Abstract
Truncated human coronavirus HCoV-229E spike glycoproteins containing amino acids 407 to 547 bound to purified, soluble virus receptor, human aminopeptidase N (hAPN). Soluble hAPN neutralized the infectivity of HCoV-229E virions at 37°C, but not 4°C. Binding of hAPN may therefore trigger conformational changes in the viral spike protein at 37°C that facilitate virus entry.
Human coronaviruses HCoV-229E in serogroup I and HCoV-OC43 in serogroup II are, after rhinoviruses, the second most important cause of the common cold (7, 13). The first step in HCoV-229E infection is binding of the trimeric 200-kDa viral spike glycoprotein (S) to human aminopeptidase N (hAPN, CD13) on human cells (26). The three-dimensional structures of the spike glycoproteins of the plus-strand RNA-containing coronaviruses are not yet known. However, the structure of S is predicted to be generally similar to that of type 1 viral fusion proteins of large negative-strand RNA viruses (12, 22, 25). The approximately 547-amino-acid (aa)-long N-terminal S1 domain of HCoV-229E spike protein has receptor-binding activity (2), and the membrane-anchored S2 domain contains several heptad repeats (3). APNs of the normal host species are also receptors for porcine, feline, and canine coronaviruses in group I (5, 6, 15, 23). The metalloprotease APN is a 150-kDa, class II glycoprotein that is expressed as a dimer on apical membranes of polarized epithelial cells, at synaptic junctions, and on antigen-presenting cells (11, 17, 19, 21).
Soluble receptors for many viruses have been used to identify critical receptor-binding sites on the viral attachment proteins, explore the specificity of virus-receptor interactions, and characterize conformational changes in the viral attachment protein that are induced by receptor binding (10, 14, 18, 20, 28). Temperature-dependent triggering of such conformational changes in a virus attachment protein by receptors on host cell membranes leads to penetration of the viral genome into the cytoplasm of the host cell. Binding of receptor to the N-terminal domain of a type 1 viral fusion protein at 37°C can trigger a conformational change in the membrane-anchored domain, leading to fusion of the viral envelope with host cell membranes (9, 10, 14, 27).
This paper reports the expression in insect cells and characterization of a soluble recombinant hAPN protein (shAPN) and interactions of shAPN with HCoV-229E virions, the S1 domain of the viral spike glycoprotein, and truncated S1 proteins.
Characterization of shAPN.
A soluble form of hAPN lacking the stalk, transmembrane, and intracellular domains (anchor/stalk-minus APN), was previously expressed in mammalian cells and shown to be enzymatically active (24). For these studies, we expressed in insect cells the 95- to 98-kDa soluble, anchor/stalk-minus protein (shAPN). The open reading frame included the BiP cleavable signal sequence (MKLCILLAVVAFVGLSGL), a linker (RS), and aa 66 to 967 of hAPN with a C-terminal V5 epitope and His tag. We purified the protein on Ni-nitrilotriacetic acid (NTA) resin (Fig. 1A). Amino acid sequencing showed that the N terminus of shAPN was RSDQSKAWN. Isoelectric focusing showed that the pI value of shAPN was 4.95. The yield of shAPN was 20 mg/liter, and the specific activity of purified shAPN was 24 U/mg.
FIG. 1.
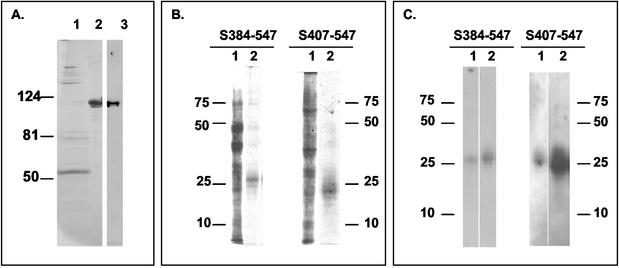
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of purified recombinant soluble receptor hAPN and truncated spike glycoproteins of HCoV-229E. (A) Molecular weight standards (lane 1), 16 μg of shAPN stained with Coomassie brilliant blue (lane 2), and 4 μg of shAPN immunoblotted with a rabbit polyclonal anti-hAPN antibody (lane 3). (B and C) Truncated HCoV-229E spike proteins in supernatant media from Sf9 cells (lanes 1) and after purification (lanes 2) are shown in silver stains (B) and in immunoblots (C) with polyclonal rabbit antibody to HCoV-229E virions.
The enzymatic activity of shAPN and its susceptibility to inhibition by a panel of anti-CD13 monoclonal antibodies (MAbs) were determined as previously described (1). Anti-hAPN antibodies were incubated at 37°C for 30 min with shAPN that had been activated with 10 mM Tris (pH 8.0), plus 1 mM CoCl2, prior to addition of the substrate l-alanine-p-nitroanalide (Sigma-Aldrich). Table 1 shows the mean A405 after 20 min from three replicate experiments. Yeager and coworkers previously showed that anti-hAPN MAbs WM15 and MY7, which block HCoV-229E infection of human cells, and WM47, which does not block infection, differ in their ability to inhibit enzyme activity of anchored hAPN (26). Here we show that MAbs WM15 and WM47and three new anti-hAPN MAbs, BB1, DW1, and Y2K, which blocked HCoV-229E infection of human cells, differed in inhibition of shAPN activity. Furthermore, when purified HCoV-229E virions were tested for their ability to inhibit APN activity, no inhibition was seen. These observations, together with previous studies of hAPN and pAPN with APN inhibitors and mutations in the catalytic site of the enzymes (4), support the conclusion that the region of hAPN to which HCoV-229E S protein binds is close to, but not identical to, the active site of the enzyme. The data also indicate that shAPN is similar in antigenicity to native hAPN.
TABLE 1.
Properties of anti-hAPN antibodiesa
| Antibody | Binds soluble hAPN | Binds anchored hAPNb | Blocks HCoV-229E infectionb | % Inhibition of enzyme activity |
|---|---|---|---|---|
| Control MAb | − | − | − | 0 |
| MAb Y2K | + | + | + | 0 |
| MAb WM47 | + | + | − | 2 |
| MAb BB1 | + | + | + | 53 |
| MAb DW1 | + | + | + | 56 |
| MAb WM15 | + | + | + | 67 |
| Polyclonal antibody P00066 | + | + | + | 42 |
Binding of anti-hAPN antibodies to immobilized shAPN was determined by ELISA. Percent inhibition of enzyme activity was determined as described in text; values are based on the mean A405 from three independent experiments, each of which was performed in duplicate.
Purification and characterization of soluble, truncated HCoV-229E S1 glycoproteins.
Two S547 proteins with C-terminal His tags and N-terminal truncations, called S384-547 and S407-547, were expressed in insect cells and purified by Ni-NTA chromatography. Truncated HCoV-229E spike proteins after purification are shown in silver stains (Fig. 1B) and in immunoblots with polyclonal rabbit antibody to HCoV-229E virions (Fig. 1C). Immobilized S384-547 and S407-547 were detected in enzyme-linked immunosorbent assay (ELISA) by goat polyclonal anti-HCoV-229E antibody and anti-HCoV-229E spike MAb 4-9H.5, but not MAb 5-11H.6 (data not shown). Immobilized S1 proteins with C-terminal truncations, S268 and S417 (2), were detected by the goat polyclonal antibody and MAb 5-11H.6, but not by MAb 4-9H.5 (data not shown). Thus, the two N-terminally-truncated S1 proteins retained the epitope previously mapped to this region on anchored hAPN (2).
Binding of truncated HCoV-229E S glycoproteins to shAPN.
The APN-binding residues on the spike glycoproteins of group I coronaviruses lie in the N-terminal S1 domain (2). We previously used flow cytometry experiments to show that although S547 bound to anchored hAPN expressed on murine cell membranes, the C-terminally-truncated S1 proteins S268 and S417 did not (2). Furthermore, antibody directed against the region of HCoV-229E S downstream of aa 417 blocked binding of S547 to anchored hAPN (2).
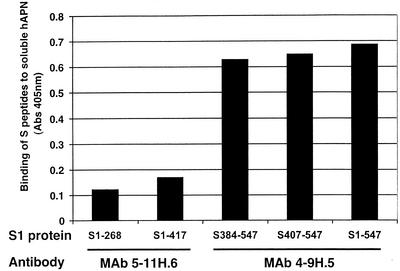
We used ELISAs to characterize binding of S1 to shAPN and to identify the smallest truncated S1 protein that could bind to immobilized shAPN. S547 bound to immobilized shAPN at room temperature (Fig. 2), 4°C, and 37°C (data not shown) and did not elute significantly during 5 h at 4 or 37°C (data not shown). Binding of S547 was specific for shAPN, and no binding to immobilized murine CEACAM1a, the receptor for murine coronavirus (28), was observed (data not shown). Both of the N-terminally-truncated S1 proteins, S384-547 and S407-547, bound to immobilized shAPN, but S268 and S417 did not (Fig. 2). These data confirm and extend the conclusions of our previous fluorescence-activated cell sorting (FACS) analysis (2) and localize the minimal receptor-binding domain of HCoV-229E to the domain of S1 between aa 407 and 547.
FIG. 2.
Identification of the receptor-binding domain on the S1 domain of the spike glycoprotein of HCoV-229E. Purified shAPN was immobilized on ELISA plates, and the S1 domain of the HCoV-229E spike protein (S547) and C- or N-terminally-truncated S1 proteins expressed in baculovirus vectors were bound at 4°C for 4 h. Binding of the S proteins to shAPN at 4°C was detected by binding of the indicated anti-S MAbs followed by peroxidase-conjugated antirabbit immunoglobulin. The y axis shows the difference between the absorbance values (A405) of each sample incubated with anti-HCoV-229E MAb and a control MAb.
A soluble truncation protein containing aa 506 to 655 of TGEV S1 coexpressed in insect cells with anchored pAPN, was found to be coimmunoprecipitated with pAPN (8). The data were interpreted as implicating the aa 506 to 655 domain of transmissible gastroenteritis virus (TGEV) S protein in receptor binding, although direct binding of purified S protein to pAPN on cell membranes was not demonstrated (8). By amino acid sequence alignment, this domain of TGEV S1 corresponds to aa 279 to 417 of HCoV-229E, which does not bind to anchored (2) or soluble hAPN. These data suggest that the receptor-binding domain of the HCoV-229E spike protein is downstream of the putative receptor-binding domain of the TGEV spike protein.
Neutralization of HCoV-229E virions by shAPN at 37°C.
To analyze the effects of shAPN upon the infectivity of HCoV-229E virions, shAPN was incubated with 2,500 PFU of HCoV-229E for 1 h at pH 7.0, at either 37 or 4°C, and the percent virus neutralization was determined by plaque assay (Fig. 3). At pH 7.0, 50% of the infectivity of HCoV-229E virions was neutralized by 0.16 nmol of shAPN at 37°C, but virus was not neutralized by 2.55 nmol of shAPN at 4°C. As a negative control for receptor specificity, soluble murine CEACAM1a did not neutralize HCoV-229E at 37°C (Fig. 3). These results show that shAPN neutralization of HCoV-229E virions is specific and concentration and temperature dependent.
FIG. 3.
Neutralization of HCoV-229E infectivity by soluble human APN. HCoV-229E virions were incubated with dilutions of shAPN at 37 or 4°C for 1 h at pH 7.0 or with sCEACAM1a as a control. Virus infectivity was quantified by plaque assay. The percentage of virus neutralization relative to that in samples without receptor is plotted against the molar concentration of purified shAPN.
We previously showed that soluble murine CEACAM1a neutralized the group II coronavirus mouse hepatitis virus (MHV) at 37°C and that virus neutralization was associated with a conformational change in S2 that made virions hydrophobic and made S2 susceptible to degradation by trypsin (27). The data in this paper suggest that at 37°C, binding of the soluble hAPN receptor to aa 407 to 547 of S1 on HCoV-229E virions may also trigger a conformational change in the spikes like that associated with membrane fusion and entry for type 1 viral fusion glycoproteins of other enveloped viruses.
Interestingly, the efficient neutralization of MHV by soluble receptor is dependent on the cleavage of the S glycoprotein (16, 27), while spike glycoproteins of group I coronaviruses do not have a proteolytic cleavage site between S1 and S2. Possibly the S proteins of group I coronaviruses are flexible enough to expose a membrane fusion peptide in response to receptor binding at 37°C without requiring protease activation.
Elucidation of the molecular mechanisms of human coronavirus receptor-mediated neutralization and virus entry may lead to new strategies for the prevention and/or treatment of upper respiratory diseases caused by human coronaviruses.
Acknowledgments
J.J.B. and I.M. contributed equally to this work.
This work was supported by NIH grants AI26075 and AI25231, Operating Grant MT-9203 from the Canadian Institutes of Health Research, the Novo Nordisk Foundation, Danish Cancer Society, and the Danish Medical Research Council. The project was part of a program under the Danish Biomembrane Research Center. J.J.B. was supported by NIH National Research Service Award AI49678.
We thank the University of Colorado Health Sciences Center Cancer Center Core (supported by NIH/NCI grant CA46934) for assistance with DNA sequencing and baculovirus expression of soluble glycoproteins. We thank Francine Lambert for technical help and David Wentworth, Brian Turner, and Larissa Thackray for many helpful discussions.
REFERENCES
- 1.Ashmun, R. A., and A. T. Look. 1990. Metalloprotease activity of CD13/aminopeptidase N on the surface of human myeloid cells. Blood 75:462-469. [PubMed] [Google Scholar]
- 2.Bonavia, A., B. D. Zelus, D. E. Wentworth, P. J. Talbot, and K. V. Holmes. 2003. Identification of the receptor binding domain of HCoV-229E spike glycoprotein. J. Virol. 77:2530-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavanagh, D. 1995. The coronavirus surface glycoprotein, p. 73-113. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, N.Y.
- 4.Delmas, B., J. Gelfi, E. Kut, H. Sjostrom, O. Noren, and H. Laude. 1994. Determinants essential for the transmissible gastroenteritis virus-receptor interaction reside within a domain of aminopeptidase-N that is distinct from the enzymatic site. J. Virol. 68:5216-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delmas, B., J. Gelfi, R. L'Haridon, L. K. Vogel, H. Sjostrom, O. Noren, and H. Laude. 1992. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature 357:417-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delmas, B. J., H. Gelfi, H. Sjostrom, O. Noren, and H. Laude. 1993. Further characterization of aminopeptidase-N as a receptor for coronaviruses. Adv. Exp. Med. Biol. 342:293-298. [DOI] [PubMed] [Google Scholar]
- 7.Denison, M. R. 1999. The common cold. Rhinoviruses and coronaviruses, p. 253-280. In R. Dolin and F. P. Wright (ed.), Viral infections of the respiratory tract. Marcel Dekker, New York, N.Y.
- 8.Godet, M., J. Grosclaude, B. Delmas, and H. Laude. 1994. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J. Virol. 68:8008-8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomez Yafal, A., G. Kaplan, V. R. Racaniello, and J. M. Hogle. 1993. Characterization of poliovirus conformational alteration mediated by soluble cell receptors. Virology 197:501-505. [DOI] [PubMed] [Google Scholar]
- 10.Greve, J. M., C. P. Forte, C. W. Marlor, A. M. Meyer, H. Hoover-Litty, D. Wunderlich, and A. McClelland. 1991. Mechanisms of receptor-mediated rhinovirus neutralization defined by two soluble forms of ICAM-1. J. Virol. 65:6015-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen, A. S., O. Noren, H. Sjostrom, and O. Werdelin. 1993. A mouse aminopeptidase N is a marker for antigen-presenting cells and appears to be co-expressed with major histocompatibility complex class II molecules. Eur. J. Immunol. 23:2358-2364. [DOI] [PubMed] [Google Scholar]
- 12.Heinz, F. X., and S. L. Allison. 2001. The machinery of flavivirus fusion with host cell membranes. Curr. Opin. Microbiol. 4:450-455. [DOI] [PubMed] [Google Scholar]
- 13.Holmes, K. V. 2001. Coronaviruses, p. 1187-1203. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Williams & Wilkins, Philadelphia, Pa.
- 14.Kaplan, G., M. S. Freistadt, and V. R. Racaniello. 1990. Neutralization of poliovirus by cell receptors expressed in insect cells. J. Virol. 64:4697-4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolb, A. F., A. Hegyi, J. Maile, A. Heister, M. Hagemann, and S. G. Siddell. 1998. Molecular analysis of the coronavirus-receptor function of aminopeptidase N. Adv. Exp. Med. Biol. 440:61-67. [DOI] [PubMed] [Google Scholar]
- 16.Lewicki, D. N., and T. M. Gallagher. 2002. Quaternary structure of coronavirus spikes in complex with carcinoembryonic antigen-related cell adhesion molecule cellular receptors. J. Biol. Chem. 277:19727-19734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noren, O., H. Sjostrom, and J. Olsen. 1997. Aminopeptidase N, p. 175-191. In A. J. B. C. M. Kenny (ed.), Cell-surface peptidases in health and disease. BIOS Scientific Publishers, Oxford, United Kingdom.
- 18.Powell, R. M., T. Ward, D. J. Evans, and J. W. Almond. 1997. Interaction between echovirus 7 and its receptor, decay-accelerating factor (CD55): evidence for a secondary cellular factor in A-particle formation. J. Virol. 71:9306-9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riemann, D., A. Kehlen, and J. Langner. 1999. CD13—not just a marker in leukemia typing. Immunol. Today 20:83-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Signoret, N., P. Poignard, D. Blanc, and Q. J. Sattentau. 1993. Human and simian immunodeficiency viruses: virus-receptor interactions. Trends Microbiol. 1:328-333. [DOI] [PubMed] [Google Scholar]
- 21.Sjostrom, H., O. Noren, and J. Olsen. 2000. Structure and function of aminopeptidase N, p. 25-34. In J. A. S. Langner (ed.), Cellular peptidases in immune functions and diseases 2. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 22.Stegmann, T. 2000. Membrane fusion mechanisms: the influenza hemagglutinin paradigm and its implications for intracellular fusion. Traffic 1:598-604. [DOI] [PubMed] [Google Scholar]
- 23.Tresnan, D. B., R. Levis, and K. V. Holmes. 1996. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J. Virol. 70:8669-8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogel, L. K., O. Noren, and H. Sjostrom. 1992. The apical sorting signal on human aminopeptidase N is not located in the stalk but in the catalytic head group. FEBS Lett. 308:14-17. [DOI] [PubMed] [Google Scholar]
- 25.Wiley, D. C., and J. J. Skehel. 1987. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 56:365-394. [DOI] [PubMed] [Google Scholar]
- 26.Yeager, C. L., R. A. Ashmun, R. K. Williams, C. B. Cardellichio, L. H. Shapiro, A. T. Look, and K. V. Holmes. 1992. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357:420-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zelus, B. D., J. H. Schickli, D. M. Blau, S. R. Weiss, and K. V. Holmes. 2003. Confirmational changes in the spike glycoprotein of murine coronavirus are induced at 37°C either by soluble murine CEACAM1 receptor glycoproteins or by pH 8. J. Virol. 77:830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zelus, B. D., D. R. Wessner, R. K. Williams, M. N. Pensiero, F. T. Phibbs, M. deSouza, G. S. Dveksler, and K. V. Holmes. 1998. Purified, soluble recombinant mouse hepatitis virus receptor, Bgp1b, and Bgp2 murine coronavirus receptors differ in mouse hepatitis virus binding and neutralizing activities. J. Virol. 72:7237-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]