Abstract
The purified recombinant African swine fever virus polyprotein processing protease cleaves the two GG-X sites in polyprotein pp62 with the same efficiency. Cleavage at the site that is first recognized in vivo is not a requisite for cleavage at the second site, suggesting the existence of mechanisms that control the ordered processing of the polyprotein during infection.
African swine fever virus (ASFV) is the causative agent of a severe hemorrhagic disease of domestic swine. It is a large double-stranded DNA icosahedral virus that infects macrophages/monocytes of swine as well as ticks of the Ornithodoros genus. Its genome is about 170 to 190 kbp, depending on the strain, and it contains more than 150 potential genes (12). ASFV replicates mainly in the cytoplasm of the infected cell, where large viral factories containing assembling particles are formed. The viral particle contains more than 30 polypeptides that are organized into several concentric domains, which are, from the inside to the outside of the virion, the DNA-containing nucleoid, the core shell, the endoplasmic reticulum-derived inner envelope, the capsid, and the outer envelope, which is acquired from the plasma membrane during virus budding. ASFV has been shown to encode two polyproteins named pp220 and pp62 that are sequentially processed during infection to produce six different structural proteins (13, 14). These are located at the core shell, the domain that bridges the nucleoid to the inner envelope. Altogether the processing products of both polyproteins account for more than 30% of the total virion protein mass, being present at an equimolecular stoichiometry (1, 3). Processing of polyproteins pp220 and pp62 takes place after the second glycine in the consensus motif GGX (9) and is catalyzed by the recently identified ASFV protease, protein pS273R, that belongs to the family of SUMO-1-processing cysteine proteinases (2). This family includes an increasing number of cellular cysteine proteases, along with the adenovirus protease and the vaccinia virus protein encoded by gene I7 (8). All of them are characterized by the presence of a conserved domain of about 90 amino acids that includes the catalytic triad formed, in the case of the ASFV protease, by H168, N187, and C232.
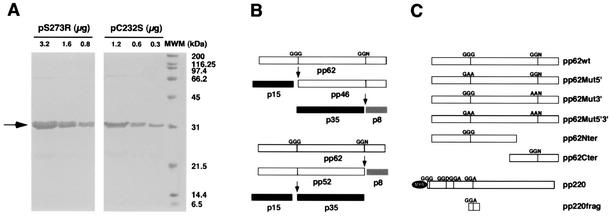
Previous work had shown that extracts containing the ASFV pS273R protein were able to correctly process polyproteins pp220 and pp62 in vitro (2). In this study we have expressed and purified the recombinant ASFV protease in order to characterize its activity in greater detail. Briefly, we have cloned the S273R gene encoding the ASFV protease into the pGEX2T vector (Amersham-Pharmacia Biotech). The recombinant glutathione S-transferase fusion protein was expressed in Escherichia coli with a yield of approximately 60 mg/liter of culture, more than 70% of which being found in a soluble form. A soluble extract of transformed bacteria was bound to glutathione S-transferase beads, and after extensive washing with phosphate-buffered saline the bound protein was processed in batch with bovine plasma thrombin. The processed recombinant protein pS273R was purified away from thrombin by using p-aminobenzamidine beads. A recombinant single-residue mutant designated protein pC232S that corresponds to a catalytically inactive form of the protease (2) was obtained in a similar way. The purity of the final protease preparations was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie blue staining (Fig. 1A) and was found to be greater than 80% for pC232S and 90% for pS273R.
FIG. 1.
(A) Analysis by PAGE of purified ASFV recombinant protease. The indicated amounts of the purified ASFV protease pS273R and the catalytic mutant pC232S were analyzed by SDS-12% PAGE and were stained with Coomassie blue. MWM, molecular size markers. (B) Representation of the processing pathway of ASFV polyprotein pp62. The upper scheme depicts the favored pathway during ASFV infection, where the first site to be cleaved is the one located closer to the N terminus of the precursor polyprotein, giving rise to protein p15 and the intermediate precursor pp46. The latter is subsequently cleaved, producing p35 and a polypeptide of about 8 kDa. The lower scheme represents a secondary pathway where the site located closer to the C terminus is first processed, releasing the p8 polypeptide along with the pp52 precursor. The pp52 protein would then be processed to produce p35 and p15. (C) Representation of the different pp62- and pp220-derived constructs used in the in vitro processing experiments. The mutations introduced at the processing sites are indicated for each case. The N-terminal fragment of pp62 spans residues M1 to L385, and the C-terminal fragment spans residues M286 to E530. The pp220-derived fragment contains the first cleavage site of pp220 used during infection and spans residues A820 to E1044 of the complete precursor.
In order to analyze the activity of the recombinant ASFV protease, we used in vitro-translated proteins that correspond to pp62- and pp220-derived constructs as substrates (Fig. 1C). Polyprotein pp62 contains two processing sites, which are sequentially cleaved during infection (14). Thus, the site located closer to the N terminus is cleaved first, releasing the final product p15 along with an intermediate precursor termed pp46. The latter is then cleaved at the second site to produce p35 and an 8-kDa polypeptide that has not been detected in infected cells. An alternative pathway could use the sites in a reverse order, releasing in the first place the p8 product and a precursor of 52 kDa that would finally be cleaved into p35 and p15. This second pathway has only been observed in in vitro reactions with extracts of transfected cells (2), and its relevance during infection has not been established. Both pathways are summarized in Fig. 1B.
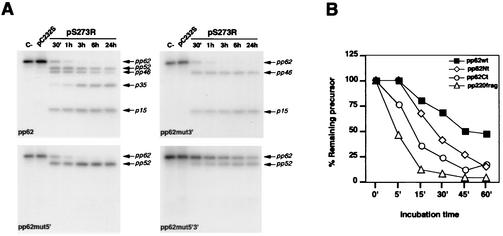
We first analyzed the in vitro processing of four different constructs corresponding to the complete polyprotein pp62 or mutated forms thereof (Fig. 1C). Thus, a complete, wild-type pp62, a pp62 with G158 and G159 (at the first cleavage site) mutated to alanine, a pp62 with G462 and G463 (at the second cleavage site) mutated to alanine, and a pp62 containing all four mutations were translated in vitro in the presence of [35S]-Met, Cys and were incubated with 0.65 μg of pS273R or pC232S/ml for different time periods at 30°C in 50 mM Tris-HCl, pH 7.5. The samples were analyzed on 20 to 10% gradient polyacrylamide gels followed by fluorography, and the autoradiograms were scanned by using a GS-710 Bio-Rad densitometer. As shown in Fig. 2A, the recombinant ASFV pS273R was able to correctly process the translated full-length pp62, as judged by the appearance of the final products p35 and p15, while the mutant protease was inactive. These results confirm that the ASFV pS273R protein is the polyprotein processing protease and show that in vitro the protease is active in the absence of any additional viral factor. After a 30-min incubation considerably more p15 than p35 is detected, as expected from the cleavage at the first site. However, the detection of the pp52 intermediate at this time indicates that the alternate pathway is also used under these conditions.
FIG. 2.
(A) In vitro processing of wild-type or mutated polyprotein pp62 forms by the ASFV protease. As a control, the translated products were incubated in the absence of protease (C−) or in the presence of the catalytically inactive protease (pC232S) for 1 h. In incubations for up to 24 h with the mutant protease, no processing of the polyprotein was observed (data not shown). The reactions were carried out for the indicated times at 30°C. The intermediate and final processing products are indicated. (B) Representation of the percentage of unprocessed precursor at the time points indicated. The pp62- and pp220-derived fragments containing only one cleavage site were compared to the full-length polyprotein pp62.
Under the same reaction conditions, mutation of the first processing site (Fig. 2A, pp62mut5′ panel) completely abrogated its cleavage, while the second site could still be efficiently used, as judged by the appearance of the pp52 intermediate. Thus, at 30 min, 70% of the precursor had been processed. This result indicates that, under these conditions, processing of the first cleavage site of polyprotein pp62 is not a necessary requisite for recognition of the second one. Similarly, mutation of the second site (Fig. 2A, pp62mut3′ panel) completely prevented its processing while the first site was still used, as the appearance of p15 and pp46 demonstrates. A densitometric analysis showed that, at 30 min, 80% of the precursor was already processed. These results confirm that in vitro the protease cleaves both sites in polyprotein pp62 with a similar efficiency, in contrast to the preferential use of the first site during the infection, which may suggest the existence in vivo of mechanisms that control the ordered processing of the polyprotein. However, the possibility that an altered folding of the polyprotein substrate produced in the in vitro translation system might affect cleavage site utilization by the protease cannot be completely ruled out.
Surprisingly, when the double mutant pp62 was assayed, a certain degree of processing into pp52 was detected. After 3 h, 30% of the precursor had been processed, a percentage that increased to 51% at 24 h. No further processing into the final products p35 and p15 was detected. Thus, the GG-X motif proposed as a recognition site for ASFV protease is not an absolute requisite for processing, at least under in vitro conditions. In relation to this, it should be noted that the substrate specificity of adenovirus protease has been defined as (M,L,I)XGG-X and (M,L,I)XGX-G (18) and that proteolytic processing of vaccinia virus core precursors takes place at AG-X motifs (17). Further studies will be needed to determine the precise sequence requirements for ASFV protease-mediated processing.
As described above, the mutated second processing site is to some extent accessible to cleavage when present on an intact pp62 polyprotein, while when exposed to the protease on the intermediate pp46 protein it is not cleaved. This result may be interpreted as the consequence of a conformational switch after cleavage at the first site in polyprotein pp62. If such a conformational switch also occurs during the proteolytic processing in infected cells, the mature products might have a conformation different from that of the corresponding regions in the precursor, which could be important for their subsequent interaction with additional proteins during virus assembly.
In order to study the activity of pS273R in greater detail, we decided to use as substrate smaller constructs derived from both polyproteins (see Fig. 1C). Thus, an N-terminal fragment of polyprotein pp62 containing the first cleavage site, a C-terminal fragment of pp62 containing the second cleavage site and a pp220-derived fragment containing the first cleavage site used in this polyprotein (13), as well as the complete polyprotein pp62 were translated in vitro and incubated as before with pS273R (Fig. 2B). The percentage of remaining precursor after the indicated reaction times was determined as a measure of the activity of the ASFV protease on the different substrates. The two pp62 fragments were processed more efficiently than the full-length pp62, which might be due to a greater conformational flexibility of the smaller proteins. On the other hand, the pp220 fragment, with more than 50% processed after the 5-min incubation, was the most efficiently cleaved substrate. At the same time point, about 25% of the C-terminal pp62 fragment was processed, while the N-terminal pp62 fragment and the full-length precursor remained essentially intact. Therefore, the C-terminal and the pp220-derived fragments were selected as substrates for subsequent analyses.
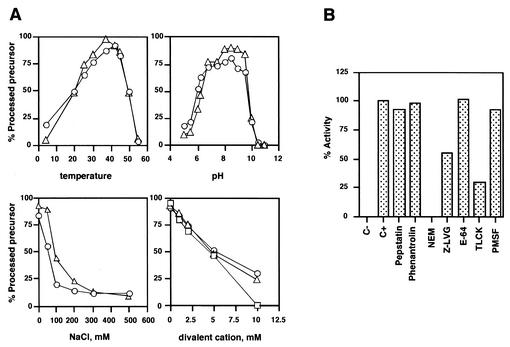
First we determined the pH, temperature, ionic strength, and divalent cation dependence of the ASFV protease activity. As shown in Fig. 3A, the ASFV protease was active over the pH 6.0 to 9.5 range, with the highest activities obtained around pH 8.5. The activity of the ASFV protease increased with increasing temperature over the range from 4 to 42°C, the highest activities being obtained around 37°C. The presence of NaCl in the reaction mixture was found to strongly inhibit pS273R activity at concentrations of 100 mM and higher. A similar inhibitory effect was obtained with KCl (data not shown). The divalent cations Mg2+, Mn2+, and Ca2+ inhibited the protease at all concentrations tested, 50% inhibition being obtained at 5 mM concentration (Fig. 3A). A greater effect was found in the case of Zn2+, which completely inhibited the protease activity at a concentration of 1 mM (data not shown). Additionally, we performed an inhibitor profiling with inhibitors of different protease classes. N-ethylmaleimide, a typical cysteine reactive compound; the LVG diazomethane, a cysteine protease inhibitor; and Nα-p-tosyl-l-lysine chloromethyl ketone, an inhibitor of trypsin-like serine proteases that also inhibits some cysteine proteases efficiently blocked pS273R-mediated proteolysis. The other inhibitors assayed did not affect protease activity, in accordance with previous results obtained by using extracts of cells expressing the protease (2). Surprisingly, the typical cysteine protease inhibitor E-64 was not able to block ASFV protease activity, as has also been described for the adenovirus protease (15, 16). This might reflect a common structural feature of this new family of SUMO-1-processing proteases. It remains to be addressed whether other members of this group are susceptible or not to E-64 action. We also found that the decapeptide MILGGADELE, which corresponds to the sequence encompassing the first processing site of polyprotein pp220, was able to inhibit the processing of the pp220 fragment by 52% at a 1 mM concentration. This effect was specific, since a control peptide with the same amino acid composition but with a randomized sequence had no effect on the protease at 1 mM concentration. This result raises the possibility of using small peptide derivatives as inhibitors of the protease activity in vitro and in vivo.
FIG. 3.
(A) pH, temperature, ionic strength, and divalent cation dependence of ASFV protease activity. To study pH dependence, the following buffers were used: 50 mM sodium acetate at pH 5.0, 5.5, and 6.0; 50 mM PIPES at pH 6.2 and 6.8; 50 mM Tris-HCl at pH 7.5, 8.0, and 8.5; 50 mM glycine-NaOH at pH 9.0, 9.5, 10.0, 10.5, and 11.0. The results for the pp62Ct (circles) and pp220frag (triangles) constructs are shown. The following divalent cations were tested with the pp220frag substrate: MgCl2 (triangles), MnCl2 (circles), and CaCl2 (squares). Controls in the presence of pC232S were included for all the conditions assayed. (B) Effect of protease inhibitors on ASFV protease activity. The inhibitors were freshly prepared and added at the following concentrations: pepstatin, 10 μM; phenanthroline, 1 mM; phenylmethylsulfonyl fluoride (PMSF), 1 mM; E-64, 100 μM; Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK), 100 μM; Z-LVG-CHN2, 100 μM; N-ethylmaleimide (NEM), 2.5 mM. They were assayed on full-length pp62 and the pp220 fragment. Activity of 100% corresponded to the processing obtained in the absence of inhibitors for each construct. The data presented are the means of two independent experiments performed on both substrates.
Finally, we asked the question of whether the ASFV protease would be able to recognize SUMO-1, the target for other proteases of this family. We assayed protein pRan-GAP, a 90-kDa protein present at the cytosolic face of the nuclear pores that is sumoylated during in vitro translation reactions (11). The ASFV protease was not able to desumoylate this substrate under the assayed conditions (data not shown). It remains to be addressed whether the ASFV protease would be able to process the SUMO-1 precursor or even possess a deubiquitinizing activity, as recently shown for the adenovirus proteinase (4).
As shown here, the recombinant ASFV protease is able to correctly and efficiently process polyprotein pp62 in vitro to the mature products p35 and p15. While this processing is observed after short incubation times, the pp62 processing products can only be detected after 3-h chase periods during infection (14). Furthermore, we have recently found that during ASFV infection polyprotein pp220 and pp62 processing occurs only on assembling particles (3), suggesting a tight regulation of proteolytic activity in the infected cells. At the present time, the specific molecular mechanisms mediating this regulation are unknown. In this connection, a remarkable feature described for the adenovirus L3 protease is its activation both by a viral peptide and DNA, which ensures that only genome-containing particles are matured into infectious virions (6, 10). With regard to the third viral member of the family of SUMO-1-processing proteases, the vaccinia virus I7 protein recently characterized as the core protein protease (5), it has been described that a thermosensitive mutant for this protein fails to process the core precursors P4a, P4b, and VP8, accumulating immature viral particles at a morphogenetic stage between the immature virion and intracellular mature virus forms (7).
Further studies on the ASFV protease and its relationship to other viral or nonviral members of the SUMO-1-specific protease family will help to establish its role during infection and the mechanisms of its regulation.
Acknowledgments
D. Rubio and A. Alejo contributed equally to this work.
We thank J. Salas for helpful discussions and critical reading of the manuscript. We also thank M. Hochstrasser for the gift of plasmid pRanGAP1.
This work was supported by grants from the Dirección General de Investigación Científica y Técnica (BMC2000-1485), the European Commission (QLRT-2000-02216), and the Comunidad Autónoma de Madrid (07G/0034/2000) and by an Institutional Grant from Fundación Ramón Areces.
REFERENCES
- 1.Andrés, G., C. Simón-Mateo, and E. Viñuela. 1997. Assembly of African swine fever virus: role of polyprotein pp220. J. Virol. 71:2331-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrés, G., A. Alejo, C. Simón-Mateo, and M. L. Salas. 2001. African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. J. Biol. Chem. 276:780-787. [DOI] [PubMed] [Google Scholar]
- 3.Andrés, G., A. Alejo, J. Salas, and M. L. Salas. 2002. African swine fever virus polyproteins pp220 and pp62 assemble into the core shell. J. Virol. 76:12473-12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balakirev, M. Y., M. Jaquinod, A. L. Haas, and J. Chroboczek. 2002. Deubiquitinating function of adenovirus proteinase. J. Virol. 76:6323-6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byrd, C. M., T. C. Bolken, and D. E. Hruby. 2002. The vaccinia virus I7L gene product is the core protein proteinase. J. Virol. 76:8973-8976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding, J., W. J. McGrath, R. M. Sweet, and W. F. Mangel. 1996. Crystal structure of the human adenovirus proteinase with its 11 amino acid cofactor. EMBO J. 15:1778-1783. [PMC free article] [PubMed] [Google Scholar]
- 7.Ericsson, M., S. Cudmore, S. Shuman, R. C. Condit, G. Griffiths, and J. Krijnse-Locker. 1995. Characterization of ts16, a temperature-sensitive mutant of vaccinia virus. J. Virol. 69:7072-7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li, S.-H., and M. Hochstrasser. 1999. A new protease required for cell-cycle progression in yeast. Nature 398:246-251. [DOI] [PubMed] [Google Scholar]
- 9.López-Otín, C., C. Simón-Mateo, L. Martínez, and E. Viñuela. 1989. Gly-Gly-X, a novel consensus sequence for the proteolytic processing of viral and cellular proteins. J. Biol. Chem. 264:9107-9110. [PubMed] [Google Scholar]
- 10.Mangel, W. F., W. J. McGrath, D. L. Toledo, and C. W. Anderson. 1993. Viral DNA and a viral peptide can act as cofactors of adenovirus virion proteinase activity. Nature 361:274-275. [DOI] [PubMed] [Google Scholar]
- 11.Matunis, M. J., J. Wu, and G. Blobel. 1998. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 140:499-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salas, J., M. L. Salas, and E. Viñuela. 1999. African swine fever virus: a missing link between poxviruses and iridoviruses?, p. 467-480. In E. Domingo, R. G. Webster, and J. J. Holland (ed.), Origin and evolution of viruses. Academic Press, London, United Kingdom.
- 13.Simón-Mateo, C., G. Andrés, and E. Viñuela. 1993. Polyprotein processing in African swine fever virus: a novel gene expression strategy for a DNA virus. EMBO J. 12:2977-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simón-Mateo, C., G. Andrés, F. Almazán, and E. Viñuela. 1997. Proteolytic processing in African swine fever virus: evidence for a new structural polyprotein, pp62. J. Virol. 71:5799-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sircar, S., A. Ruzindana-Umunyana, W. Neugebauer, and J. M. Weber. 1998. Adenovirus endopeptidase and papain are inhibited by the same agents. Antivir. Res. 40:45-51. [DOI] [PubMed] [Google Scholar]
- 16.Tihanyi, K., M. Bourbonniere, A. Houde, C. Rancourt, and J. M. Weber. 1993. Isolation and properties of adenovirus type 2 proteinase. J. Biol. Chem. 268:1780-1785. [PubMed] [Google Scholar]
- 17.Vanslyke, J. K., S. S. Whitehead, E. M. Wilson, and D. E. Hruby. 1991. The multistep proteolytic maturation pathway utilized by vaccinia virus P4a protein: a degenerate conserved cleavage motif within core proteins. Virology 183:467-478. [DOI] [PubMed] [Google Scholar]
- 18.Webster, A., S. Russell, P. Talbot, W. C. Russell, and G. D. Kemp. 1989. Characterization of the adenovirus proteinase: substrate specificity. J. Gen. Virol. 70:3225-3234. [DOI] [PubMed] [Google Scholar]