dye sublimation printer (Codonics, Middleburg Heights, OH).
Mutagenesis
Various coilin mutants used throughout this study were obtained using the Quick Change Mutagenesis kit (Stratagene, La Jolla, CA). GFP-coilin(1–248) and GFP-coilin(1–315) were generated by the introduction of stop codons. Constructs with single coilin amino acid changes, such as GFP-coilin(S184A), were generated by point mutagenesis. All constructs, including those generated by standard molecular biological techniques, were verified by sequencing and Western blotting.
In Vitro Kinase Assay
Coilin used as a substrate for in vitro kinase assays was obtained by coupled in vitro transcription/translation (Promega, Madison, WI), followed by immunoprecipitation with anticoilin antibodies (R288) and adsorption onto Protein A Sepharose beads (Pharmacia). The in vitro kinase assay was conducted essentially as described previously (Liu et al., 2000), except that 2 μl of casein kinase 2 (Upstate Biotechnology, Lake Placid, NY) was used.
RESULTS
Coilin Is a Self-Interacting Protein
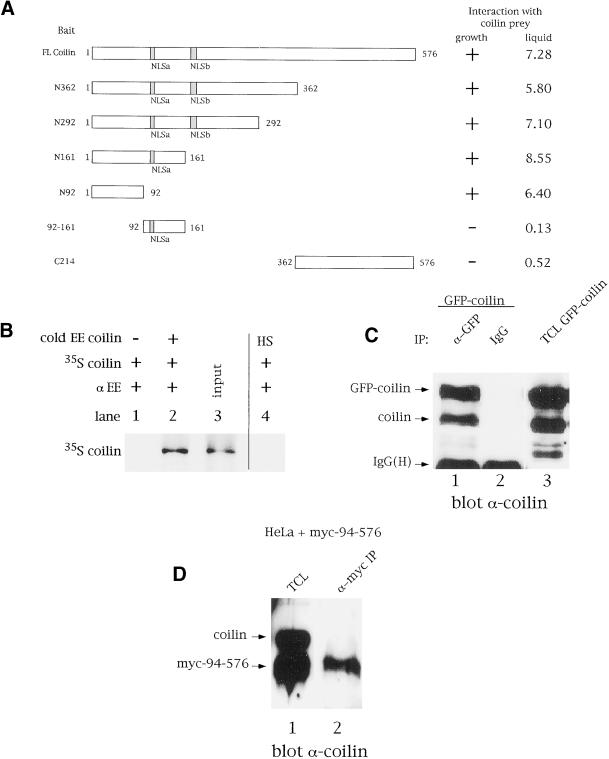
Recent data have shown that the signature proteins of several different nuclear bodies can self-oligomerize (e.g., SMN, [Lorson et al., 1998]; Sam68, [Chen et al., 1999]; PML, [Ishov et al., 1999]). Thus, it seemed plausible that coilin might also self-interact. Using a directed two-hybrid screen with coilin as both bait and prey, we found that, indeed, coilin self-associates (Figure 1A). Furthermore, the use of coilin bait truncations localized the self-interaction domain to within the N-terminal 92 aa of the protein (Figure 1A). To confirm this interaction, coimmunoprecipitations were conducted with in vitro translated coilin. As shown in Figure 1B, immunoprecipitation of cold EE epitope-tagged coilin with anti-EE antibodies specifically recovered radiolabeled coilin (lane 2). In the absence of EE-coilin (lane 1) or the presence of a heterologous EE-tagged protein (lane 4), no 35S-labeled coilin was precipitated (Figure 1B). Coilin self-interaction in vivo was also demonstrated using transfected cell extracts (Figure 1C). In this experiment, HeLa cells were transiently transfected with a construct expressing GFP-coilin, followed by lysis and immunoprecipitation with anti-GFP antibodies. Endogenous coilin coimmunopreciptiated with anti-GFP, but not with normal rabbit serum (Figure 1C, lanes 1 and 2). As a negative control, no coilin was detected after immunoprecipitation with anti-GFP using untransfected cell extracts or cells transfected with GFP alone (our unpublished results).
Figure 1.
The N-terminus of coilin mediates self-interaction. Coilin self-interacts in the yeast two-hybrid system (A), as well as by coimmunoprecipitation in vitro (B) and from cell extracts (C). In panel A, various coilin baits, indicated on the left, were cotransformed into the yeast strain PJ69–2A with coilin prey. Interaction was defined as the ability of PJ69–2A containing both bait and prey to grow on selective medium within 3 days. The (+) sign denotes robust growth after 3 days incubation, whereas the ({minus}) sign represents no growth. The lack of coilin prey interaction with aa 92–161 or the C-terminal 214 aa of coilin verified the specificity of the interaction. The interactions were quantified by performing liquid β-galactosidase assays, as described in MATERIALS AND METHODS. Values are reported in Miller units and varied by no more than 20%. (B) Coilin and EE-coilin were in vitro translated with or without [35S]methionine, respectively. Cold EE-coilin (5 μl) was mixed with 5 μl 35S-labeled coilin and immunoprecipitated with anti-EE antibodies (lane 2). Lane 3 contains 1/15th the amount of 35S-labeled coilin used in the reactions. No 35S-labeled coilin was recovered when immunoprecipitated in the absence of EE-coilin (lane 1) or in the presence of an EE-tagged heterologous sequence (HS, lane 4). (C) Cell extracts from GFP-coilin transfected HeLa cells were subjected to immunoprecipitation with anti-GFP antibodies, followed by SDS-PAGE and western blotting with antibodies against coilin. Normal rabbit serum was used as a negative control for immunoprecipitation. Lane 3 contains 10 μl of total cell lysate (TCL) from GFP-coilin transfected cells. IgG(H) is the immunoglobulin heavy chain. (D) Cell extracts from myc-coilin(94–576) transfected HeLa cells were immunoprecipitated with antimyc antibodies, followed by SDS-PAGE and western blotting with antibodies against coilin. No endogenous coilin is observed in lane 2, corresponding to the IP using antimyc antibodies. Lane 1 contains 10 μl of total cell lysate (TCL) from myc-coilin(94–576) transfected cells. Normal mouse serum was used as a negative control for immunoprecipitation (our unpublished results).
To demonstrate that the N-terminus of coilin is required for its self-interaction, as suggested by our two-hybrid data, we transfected HeLa cells with a myc-tagged coilin construct lacking the first 93 aa of the protein and coimmunoprecipitated with antimyc antibodies, (Figure 1D). No endogenous coilin was observed upon immunoprecipitation with coilin(94–576). Thus the amino terminus of coilin is required for its self-interaction.
The Self-Interaction Domain Is Sufficient for CB Localization
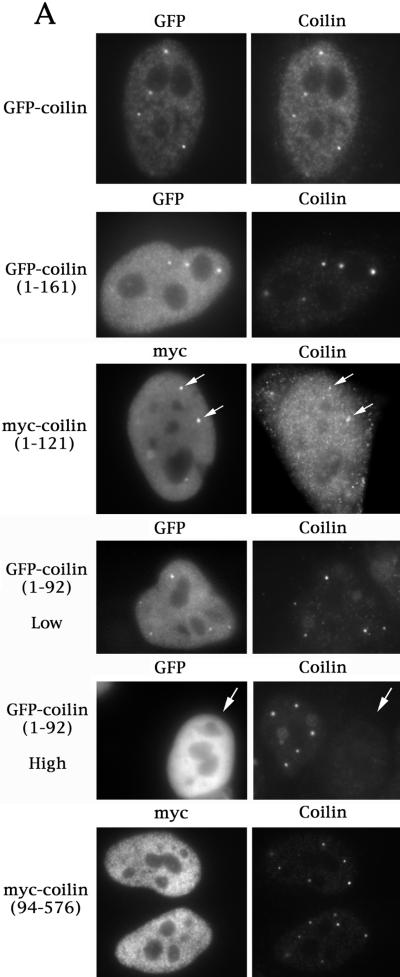
Previous work by Bohmann et al. (1995) demonstrated that various coilin truncation mutations affect its subnuclear localization, but no Cajal body localization domain was identified. In contrast, Gall and coworkers (Wu et al., 1994) showed that a construct containing only the N-terminal 102 aa (along with exogenous myc and NLS tags) of human coilin correctly localized to CBs in Xenopus oocytes. Since we had mapped the coilin self-interaction domain to within the first 92 aa of the protein (Figure 1), it seemed plausible that N-terminal constructs of coilin might also localize to CBs in somatic cells. Therefore, we decided to reinvestigate the issue of CB localization in HeLa cells. Three differentially tagged N-terminal coilin constructs were employed: GFP-coilin(1–161), myc-coilin(1–121) and GFP-myc-NLS-coilin(1–92). Cells expressing lower levels of each of the constructs tended to localize both within CBs as well as diffusely throughout the nucleoplasm (Figure 2A). However, higher levels of expression resulted in an increase in the diffuse, nucleoplasmic component and a concomitant loss of both the mutant and endogenous coilin from CBs (Figure 2A). The presence of an exogenous NLS was important for the localization to CBs, as expression of myc-coilin(1–92) was diffuse throughout the nucleus and the cytoplasm (Bohmann et al., 1995, and our unpublished results). We were able to monitor the effects of overexpression on the endogenous protein using an antibody to the coilin C-terminus that does not recognize the various N-terminal fragments (Figure 2A). The finding, that these N-terminal constructs can localize to CBs, demonstrates that the self-interaction domain, along with an NLS, is sufficient for localization to CBs. In support of this finding, the coilin(94–576) construct does not localize to CBs (Figure 2A; Bohmann et al., 1995).
Figure 2.
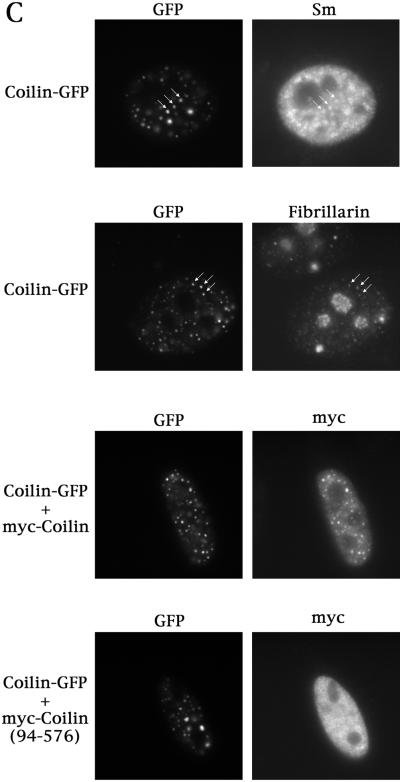
(A) The self-interaction domain of coilin is necessary and sufficient for CB localization. HeLa cells transfected with the indicated constructs were assayed for coilin localization by immunofluorescence using anticoilin antibody R508. The myc-coilin(1–121) and myc-coilin(94–576) constructs (Bohmann et al., 1995) were localized using antimyc antibodies. GFP-myc-NLS-coilin(1–92), labeled GFP-coilin(1–92), is shown at both high and low expression levels. The highly expressing GFP-myc-NLS-coilin(1–92) cell is indicated by an arrow. GFP-coilin(1–161) and myc-coilin(1–121) are shown at low expression levels. Facing page: (B) Overexpression of GFP-coilin disrupts CBs and results in the accumulation of GFP-coilin in thenucleolus. GFP-coilin was transfected into HeLa cells and immunofluorescence with coilin, SMN, and PML antibodies was conducted. (C) Fusion of GFP to the C-terminus of coilin apparently increases the number of CBs. Coilin was cloned into pEGFP-N3 (Clontech) and transfected into HeLa cells alone or in the presence of myc-coilin or myc-coilin (94–576). Immunofluorescence with antibodies to Sm (for snRNP localization), fibrillarin (for snoRNPs) or myc was conducted. Coilin-GFP localizes to numerous small foci which colocalize with Sm, fibrillarin, and ectopically expressed coilin, some of which are indicated by arrows. No colocalization was observed for coilin-GFP with myc-coilin(94–576).
To determine whether the observed loss of endogenous protein from CBs upon overexpression was due to a dominant negative effect of the coilin N-terminus, we decided to test whether overexpression of full-length GFP-coilin might have the same effect. Similarly, cells expressing low levels of GFP-coilin display CBs (Figure 2A). However, high levels of expression result in the apparent disruption of CBs and accumulation of GFP-coilin in the nucleolus (Figure 2B). Because we could not discriminate endogenous coilin protein from GFP-coilin in these experiments, we monitored the expression of other CB marker proteins in cells overexpressing the GFP constructs. As shown in Figure 2B, SMN foci are not detected in the nuclei of high-expressing cells. Similar results were obtained when cells were transfected with the N-terminal coilin fragments (our unpublished results). Likewise, nucleolar proteins Nopp140, fibrillarin, and GAR1 were readily detectable in the nucleoli of transfected cells, but CB-sized foci were not observed (our unpublished results). As a negative control, the localization of PML protein was monitored (Figure 2B) and found to be normal (i.e. within PML bodies). Thus the disruption of CBs in cells expressing high levels of coilin is not simply due to a general disorganization of the nucleoplasmic architecture.
The results presented above indicate that overexpression of the coilin self-interaction domain or the full-length coilin protein can alter endogenous coilin localization. Unlike our N-terminal coilin fusions, we have found that fusion of GFP to the C-terminus (coilin-GFP) results in an expression pattern that is quite distinct. Specifically, coilin-GFP localized to numerous foci, in contrast to the normal CB pattern (compare Figure 2, panels A and C). In particular, cells expressing coilin-GFP lack the diffuse nucleoplasmic staining component (Figure 2C) that is readily apparent in untransfected cells or those expressing N-terminally tagged coilin. Thus, fusion of GFP at the C-terminus of coilin might disrupt the normal trafficking between the nucleoplasmic pool and the CB, possibly due to improper folding of coilin-GFP. Intriguingly, these multiple coilin-GFP foci contain other antigens typically found in CBs, such as Sm and fibrillarin (Figure 2C), suggesting the presence of snRNPs and snoRNPs. Additionally, cotransfection experiments with coilin-GFP + myc-coilin demonstrate that these multiple foci also contain myc-coilin (Figure 2C), which, when expressed alone, correctly localizes to CBs. Strikingly, cotransfection of coilin-GFP with myc-coilin(94–576) reveals no change in the localization pattern of the N-terminal truncation product (Figure 2C). Thus coilin-GFP can recruit myc-tagged coilin to the multiple CB-like structures only in the presence of the N-terminus, supporting our assertion that the N-terminus is responsible for the self-interaction.
Identification of Coilin Mutants that Localize in the Nucleolus
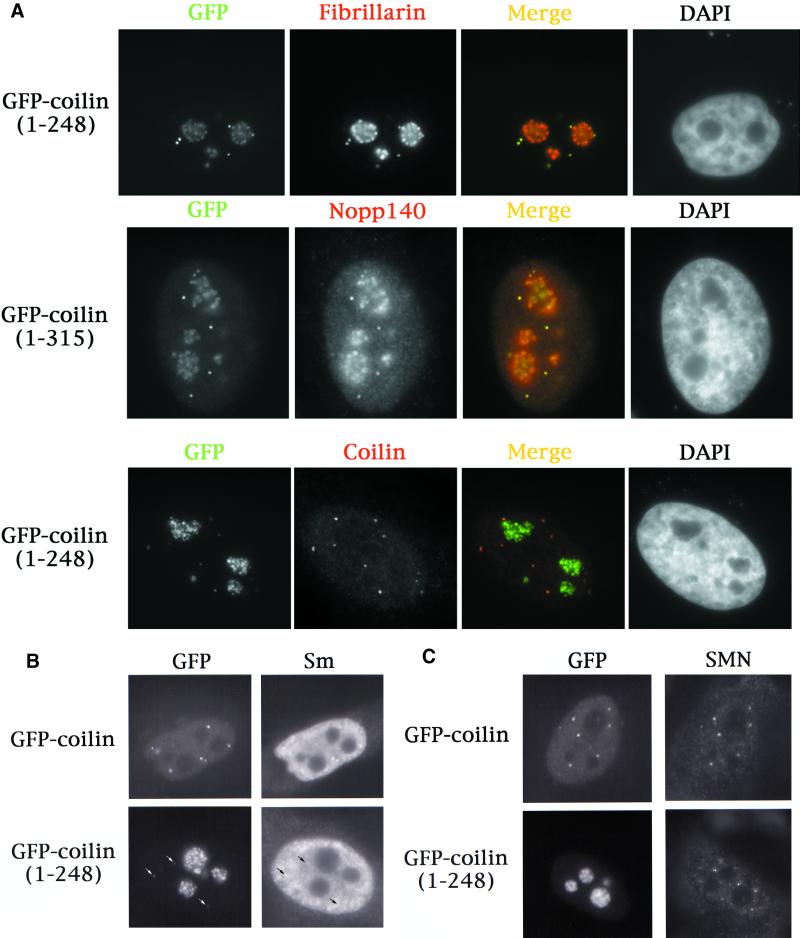
In the course of our studies, we generated two new GFP-coilin truncation mutants that have interesting expression patterns. GFP-coilin(1–248) and GFP-coilin(1–315) localize primarily to the nucleolus, but also accumulate within CBs (Figure 3A). The distribution pattern of the mutant proteins is indistinguishable from those of fibrillarin and Nopp140 (Figure 3A). However, unlike the full-length fusions or N-terminal fragments described above, these constructs do not display such variable phenotypes. Additionally, the nucleolar accumulations are not at all like those observed in high-expressing GFP-coilin cells, which display focal accumulations similar in appearance to fibrillar centers (Figure 2B). Instead, the nucleolar signals in GFP-coilin(1–248) and GFP-coilin(1–315) cells are more reminiscent of the dense fibrillar component (Figure 3A); indeed, the nucleolar distributions of fibrillarin and Nopp140 are virtually identical in transfected and untransfected cells. Strikingly, endogenous coilin staining is undisturbed by expression of these mutants (Figure 3A). Therefore, both truncations properly localize in CBs, with additional concentrations present in the nucleolus.
Figure 3.
Two coilin truncation mutants localize to the nucleolus and CBs, but do not mis-localize endogenous coilin, snRNPs, or SMN to the nucleolus. (A) GFP-coilin(1–248) and GFP-coilin(1–315) mutants, upon transfection into HeLa, were found to have identical nucleolar and CB expression profiles. Antibodies to fibrillarin and Nopp140, which normally localize in nucleoli and CBs, reveal complete colocalization with these proteins. Endogenous coilin localization was determined in GFP-coilin(1–248) transfected cells using anticoilin antibodies that recognize epitopes downstream of the truncation. Note that endogenous coilin is not mislocalized to the nucleolus in GFP-coilin(1–248)transfected cells. (B and C) GFP-coilin(1–248) transfected cells were assessed for the localization of Sm proteins (B) and SMN (C). No nucleolar accumulations of snRNPs or SMN were observed. As expected, accumulations of snRNPs could be detected in the CBs (arrows). Likewise, SMN is present in the CB foci of GFP-coilin(1–248) transfected cells (our unpublished results). However, a small fraction of GFP-coilin(1–248) cells did not display CBs, yet SMN foci, or gems, are present in these cells (C).
To determine if other antigens normally found in CBs are also mislocalized to nucleoli of cells transfected with GFP-coilin(1–248), we tested for the presence of snRNPs (using an antibody to Sm proteins) and SMN. Figure 3, panels B and C, demonstrates that while GFP-coilin(1–248) is found in the nucleolus, these other epitopes are not mislocalized. Localization of snRNPs appears to be normal in GFP-coilin(1–248) transfected cells (Figure 3B). Likewise, SMN shows typical localization in gems and CBs. As shown in Figure 3C, some cells transfected with GFP-coilin(1–248) do not display CBs, while others do (Figure 3, A and B). Nevertheless, it is clear that SMN and Sm proteins are not translocated to the nucleolus along with the mutant coilin.
Identification of a Cryptic Nucleolar Localization Signal in Coilin
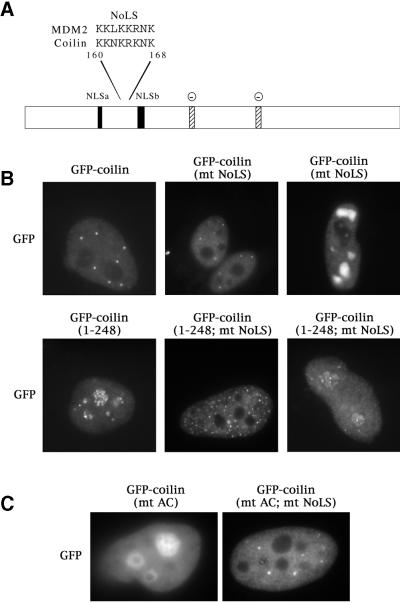
The finding that two new coilin mutations result in the mislocalization of coilin to the nucleolus led us to speculate about the presence of a nucleolar localization signal (NoLS) in coilin. The connection between CBs and the nucleolus is a time-honored one and has been suggested by numerous studies (e.g., Cajal, 1903; Hardin et al., 1969; Lafarga et al., 1983; Malatesta et al., 1994; Bohmann et al. 1995; Isaac et al., 1998). Why do the mutant coilin proteins accumulate in nucleoli? Is there a sequence in coilin that is buried in the normal protein's tertiary structure, but free to interact in the mutants? Recent work by Lohrum et al. (2000) provides a clue. These investigators showed that MDM2, a regulator of p53 function and stability, contains a basic stretch of 8 aa that is required to localize the protein in the nucleolus when stimulated by cell stress (Lohrum et al., 2000). The MDM2 NoLS does not function in unstressed cells but works in concert with its binding partner p14ARF to relocalize both proteins to the nucleolus upon receiving the proper cellular signals (Weber et al., 1999). Thus this sequence is not a constitutive element and has been dubbed a “cryptic” NoLS.
Alignment of the MDM2 NoLS against coilin revealed a nearly identical sequence (Figure 4A). We next mutated the putative coilin NoLS by changing the wild-type basic sequence KKNKRKNK to the neutral IINNIINI. This mutation was generated on both the GFP-coilin and GFP-coilin(1–248) backgrounds. As seen for the MDM2 protein, mutation of the NoLS in the full-length coilin construct did not alter its localization pattern in most transfected cells (Figure 4B, top middle panel). A few cells did have what looked to be large nucleoplasmic inclusions that were not normally observed in cells transfected with GFP-coilin (Figure 4B, top right). Importantly, mutation of the NoLS in the GFP-coilin(1–248) background, in which all transfected cells showed the nucleolar phenotype, resulted in complete exclusion of coilin from the nucleolus (Figure 4B, bottom middle). This pattern was observed in roughly one-third of the cells, however, even cells that retained some nucleolar GFP-coilin(1–248; mt NoLS) signal showed a lower level of nucleolar fluorescence and a correspondingly brighter nucleoplasm (Figure 4B, bottom right). Thus mutation of the NoLS in the GFP-coilin(1–248) or-coilin(1–315) backgrounds results in a decrease, often a dramatic one, in nucleolar coilin accumulation.
Figure 4.
Identification and mutation of a nucleolar localization signal (NoLS) in coilin. (A) Coilin contains a sequence, located between the two putative NLSs, that is nearly identical to the NoLS of MDM2 (Lohrum et al., 2000). Stippled boxes indicate potentially-acidic serine patches (AC) spanning amino acids 242–259 and 312–325. Facing page: (B) The putative coilin NoLS was mutated from KKNKRKNK to IINNIINI (mt NoLS) in both the GFP-coilin and GFP-coilin(1–248) backgrounds. Note that, while the truncation alone localized to nucleoli and CBs, mutation of the NoLS depleted coilin from nucleoli. (C) Similarly, block substitution of the two acidic patches (S245–252A and S314–320A, termed GFP-coilin(mt AC)) also resulted in nucleolar accumulation. Additional localization in CBs was occasionally observed (our unpublished results). However, mutant proteins bearing substitutions in both the NoLS and the acidic patches (GFP-coilin(mt AC; mt NoLS)) did not accumulate in nucleoli, resulting in a distribution pattern indistinguishable from that of wild-type coilin.
We hypothesized that the potentially acidic serine patches, located downstream of the NoLS and deleted by the coilin(1–248) truncation (AC in Figure 4A), might be responsible for sequestering the NoLS in the full-length protein. Consistent with this interpretation, block substitution of the serine patches to alanines resulted in nucleolar localization of coilin (Figure 4C). DAPI and fibrillarin costaining confirmed the nucleolar localization (our unpublished results). Importantly, mutation of the NoLS in the background of the block substitutions released the protein from the nucleolus, resulting in an essentially normal distribution pattern (Figure 4C). Unlike the case with truncated double mutants (e.g., coilin[1–248; mt NoLS]), where only a third of the cells had vacant nucleoli, there was very little variability in the phenotype of the block-substitution double mutant. Thus mutation of the NoLS in the background of the serine substitutions, GFP-coilin(mt AC; mt NoLS), reversed the nucleolar phenotype. Together, these data strongly support the idea that coilin can adopt alternatively folded structures that can interact with nucleolar epitopes.
Mutation of Another Putative Phosphoserine Residue Affects Coilin's Subcellular Localization
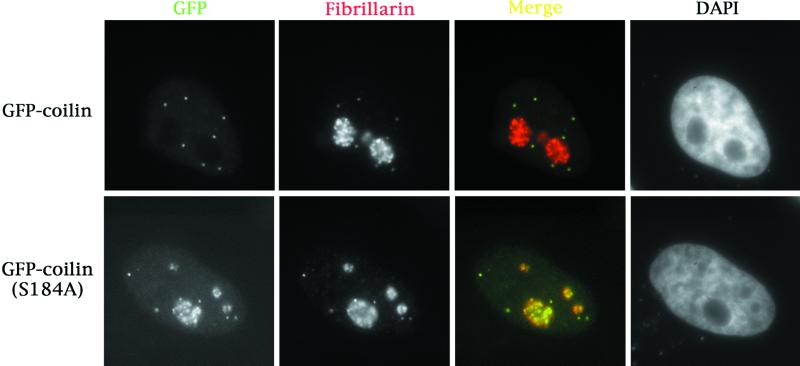
In addition to the serine patches described above, there are numerous other potential phosphorylation sites. In particular, we have recently found that the CDK2/cyclin E complex can phosphorylate coilin in vitro (Liu et al., 2000). Because coilin is only phosphorylated on serines (Carmo-Fonseca et al., 1993), there is but a single consensus CDK2/cyclin E phosphorylation site in coilin, located at serine 184. Mutation of this residue to aspartate (S184D), mimicking a constitutively phosphorylated state, had no affect on the localization of the protein (our unpublished results). However, when serine 184 was mutated to an alanine (S184A), mimicking a dephosphorylated state, a nucleolar mislocalization similar to that of GFP-coilin(1–248) was observed (Figure 5). The localization pattern is identical to that seen in cells transfected with GFP-coilin(1–248) and GFP-coilin(1–315) with one major exception—only around 30% of GFP-coilin(S184A) transfected cells displayed this phenotype; the remaining cells appeared normal. In contrast, 100% of the GFP-coilin(1–248) or GFP-coilin(1–315) showed this nucleolar staining pattern (Figure 3A).
Figure 5.
Mutation of the CDK2/cyclin E phosphorylation site in coilin at S184 to alanine results in nucleolar and CB localization. GFP-coilin(S184A) was transfected into HeLa cells. Immunofluorescence with antibodies to fibrillarin showed complete colocalization with GFP-coilin(S184A). Approximately 30% of transfected cells show this phenotype; the rest of the cells appear normal.
It is possible that the relative expression levels of GFP-coilin(S184A) may alter its localization, as we observed an increase in the number of cells showing the mutant phenotype at longer times posttransfection (our unpublished results). Perhaps cell cycle-specific (de)phosphorylation events effect a conformational change in coilin such that the cryptic NoLS is revealed. Consistent with these observations, mutation of the NoLS in the GFP-coilin(S184A) background reduces the amount of nucleolar staining (our unpublished results). These results demonstrate that the CDK2/cyclin E consensus phosphorylation site at S184 can play a role in the subnuclear localization of coilin.
Coilin Hyperphosphorylation Reduces Its Self-Interaction
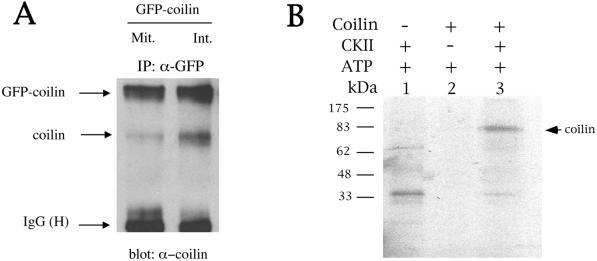
CBs are dynamic organelles; they assemble in mid- to late-G1 and disassemble during M phase of the cell cycle (Andrade et al., 1993). Like many mitotic processes, CB disassembly may be signaled by general protein hyperphosphorylation (Carmo-Fonseca et al., 1993). Thus if coilin self-interaction were important for CB formation, then hyperphosphorylation of coilin during mitosis might plausibly reduce this self-interaction, leading to the disassembly of CBs. We decided to test this hypothesis by a coimmunoprecipitation (coIP) assay using cells transfected with GFP-coilin. CoIPs were conducted on transfected cells that had been either untreated or treated with nocodazole, which enriches cells for hyperphosphorylated coilin (Andrade et al., 1993). As shown in Figure 6A, comparison of the amount of coilin immunoprecipitated in treated versus nontreated cells reveals that there is a reduction in the amount of endogenous coilin immunoprecipitated in mitotic versus interphase lysates. Thus, the increase in coilin phosphorylation correlates with a reduction in coilin self-interaction.
Figure 6.
(A) Hyperphosphorylation of coilin reduces coilin self-interaction. GFP-coilin was transfected into HeLa and cells were treated with nocodazole to enrich for mitotic cells (Mit.) or left untreated for interphase cells (Int.). Extracts were subjected to immunoprecipitation with anti-GFP antibodies, followed by SDS-PAGE and western blotting with antibodies against coilin. Equivalent amounts of protein were used for each reaction. (B) Coilin was phosphorylated by casein kinase 2 in vitro. Reactions were conducted as described in the MATERIALS AND METHODS section. Phosphorylation of coilin was dependent on the presence of casein kinase 2 (CKII). Radiolabeled bands present in lane 1 and below the coilin radioactive band in lane 3 are most likely CKII autophosphorylation products.
What kinases are responsible for coilin (hyper)phosphorylation? We previously demonstrated that CDK2/cyclin E may be responsible for the phosphorylation of coilin at one site (S184), and mutation of this residue to an alanine redistributes coilin to the nucleolus (Figure 5). However, in separate experiments, we did not observe any changes in the amount of endogenous coilin coimmunoprecipitated in cells transfected with coilin mutants S184A or S184D (our unpublished results). Furthermore, no significant changes in the coimmunoprecipitation profiles of endogenous coilin were observed from cell lysates transfected with coilin that had 4 serines mutated to alanines or aspartates (S184, S202, S248, S315). We do not currently know which coilin serine residues are phosphorylated in vivo, although labeling experiments reveal ∼12–15 phosphoserine positive tryptic peptides in interphase, and two more in mitosis (Carmo-Fonseca et al., 1993). Therefore, it is likely that additional residues need to be phosphorylated in order to account for the observed reduction in coilin self-interaction from mitotic cell lysates. Clearly, this will be a subject of future investigation.
However, one candidate coilin kinase is casein kinase 2 (CK2). Isaac et al. (1998) have shown that Nopp140 interacts with coilin in vitro and by the yeast dihybrid system. Other experiments reveal that Nopp140 tightly interacts with CK2 (Meier, 1996). Because CDK2/cyclin E has only one potential phosphorylation site in coilin (S184), we wanted to test if CK2 could phosphorylate coilin in vitro. There are numerous consensus CK2 phosphorylation sites (S/TXXD/E) present in coilin, including several within the serine patches shown in Figure 4A. As Figure 6B demonstrates, CK2 can indeed phosphorylate coilin in vitro, thus providing a mechanism by which coilin function may be modulated by interaction with Nopp140 and CK2.
DISCUSSION
We have found that coilin is a self-interacting protein, and we mapped the responsible domain to the coilin N-terminus. Together with an exogenously added NLS, this self-interaction domain is necessary and sufficient for localization to CBs in HeLa cells. Overexpression of various wild-type and mutant coilin constructs results in the disruption of CBs and SMN gems, but not PML bodies. Mitotic hyperphosphorylation of coilin, which coincides with CB disassembly, was correlated with a reduction in coilin self-interaction.
Additionally, we have identified two truncation mutants that indicate the presence of a cryptic nucleolar localization signal (NoLS) within the coilin protein. These mutants localize to both CBs as well as the nucleolus and provide further evidence for a functional link between these two nuclear suborganelles. Mutation of the NoLS in the nucleolar background, but not the wild-type, resulted in a dramatic reduction of nucleolar accumulation. A point mutation in the consensus CDK2-cyclin E phosphorylation site in coilin (S184A) results in the same phenotype as observed for the truncations, suggesting that the coilin NoLS is exposed in specific coilin phospho-isoforms. Similarly, block substitution of two serine-rich patches near the middle of the protein also resulted in nucleolar accumulation. Therefore, coilin contains a cryptic NoLS, which may function in response to specific cellular signals that are regulated by phosphorylation. Other kinases that may be responsible for coilin phosphorylation include casein kinase 2 (CK2), which interacts with the nucleolar shuttling protein Nopp140. We showed that coilin can be phosphorylated by CK2 in vitro. Since Nopp140 also interacts with coilin (Isaac et al., 1998), these data provide a mechanism by which coilin localization and function may be modulated by binding to Nopp140 and CK2.
Mutational Analysis of Coilin Strengthens the Link between CBs and Nucleoli
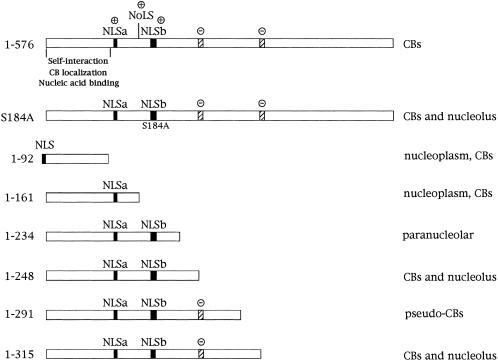
The nucleolus and Cajal body share many protein constituents, such as Nopp140, GAR1, and fibrillarin. Nopp140 is interesting in this regard because it has been shown to interact with coilin by two-hybrid and coimmunoprecipitation assays (Isaac et al., 1998) and to shuttle between the cytoplasm, the nucleolus, and the CB (Meier and Blobel, 1992). Furthermore, mutations of Nopp140 affect coilin localization and CB formation in a dominant negative manner (Isaac et al., 1998). We have found that expression of two coilin truncations, 1–248 and 1–315, results in the complete colocalization of the mutant proteins with fibrillarin and Nopp140 in the nucleoli and CBs (Figure 3A). Unlike the overexpression of full-length GFP-coilin, which disassembles CBs and SMN gems (Figure 2B), GFP-coilin(1–248) and -coilin(1–315) do not have effects on localization of the endogenous coilin protein, SMN or Sm snRNPs (Figures 3B, 3C). Together with two additional coilin truncation mutants, myc-coilin(1–234) and -coilin(1–291), described by Bohmann et al. (1995), our data reveal that the region between amino acids 234 and 315 is very sensitive to mutation, perhaps reflecting differential binding affinities to Nopp140. As summarized in Figure 7, there is a striking difference in the phenotypes of coilin(1–234) and coilin(1–248); when coilin is truncated just 14 aa upstream of position 248, the mutant protein is localized in a ring around the outer edge of the nucleolus. No internal nucleolar fluorescence is detected (Bohmann et al., 1995 and our unpublished results). Another interesting pair of mutations are coilin(1–291) and coilin(1–315). Both constructs colocalize completely with Nopp140, however, while coilin(1–315) accumulates in both CBs and nucleoli, coilin(1–291) expression gives rise to structures called pseudoCBs (Bohmann et al., 1995). PseudoCBs resemble large, electron-dense inclusions and have a very distinct morphology from either CBs or nucleoli. In fact, coilin(1–291) expression tends to remove Nopp140 from the nucleoli, which remain positively stained by fibrillarin (Bohmann et al., 1995). Taken together, these results point to the importance of the region between amino acids 234 and 315 of the coilin protein for its subnuclear localization. However, as discussed below, we do not believe that this region functions in a positive manner but may instead be important for the proper folding of the protein, exposing or sequestering regions that interact with nucleolar factors. This idea is further supported by our findings that point mutations outside of this domain (e.g., S184A) also display the same nucleolar and CB phenotype (Figure 7).
Figure 7.
Coilin mutational summary. The localization of full-length coilin (1–576) and coilin mutants generated in this study are described. The putative coilin nucleolar localization signal (NoLS), nuclear localization signals (NLS, dark boxes), self-interaction, CB localization, and nucleic acid binding domains are indicated. The NLS in the coilin(1–92) construct was added exogneously, along with GFP- and myc-tags. Positive signs (+) denote basic regions of coilin, whereas negative signs (−) indicate potentially-acidic serine patches from amino acids 242–259 and 312–325 (stippled boxes). Two additional coilin mutants, myc-coilin(1–234) and myc-coilin(1–291) were generated by Bohmann et al. (1995).
Does Coilin Traffic through the Nucleolus?
Studies linking CBs and nucleoli (Malatesta et al., 1994; Bohmann et al., 1995; Lyon et al., 1997; Isaac et al., 1998; Sleeman et al., 1998) raise the possibility that coilin transits through the nucleolus during the protein's normal life cycle. The identification of a cryptic NoLS within coilin, nearly identical to that of the MDM2 protein (Lohrum et al., 2000), further supports this idea. The coilin NoLS (Figure 7) lies within a highly basic region of the molecule that spans residues 107–198. Just as in the case of MDM2, mutation of the NoLS in the wild-type background has no effect on the localization of coilin. The MDM2 protein is translocated to the nucleolus only when cells are stressed (Lohrum et al., 2000). We simply have yet to identify the cellular signals that naturally cover/uncover the coilin NoLS. However, also like MDM2, mutations such as coilin(1–248) can cause the protein to localize in the nucleolus (Figure 3A). Double mutants, such as coilin(1–248; mt NoLS), result in a dramatic reduction of the nucleolar signal (Figure 4B).
We therefore hypothesized that the potentially acidic serine patches surrounding positions 248 and 315 of coilin (Figure 7) can form electrostatic interactions with the basic residues surrounding the NoLS. Consistent with this idea, block substitution mutants (S245–252A and S314–320A, respectively) that cannot form these putative electrostatic bonds also accumulate in nucleoli (Figure 4C). Mutation of the NoLS, in addition to the block substitutions, restored the localization to CBs (Figure 4C). Thus the serine patches are likely to be important for folding of the coilin protein to either expose or conceal the basic residues surrounding the NoLS. Similar intramolecular interactions between basic and acidic patches have been important for function of the retinoblastoma protein (Harbor et al., 1999). In the case of coilin, such interactions may play a role in the release from binding to Nopp140, which is thought to bind in the basic region of coilin (Isaac et al., 1998, and our unpublished results). This speculation is supported by the finding that coilin truncations (1–248) and (1–315) colocalize with Nopp140. Furthermore, this same phenotype is observed in coilin(S184A). Thus it is plausible that phosphorylation controls aspects of coilin function by altering its folding and changing its subcellular localization.
Self-Interaction and Nuclear Body Formation
We have seen that changes in the phosphorylation status of coilin are important molecular switches that can govern the subcellular localization of the protein (Figure 5) as well as its self-interaction (Figure 6A). We have mapped the self-interaction domain to the coilin amino terminus (Figures 1 and 7). In agreement with Wu et al. (1994), our data reveal that the N-terminal 92 amino acids of coilin are required for localization of the protein to CBs (Figure 7). It should be noted that, while most cells transfected with GFP-coilin(1–161), myc-coilin(1–121), or GFP-myc-NLS-coilin(1–92) constructs showed nucleoplasmic labeling without detectable coilin foci, some cells did display CBs (Figure 2A). We cannot, at present, exclude the possibility that endogenous coilin is required for these N-terminal constructs to localize within CBs. However, it is clear that high levels of expression of the coilin N-terminal fragments resulted in a significant decrease in the amount of endogenous coilin in the CB compartment (Figure 2A). We also observed that high levels of full-length coilin expression disrupted endogenous CBs. We speculate that high levels of ectopically expressed coilin alter CBs due to a titration of other factors, such as kinases or phosphatases, which may facilitate CB formation.
The Cajal body is one of several different nuclear body subtypes present in the interphase mammalian nucleus (reviewed in Matera, 1999). Interestingly, each of these other nuclear bodies contain marker proteins that also self-associate—Sam68, (Chen et al., 1999), PML (Ishov et al., 1999), and SMN (Lorson et al., 1998). In the case of SMN gems, self-oligomerization is the key to creation of a high-affinity binding site for the Sm proteins (Pellizzoni et al., 1999). Notably, the self-association domain of SMN also overlaps with regions of the protein shown to interact with the E2 transcriptional activator (Strasswimmer et al., 1999) and profilin (Giesemann et al., 1999). The self-interaction domain of coilin also has other activities, including nucleic acid binding (Bellini and Gall, 1998) and CB localization (Wu et al., 1994; this work). It is possible that these overlapping activities may be related or even antagonistic. In the case of Cajal bodies, we do not actually know if the coilin molecules that self-associate are in the nucleoplasm, the CBs, or both compartments; additional experiments will be required.
Thus formation of nuclear bodies may well be dependent on marker protein self-association. Such interactions may provide a scaffold upon which other components of the nuclear body can then coalesce. Recent work with PML knockout cells showed that PML bodies require the PML protein in order to form (Ishov et al., 1999; Zhong et al., 2000). Modification of PML by SUMO-1 is important for PML body assembly (Zhong et al., 2000), but it remains unclear whether PML self-association is also required. Thus, in the same way that PML is important for PML body formation, coilin may be required for Cajal body formation. Alternatively, coilin may simply be a passenger protein, accumulating in CBs by virtue of interaction with other cellular factors. We are currently generating a genetic model system, lacking endogenous coilin, to test this hypothesis.
ACKNOWLEDGMENTS
We thank M. Frey, P. Szymczyk, and J. Ospina for scientific discussions and for help with some of the subcloning. We also thank the following groups for generously providing reagents used in this study: A. Lamond (University of Dundee, Dundee, UK) for coilin mutant constructs; E. Chan (The Scripps Research Institute, La Jolla, California) for anticoilin and antifibrillarin antibodies; T. Meier (Albert Einstein College of Medicine of Yeshiva University, Bronx, New York) for anti-Nopp140; J. Steitz (Yale University, New Haven, CT) for anti-Sm; F. Dragon (Friedrich Miescher Institute, Basel, Switzerland) for anti-GAR1; G. Dreyfuss (University of Pennsylvania, Philadelphia, Pennsylvania) for anti-SMN; and L. de Jong (E.C. Slater Institute, Amsterdam) for anti-PML. M.D.H. was supported in part by NIH Postdoctoral Training Grant T32-HD07518. This work was supported by grants to A.G.M. from the NIH (GM-53034) and from the Muscular Dystrophy Association.
REFERENCES
- Andrade LEC, Chan EKL, Raska I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: Immunological characterization and cDNA cloning of p80 coilin. J Exp Med. 1991;173:1407–1419. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade LEC, Tan EM, Chan EKL. Immunocytochemical analysis of the coiled body in the cell cycle and during cell proliferation. Proc Natl Acad Sci USA. 1993;90:1947–1951. doi: 10.1073/pnas.90.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini M, Gall JG. Coilin can form a complex with the U7 small nuclear ribonucleoprotein. Mol Biol Cell. 1998;9:2987–3001. doi: 10.1091/mbc.9.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann K, Ferreira J, Lamond A. Mutational analysis of p80 coilin indicates a functional interaction between coiled bodies and the nucleolus. J Cell Biol. 1995;131:817–831. doi: 10.1083/jcb.131.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajal SRy. Un sencillo metodo de coloracion selectiva del reticulo protoplasmico y sus efectos en los diversos organos nerviosos de vertebrados y invertebrados. Trab Lab Invest Biol (Madrid) 1903;2:129–221. [Google Scholar]
- Carmo-Fonseca M, Ferreira J, Lamond AI. Assembly of snRNP-containing Coiled Bodies Is Regulated in Interphase and Mitosis - Evidence that the Coiled Body Is a Kinetic Nuclear Structure. J Cell Biol. 1993;120:841–852. doi: 10.1083/jcb.120.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho T, Almeida F, Calapez A, Lafarga M, Berciano MT, Carmo-Fonseca M. The spinal muscular atrophy disease gene product, SMN: A link between snRNP biogenesis and the Cajal (coiled) body. J Cell Biol. 1999;147:715–728. doi: 10.1083/jcb.147.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EKL, Takano S, Andrade LEC, Hamel JC, Matera AG. Structure, expression and chromosomal localization of the human p80-coilin gene. Nucleic Acids Res. 1994;22:4462–4469. doi: 10.1093/nar/22.21.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charroux B, Pellizzoni L, Perkinson RA, Shevchenko A, Mann M, Dreyfuss G. Gemin3: A novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J Cell Biol. 1999;147:1181–1194. doi: 10.1083/jcb.147.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charroux B, Pellizzoni L, Perkinson RA, Yong J, Shevchenko A, Mann M, Dreyfuss G. Gemin4: A novel component of the SMN complex that is found in both gems and nucleoli. J Cell Biol. 2000;148:1177–1186. doi: 10.1083/jcb.148.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Boisvert FM, Bazett-Jones DP, Richard S. A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines. Mol Biol Cell. 1999;10:3015–3033. doi: 10.1091/mbc.10.9.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase cells. Proc Natl Acad Sci USA. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesemann T, Rathke-Hartlieb S, Rothkegel M, Bartsch JW, Buchmeier S, Jockusch BM, Jockusch H. A role for polyproline motifs in the spinal muscular atrophy protein SMN. Profilins bind to and colocalize with SMN in nuclear gems. J Biol Chem. 1999;274:37908–37914. doi: 10.1074/jbc.274.53.37908. [DOI] [PubMed] [Google Scholar]
- Harbor JW, Luo RX, Dei Santi A, Postigo AA, Dean DC. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98:859–869. doi: 10.1016/s0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- Hardin JH, Spicer SS, Greene WB. The paranucleolar structure, accessory body of Cajal, sex chromatin and related structures in nuclei of rat trigeminal neurons: a cytochemical and ultrastructural study. Anat Rec. 1969;164:403–432. doi: 10.1002/ar.1091640403. [DOI] [PubMed] [Google Scholar]
- Horton LE, Templeton DJ. The cyclin box and C-terminus of cyclins A and E specify CDK activation and substrate specificity. Oncogene. 1997;14:491–498. doi: 10.1038/sj.onc.1200851. [DOI] [PubMed] [Google Scholar]
- Isaac C, Yang Y, Meier UT. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J Cell Biol. 1998;142:407–417. doi: 10.1083/jcb.142.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss J F, 3rd, Maul GG. PML Is Critical for ND10 Formation and Recruits the PML-interacting Protein Daxx to this Nuclear Structure When Modified by SUMO-1. J Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafarga M, Hervas JP, Santa-Cruz MC, Villegas J, Crespo D. The “acccessory body”of Cajal in the neuronal nucleus: a light and electron microscopic approach Anat. Embryol. 1983;166:19–30. doi: 10.1007/BF00317942. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, Le Pasilier D, Frezal J, Cohen D, Weissenbach J, Munnich A, Melki J. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA, Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular components: Probes for molecular biology and autoimmune disease. Proc Natl Acad Sci USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JL, Hebert MD, Ye Y, Kung HJ, Matera AG. Cell cycle-dependent localization of the CDK2-cyclin E complex in Cajal (coiled) bodies. J Cell Sci. 2000;113:1543–1552. doi: 10.1242/jcs.113.9.1543. [DOI] [PubMed] [Google Scholar]
- Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- Lohrum MA, Ashcroft M, Kubbutat MH, Vousden KH. Identification of a cryptic nucleolar-localization signal in MDM2. Nat Cell Biol. 2000;2:179–181. doi: 10.1038/35004057. [DOI] [PubMed] [Google Scholar]
- Lorson CL, Strasswimmer J, Yao JM, Baleja JD, Hahnen E, Wirth B, Le T, Burghes AH, Androphy EJ. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nature Genet. 1998;19:63–66. doi: 10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- Lyon CE, Bohmann K, Sleeman J, Lamond AI. Inhibition of protein dephosphorylation results in the accumulation of splicing snRNPs and coiled bodies within the nucleolus. Exp Cell Res. 1997;230:84–93. doi: 10.1006/excr.1996.3380. [DOI] [PubMed] [Google Scholar]
- Malatesta M, Zancanaro C, Martin T, Chan E, Amalric FL, R, Vogel P, Fakan S. Is the coiled body involved in nucleolar functions? Exp Cell Res. 1994;211:415–419. doi: 10.1006/excr.1994.1106. [DOI] [PubMed] [Google Scholar]
- Matera AG. Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol. 1999;9:302–309. doi: 10.1016/s0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- Matera AG, Frey MR. Coiled bodies and gems: Janus or Gemini? Am J Hum Genet. 1998;63:317–321. doi: 10.1086/301992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier UT. Comparison of the rat nucleolar protein nopp140 with its yeast homolog SRP40. Differential phosphorylation in vertebrates and yeast. J Biol Chem. 1996;271:19376–19384. [PubMed] [Google Scholar]
- Meier UT, Blobel G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell. 1992;70:127–138. doi: 10.1016/0092-8674(92)90539-o. [DOI] [PubMed] [Google Scholar]
- Misteli T, Spector DL. The cellular organization of gene expression. Curr Opin Cell Biol. 1998;10:323–331. doi: 10.1016/s0955-0674(98)80007-0. [DOI] [PubMed] [Google Scholar]
- Moss B, Elroy-Stein O, Mizukami T, Alexander WA, Fuerst TR. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Charroux B, Dreyfuss G. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc Natl Acad Sci USA. 1999;96:11167–11172. doi: 10.1073/pnas.96.20.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I, Andrade LEC, Ochs RL, Chan EKL, Chang C-M, Roos G, Tan EM. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp Cell Res. 1991;195:27–37. doi: 10.1016/0014-4827(91)90496-h. [DOI] [PubMed] [Google Scholar]
- Sleeman J, Lyon CE, Platani M, Kreivi J-P, Lamond AI. Dynamic interactions between splicing snRNPs, coiled bodies and nucleoli revealed using snRNP protein fusions to the green fluorescent protein. Exp Cell Res. 1998;243:290–304. doi: 10.1006/excr.1998.4135. [DOI] [PubMed] [Google Scholar]
- Strasswimmer J, Lorson CL, Breiding DE, Chen JJ, Le T, Burghes AH, Androphy EJ. Identification of survival motor neuron as a transcriptional activator- binding protein. Hum Mol Genet. 1999;8:1219–1226. doi: 10.1093/hmg/8.7.1219. [DOI] [PubMed] [Google Scholar]
- Stuurman N, de Graaf A, Floore A, Josso A, Humbel B, de Jong L, van Driel R. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J Cell Sci. 1992;101:773–784. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- Wu Z, Murphy C, Gall JG. Human p80-coilin is targeted to sphere organelles in the amphibian germinal vesicle. Mol Biol Cell. 1994;5:1119–1127. doi: 10.1091/mbc.5.10.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young PJ, Le TT, thi Man N, Burghes AH, Morris GE. The relationship between SMN, the spinal muscular atrophy protein, and nuclear coiled bodies in differentiated tissues and cultured cells. Exp Cell Res. 2000;256:365–374. doi: 10.1006/excr.2000.4858. [DOI] [PubMed] [Google Scholar]
- Zhong S, Muller S, Ronchetti S, Freemont PS, Dejean A, Pandolfi PP. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95:2748–2752. [PubMed] [Google Scholar]