Abstract
Protein-protein interactions drive the assembly of the herpes simplex virus type 1 (HSV-1) capsid. A key interaction occurs between the C-terminal tail of the scaffold protein (pre-22a) and the major capsid protein (VP5). Previously (Z. Hong, M. Beaudet-Miller, J. Durkin, R. Zhang, and A. D. Kwong, J. Virol. 70:533-540, 1996) it was shown that the minimal domain in the scaffold protein necessary for this interaction was composed of a hydrophobic amphipathic helix. The goal of this study was to identify the hydrophobic residues in VP5 important for this bimolecular interaction. Results from the genetic analysis of second-site revertant virus mutants identified the importance of the N terminus of VP5 for the interaction with the scaffold protein. This allowed us to focus our efforts on a small region of this large polypeptide. Twenty-four hydrophobic residues, starting at L23 and ending at F84, were mutated to alanine. All the mutants were first screened for interaction with pre-22a in the yeast two-hybrid assay. From this in vitro assay, seven residues, I27, L35, F39, L58, L65, L67, and L71, that eliminated the interaction when mutated were identified. All 24 mutants were introduced into the virus genome with a genetic marker rescue/marker transfer system. For this system, viruses and cell lines that greatly facilitated the introduction of the mutants into the genome were made. The same seven mutants that abolished interaction of VP5 with pre-22a resulted in an absolute requirement for wild-type VP5 for growth of the viruses. The viruses encoding these mutations in VP5 were capable of forming capsid shells comprised of VP5, VP19C, VP23, and VP26, but the closure of these shells into an icosahedral structure was prevented. Mutation at L75 did not affect the ability of this protein to interact with pre-22a, as judged from the in vitro assay, but this mutation specified a lethal effect for virus growth and abolished the formation of any detectable assembled structure. Thus, it appears that the L75 residue is important for another essential interaction of VP5 with the capsid shell proteins. The congruence of the data from the previous and present studies demonstrates the key roles of two regions in the N terminus of this large protein that are crucial for this bimolecular interaction. Thus, residues I27, L35, and F39 comprise the first subdomain and residues L58, L65, L67 and L71 comprise a second subdomain of VP5. These seven hydrophobic residues are important for the interaction of VP5 with the scaffold protein and consequently the formation of an icosahedral shell structure that encloses the viral genome.
The construction of a protein shell that encloses the viral genome is an essential step in the morphogenesis of phage and animal viruses. The assembly process of the herpes simplex virus type 1 (HSV-1) particle results in the production of three sedimentable capsid particles from lysates of infected cells. These are designated A, B, and C capsids depending on the distance sedimented in sucrose gradients (13). C capsids, which sediment farthest, contain viral DNA and are the capsids that mature into infectious virions. B capsids do not contain DNA but do contain the scaffold proteins, whereas A capsids are empty, containing neither DNA nor scaffold proteins. These are thought to be capsids that failed in the DNA packaging process (reviewed in references 24, 27, and 31). Another fourth capsid type that cannot be isolated in sucrose gradients but has been observed by ultrastructural analysis of infected cells is the procapsid, which is thought to be the earliest spherical structure formed in HSV-1-infected cells (19, 26).
There are seven proteins in B capsids. They and the genes encoding them are VP5 (UL19), VP19C (UL38), 21 (UL26), 22a (UL26.5), VP23 (UL18), VP24 (UL26), and VP26 (UL35). The capsid shell comprises four proteins, VP5, VP19C, VP23, and VP26 (reviewed in references 24, 27, and 31). During the capsid maturation process the scaffold proteins (22a and 21) occupy the internal space of the capsid, and the shell achieves icosahedral symmetry due to the presence of these scaffold proteins (8, 33, 34). A key interaction for assembly of the capsid is that between the scaffold proteins and the major capsid protein VP5. This interaction has been detected in vitro with the yeast-two hybrid assay, coimmunoprecipitation, colocalization, and protein-binding assays (9, 15, 20, 22, 25, 34). Several studies have shown that the C-terminal amino acids of the scaffold protein are necessary and sufficient for the interaction with the major capsid protein (2, 9, 15, 16, 17, 21, 22, 34, 38). A study by Hong et al. (15) showed that the C-terminal tail of pre-22a that interacts with VP5 conforms to an alpha-helical structure. The hydrophobic residues that are exposed on this amphipathic helix are key for the interaction with VP5. In particular, a highly conserved phenylalanine was important for this interaction (15).
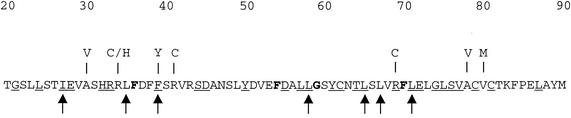
Before or concurrent with packaging the DNA, the scaffold proteins are released from the capsid. Cleavage of the scaffold proteins at the maturation site by the virally encoded protease (VP24) results in the loss of proteins 22a and 21 from the interior of the capsid. This space is subsequently occupied by the viral genome (reviewed in references 24, 27, and 31). We recently identified a small region at the N terminus of VP5 as being critical for its interaction with pre-22a. These studies utilized a virus in which the maturation cleavage site in the scaffold proteins was blocked by mutation (23). This mutation is lethal because it prevents the release of the scaffold molecules from the capsid structure, and thus these capsids failed to acquire a genome; however, spontaneous revertants of this mutant virus were readily isolated on noncomplementing cells (11). These revertants had second-site mutations, and subsequent mapping and sequence analysis revealed that the majority (21 out of 33) of these mutations were localized at the N terminus of VP5, between residues 30 and 80 (see Fig. 1) (36). The mutations occurred in two 12-amino-acid stretches; five were at amino acid 34, and 10 were at amino acid 78 (see Fig. 1) (36).
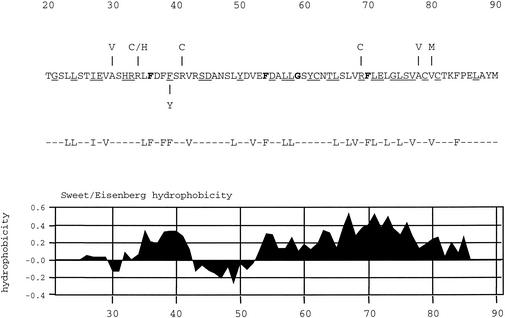
FIG. 1.
Amino acid sequence of the N terminus of VP5, which is important for interaction with the scaffold protein. The amino acid sequence of VP5 (strain KOS) from residues 20 to 90 is shown. Displayed above the sequence are the mutations identified in the revertant viruses that overcame the blocked maturation cleavage site in the scaffold proteins (36). Also illustrated below the sequence is the most frequent mutation (F39→Y) discovered in wild-type VP5, which reduced the VP5-pre-22a interaction in the yeast two-hybrid assay (37). The 24 hydrophobic residues targeted for mutation are shown below this figure. All the residues were changed to alanine. Displayed below the targeted amino acids is a plot of the hydrophobicities of the amino acids in this sequence, as determined with the Sweet-Eisenberg computer program (32). Residues in bold are conserved in all herpesviruses examined (herpes simplex viruses type 1 and type 2, pseudorabies virus, equine herpesvirus 1, bovine herpesvirus 1, varicella-zoster virus, turkey herpesvirus 2, human herpesvirus 8, equine herpesvirus 2, Epstein-Barr virus, murine herpesvirus 68, alcelaphine herpesvirus 1, human herpes virus 6, human herpesvirus 7, human cytomegalovirus, simian cytomegalovirus, and murine cytomegalovirus. The residues that are underlined are conserved in the alphaherpesviruses (herpes simplex viruses type 1 and type 2, pseudorabies virus, equine herpesvirus 1, bovine herpesvirus 1, turkey herpesvirus 2, and varicella-zoster virus).
These second-site mutations are suggested to act by weakening the interaction between VP5 and pre-22a once the capsid has formed, thereby allowing the release of the scaffold proteins from the maturing capsid in the absence of cleavage (36). With the in vitro yeast two-hybrid assay, mutants of KOS (wild-type) VP5 that result in loss of activity in this assay were selected following random mutagenesis. Several of the mutations were mapped to residue F39 (see Fig. 1) (37), indicating the importance of this hydrophobic residue for interaction with pre-22a.
The goal of the present study was to target residues of VP5 that directly participate in this interaction for mutagenesis. The study by Hong et al. (15) showed that the interaction is hydrophobic in nature by virtue of the structural data, the mutagenesis experiments, and the biochemical analysis of this interaction. To this end, we mutagenized the hydrophobic residues of VP5 that are found in the N terminus spanning residues 23 to 84. In the study described below, we have identified single amino acids of this large protein that are important for interaction with the scaffold protein, as judged from the yeast two-hybrid assay, and that are essential for the closure of HSV-1 capsid shells in infected cells.
MATERIALS AND METHODS
Cells and viruses.
Vero cells and transformed Vero cell lines were grown in minimum essential medium-alpha medium supplemented with 10% fetal calf serum (Gibco-Invitrogen) and passaged as described by Desai et al. (10). Virus stocks of KOS (HSV-1) and the mutant viruses were prepared as previously described (10).
Plasmids.
For this study, the KpnII genomic fragment derived from strain KOS was isolated and transferred into the KpnI site of pBSII-KS in which the HindIII site in the multiple cloning site was destroyed by a fill-in reaction. This plasmid was designated pKKI (see Fig. 2). A subclone of the KpnII fragment which encodes just UL18 and UL19, designated pKUL18-UL19, has been described previously (23) (see Fig. 2).
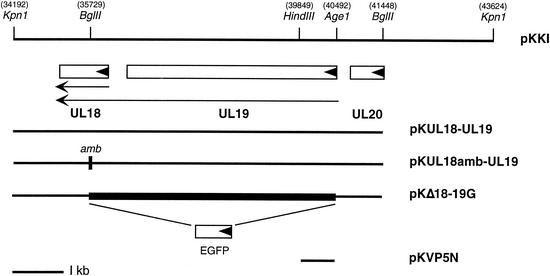
FIG. 2.
Schematic of the KpnI I region of HSV-1 strain KOS. The KpnI I fragment of HSV-1 spans from nucleotides 34192 to 43624 (18) and encodes genes UL18, UL19, and UL20. A subclone of the KpnI I fragment designated pKUL18-UL19, which encodes just the UL18 and UL19 genes, extends from the 5′ KpnI site (34192) to the BglII site (41448). In plasmid pKUL18amb-UL19, an oligonucleotide which specified translation termination in all three reading frames and an SpeI restriction site were inserted into the BglII restriction site in UL18 (35729). A deletion (filled box) that spans UL19 and UL18 (pKΔ18-19G) was made by eliminating the sequences between the AgeI (40492) and BglII (35729) sites and replacing it with an EGFP reporter gene. The template for the QuikChange mutagenesis was a small fragment (pKVP5N) encoding the N-terminal 226 residues of VP5. The directions of translation are indicated by the solid arrowheads in the open boxes, and the direction and extent of the UL18 and UL19 transcripts are shown by the arrows. The 1-kb size marker is displayed at the bottom.
To derive a fragment that encodes UL19 but not UL18, an amber codon was inserted into UL18. Plasmid pKUL18-UL19 was digested with BglII, which cuts in UL18, and an oligonucleotide (GATCTCTAGACTAGTCTAGA) was inserted into this site. The oligonucleotide specified translation termination codons in all six reading frames and also a SpeI restriction enzyme site. This plasmid was designated pKUL18amb-UL19 (see Fig. 2). Digesting pKUL18-UL19 with AgeI and BglII generated a deletion that spans the open reading frames of UL18 and UL19. The AgeI site spans codons 11 to 13 of UL19, and the BglII site spans residues 106 to 108 of UL18 (18). The enhanced green fluorescent protein (EGFP) open reading frame produced by PCR with Pfu Turbo (Stratagene) was cloned into the site of the deletion so that it was synthesized in frame with the first 10 codons of UL19 (see Fig. 2). The template for the mutagenesis of the VP5 N terminus was derived from the yeast two-hybrid vector pGAD424-VP5 (9) as an EcoRI-HindIII fragment. This 678-bp fragment encoding the N-terminal 226 residues was cloned into pUC19 that had been digested with EcoRI and HindIII, and the resulting plasmid was designated pKVP5N (see Fig. 2).
Mutagenesis.
Site-directed mutations were created by the QuikChange mutagenesis procedure (Stratagene). This method involves the use of complementary primers containing the mutation. Primers containing 15 nucleotides on either side of the mutation were made. Following PCR assays according to the manufacturer's protocol, the DNA was digested with DpnI and transformed into Escherichia coli. Positive clones were isolated, and the introduction of the mutation was confirmed by sequence analysis. The mutant sequence was then transferred into the yeast-two hybrid vector containing VP5 (pGBT9-VP5) as an EcoRI-HindIII fragment and into pKKI as an AgeI-HindIII fragment, that is, the wild-type sequences were replaced with the sequence containing the site-directed mutation.
Yeast two-hybrid assays.
The yeast two-hybrid assays were performed essentially as described by Desai and Person (9). Plasmid pGBT9-VP5, in which VP5 is fused to the Gal4 DNA-binding domain, was cotransformed into SFY526 cells together with pGAD424-22a, in which the scaffold protein is fused to the Gal4 transactivation domain (9). The pGBT9-VP5 mutant clones were cotransformed with pGAD424-22a into SFY526, and the transformants were allowed to grow on filter paper in order to screen each mutation with the 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) filter assay (3). Similar transformations were also performed, and the colonies were allowed to grow on the selective medium plates. These cells were then used for the o-nitrophenyl-β-d-galactopyranoside (ONPG) liquid assay in order to quantitate the levels of β-galactosidase expression. Quantitation was performed as described by Rose et al. (28) and Desai and Person (9).
Construction of transformed Vero cell lines.
The procedure of DeLuca et al. (5) was followed for transformation of Vero cells and is also described by Desai et al. (6). Subconfluent monolayers of Vero cells were cotransfected with pSV2-neo (1.0 μg) (30) and a molar three- or fivefold excess of the plasmid encoding the UL19 gene (pKUL18amb-UL19; see Fig. 2). Sixty-one G418-resistant colonies were tested for their ability to support the replication of K5ΔZ, a null mutant of the UL19 open reading frame (6). Nine out of the 61 were able to complement K5ΔZ. These cell lines were also tested for their ability to complement the growth of K23Z, a null mutant virus of VP23 (6), and as expected, all the cell lines that complemented K5ΔZ failed to complement K23Z. One such cell line, designated 31, was chosen for further analysis and subsequently for isolation of the VP5 mutants.
Marker transfer and marker rescue.
Marker transfer of the UL18-UL19 null mutation was carried out essentially as described by Person and Desai (23). Subconfluent monolayers of C32 cells (0.75 × 106) in 60-mm dishes were cotransfected with 2 μg of linearized plasmid and 5 to 10 μg of infected cell DNA. When foci were observed (48 h after transfection), the cell monolayers were harvested, freeze-thawed once, and sonicated, and titers of total virus progeny were determined. Single-plaque isolates were screened in the fluorescence microscope for green fluorescence. Isolates that exhibited a green fluorescence phenotype were plaque purified three times prior to further characterization. This virus was designated KΔ18-19G. For marker rescue/marker transfer assays, C32 cells were cotransfected with KΔ18-19G CsCl-purified virus DNA (5 μg) and linearized pKKI mutant plasmid DNA (1 μg). The transfection progeny were counted on Vero, C32, and 31 cells to determine the phenotype of the mutation.
Southern blot hybridization.
Southern blot analysis was performed as described by Desai et al. (6).
Radiolabeling and SDS-PAGE.
Radiolabeling of infected cells and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis were performed as described by Person and Desai (23).
Sedimentation analysis of capsids.
Sedimentation analysis of capsids from infected cells was performed as described by Desai et al. (6) and Person and Desai (23). All gradients were made with a BioComp Gradient Mate (BioComp). Generally, lysates for sedimentation were prepared from cells (107) in 100-mm dishes. Infected cells were harvested by scraping into phosphate-buffered saline and pelleted, washed once in phosphate-buffered saline, and repelleted. The cell pellet was resuspended in 0.5× capsid lysis buffer (2× capsid lysis buffer is 2% Triton X-100, 2 M NaCl, 10 mM Tris [pH 7.5], and 2 mM EDTA), and the nuclear fraction was pelleted at 3,500 rpm for 20 min (Beckman, GS-6R). The nuclear pellet was lysed in 2× capsid lysis buffer, and the lysate was sonicated prior to sedimentation.
Electron microscopy.
Vero cells (107) in 100-mm dishes were infected at a multiplicity of infection of 10 PFU/cell. The samples were processed for transmission electron microscopy as described by Hendricks et al. (14). The cells were fixed for 1 h at room temperature in a solution containing 2.5% glutaraldehyde in 100 mM cacodylate (pH 7.4) and 2.5% sucrose. The cells were then lifted off the dish, pelleted, and subsequently treated with Palade's OsO4 (1 h at 4°C). The pellet was then washed three times in 100 mM cacodylate, pH 7.4, washed three times in double-distilled H2O, and incubated overnight in Kellenberger's uranyl acetate (14). The pellet was then dehydrated through a graded series of ethanol and embedded in Embed-812. Sections were cut on a Leica Ultracut UCT ultramicrotome, collected onto 400-mesh nickel grids, poststained in uranyl acetate and lead citrate, and observed in a Philips EM420 transmission electron microscope.
For negative-staining analysis, 400-mesh nickel grids coated with Formvar were floated on sucrose gradient fractions and then stained with 2% uranyl acetate. The samples were then air dried and observed with a Philips EM420 transmission electron microscope with Soft Imaging System MegaView III camera and AnalySIS software.
Data and figure preparation.
For figure preparation, autoradiographs were scanned at 600 dots per inch in Adobe Photoshop. Electron micrographs were captured as 12-bit images (16-bit tiff files) and then exported as 8-bit tiff files, which were adjusted for brightness and contrast and resized in Adobe Photoshop. Figures were compiled with Adobe Illustrator.
RESULTS
Mutagenesis of hydrophobic residues of the N terminus of VP5.
The 25 C-terminal amino acids of the scaffold protein (pre-22a) have been shown to be required for interaction with VP5 (9, 15, 16, 17, 22, 34), while N-terminal residues 27 to 80 of VP5 are important for this interaction (11, 36, 37). For pre-22a, hydrophobic residues in the C terminus were shown to be key for its interaction with VP5 (15). This finding was the rationale for mutation of the hydrophobic residues of VP5.
Analysis of the sequence of VP5 with the Sweet-Eisenberg hydrophobicity plot (32) revealed two hydrophobic stretches in the N terminus of VP5, a small region spanning residues 35 to 39 and the largest hydrophobic region in the whole protein spanning residues 52 to 86 (see Fig. 1). The confluence of evidence pointed to the hydrophobic residues in the N terminus, and thus these were sequentially mutated with the QuikChange mutagenesis protocol (Stratagene). The residues targeted are shown in Fig. 1, and all were mutated to alanine, which minimally perturbs secondary structure. The template for mutagenesis of VP5 was a subclone of pGAD424-VP5 which spans from the start of VP5 (EcoRI) to residue 226 (HindIII) (see Fig. 2). Following QuikChange mutagenesis, the introduction of the mutation was confirmed by sequencing and for the authentic amplification of the rest of the sequence. The mutation was transferred into the yeast two-hybrid vector, which contains VP5 (pGBT9-VP5), as an EcoRI-HindIII fragment for analysis in the yeast two-hybrid assay or into the genomic clone pKKI as an AgeI-HindIII fragment for introduction into the virus genome.
Yeast two-hybrid analysis of the VP5 mutations.
The mutations were analyzed with the yeast two-hybrid assay (1, 4, 12) to determine the effect of these changes on the interaction with the scaffold protein. Previously, cotransformation of the SFY526 yeast strain (12) with the DNA encoding the scaffold protein fused to the Gal4 transactivation domain (pGAD424-22a) and the DNA encoding VP5 fused to the Gal4 DNA binding domain (pGBT9-VP5) gave colonies that turned blue in the X-Gal filter assay due to the expression of β-galactosidase, indicative of protein-protein interaction (9). The colonies consistently began to attain a blue color approximately 20 min after the initiation of the assay. Quantitative analysis of this interaction was determined by enumeration of the β-galactosidase enzyme activity with a liquid assay with o-nitrophenyl-β-d-galactopyranoside (ONPG) as the substrate (28). The VP5 mutants were thus analyzed with the X-Gal filter assay, and a quantitation of the enzyme activity was determined from separate experiments (see Table 1).
TABLE 1.
Yeast two-hybrid assay: interaction of VP5 mutations with the scaffold proteina
| VP5 | X-Gal filter assay | β-Galactosidase activity (% of wild-type activity) |
|---|---|---|
| Wild type | +++ | 100.00 |
| L23A | +++ | 83.16 |
| L24A | +++ | 73.36 |
| I27A | − | 0.84 |
| V29A | +++ | 338.19 |
| L35A | − | 1.19 |
| F36A | ++ | 1.68 |
| F38A | ++ | 1.85 |
| F39A | − | 0.95 |
| V42A | +++ | 86.74 |
| L49A | +++ | 25.37 |
| V52A | +++ | 138.54 |
| F54A | +++ | 34.54 |
| L57A | ++ | 4.77 |
| L58A | − | 1.15 |
| L65A | − | 0.87 |
| L67A | + | 4.01 |
| V68A | +++ | 24.58 |
| F70A | +++ | 55.86 |
| L71A | − | 0.74 |
| L73A | +++ | 62.88 |
| L75A | +++ | 17.91 |
| V77A | +++ | 27.58 |
| V80A | ++ | 3.82 |
| F84A | +++ | 127.66 |
| None (vector alone) | − | 1.16 |
In the X-Gal filter assay, the time it took for the appearance of blue colonies was determined. +++, wild-type kinetics of blue color formation (that is, within 30 min of the start of the assay); ++, 30 to 120 min for the appearance of blue color; +, greater than 2 h for blue color development; −, colonies remained clear even after prolonged incubation. The data for both the X-Gal filter assay and the β-galactosidase liquid assay were derived from two independent transformations.
The mutations fell into three classes of phenotypes: those that interacted with pre-22a seemingly with wild-type kinetics of blue colony appearance (+++), those that showed a decreased interaction with pre-22a, as judged from the slower appearance of the blue colonies (++ and +), and those that failed to interact with pre-22a (−), as judged from the absence of blue color of the colonies (see Table 1). Thirteen of the 24 mutants all interacted with pre-22a in a manner analogous to wild-type VP5, as judged from the appearance of blue colonies within 20 to 30 min of the start of the assay.
Measurement of the enzyme activity revealed some interesting differences. For the L23A, L24A, V42A, L49A, F54A, V68A, F70A, L73A, L75A, and V77A mutants, the enzyme activity was anywhere between 20 and 85% of wild-type activity. However, for three mutants, V29A, V52A, and F84A, the enzyme activity was increased over wild-type activity in some cases by threefold, indicating that the interaction of the VP5 polypeptide expressed by these mutants with pre-22a was enhanced. This result may suggest that these mutants offer a conformationally better fit with pre-22a in this assay. Mutations in five residues, F36, F38, L57, L67, and V80, caused a decreased ability to interact with pre-22a. This was reflected in the slower appearance of blue colonies, anywhere from 30 min to 6 h for mutant L67A, and by the reduced levels of β-galactosidase enzyme activity observed, typically at or below 5% of the wild-type level. Even though mutants F36A and F38A gave rise to blue colonies in the filter assay, the amounts of β-galactosidase enzyme produced by their interaction with pre-22a were similar to those of mutants carrying a mutation in the group of hydrophobic residues spanning from L35 to F39 (Table 1). Finally, six mutations in residues I27, L35, F39, L58, L65, and L71 abolished interaction with the scaffold proteins. The yeast cotransformants did not turn blue in the X-Gal filter assay, and the enzyme activity detected was at background levels (at or below 1% of wild-type activity). The background level was the enzyme activity obtained with no insert in the pGBT9 plasmid. Thus, these six hydrophobic residues in VP5 are essential for the interaction with scaffold proteins.
Introduction of VP5 mutations into the virus genome.
The yeast two-hybrid assay results were indicative of the ability of the different mutants to interact with the scaffold proteins. In order to correlate the different phenotypes observed in this assay with the effect of the mutations on virus replication and capsid assembly, we decided to introduce all 24 mutations into the virus genome. To facilitate the rapid introduction and selection of the virus mutants, especially for such a large number of mutants, we decided to use the marker rescue/marker transfer assay that we had utilized previously for the introduction of mutations in glycoprotein B (7) and the UL26 open reading frame (23) into the KOS genome. In this genetic system, virus and plasmid constructs are generated so that a mutation in the recipient viral genome is rescued and the desired marker in an adjacent gene is transferred to the viral genome in the same homologous recombination event. The assay requires that the two adjacent genes be essential for viral replication and that transformed cell lines that express both genes and the individual genes be available. The UL18 (VP23) and UL19 (VP5) genes fit these criteria, and this genetic system was used to introduce UL19 mutations into the virus.
For this procedure, a double mutant virus with a deletion in most of the UL19 gene and extending partway into the adjacent UL18 gene was constructed. The deletion spans from the tenth codon of VP5 to the 107th residue of VP23 (total number of VP23 residues = 318) (see Fig. 2). The deletion of all but the first 10 residues of VP5 was necessary because the mutations to be introduced are at the N terminus of VP5; this deletion therefore ensures that homologous recombination will introduce the mutation into the UL19 genetic locus. The open reading frame encoding the enhanced green fluorescent protein (EGFP) was introduced into the site of the deletion so that the first 10 codons of UL19 were fused in frame to the start of EGFP (see Fig. 2). Expression of the reporter cassette was under the control of the UL19 promoter. The GFP reporter was used to identify virus recombinants that contained the specified double mutation. The mutation was transferred into the virus with standard marker transfer transfection experiments.
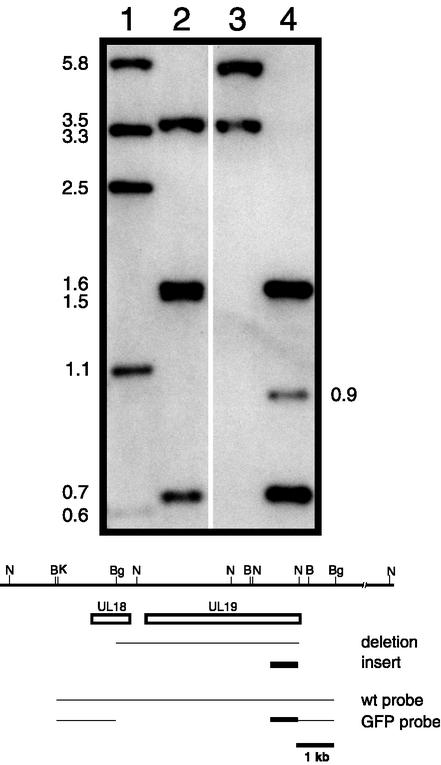
Viruses that formed green fluorescent plaques on C32 cells (expresses both VP23 and VP5) (see Table 2) were picked, purified, and characterized further. The genome of this virus, designated KΔ18-19G, was examined by Southern blot analysis. Infected-cell DNA was digested and examined with radiolabeled probes corresponding to both the pKUL18-UL19 sequence and plasmid pKΔ18-19G for the EGFP sequence probe. Shown in Fig. 3 is the result of this blot. When DNA from KOS-infected cells was digested with BamHI and BglII, the probe hybridized to fragments of 3.5, 1.6, 1.5, and 0.67 kb (Fig. 3, lane 2). The probe hybridized to only the 1.6- and 0.67-kb fragments (see Fig. 3, lane 4) in DNA extracted from KΔ18-19G-infected cells because the deletion eliminated the sequences that would give rise to the 3.5- and 1.5-kb fragments (see cartoon below blot in Fig. 3). The probe also hybridized to a novel 0.9-kb band which corresponds to the GFP open reading frame (0.65 kb) fused to upstream sequences from the tenth UL19 codon to the BamHI site in the UL19 promoter (Fig. 3, lane 4). In KOS DNA digested with NcoI, the probes hybridized to fragments of 5.8, 3.3, 2.5, 1.1, and 0.6 kb (Fig. 3, lane 1), of which only the 5.8-kb fragment was unaltered by the deletion in the mutant virus. The 3.3-kb fragment was shorter in the mutant DNA, but because of the fusion with GFP, it increased in size to 3.5 kb (Fig. 3, lane 3). This analysis confirmed the genotype of the KΔ18-19G virus.
TABLE 2.
Genotype and phenotype of the mutant viruses and complementing cell lines as determined by the plating efficiency assay
| Virus | Plating efficiency (PFU/ml) of wild-type (KOS) and mutant viruses on:
|
||
|---|---|---|---|
| Vero cells | C32 cells | 31 cells | |
| KOS | 8.8 × 109 | 9.3 × 109 | 6.8 × 109 |
| K5ΔZ | 3 × 105 | 2.10 × 109 | 2.29 × 109 |
| K23Z | 1 × 105 | 2.63 × 109 | 4 × 105 |
| KΔ18-19G | <104 | 3.8 × 108 | <104 |
FIG. 3.
Southern blot analysis of the KΔ18-19G genome. We digested 2 μg of KOS-infected cell DNA (lanes 1 and 2) and KΔ18-19G-infected cell DNA (lanes 3 and 4) with NcoI (lanes 1 and 3) or BamHI and BglII (lanes 2 and 4) and resolved the DNAs by agarose gel electrophoresis prior to analysis by Southern blot hybridization. Filters were probed with a 32P-labeled DNA probe corresponding to both the wild-type pKUL18-UL19 sequence (wt probe) and pKΔ18-19G (GFP probe) sequence. The size of hybridized fragments (in kilobases) are indicated at the sides of the gel. The cartoon at the bottom of the figure shows the region of HSV-1 from genome nucleotides 32576 to 46293. The open box depicts the locations of the UL18 and UL19 open reading frames, the line below shows the extent of the Δ18-19 deletion, and the solid box depicts the GFP insert. The wild-type and Δ18-19GFP probes used are shown below this. Relevant restriction enzyme sites are shown at the top (B, BamHI; Bg, BglII; N, NcoI; and K, KpnI), and the 1-kb size marker is at the bottom.
The other requisite for this genetic transfer system was transformed cell lines that express UL18 and UL19. Cell line C32 (23), which expresses both genes, was already available and was used to propagate this double mutant virus (see Table 2). A cell line was also needed that expressed just UL19. This was achieved by inserting an amber codon into the UL18 gene. Again, plasmid pUL18-UL19 was used; it was digested with BglII, and an SpeI linker that specified translation termination in all three reading frames was inserted into the BglII site (see Fig. 2). Vero cell lines were cotransformed with this plasmid, pKUL18amb-UL19, and pSV2neo. G418-resistant cell lines were screened for their ability to plaque the VP5 null mutant K5ΔZ (6). Several cell lines that exhibited this phenotype were isolated. These cell lines were also screened for their ability to plaque the VP23 null mutant K23Z (6). All the cell lines failed to grow K23Z, as expected, indicative of translation ablation of VP23. One cell line, designated 31, was chosen for further analysis. This cell line was capable of plaque formation of K5ΔZ but not of K23Z (see Table 2). Growth of K5ΔZ in cell line 31 was comparable to its growth in C32 cells, as judged from burst size yields (data not shown).
For transfer of the mutations into the virus, all the mutations were derived as AgeI-HindIII fragments, and this fragment cloned into pKKI. Plasmid pKKI specifies sufficient flanking sequences on either side of the UL18-UL19 deletion (see Fig. 2). To initially test the genetic transfer system and also to derive a rescued virus of this mutation, C32 cells were cotransfected with wild-type pKKI and viral DNA derived from KΔ18-19G. When the transfection progeny virus was plated on Vero cells, plaques were readily observed. Thus, the system was capable of easily transferring a VP5 marker. This virus was purified further and designated K5R.
The VP5 mutants were transferred into the virus genome with similar cotransfection experiments. For each mutant, the transfection progeny were plated onto C32, Vero, and 31 cell lines. The viruses that formed plaques on C32 cells were predominantly the parental virus KΔ18-19G. Viruses that formed plaques on 31 cells were viruses in which the UL18 mutation was rescued and the UL19 mutation was transferred into the UL19 locus. In many cases, viruses were also observed to form plaques on Vero cells, indicating that the mutation was not lethal. We typically obtained marker transfer/marker rescue efficiencies of 2 to 20%.
All the mutant viruses were plaque purified prior to characterization. Most of the VP5 mutants were capable of growth on noncomplementing Vero cells. Mutations of residues I27, L35, F39, L58, L65, L67, F70, L71, and L75 were lethal for virus replication, as judged from the requirement of the 31 cell line for the propagation of these viruses. For these viruses, a PCR product which spans from the start of the VP5 gene to residue 153 was amplified with the virus DNA and then sequenced. This confirmed the introduction of each of the lethal mutations into the virus genome.
Single-step growth of the VP5 mutants.
Even though the majority of the mutants formed plaques and replicated on Vero cells, there were some mutants that formed significantly smaller plaques. In order to obtain quantitative measurement of the growth properties of the mutant viruses, single-step growth assays were performed to determine the burst size for each virus. Confluent monolayers of Vero cells were infected with the mutants, and the progeny were harvested 24 h following infection. The yield of virus was determined by titration on Vero (or C32) cells, and the data are shown in Table 3.
TABLE 3.
Single-step growth of the VP5 mutantsa
| Virus | Burst size (% of wild-type burst size) |
|---|---|
| KOS (wild type) | 100 |
| K5R | 74.4 |
| L23A | 43.5 |
| L24A | 46.5 |
| I27A | 0.04 |
| V29A | 44.0 |
| L35A | 0.15 |
| F36A | 41.8 |
| F38A | 48.4 |
| F39A | 0.10 |
| V42A | 59.9 |
| L49A | 61.8 |
| V52A | 68.0 |
| F54A | 57.7 |
| L57A | 38.1 |
| L58A | 0.07 |
| L65A | 0.06 |
| L67A | 0.01 |
| V68A | 56.3 |
| F70A | 0.07 |
| L71A | 0.04 |
| L73A | 42.1 |
| L75A | 0.03 |
| V77A | 52.8 |
| V80A | 36.5 |
| F84A | 7.4 |
Data were derived from three independent infections.
Most of the VP5 mutants gave a burst size of between 36 and 68% of the wild-type burst size. The only exception was mutant F84A, which gave a mean burst size of 7.4% of the wild-type burst size. This virus formed very small plaques on Vero cells, and we attribute this to a temperature-sensitive phenotype because plaques of this virus could not form at the higher temperature (39.5°C; data not shown). The mutants carrying mutations that resulted in lethal phenotypes (I27A, L35A, F39A, L58A, L65A, L67A, F70A, L71A, and L75A) were incapable of growth in Vero cells; they typically gave a burst size of less than 0.1% of the wild-type burst size. Any growth observed for these viruses was most likely due to background wild-type virus present in these stocks, which ranged from 0.3 to 0.001%. This background virus arises following recombination with the endogenous DNA sequence present in the cell line.
From these data, there was a good correlation between the results of the yeast two-hybrid assay and the ability of the mutants to replicate. Thus, mutants at residues I27, L35, F39, L58, L65, and L71, which failed to interact with pre-22a, were also not capable of growth in the absence of helper function. Mutant L67A exhibited an extremely weak interaction with pre-22a, which could explain why this mutation was lethal for virus growth. The surprising result was that observed with mutants F70A and L75A; both interacted with pre-22a in the yeast assay, but when the mutations were transferred into the virus, they resulted in a lethal growth phenotype.
Synthesis and accumulation of ICP5 (VP5) polypeptide in mutant-infected cells.
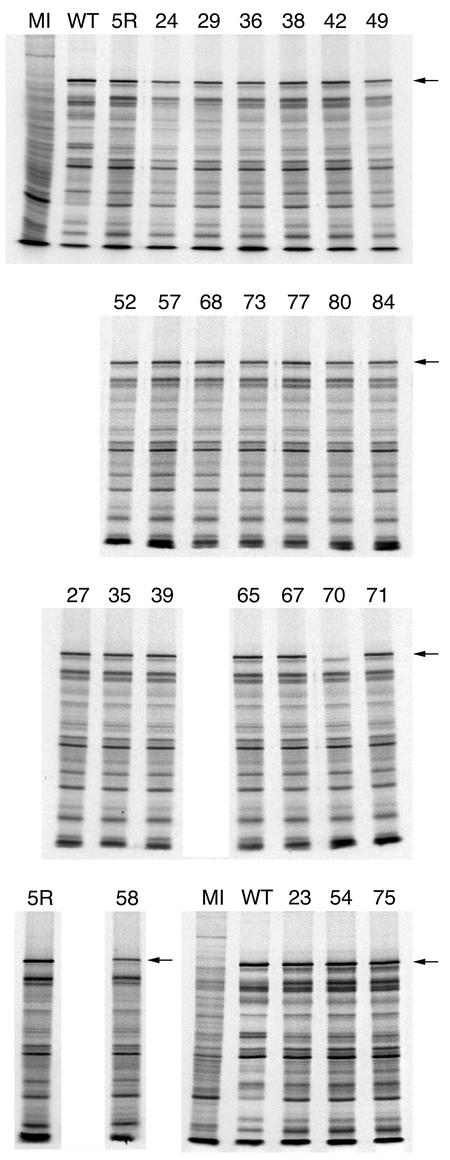
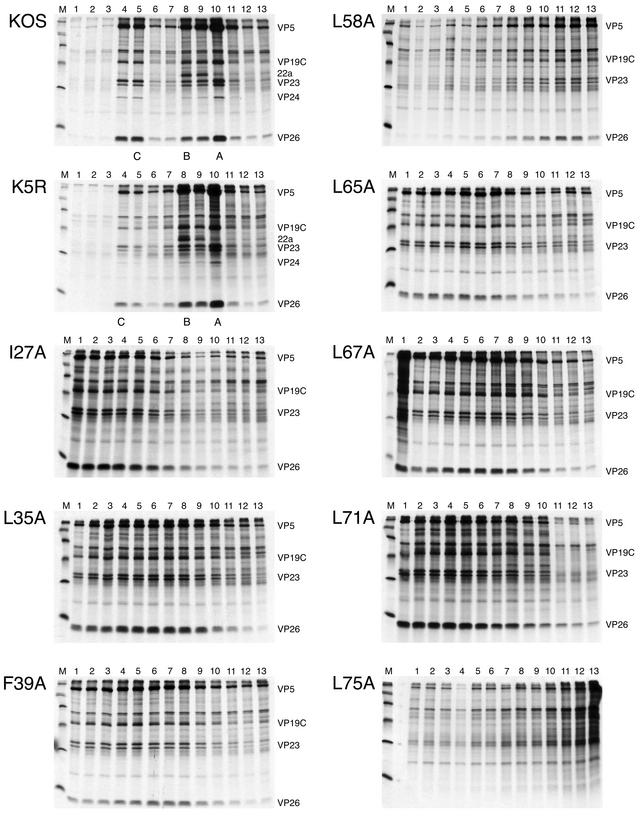
The effect of the mutations on the expression and accumulation of the ICP5 polypeptide was examined by SDS-PAGE of radiolabeled infected cell lysates. Vero cells were infected with the wild-type, marker-rescued, and mutant viruses and pulse-labeled with [35S]methionine from 7 to 9 h postinfection or labeled with [35S]methionine from 9 to 24 h after infection. The protein lysates were resolved by SDS-PAGE, and the results of the overnight labeling experiment are shown in Fig. 4. The majority of the mutant viruses accumulated appreciable amounts of the ICP5 polypeptide. The mutant L58A appeared to accumulate slightly reduced amounts of the ICP5 polypeptide (see Fig. 4, lane 58). The results with the pulse-labeled lysates revealed similar synthesis of the ICP5 polypeptide for most of the mutant viruses (data not shown). The only exception was observed in the lysate derived from mutant F70A-infected cells; the ICP5 polypeptide was absent in the lysate, as judged from the absence of radioactivity in the gel (see Fig. 4, lane labeled 70). This result was unexpected but would explain why this mutation caused a lethal phenotype when introduced into the virus. Sequence analysis of this mutation in the original plasmid, in the virus genome, and in the yeast two-hybrid vector showed only the presence of the generated mutation. Thus, the mutation at F70 must have an effect on the stability of the ICP5 polypeptide.
FIG. 4.
Accumulation of ICP5 in the mutant-infected cells. Vero cell monolayers (2.5 × 105 cells) were either mock infected (MI) or infected with wild-type KOS (WT), the marker-rescued virus K5R (5R), or the mutant viruses at a multiplicity of infection of 10 PFU/cell. Infected cells were radiolabeled with [35S]methionine from 9 to 24 h postinfection. The cells were lysed in Laemmli sample buffer, and the proteins were analyzed by SDS-PAGE (9% acrylamide). The autoradiographs obtained following exposure of the dried gels to X-ray film are shown in the figure. The numbers above each lane refer to the mutated residue. The position of the ICP5 polypeptide is shown on the right of the panels (see arrow).
Ultrastructural analysis of infected cells.
Conventional transmission electron microscopy examination of mutant-infected cells was carried out to determine the phenotype of the VP5 point mutants. This analysis was limited to just the mutant viruses that contained a lethal VP5 mutation and thus were not capable of growth in cells that did not provide VP5 in trans. Vero cells were infected with the VP5 mutant viruses and with wild-type virus and the marker-rescued virus K5R. The infected cells were harvested 18 h following infection and fixed, and the cell pellets were sectioned. The thin sections were processed for electron microscopy analysis, the results of which are shown in Fig. 5 and Fig. 6.
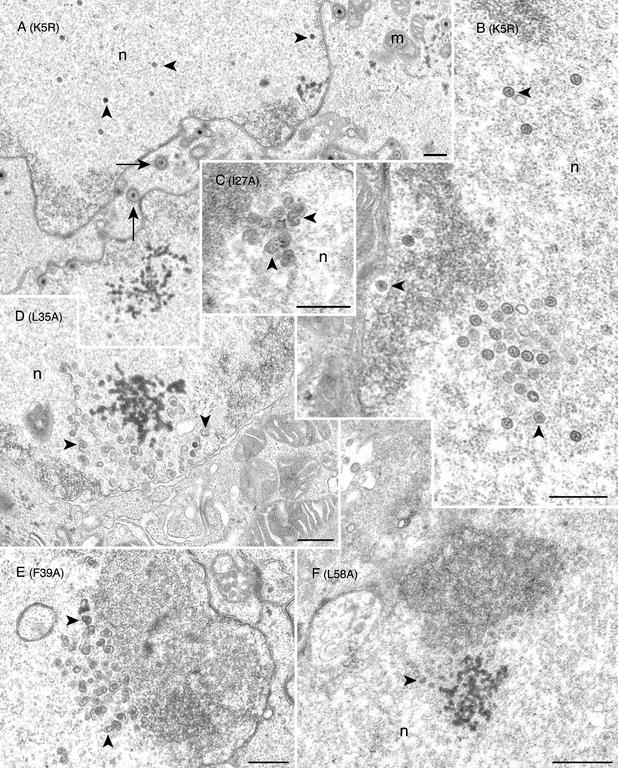
FIG. 5.
Conventional transmission electron microscopy of the mutant-infected cells reveals open shells in the nucleus. Vero cells were infected with K5R (A and B) or with the mutant viruses I27A(C), L35A (D), F39A (E), and L58A (F). Infected cells were harvested 18 h following infection and processed for transmission electron microscopy. Mature capsids (marked by arrowheads) were seen in the nucleus (n), and enveloped virions (marked by arrows) were evident in K5R-infected cells. In the mutant-infected cells (mutation shown in parentheses), numerous large open capsid shell structures (marked by arrowheads) were detected in the nuclei (n). In L58A-infected cell nuclei, smaller capsid shells (see arrowhead) were observed (panel F). Bars, 0.5 μm. Mitochondria (m) are indicated.
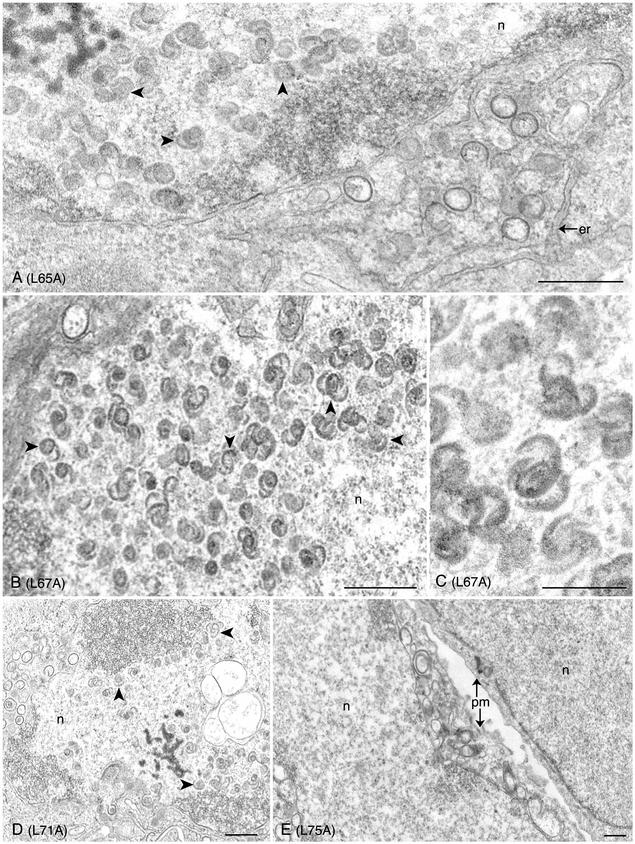
FIG. 6.
Ultrastructural analysis of the L65A, L67A, and L71A VP5 mutants reveals capsid shells in the infected cell. Infected cells were processed for conventional electron microscopy as described for Fig. 5. Capsid shells (marked by arrowheads) that have the characteristic 6s and 9s configuration (see panel C) were observed in the nuclei (n) of cells infected with mutants L65A (A), L67A (B and C), and L71A (D). Shown in panel E are cells infected with mutant L75A. No assembled structures were evident in these cells. Plasma membrane (pm) and endoplasmic reticulum (er) are indicated. Bars: (A, B, D, and E) 0.5 μm; (C) 0.25 μm.
In cells infected with the marker-rescued virus (K5R), both mature capsids (Fig. 5B) and enveloped virions at the nuclear envelope were detected, as expected (see Fig. 5A). In wild-type virus-infected cells, mature capsids and enveloped virions were also observed (data not shown). In cells that were infected with mutants I27A (Fig. 5C), L35A (Fig. 5D), F39A (Fig. 5E), L58A (Fig. 5F), L65A (Fig. 6A), L67A (Fig. 6B-C), and L71A (Fig. 6D), fully matured capsid structures were not observed in the nuclei of these infected cells. Rather, structures that resemble capsid shells were detected in the nucleus (see Fig. 6C). These structures aggregated in the nucleus and were sometimes adjacent to the nuclear envelope (see Fig. 6A). These structures have the characteristic appearance of open shells termed 6s and 9s (see Fig. 6C) and are similar to the shells observed in cells infected with the scaffold null mutant virus (8) (data not shown).
The shell structures observed for the majority of the mutants were very similar in appearance, and their features are evident in the high magnification shown in Fig. 6C. However, the shells observed in cells infected with L58A were harder to discern due to their relatively smaller size (see arrowhead in Fig. 5F). A thorough examination of many cells did not reveal any closed icosahedral capsid structures in any of the mutant-infected cells. The results from this ultrastructural analysis were consistent with the yeast two-hybrid data, which indicated an abolishment of scaffold-protein interaction which in infected cells resulted in the absence of growth and an accumulation of capsid shells. For mutant L75A (see Fig. 6E), no discernible structures were observed in the nucleus of these infected cells. Many infected cells were scanned by this analysis. This was surprising for L75A, since the yeast two-hybrid result indicated that this mutation did not affect interaction with the scaffold proteins. We also analyzed cells infected with the mutant F70A by transmission electron microscopy. As expected, we did not detect any capsid structures in the nuclei of these cells (data not shown). The absence of the ICP5 polypeptide in cells infected with this mutant would prevent any capsid structure formation.
Sedimentation analysis of infected cell lysates.
In order to confirm the data obtained from the ultrastructural analysis of infected cells and to determine the polypeptide composition of the shell structures, sedimentation of radiolabeled lysates derived from mutant-infected cells was performed. Vero cell monolayers were infected with the mutant viruses and also with wild-type and marker-rescued viruses. The cells were metabolically labeled with [35S]methionine. The infected cells were harvested 24 h after infection, and nuclear lysates were prepared, which were then sedimented in sucrose gradients. The gradients were fractionated, and the protein composition of the fractions were determined by SDS-PAGE. The results of this analysis are shown in Fig. 7.
FIG. 7.
Sedimentation analysis of the VP5 hydrophobic point mutants. Vero cell monolayers (107 cells) were infected with KOS, K5R, and the mutant viruses at a multiplicity of infection of 10 PFU/cell and labeled with 35[S]methionine from 8 to 24 h postinfection. Nuclear extracts were prepared and layered onto 20 to 50% sucrose gradients. Following sedimentation, fractions were analyzed by SDS-PAGE (17% acrylamide). The autoradiographs obtained following exposure of the dried gels to X-ray film are shown in the figure. The direction of sedimentation was from right to left. The positions in the gradient where capsids sediment are indicated at the bottom of the panel for KOS and K5R. The positions of the capsid proteins are indicated on the right of each panel except for L75A, for which there was no evident cosedimentation of capsid proteins. Molecular size standards are shown in lane M and are 220, 97.4, 66, 46, 30, and 14.3 kDa.
Fraction 1 corresponds to the bottom of the centrifuge tube. For KOS- and K5R-infected cell nuclear lysates, three peaks of radioactivity and consequently of capsid proteins were observed. These correspond to the positions where A, B, and C capsids sediment (labeled at the bottom of the panels). In these gradients, A capsids were the predominant species because viral DNA was occasionally lost from the C capsid, resulting in its conversion into an A (empty) capsid, during the preparation of the lysates. All three capsid types contain VP5, VP19C, VP23, VP24, and VP26. B capsids contain the abundant scaffold protein 22a and lesser amounts of protein 21 (not labeled). In our analysis, VP23 was always observed as a doublet polypeptide species (confirmed by Western blot analysis with antiserum to VP23).
The sedimentation of lysates derived from the mutants that formed the open shells (as determined by transmission electron microscopy) showed cosedimentation of four of the shell proteins, VP5, VP19C, VP23, and VP26. These cosedimenting proteins were observed in a number of the fractions but were generally more abundant in the lower portion of the gradients (fractions 1 to 10), as judged from the peak radioactivities corresponding to the capsid proteins. The shell structures formed by these mutants were most probably of different sizes and shapes, which would account for the distribution of these structures in the gradients. The result obtained with the lysate derived from cells infected with the mutant L58A differed from those with the lysates of the other mutants in that the four cosedimenting shell proteins were generally detected in the upper part of the gradient (fractions 10 to 13). This would indicate that the shell structures formed in cells infected with this mutant were smaller than those formed in the cells infected with the other mutants. This was consistently observed in repeated experiments and when a second independent isolate of L58A was examined in this assay.
Sedimentation analysis of mutants F70A (data not shown) and L75A did not reveal any cosedimenting capsid proteins in any of the fractions, indicating that these mutations prevented any assembly of a capsid precursor. The individual capsid proteins in these lysates were presumably at the top of the gradient, where the majority of unassembled and monomeric proteins are found. These data were consistent with the electron microscopy results of these two mutants, which also did not reveal an assembled structure in the infected cell nucleus.
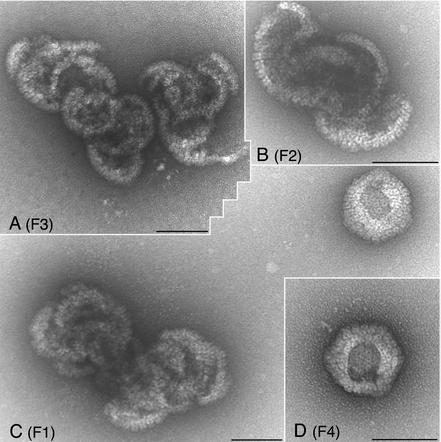
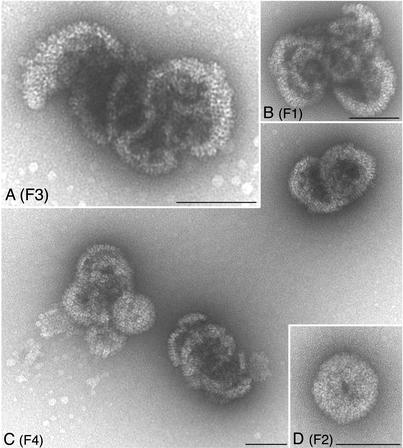
In order to analyze the sedimenting structures in detail, we performed an electron microscopy analysis of fractions from sucrose gradients. Vero cell monolayers were infected with KOS, I27A, and L65A. The cells were not labeled for this experiment. The infected cells were harvested, and nuclear lysates were sedimented through sucrose gradients as described above. KOS material in fractions where C capsids (fraction 4) and A/B capsids (fraction 9) sediment was analyzed by electron microscopy. For the two mutants (I27A and L65A), material from fractions 1 to 4, representing the bottom one third of the gradient, was assayed. A small amount of the fraction was loaded onto electron microscopy grids, negatively stained, and examined in the electron-microscope. The results of this analysis are shown in Fig. 8 to 10.
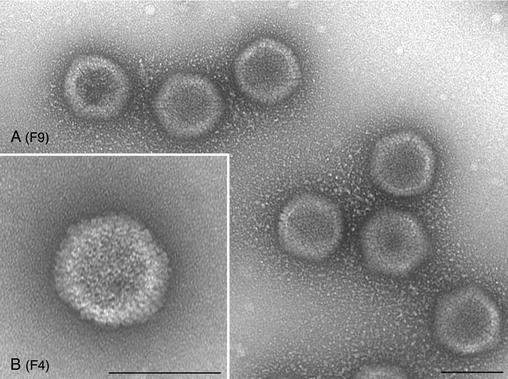
FIG. 8.
Electron micrographs of negatively stained KOS capsids. Infected-cell lysates were sedimented through sucrose gradients, and the capsids in the fractions were stained prior to electron microscopy. The gradients were fractioned from the bottom up. Shown in panel A are capsids (mixture of A and B capsids) present in fraction 9 (F9) of the gradient. C capsids (B) were present in fraction 4 (F4) of the gradient. Bars, 100 nm.
FIG. 10.
Negatively stained capsid shell particles observed in lysates of VP5 mutant L65A. Lysates derived from L65A-infected cells were processed as described for Fig. 9. Four fractions (F1 to F4) closest to the bottom of the tube were examined. Large open shell structures were evident in all fractions, with some nearing closure but never becoming completely closed. Bars, 100 nm.
In the KOS (Fig. 8) samples, icosahedral capsids that correspond to C capsids (F4) or A/B capsids (F9) were observed. In the mutant samples, I27A (Fig. 9) and L65A (Fig. 10), all the fractions contained large open capsid shell structures in which the capsomeres were clearly visible. There was no discernible size distribution of these structures relative to the fraction analyzed. The structures typically observed were large elaborate shells that appeared to twine around each other. Occasionally shell structures that appeared to be forming a closed spherical structure were observed, but a hole was still present in the structure (see Fig. 9D, F2, and Fig. 10C, F1, and 10D, F4). This could be an artifact of the staining process; however, the absence of such structures in the wild-type samples could indicate that these spherical structures are truly unclosed particles. Nevertheless, such structures were rarely observed, and the majority of the structures detected were the large open shells. This analysis visually demonstrates the importance of the VP5-scaffold protein interaction for the closure of the shell into an icosahedral capsid.
FIG. 9.
Negatively stained structures observed in I27A mutant lysates. Nuclear lysates obtained from I27A-infected cells were sedimented as described in the Fig. 8 legend. Material present in fractions F1 to F4 that represented the lower portion of the tube was analyzed. Large open shell structures were observed in fractions 1, 3, and 4. Bars, 100 nm.
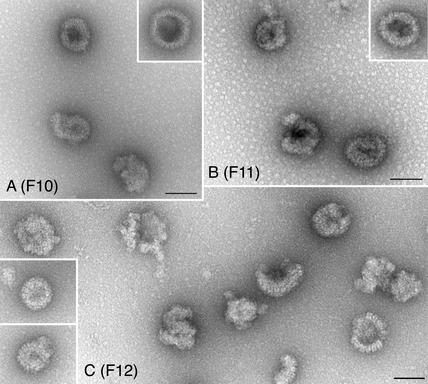
We also analyzed the shell structures formed in cells infected with L58A in similar experiments. For this mutant, the upper fractions of the sucrose gradients were analyzed (fractions 15 to 10) because the sedimentation results indicated that shells were more evident in these fractions (see Fig. 7, L58A). As shown in Fig. 11, shells were observed in these fractions (10 to 12); however, unlike those seen with the L65A and I27A mutants, these structures were much smaller. We did not observe the large intertwined shell structures in any of the fractions examined. Thus, it appears that although L58A is capable of forming capsid shells, these shells do not become larger in size nor do they form the elaborate structures typically seen and termed 6s and 9s.
FIG. 11.
Small shells observed in lysates of VP5 mutant L58A. Capsid structures in sucrose gradient fractions were negatively stained prior to electron microscopy analysis. The fractions analyzed were fractions 10 to 12 (F10 through F12) from the top half of the gradient. Small shell structures were evident in the fractions. Bars, 100 nm.
In summary, 24 mutations were made in the N-terminal region of VP5, spanning from residues L23 to F84. All 24 residues targeted specified hydrophobic side chains. Out of the 24, 7 were not able to interact with pre-22a, as judged from the yeast two-hybrid assay (see Table 4). When the mutations in these residues were transferred into the virus, all seven mutations specified a phenotype lethal for virus growth. Thus, seven key residues that are essential for pre-22a interaction and hence closure of the capsid shell into an icosahedral structure were identified (see Table 4). The leucine residue at position 58 of VP5 may also be important for capsomere-capsomere interactions that allow the formation of a large capsid shell. Mutation at residue F70 results in absence of the ICP5 protein in infected cells. Mutation at residue L75 of VP5 may affect another capsid protein interaction, as judged from the absence of an assembled structure in infected cells (see Table 4).
TABLE 4.
Summary of the residues of VP5 important for scaffold protein interaction
| Phenotype | VP5 residue |
|---|---|
| Loss of interaction with pre-22a in yeast two-hybrid assay | I27, L35, F39, L58, L65, L67, and L71 |
| Interaction with pre-22a in yeast two-hybrid assay | F70 and L75 |
| Formation of open capsid shells in infected cells | I27, L35, F39, L58, L65, L67, and L71 |
| No assembled structure in infected cells | F70 and L75 |
DISCUSSION
Previously we had derived a number of second-site revertants (PR mutants) of a mutant virus that contained a blocked maturation cleavage site in the scaffold protein (36). All but one of these revertant viruses contained a second-site mutation in the open reading frame encoding VP5, the interactive partner of the scaffold protein (pre-22a). The majority of these mutations mapped to the N terminus of this large polypeptide. The present study was aided by the analysis of the second-site mutations in VP5, which allowed us to focus our efforts on a small specific region of this large polypeptide. Although the PR mutants and mutants that were selected for their altered interaction with pre-22a following random mutagenesis of the VP5 N terminus (37) provided an extensive map of important VP5 residues, we did not have the identity of the key interactive residues.
In an important study, Hong and colleagues (15) determined the key interactive residues in the C-terminal tail of pre-22a and showed that an amphipathic helix which projected hydrophobic residues to the interactive surface was important. The hydrophobic residues of VP5 were thus likely candidates for this interaction, and so we mutated each hydrophobic residue starting at L23 and ending at F84. We made 24 mutations in this region, and all were changes to alanine to determine the contribution of the hydrophobic side chain. All the mutants were analyzed for interaction with pre-22a in the yeast two-hybrid system. Of the 24 mutations, only 7 abolished interaction with the scaffold protein; they were I27A, L35A, F39A, L58A, L65A, L67A, and L71A. These mutations were clustered in essentially two regions of the N terminus, and these regions correlate closely with the two regions in which the PR mutations mapped (see Fig. 12). Residue 39 was also shown from previous studies to be important for the interaction between these proteins in the yeast two-hybrid assay (37).
FIG. 12.
Residues in the N terminus of VP5 that are important for its interaction with the scaffold protein. The amino acid sequence of VP5 from residues 20 to 90 is shown. Above this sequence are the mutations identified in the revertant viruses and in the in vitro yeast two-hybrid assay. Shown below the sequence identified by solid arrows are the key hydrophobic residues identified in this study that abolish interaction with pre-22a when mutated.
To determine the effect of these mutations on virus replication and capsid assembly, all 24 mutations were introduced into the virus genome. We developed a genetic marker rescue/marker transfer procedure for the easy and rapid introduction of these mutations into the UL19 locus encoding VP5. This genetic approach facilitated the rapid generation of these mutant viruses and thus permitted us to analyze all the mutants in infected cells. This was important because mutations that had additional phenotypes were revealed only in the context of the virus replication cycle. There was an almost complete correlation between the inability of the mutant VP5 to interact with pre-22a and its ability to support virus replication. Thus, the same mutants which did not interact with the scaffold protein, I27A, L35A, F39A, L58A, L65A, L67A and L71A, were not capable of growth on Vero cells. The remaining mutant viruses grew on Vero cells with varied efficiencies. It was interesting that mutations adjacent to the lethal mutations listed above, for example, at F36, F38, and L57, also caused a decrease in the ability to interact with the scaffold protein in the yeast two-hybrid assay (see Table 1). However, when the same mutations were analyzed in the context of the virus replication cycle, they had a minimal effect on virus production (see Table 3).
Two mutants, F70A and L75A, did not fall into this pattern. The mutation at F70, when fused to the Gal4 DNA binding domain, was capable of the bimolecular interaction. However, the mutant polypeptide expressed from the virus genome was unstable, and no accumulation was detected. This explained the absence of growth of this mutant virus in Vero cells. The result of the mutation at residue L75 was unexpected because this mutant also interacted with pre-22a in the yeast two-hybrid assay, but the mutation was lethal for virus growth. The ICP5 polypeptide accumulates at levels comparable to the other viruses, and ultrastructural and sedimentation analysis did not reveal any capsid structure or precursor in the mutant-infected cells. Thus, if mutations of L75 do not result in a misfolded protein, this residue may be important for interaction of VP5 with other capsid proteins that are essential for assembly or capsomere formation.
Ultrastuctural analysis of the mutations that specified a lethal phenotype for virus growth visually revealed the effect of disrupting the VP5-pre-22a interaction. In cells infected with mutant viruses encoding changes at residues I27, L35, F39, L65, L67, and L71, large structures of capsid shells were observed. These shells, which were composed of numerous capsomeres and contained all the essential shell proteins (VP5, VP19C, and VP23) and also VP26, could not close into an icosahedral structure. These elaborate structures are presumably formed due to uncontrolled accretion of capsomeres with each other. The controlling factor in wild-type-infected cells is the scaffold protein, which “catalyzes” the synthesis of the icosahedral capsid. This phenotype was similar to that seen in cells infected with a virus that carries a null mutation in the gene encoding the scaffold protein (8).
The shell structures formed in cells infected with the L58A mutant were smaller, as shown by ultrastructural examination of infected cells, sedimentation analysis of lysates through sucrose gradients, and finally examination of these shells with electron microscopy. These shells were unable to continue accretion of the capsomeres to form large elaborate capsid shells. It is thus possible that residue L58 is important for capsomere-capsomere interactions that allow the formation of a large capsid shell necessary for enclosing the genome of this virus. It is also possible that structural perturbations of VP5 may explain this result; however, the mutant L58A polypeptide was capable of forming interactions with the remaining proteins to yield assembled shells.
Although the mutations were relatively conservative changes, some of the changes could have an affect on the correct folding of the VP5 molecule in that region or could have global effects on secondary structure that disrupt protein interactions. For mutants I27, L35, F39, L65, L67, and L71, this was not the case, as the mutant ICP5 polypeptide was still capable of the interactions that give rise to the capsid shells; these interactions allowed the formation of the capsomeres and the association with the triplex proteins to yield large sheets of the capsid shell. The mutation at residue L58 did result in slightly reduced levels of ICP5 accumulation. This was also observed in a second independent isolate of L58A. Both L58A isolates formed capsid shells, but these structures were less elaborate than the shells synthesized by the other mutant viruses. We do not think this is a consequence of the slightly lower accumulation of ICP5 in L58A mutant-infected cells, because if the mutant was capable of synthesizing large shells, they would still form, but fewer shells would be seen because of the reduced amounts of ICP5.
Mutation F70A did have an effect on protein stability. The mutation at residue 70 was puzzling because the mutant VP5 polypeptide fused to the Gal4 DNA binding domain interacts with pre-22a very well, but when the mutation was incorporated into the genetic locus encoding VP5, the ICP5 polypeptide was absent. We confirmed in the yeast assay that activation of the reporter gene was only seen when both VP5 F70A and pre-22a were present, that is, the F70A mutation in VP5 did not cause it to activate the reporter gene by itself. We recloned the sequenced F70A mutant fragment into the virus transfer plasmid and isolated two additional independent isolates. Both of these isolates required the VP5 complementing cell line for growth, and the ICP5 polypeptide was absent in cells infected with these viruses. Thus, we got the same result with three independent mutant isolations, which indicates that this phenotype could not be due to a second-site mutation. The simplest explanation for this result is that the mutation destabilizes ICP5 accumulation in infected cells, but fusion of the polypeptide to another protein domain overcomes this destabilization.
All the mutations generated in this study targeted hydrophobic residues, and the mutations were generally conservative changes to alanine. The methyl group of alanine maintains a hydrophobic group at the altered residues, while the smaller size of the methyl group should not cause physical disruption of VP5 structure. The change to alanine, resulting in abrogation of protein interaction, could indicate the requirement of a specific hydrophobic side chain for interaction, or it could indicate a change in the structural positioning of the binding domain, because leucines that are frequently encountered in alpha-helices were shown to be important for interaction. Although not much is known about the secondary structure in this region, cryoelectron microscopy of the HSV capsid has revealed that there are two long alpha-helices in the region of VP5 that is closest to the interior of the capsid, which is presumably the region that interacts with the scaffold proteins (39). The second-site revertants from the previous studies were located in an alpha-helix and a beta-sheet conformation of the protein, as predicted with a computer-generated program (29). The majority of the mutations from this experiment are also located in two alpha-helices, spanning residues 22 to 42 and residues 58 to 72, as judged from the same protein structure prediction methods. Structural analysis of the N terminus of VP5 complexed with the C terminus of the scaffold protein should reveal the locations and positions of the seven key residues identified in this study in this complex and establish how these residues contact the amphipathic helix in the C-terminal tail of the scaffold protein. This type of analysis would also identify the nature of the hydrophobic interactive surface in VP5.
The data presented here show again the importance of the N terminus of this large polypeptide for this bimolecular interaction, and now the specific residues of VP5 that participate in this interaction have been identified. Mutation of just one of these residues completely abolishes capsid assembly and virus replication. This has significant implications for developing an antiviral compound that targets an essential protein-protein interaction. The results of this study will enable one to design peptides that can effectively compete for interaction with the tail of the scaffold protein and determine the antiviral potential of these interactions. Alignment of the open reading frames of the major capsid proteins from the three herpesvirus families revealed four absolutely conserved residues in this region, F36, F54, G59, and F70. Mutations at positions F36 and F54 did not affect the VP5-pre-22a interaction. Mutation at residue F70 resulted in a defect in accumulation of the ICP5 polypeptide. This analysis also revealed that there is an absolute conservation of the hydrophobic side chain at residues 27, 35, 39, and 71. The potential benefits of this information could be the development of novel antiviral compounds that could block an essential step in herpesvirus replication.
These studies have revealed three important phenotypes associated with the major capsid protein and the residues that impart these phenotypes. Thus, closure of the capsid shell into an icosahedral structure by interaction with the scaffold protein is mediated through residues in two small domains of this large protein. Accretion of capsomeres during the synthesis of these shells may involve residue L58, and assembly of a multiprotein complex appears to require the participation of residue L75. These interpretations could and should be confirmed by high-resolution structural analysis of the protein assemblies. Nevertheless, this extensive analysis revealed that out of the 24 mutations that we made, only 7 resulted in loss of interaction with pre-22a in the yeast two-hybrid assay. All seven were essential for virus replication in the cell. The mutant viruses encoding these mutations formed the structures termed 6s and 9s (35), which demonstrates the importance of this interaction for capsid assembly. Thus, we have identified the hydrophobic residues that constitute the interactive domain in VP5 that binds to the C terminus of the scaffold protein.
Acknowledgments
This work was supported by National Institutes of Health grant AI33077 and by National Science Foundation grant NSF-DBI-0099706 to J.M.M.
We thank Stanley Person for his enduring enthusiasm for this work, insight into the data, and critical review of the manuscript. We also thank Wade Gibson for support and discussions of the data.
REFERENCES
- 1.Bartel, P. L., C-T. Chien, R. Sternglanz, and S. Fields. 1993. With the two-hybrid system to detect protein-protein interactions, p. 153-179. In D. A. Hartley (ed.), Cellular interactions in development: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 2.Beaudet-Miller, M., R. Zhang, J. Durkin, W. Gibson, A. D. Kwong, and Z. Hong. 1996. Virus-specific interaction between the human cytomegalovirus major capsid protein and the C terminus of the assembly protein precursor. J. Virol. 70:8081-8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breeden, L., and K. Nasmyth. 1985. Regulation of the yeast HO gene. Cold Spring Harb. Symp. Quant. Biol. 50: 643-650. [DOI] [PubMed] [Google Scholar]
- 4.Chien, C.-T., P. L. Bartel, R. Sternglanz, and S. Fields. 1991. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl. Acad. Sci. USA 88:9578-9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeLuca, N. A., A. M. McCarthy, and P. A. Schaffer. 1985. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J. Virol. 56:558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desai, P., N. A. DeLuca, J. C. Glorioso, and S. Person. 1993. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 inhibit capsid formation and cleavage of replicated DNA. J. Virol. 67:1357-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desai, P., F. L. Homa, S. Person, and J. C. Glorioso. 1994. A genetic selection method for the transfer of HSV-1 glycoprotein B mutations from plasmid to the viral genome: preliminary characterization of transdominance and entry kinetics of mutant viruses. Virology 204:312-322. [DOI] [PubMed] [Google Scholar]
- 8.Desai, P., S. C. Watkins, and S. Person. 1994. The size and symmetry of B capsids of herpes simplex virus type 1 are determined by the gene products of the UL26 open reading frame. J. Virol. 68:5365-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desai, P., and S. Person. 1996. Molecular interactions between the HSV-1 capsid proteins as measured by the yeast two-hybrid system. Virology 220:516-521. [DOI] [PubMed] [Google Scholar]
- 10.Desai, P., N. A. DeLuca, and S. Person. 1998. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology 247:115-124. [DOI] [PubMed] [Google Scholar]
- 11.Desai, P., and S. Person. 1999. Second site mutations in the N terminus of the major capsid protein (VP5) overcome a block at the maturation cleavage site of the capsid scaffold proteins of herpes simplex virus type 1. Virology 261:357-366. [DOI] [PubMed] [Google Scholar]
- 12.Fields, S., and O.-K. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 13.Gibson, W., and B. Roizman. 1972. Proteins specified by herpes simplex virus. VIII. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 10:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendricks, L. C., J. M. McCaffery, G. E. Palade, and M. G. Farquhar. 1993. Disruption of ER to Golgi transport leads to the accumulation of large aggregates containing β-COP in pancreatic acinar cells. Mol. Biol. Cell 4:413-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong, Z., M. Beaudet-Miller, J. Durkin, R. Zhang, and A. D. Kwong. 1996. Identification of a minimal hydrophobic domain in the HSV-1 scaffolding protein which is required for its interaction with the major capsid protein. J. Virol. 70:533-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kennard, J., F. J. Rixon, I. M. McDougall, J. D. Tatman, and V. G. Preston. 1995. The 25 amino acid residues at the carboxy terminus of the herpes simplex virus type 1 UL26.5 protein are required for the formation of the capsid shell around the scaffold. J. Gen. Virol. 76:1611-1621. [DOI] [PubMed] [Google Scholar]
- 17.Matusick-Kumar, L., W. W. Newcomb, J. C. Brown, P. J. McCann, I. I. I., W. Hurlburt, S. P. Weinheimer, and M. Gao. 1995. The C-terminal 25 amino acids of the protease and its substrate ICP35 of herpes simplex virus type 1 are involved in the formation of sealed capsids. J. Virol. 69:4347-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 19.Newcomb, W. W., B. L. Trus, N. Cheng, A. C. Steven, A. K. Sheaffer, D. J. Tenney, S. K. Weller, and J. C. Brown. 2000. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J. Virol. 74:1663-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicholson, P., C. Addison, A. M. Cross, J. Kennard, V. G. Preston, and F. J. Rixon. 1994. Localization of the herpes simplex virus type 1 major capsid protein VP5 to the cell nucleus requires the abundant scaffolding protein VP22a. J. Gen. Virol. 75:1091-1099. [DOI] [PubMed] [Google Scholar]
- 21.Oien, N. L., D. R. Thomsen, M. W. Wathen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1997. Assembly of herpes simplex virus capsids with the human cytomegalovirus scaffold protein: critical role of the C terminus. J. Virol. 71:1281-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pelletier, A., F. Do, J. J. Brisebois, L. Lagace, and M. G. Cordingly. 1997. Self-association of herpes simplex virus type 1 ICP35 is via coiled-coil interactions and promotes stable interaction with the major capsid protein. J. Virol. 71:5197-5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Person, S., and P. Desai. 1998. Capsids are formed in a mutant virus blocked at the maturation site of the UL26 and UL26.5 open reading frame of HSV-1 but are not formed in a null mutant of UL38 (VP19C). Virology 242:193-203. [DOI] [PubMed] [Google Scholar]
- 24.Rixon, F. J. 1993. Structure and assembly of herpesviruses. Semin. Virol. 4:135-144. [Google Scholar]
- 25.Rixon, F. J., C. Addison, A. McGregor, S. J. McNab, P. Nicholson, V. G. Preston, and J. D. Tatman. 1996. Multiple interactions control the intracellular localization of the herpes simplex virus type 1 capsid proteins. J. Gen. Virol. 77:2251-2260. [DOI] [PubMed] [Google Scholar]
- 26.Rixon, F. J., and D. McNab. 1999. Packaging-competent capsids of a herpes simplex virus temperature-sensitive mutant have properties similar to those of in vitro-assembled procapsids. J. Virol. 73:5714-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roizman, B., and A. Sears. 1996. Herpes simplex viruses and their replication, p. 2223-2295. In B. N. Fields, D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology. Lippincott-Raven, Philadelphia, Pa.
- 28.Rose, M., M. J. Casadaban, and D. Botstein. 1981. Yeast genes fused to β-galactosidase in Escherichia coli can be expressed normally in yeast. Proc. Natl. Acad. Sci. USA 78:2460-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rost, B., and C. Sander. 1994. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins 19:55-72. [DOI] [PubMed] [Google Scholar]
- 30.Southern, P. J., and P. Berg. 1982. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under the control of the SV40 early region promoter. J. Mol. Appl. Genet. 1:327-341. [PubMed] [Google Scholar]
- 31.Steven, A. C., and P. G. Spear. 1996. Herpesvirus capsid assembly and envelopment, p. 312-351. In R. Burnett et al. (ed.), Structural biology of viruses. Oxford University Press, New York, N.Y.
- 32.Sweet, R. M., and D. Eisenberg. 1983. Correlation of sequence hydrophobicities measures similarity in three-dimensional protein structure. J. Mol. Biol. 171:479-488. [DOI] [PubMed] [Google Scholar]
- 33.Tatman, J. D., V. G. Preston, P. Nicholson, R. M. Elliot, and F. J. Rixon. 1994. Assembly of herpes simplex virus type 1 capsids with a panel of recombinant baculoviruses. J. Gen. Virol. 75:1101-1113. [DOI] [PubMed] [Google Scholar]
- 34.Thomsen, D. R., W. W. Newcomb, J. C. Brown, and F. L. Homa. 1995. Assembly of herpes simplex virus capsid: requirement for the carboxy-terminal twenty-five amino acids of the proteins encoded by the UL26 and UL26.5 genes. J. Virol. 69:3690-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomsen, D. R., L. L. Roof, and F. L. Homa. 1994. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J. Virol. 68:2442-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warner, S. C., P. Desai, and S. Person. 2000. Second site mutations encoding residues 34 and 78 of the major capsid protein (VP5) of herpes simplex virus type 1 are important for overcoming a blocked maturation cleavage site of the capsid scaffold proteins. Virology 278:217-226. [DOI] [PubMed] [Google Scholar]
- 37.Warner, S. C., G. Chytrova, P. Desai, and S. Person. 2001. Mutations in the N terminus of VP5 alter its interaction with the scaffold proteins of herpes simplex virus type 1. Virology 284:308-316. [DOI] [PubMed] [Google Scholar]
- 38.Wood, L. J., M. K. Baxter, S. M. Plafker, and W. Gibson. 1997. Human cytomegalovirus capsid assembly protein precursor (pUL80.5) interacts with itself and with the major capsid protein (pUL86) through two different domains. J. Virol. 71:179-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou, Z. H., M. Dougherty, J. Jakana, J. He, F. J. Rixon, and W. Chiu. 2000. Seeing the herpesvirus capsid at 8.5A. Science 288:877-880. [DOI] [PubMed] [Google Scholar]