Abstract
This study shows a mechanism for the increase of signal transducer and activator of transcription 1 (STAT1) in Epstein-Barr virus-immortalized cells. Latent membrane protein 1 (LMP-1) expression was sufficient to induce STAT1 expression, DNA binding, and transcriptional activity. LMP-1-expressing cells can induce an increase in STAT1 expression in LMP-negative cells in the same culture, suggesting an indirect regulation of STAT1 expression. The increase in STAT1 expression is mediated by the C-terminal activating region 1 (CTAR-1) and/or CTAR-2 domains of LMP-1 and is inhibited by mutant IκB, demonstrating a role for NFκB in LMP-1-mediated STAT1 expression.
Constitutive STAT activation has been detected in a wide variety of human cancers, including Epstein-Barr virus (EBV)-associated tumors, implicating these molecules in tumor formation and progression. Immunohistochemical analysis has demonstrated the constitutive activation of STAT members 1, 3, and 5 in nasopharyngeal carcinoma tumors (10). Furthermore, a recent study has demonstrated constitutive STAT3 activation in the malignant cells of Hodgkin's disease (22). STAT proteins might promote oncogenesis through regulating the expression of genes encoding proteins involved in cell cycle progression and/or apoptosis—Bcl-XL, cyclin D1, and c-Myc (4). For example, interfering with STAT3 activation by inhibition of the interleukin-6 signaling pathway decreases the expression of Bcl-XL and leads to apoptosis (9). Dominant-negative forms of STAT3 protein have been shown to block transformation of fibroblasts by the Src oncoprotein (7) and to significantly inhibit tumor growth and tumor regression in syngeneic mice (24). The observation that STATs 1 and 3 are constitutively activated in EBV-positive lymphoblastoid cell lines (LCLs), but not in EBV-negative Burkitt's lymphoma (BL) cell lines (28), implies that EBV genes contribute to constitutive STAT activation. Recent studies suggest that STAT1 actually has growth-inhibitory effects (6, 8, 11). However, in cells in which STATs 1, 3, and 5 are simultaneously activated, the proproliferative properties of STATs 3 and 5 potentially predominate the antiproliferative activity of STAT1 (4). Relatively little is known about the mechanisms regulating STAT proteins in lymphoid tumors associated with EBV infection.
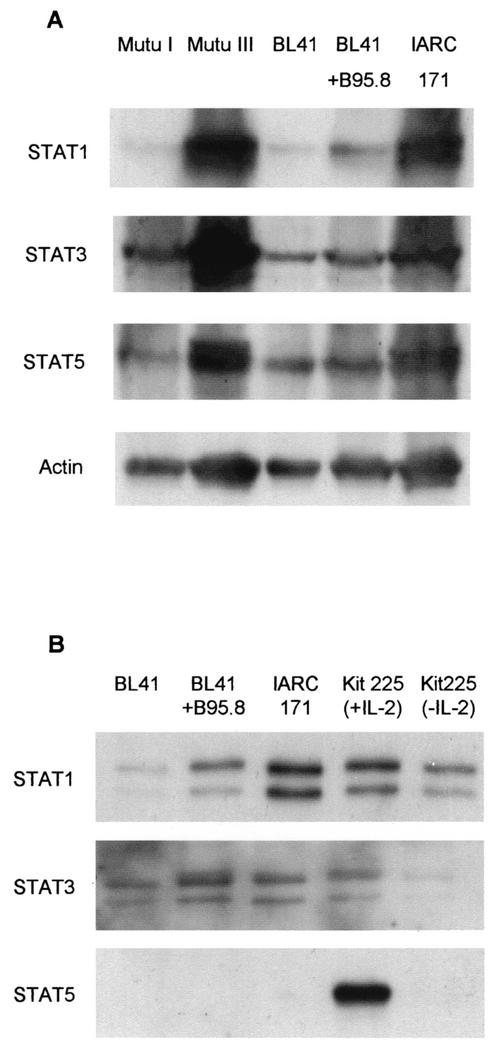
Two sets of B-cell lines were chosen in which to analyze STAT proteins. The two sets are essentially genetically identical and differ primarily in their patterns of EBV gene expression. The first set consists of BL41, BL41+B95.8, and IARC-171 (26). BL41 is an EBV-negative BL line. BL41+B95.8 is the same line after infection with the B95.8 strain of EBV. IARC-171 is an EBV-immortalized LCL derived from the same patient as BL41. The second set consists of Mutu I and Mutu III (16). These are two sublines from an EBV-positive BL tumor. Mutu I expresses only the EBNA1 viral protein, a limited viral protein expression termed latency I. Mutu III, like BL41+B95.8 and IARC-171, expresses the full array of EBV latent genes—EBNA1, EBNA2, EBNA 3A, EBNA 3B, EBNA 3C, EBNA-LP, latent membrane protein 1 (LMP-1), and LMP-2. This expression is termed latency III (25). We investigated STAT1, -3, and -5 protein levels. Protein expression was investigated by resolution of total cell lysates on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blotting and antibody detection. A representative result is shown in Fig. 1A, illustrating STAT1, -3, and -5 expression in the two sets of lines. Higher levels of expression of all three STATs can be seen in Mutu III than in Mutu I. STAT1 expression was increased in BL41+B95.8 and IARC-171 compared to that in BL41 itself. This suggests that latency III viral gene expression promotes expression of STAT proteins.
FIG. 1.
EBV latent genes increase STAT protein expression and STAT1 nuclear DNA binding. (A) Total lysates of five B-cell lines with various EBV statuses were prepared and analyzed by SDS-PAGE and Western blotting. Typically, the lysates from 4 × 105 cells were applied to each track of the gel. STAT proteins were detected using antibodies specific to STAT1 (sc-346; Santa Cruz Biotechnology [SCB]), STAT3 (sc-7179; SCB), and STAT5 (610191; BD Transduction Laboratories). Protein loading was monitored by actin protein detection using an actin-specific antibody (A-2066; Sigma). (B) Using a DNA affinity precipitation assay, STAT DNA binding was measured from nuclear extracts of the BL41, BL41+B95.8, IARC-171, and Kit225 cell lines. Typically, nuclear extracts from 20 million cells were used for these experiments. Nuclear proteins were incubated (in conditions of 4°C and 80 min with rotation) with 1 μg of 5′ biotinylated, double-stranded oligonucleotide derived from the FcγR1-GAS (GRR) (GTATTTC CCAGAAAAGGAAC) and 30 μl of prewashed streptavidin-conjugated agarose beads (50% slurry in phosphate-buffered saline, 0.05% sodium azide) (s-1638; Sigma) to identify DNA binding proteins. The underlined portion of the oligonucleotide represents the STAT consensus sequence. DNA binding proteins were separated by SDS-PAGE and analyzed by Western blotting using antibodies specific to STAT1, STAT3, and STAT5 proteins.
STAT activity is regulated by changes in STAT DNA binding. We investigated STAT DNA binding in nuclear extracts from the BL41, BL41+B95.8, and IARC-171 cell lines by affinity purification. This is a nonradioactive measurement of STAT DNA binding. Nuclear extracts were prepared, and the STAT DNA binding proteins were precipitated using a biotinylated, double-stranded FcγR1-GAS (GRR) oligonucleotide, which corresponds to a STAT consensus sequence, and avidin agarose beads (1). After incubation of the nuclear extracts with the oligonucleotide coupled to the beads, the DNA binding proteins were precipitated by centrifugation. The beads were then washed to remove nonspecific interactions. The DNA binding proteins were removed from the beads by boiling in SDS loading buffer and analyzed by SDS-PAGE and Western blotting. Experiments were performed to establish the binding specificity of STAT proteins to the GRR DNA sequence (data not shown).
Figure 1B illustrates a representative result from this experiment. Analysis of DNA binding levels in BL41+B95.8 and IARC-171 reveals higher levels of STAT1 nuclear DNA binding than those seen with the EBV-negative BL41 cell line. STAT1 DNA binding patterns, similar to that observed in the IARC-171 cell line, were also observed in two other LCLs and the Mutu III cell line (data not shown). Subtle differences in STAT3 DNA binding were detected, but these were not as large as the differences in that of STAT1. STAT5 DNA binding was not observed in the BL41, BL41+B95.8, or IARC-171 cell lines but was observed in the Kit225 cells that were used as a positive control. A recent study has shown that STAT5 is not constitutively activated in spontaneous LCLs derived from posttransplant lymphoproliferative disease patients (23a). This is consistent with the lack of nuclear STAT5 binding to DNA in the BL41+B95.8 and IARC-171 cell lines. The DNA affinity precipitation results demonstrate high levels of STAT1 DNA binding in latency III cell lines, indicative of higher levels of STAT1 transcriptional activity in latency III cell lines.
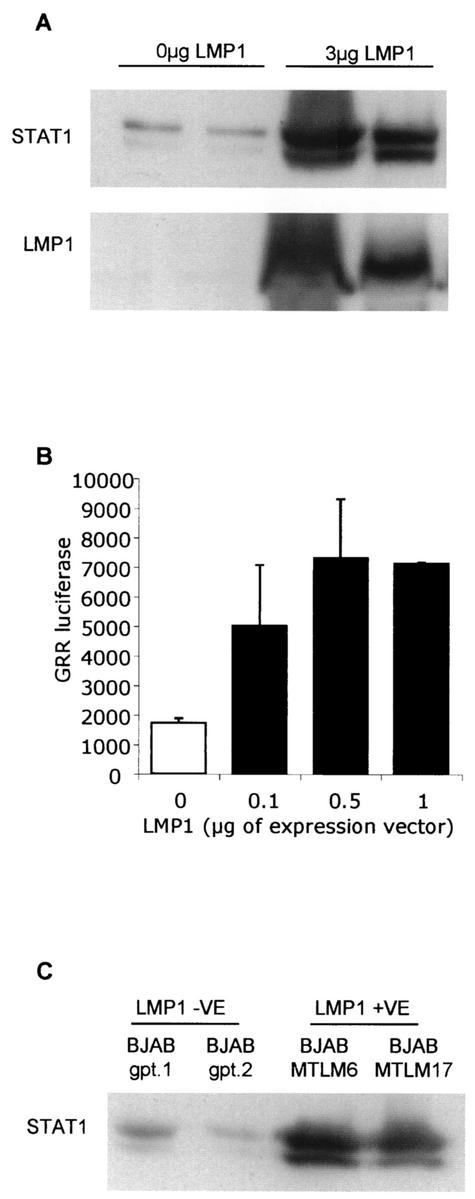
One of the critical proteins expressed during EBV latency is LMP-1, which has been shown to be essential for EBV-induced transformation of primary B cells (18, 21). We sought to determine whether EBV latent gene LMP-1 can modulate STAT1 expression and STAT1 nuclear DNA binding. An EBV-negative tumor B-cell line, DG75 (2), was transiently transfected with 3 μg of LMP-1 expression vector (17). At 48 h posttransfection, total lysates were generated and analyzed for STAT1 protein levels. DG75 cells transfected with 3 μg of LMP-1 demonstrated elevated levels of STAT1 expression compared to those seen with control transfections (Fig. 2A). The expression levels of LMP-1 following the transfection were determined (Fig. 2A, lower panel). STAT activity was also investigated following transient transfection. A synthetic STAT reporter was cotransfected with various amounts of an LMP-1 expression vector. The STAT reporter used contained five copies of the FcγR-Gas (GRR) STAT consensus sequence, the same sequence utilized for the DNA affinity precipitation, upstream of a luciferase gene. After 18 h, cells were harvested and lysed and luciferase levels were assayed. Figure 2B shows that the presence of LMP-1 was sufficient to induce STAT activity in DG75 cells. This finding is similar to data seen with other cell systems (5, 14).
FIG. 2.
EBV latent gene LMP-1 increases STAT1 protein expression, STAT transcriptional activity, and STAT1 nuclear DNA binding. (A) DG75 cells were transfected with 3 μg of LMP-1 expression vector by electroporation using a Bio-Rad Genepulser II electroporator set to 270 V and 950 μF. Typically, 10 million DG75 cells were used per transfection. The total amount of DNA used in each transfection was kept constant by the addition of an appropriate amount of pSG5 empty vector. At 48 h posttransfection, total lysates were prepared and analyzed by SDS-PAGE and Western blotting using antibodies specific to STAT1 and LMP-1 proteins. The results shown are representative of four separate experiments. (B) DG75 cells were transfected with various amounts of an LMP-1 expression vector and a STAT reporter containing five copies of the FcγR1-GAS (GRR) GTATTTCCCAGAAAAGGAAC consensus sequence upstream of a luciferase gene. After 18 h, cells were harvested, lysed, and assayed for levels of luciferase. (C) Using the agarose-coupled GRR oligonucleotide, DNA binding complexes were affinity precipitated from nuclear extracts of the two BJAB cell lines stably expressing LMP-1 and two control cell lines. DNA binding proteins were separated by SDS-PAGE and analyzed by Western blotting using an antibody specific to STAT1 protein. The results shown are representative of two separate experiments.
The ability of LMP-1 to regulate STAT1 activity, as measured by nuclear DNA binding, was also tested using two LMP-1-expressing stable transfectants of BJAB, an EBV-negative B-cell lymphoma cell line (27) (Fig. 2C). The BJAB cell lines stably expressing LMP-1 demonstrated increased nuclear STAT1 binding to DNA compared to control cell lines. Further studies using DG75 cells with tetracycline-regulatable LMP-1 expression (15) also displayed elevated STAT1 nuclear DNA binding in the presence of LMP-1 expression (data not shown). The data from this set of experiments demonstrate that LMP-1 is sufficient alone to cause an increase in STAT1 expression, STAT activity, and STAT1 DNA binding.
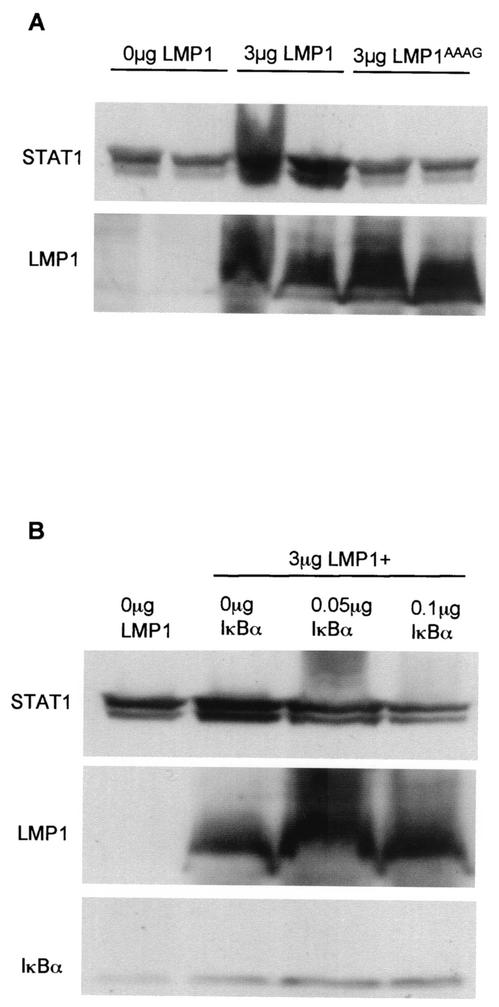
The cytoplasmic tail of LMP-1 contains two effector domains, termed C-terminal activating region 1 (CTAR-1) and CTAR-2, through which the majority of known LMP-1 signaling is mediated (13). LMP-1AAAG contains four point mutations, three in the CTAR-1 domain and one in the CTAR-2 domain, which render it defective in activating nuclear signaling pathways (5). DG75 cells were transiently transfected with either 3 μg of LMP-1 or 3 μg of LMP-1AAAG and were analyzed for STAT1 protein levels at 48 h posttransfection (Fig. 3A, upper panel). LMP-1AAAG was unable to increase STAT1 expression. The LMP-1 blot analysis revealed approximately equal quantities of LMP-1 and LMP-1AAAG in duplicate transfections (Fig. 3A, lower panel). From these data, we can conclude that LMP-1 increases STAT1 expression via CTAR-1 and/or CTAR-2 signaling pathways.
FIG. 3.
LMP-1 regulates STAT1 expression through CTAR-1 and/or CTAR-2 signaling domains via an NFκB-mediated pathway. (A) DG75 cells were transfected with 3 μg of LMP-1 expression vector or 3 μg of LMP-1AAAG expression vector. The total amount of DNA used in each transfection was kept to a constant 3 μg by the addition of an appropriate amount of pSG5 empty vector. Transfections were performed in duplicate, and control transfections are shown in lanes 1 and 2 from the left. At 48 h posttransfection, total lysates were prepared and analyzed by SDS-PAGE and Western blotting using antibodies specific to STAT1 and LMP-1 proteins. Similar data were seen in three separate experiments. (B) DG75 cells were transfected with 3 μg of LMP-1 expression vector and two different amounts of IκBαΔN expression vector (0.05 and 0.1 μg IκBαΔN). The total amount of DNA used in each transfection was kept constant by the addition of an appropriate amount of pSG5 empty vector. Control transfection is shown in the first lane from the left. At 48 h posttransfection, total lysates were prepared and analyzed by SDS-PAGE and Western blotting using antibodies specific to STAT1, LMP-1, and IκBα (sc-371; SCB) proteins. Similar results were obtained from three different experiments.
The NFκB transcription factor pathway is one of the major signaling pathways activated by the CTAR-1 and CTAR-2 domains of LMP-1 (17). To determine whether NFκB played a role in the regulation of STAT1 expression by LMP-1, we utilized IκBαΔN (kindly provided by Dean W. Ballard, Howard Hughes Medical Institute, Nashville, Tenn.), a potent inhibitor of NFκB activation. DG75 cells transfected with LMP-1 alone contained higher STAT1 levels than the control transfection (Fig. 3B). Analysis of DG75 cells transfected with LMP-1 plus 0.1 μg of IκBαΔN revealed that the inhibition of NFκB prevented LMP-1 from increasing STAT1 protein expression (Fig. 3B). Figure 3 also shows the results of an LMP-1 blot analysis, which reveals that the 0.1-μg concentration of IκBαΔN did not diminish LMP-1 levels. This dose of IκBαΔN was sufficient to inhibit LMP-1-induced NFκB activity, as measured by an NFκB reporter assay (data not shown). These observations provide evidence that LMP-1 increases STAT1 protein expression via LMP-1-mediated NFκB signaling.
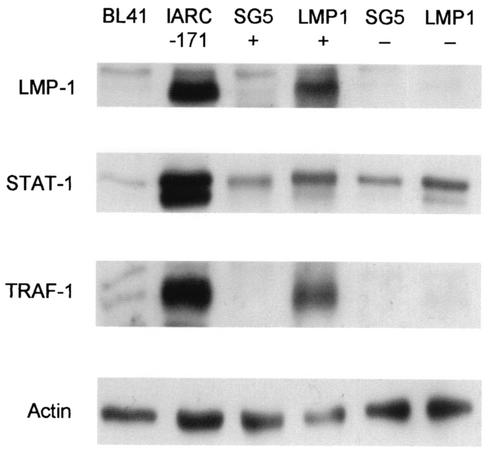
To test the method of regulation of STAT1 expression by LMP-1, we magnetically sorted transiently transfected DG75 cells. At 48 h posttransfection, DG75 cultures (transfected with 3 μg of rCD2-enhanced green fluorescent protein [EGFP] and 3 μg of pSG5 empty vector or 3 μg of LMP-1 expression vector) were incubated first with OX34 anti-rat CD2 hybridoma supernatant (19) and then with Rat anti-mouse IgG2a+b microbeads (Miltenyi Biotech). Transfected cells (positive fractions) and nontransfected cells (negative fractions) were obtained using MS+ separation columns (Miltenyi Biotech). Total lysates of the positive and negative fractions were prepared and analyzed for LMP-1, STAT1, and tumor necrosis factor receptor-associated factor 1 (TRAF-1) protein levels. Total lysates of the BL41 and IARC-171 cell lines were included in the Western blotting experiment as negative and positive controls, respectively. Figure 4 illustrates a representative result from this experiment. The results of the LMP-1 blot analysis demonstrate the purity of the sort, which was also monitored by flow cytometric analysis of gated live cells for rCD2-EGFP (20) expression, demonstrating approximately 70% sort purity for transfected cells and more than 95% sort purity for nontransfected cells (data not shown). Both the SG5-positive and -negative DG75 sorted populations contained equal, basal quantities of STAT1. The LMP-1-positive DG75 sorted population displayed an increase in STAT1 protein expression compared to the SG5-positive DG75 sorted population. Of particular interest was the LMP-1-negative DG75 sorted population, which also displayed elevated STAT1 expression. This observation provides evidence that LMP-1 increases STAT1 expression via indirect mechanisms. The TRAF-1 blot was included in this experiment as an important control. TRAF-1 has previously been shown to be upregulated via LMP-1-mediated NFκB activation (12). This experiment shows that TRAF-1 gene expression is regulated directly. This is in contrast to LMP-1 upregulation of STAT1 expression. From this experiment we can deduce that LMP-1-expressing cells can induce LMP-1-negative cells in the same culture to increase STAT1 protein expression.
FIG. 4.
LMP-1-expressing cells upregulate STAT1 expression in cocultured LMP-1-negative cells. DG75 cells were transfected with 3 μg of pSG5 empty vector or 3 μg of LMP-1 expression vector and 3 μg of rCD2-EGFP expression vector. At 48 h posttransfection, transfected cells were magnetically selected using a monoclonal antibody against rCD2. Total lysates of the transfected (+) and nontransfected (−) cell populations were prepared and analyzed by SDS-PAGE and Western blotting using antibodies specific to LMP-1, STAT1, TRAF-1 (sc-875; SCB), and actin. BL41 and IARC-171 total lysates were included as negative and positive controls, respectively. The results shown are representative of three separate experiments.
We have performed coculture studies in which LMP-1-positive cells were physically separated from LMP-1-negative cells, allowing only the passage of soluble factors such as cytokines between the two populations. Alpha interferon was used as a control for the experiment, and it strongly induced STAT1 expression in LMP-1-negative cells (data not shown). However, LMP-1-expressing cells were not able to upregulate STAT1 protein levels in the LMP-1-negative population when they were physically separated. Although it is possible that local production of cytokines is involved in the increased STAT1 expression, these data suggest that a physical cell-cell interaction is required. LMP-1 increases the expression of various cell surface molecules in an NFκB-dependent fashion (12, 23). However, the identity of the molecules that regulate this indirect STAT1 expression remains to be elucidated.
The powerful effects of the presence of LMP-1 on STAT1 expression suggest that an investigation of STAT1 function during EBV immortalization is an important next step in elucidating the role of this transcription factor in EBV-associated malignancy. To continue the work performed in this study, a dominant-negative STAT1 protein is to be used to investigate STAT1 effects on LMP-1-regulated genes. Some genes known to be regulated by LMP-1, such as HLA, are also regulated by gamma interferon, which activates STAT1 (3). Identification of STAT-dependent LMP-1-regulated genes might pinpoint mechanisms by which STAT proteins influence oncogenesis in EBV-associated tumors.
Acknowledgments
C.R. is supported by a Gordon Pillar Studentship from the Leukemia Research Fund. We thank the Leukemia Research Fund for all their generous support for the laboratory. C.F. is supported by the Leverhulme Trust.
REFERENCES
- 1.Beadling, C., J. Ng, J. W. Babbage, and D. A. Cantrell. 1996. Interleukin-2 activation of STAT5 requires the convergent action of tyrosine kinases and a serine/threonine kinase pathway distinct from the Raf1/ERK2 MAP kinase pathway. EMBO J. 15:1902-1913. [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Bassat, H. N., S. Goldblum, T. Mitrani, J. M. Goldblum, M. M. Yoffey, Z. Cohen, B. Bentwich, E. Ramot, E. Klein, and G. Klein. 1977. Establishment in continuous culture of a new type of lymphocyte from a “Burkitt like” malignant lymphoma (line D.G.-75). Int. J. Cancer 19:27-33. [DOI] [PubMed] [Google Scholar]
- 3.Boehm, U., T. Klamp, M. Groot, and J. C. Howard. 1997. Cellular responses to interferon-gamma. Annu. Rev. Immunol. 15:749-795. [DOI] [PubMed] [Google Scholar]
- 4.Bowman, T., R. Garcia, J. Turkson, and R. Jove. 2000. STATs in oncogenesis. Oncogene 19:2474-2488. [DOI] [PubMed] [Google Scholar]
- 5.Brennan, P., J. E. Floettmann, A. Mehl, M. Jones, and M. Rowe. 2001. Mechanism of action of a novel latent membrane protein-1 dominant negative. J. Biol. Chem. 276:1195-1203. [DOI] [PubMed] [Google Scholar]
- 6.Bromberg, J. F., Z. Fan, C. Brown, J. Mendelsohn, and J. E. Darnell, Jr. 1998. Epidermal growth factor-induced growth inhibition requires Stat1 activation. Cell Growth Differ. 9:505-512. [PubMed] [Google Scholar]
- 7.Bromberg, J. F., C. M. Horvath, D. Besser, W. W. Lathem, and J. E. Darnell, Jr. 1998. Stat3 activation is required for cellular transformation by v-src. Mol. Cell. Biol. 18:2553-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bromberg, J. F., C. M. Horvath, Z. Wen, R. D. Schreiber, and J. E. Darnell, Jr. 1996. Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc. Natl. Acad. Sci. USA 93:7673-7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catlett-Falcone, R., T. H. Landowski, M. M. Oshiro, J. Turkson, A. Levitzki, R. Savino, G. Ciliberto, L. Moscinski, J. L. Fernandez-Luna, G. Nunez, W. S. Dalton, and R. Jove. 1999. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity 10:105-115. [DOI] [PubMed] [Google Scholar]
- 10.Chen, H., J. M. Lee, Y. Zong, M. Borowitz, M. H. Ng, R. F. Ambinder, and S. D. Hayward. 2001. Linkage between STAT regulation and Epstein-Barr virus gene expression in tumors. J. Virol. 75:2929-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chin, Y. E., M. Kitagawa, W. C. Su, Z. H. You, Y. Iwamoto, and X. Y. Fu. 1996. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21 WAF1/CIP1 mediated by STAT1. Science 272:719-722. [DOI] [PubMed] [Google Scholar]
- 12.Devergne, O., E. C. McFarland, G. Mosialos, K. M. Izumi, C. F. Ware, and E. Kieff. 1998. Role of the TRAF binding site and NF-κB activation in Epstein-Barr virus latent membrane protein 1-induced cell gene expression. J. Virol. 72:7900-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eliopoulos, A. G., and L. S. Young. 2001. LMP1 structure and signal transduction. Semin. Cancer Biol. 11:435-444. [DOI] [PubMed] [Google Scholar]
- 14.Fielding, C. A., K. Sandvej, A. Mehl, P. Brennan, M. Jones, and M. Rowe. 2001. Epstein-Barr virus LMP-1 natural sequence variants differ in their potential to activate cellular signaling pathways. J. Virol. 75:9129-9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Floettmann, J. E., K. Ward, A. B. Rickinson, and M. Rowe. 1996. Cytostatic effect of Epstein-Barr virus latent membrane protein-1 analyzed using tetracycline-regulated expression in B cell lines. Virology 223:29-40. [DOI] [PubMed] [Google Scholar]
- 16.Gregory, C. D., M. Rowe, and A. B. Rickinson. 1990. Different Epstein-Barr virus-B cell interactions in phenotypically distinct clones of a Burkitt's lymphoma cell line. J. Gen. Virol. 71:1481-1495. [DOI] [PubMed] [Google Scholar]
- 17.Huen, D. S., S. A. Henderson, D. Croom-Carter, and M. Rowe. 1995. The Epstein-Barr virus latent membrane protein-1 (LMP1) mediates activation of NF-kappa B and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene 10:549-560. [PubMed] [Google Scholar]
- 18.Izumi, K. M. 2001. Identification of EBV transforming genes by recombinant EBV technology. Semin. Cancer Biol. 11:407-414. [DOI] [PubMed] [Google Scholar]
- 19.Jefferies, W. A., J. R. Green, and A. F. Williams. 1985. Authentic T helper CD4 (W3/25) antigen on rat peritoneal macrophages. J. Exp. Med. 162:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keating, S., S. Prince, M. Jones, and M. Rowe. 2002. The lytic cycle of Epstein-Barr virus is associated with decreased expression of cell surface major histocompatibility complex class I and class II molecules. J. Virol. 76:8179-8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieff, E. 1996. Epstein-Barr virus and its replication, p. 2343-2396. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 22.Kube, D., U. Holtick, M. Vockerodt, T. Ahmadi, B. Haier, I. Behrmann, P. C. Heinrich, V. Diehl, and H. Tesch. 2001. STAT3 is constitutively activated in Hodgkin cell lines. Blood 98:762-770. [DOI] [PubMed] [Google Scholar]
- 23.Mehl, A. M., J. E. Floettmann, M. Jones, P. Brennan, and M. Rowe. 2001. Characterization of intercellular adhesion molecule-1 regulation by Epstein-Barr virus-encoded latent membrane protein-1 identifies pathways that cooperate with nuclear factor kappa B to activate transcription. J. Biol. Chem. 276:984-992. [DOI] [PubMed] [Google Scholar]
- 23a.Nepomuceno, R. R., A. L. Snow, P. R. Beatty, S. M. Krams, and O. M. Martinez. 2002. Constitutive activation of Jak/STAT proteins in Epstein-Barr virus-infected B-cell lines from patients with posttransplant lymphoproliferative disorder. Transplantation 74:396-402. [DOI] [PubMed] [Google Scholar]
- 24.Niu, G., R. Heller, R. Catlett-Falcone, D. Coppola, M. Jaroszeski, W. Dalton, R. Jove, and H. Yu. 1999. Gene therapy with dominant-negative Stat3 suppresses growth of the murine melanoma B16 tumor in vivo. Cancer Res. 59:5059-5063. [PubMed] [Google Scholar]
- 25.Rowe, M., A. L. Lear, D. Croom-Carter, A. H. Davies, and A. B. Rickinson. 1992. Three pathways of Epstein-Barr virus gene activation from EBNA1-positive latency in B lymphocytes. J. Virol. 66:122-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rowe, M., C. M. Rooney, C. F. Edwards, G. M. Lenoir, and A. B. Rickinson. 1986. Epstein-Barr virus status and tumour cell phenotype in sporadic Burkitt's lymphoma. Int. J. Cancer 37:367-373. [DOI] [PubMed] [Google Scholar]
- 27.Wang, F., C. Gregory, C. Sample, M. Rowe, D. Liebowitz, R. Murray, A. Rickinson, and E. Kieff. 1990. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J. Virol. 64:2309-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weber-Nordt, R. M., C. Egen, J. Wehinger, W. Ludwig, V. Gouilleux-Gruart, R. Mertelsmann, and J. Finke. 1996. Constitutive activation of STAT proteins in primary lymphoid and myeloid leukemia cells and in Epstein-Barr virus (EBV)-related lymphoma cell lines. Blood 88:809-816. [PubMed] [Google Scholar]