Abstract
Microtubule nucleation on the centrosome and the fungal equivalent, the spindle pole body (SPB), is activated at the onset of mitosis. We previously reported that mitotic extracts prepared from Xenopus unfertilized eggs convert the interphase SPB of fission yeast into a competent state for microtubule nucleation. In this study, we have purified an 85-kDa SPB activator from the extracts and identified it as the ribonucleotide reductase large subunit R1. We further confirmed that recombinant mouse R1 protein was also effective for SPB activation. On the other hand, another essential subunit of ribonucleotide reductase, R2 protein, was not required for SPB activation. SPB activation by R1 protein was suppressed in the presence of anti-R1 antibodies or a partial oligopeptide of R1; the oligopeptide also inhibited aster formation on Xenopus sperm centrosomes. In accordance, R1 was detected in animal centrosomes by immunofluorescence and immunoblotting with anti-R1 antibodies. In addition, recombinant mouse R1 protein bound to γ- and α/β-tubulin in vitro. These results suggest that R1 is a bifunctional protein that acts on both ribonucleotide reduction and centrosome/SPB activation.
INTRODUCTION
The centrosome is the major microtubule-organizing center (MTOC) in animal cells. It originates a microtubule radial array in interphase and establishes cellular polarity by defining cell shape, nuclear position, and the direction of vesicle/organelle transport mediated by microtubule-dependent motors (reviewed by Kellogg et al., 1994; Reinsch and Gonczy, 1998). The centrosome duplicates once each cell cycle, and the duplicated centrosomes function as spindle poles in mitosis, establishing the bipolar spindle, which specifies the site of the cleavage furrow plane and effects balanced segregation of chromosomes (reviewed by Kellogg et al., 1994; Pereira and Schiebel, 1997; Waters and Salmon, 1997). Several reports suggest that the integrity of centrosomal dynamics and function through the cell cycle are essential for proper cell cycle progression and genetic stability: deregulated duplication of the centrosome is implicated in multipolar spindle assembly and abnormal chromosome segregation, resulting in the aneuploidy observed in many tumor cell types (reviewed by Brinkley and Goepfert, 1998). Several proteins involved in cell cycle progression localize on the centrosome; they include CDK1/cyclins (Bailly et al., 1989, 1992; Gallant and Nigg, 1992; Maldonado-Codina and Glover, 1992), components of anaphase promoting complex/cyclosome (APC/C) (Tugendreich et al., 1995) and the proteasome machinery (Wigley et al., 1999), and the p53 tumor suppressor protein (Brown et al., 1994). These observations suggest that the centrosome may be involved in signaling pathways regulating cell cycle progression.
The spindle pole body (SPB) is the fungal equivalent of the centrosome. The centrosomes and the SPBs differ structurally, yet their functions are conserved (reviewed by Hagan et al., 1998). Microtubule nucleation at the centrosomes and the SPBs is markedly enhanced at the onset of mitosis (Snyder and McIntosh, 1975; Kuriyama and Borisy, 1981; Hagan and Hyams, 1988; Masuda et al., 1992). Activation of microtubule nucleation, altered microtubule dynamics, and microtubule-based motor proteins are all required for separation of the duplicated centrosomes and proper formation of the mitotic spindle (reviewed by Vernos and Karsenti, 1996; Pereira and Schiebel, 1997; Waters and Salmon, 1997). In spite of the many studies that have identified various proteins associated with the centrosomes or the SPBs (reviewed by Kalt and Schliwa, 1993; Hagan et al., 1998), the molecular mechanism of microtubule nucleation is still unclear.
Of the candidate molecules required for microtubule nucleation at the MTOC, γ-tubulin is the most prominent: deletion of γ-tubulin function, by gene disruption or by injection of γ-tubulin antibodies, causes defects in microtubule organization in a diverse array of organisms (Oakley et al., 1990; Horio et al., 1991; Joshi et al., 1992; Sobel and Snyder, 1995; Sunkel et al., 1995; Marschall et al., 1996; Spang et al., 1996). It has been proposed that multiprotein complexes containing γ-tubulin are recruited from the cytoplasm to the centrosome/SPB and participate in microtubule nucleation (Felix et al., 1994; Stearns and Kirschner, 1994; Moritz et al., 1995; Zheng et al., 1995; Moudjou et al., 1996; Vogel et al., 1997; Martin et al., 1998; Moritz et al., 1998; Murphy et al., 1998; Pereira et al., 1998; Tassin et al., 1998; Fava et al., 1999) and that the GTP-bound form of Ran may regulate the recruitment through the action of RanBPM (Nakamura et al., 1998; reviewed in Pennisi, 1999). It remains largely unknown, however, how the microtubule-nucleating activity of γ-tubulin complexes is regulated.
Because the Schizosaccharomyces pombe SPB is the clearest example of an MTOC displaying mitosis-specific activation of existing nucleation sites already containing γ-tubulin, we have used this organism as a model system for studying mechanisms of microtubule nucleation activation (Masuda et al., 1992; Masuda and Shibata, 1996). In vitro assays of microtubule polymerization from S. pombe SPBs using lysed cells revealed that the nucleating capacity of the SPB is low during interphase and increases markedly with entry into mitosis (Masuda et al., 1992). γ-Tubulin localizes on the osmiophilic material that lies near the inner surface of the nuclear envelope immediately adjacent to the SPB (Ding et al., 1997), but because the level of this γ-tubulin does not change significantly as the cell enters mitosis (Horio et al., 1991; Masuda et al., 1992, Ding et al., 1997), simple localization of γ-tubulin to the SPB appears to be insufficient to promote microtubule assembly. We previously showed that Xenopus egg mitotic extracts convert the interphase SPB into a competent state for microtubule nucleation and that the conversion occurs downstream of CDK1/cyclin B (Masuda et al., 1992). We also showed that SPB activation probably occurs through direct interaction of an activator present in the mitotic extract with the γ-tubulin located on the SPB (Masuda and Shibata, 1996).
In this study, we have purified an SPB activator from Xenopus egg mitotic extracts. It turned out to be the large subunit (R1) of ribonucleotide reductase (RNR), which is an essential enzyme required for DNA replication and DNA repair (reviewed by Thelander and Reichard, 1979; Reichard, 1988; Elledge et al., 1993). In both Escherichia coli and higher organisms, the enzyme consists of two nonidentical subunits, a dimer of an 85-kDa protein, R1, and a dimer of a 45-kDa protein, R2. Both subunits are essential for RNR enzyme activity (Thelander et al., 1980; 1985; reviewed by Reichard, 1993). Here, we present evidence that the R1 protein also functions in MTOC activation. We propose that R1 may be involved in coordination of DNA replication/repair and mitotic spindle formation.
MATERIALS AND METHODS
Xenopus Egg Extracts
High speed extracts (HSEs) were prepared from Xenopus laevis unfertilized eggs using XB/EB buffer (10 mM HEPES, 70 mM KCl, 5.9 mM MgCl2, 9.5 mM EGTA, 24 mM β-glycerophosphate, 35 mM sucrose, 0.1 mM trolox, pH 7.6) supplemented with protease inhibitors and energy mixture (7.5 mM creatine phosphate, 1 mM ATP, 0.1 mM EGTA, 1 mM MgCl2, pH 7.7) as described previously (Murray, 1991; Masuda and Shibata, 1996). For the sperm aster formation assay, HSEs were prepared in a similar manner, except that XB buffer (10 mM HEPES, 100 mM KCl, 2 mM MgCl2, 0.1 mM CaCl2, 5 mM EGTA, and 50 mM sucrose, pH 7.7) supplemented with protease inhibitors and the energy mixture was used and the extracts were centrifuged at 80,000 rpm for only 30 min.
In Vitro SPB Activation Assay
The in vitro SPB activation assay was performed as described previously (Masuda et al., 1992; Masuda and Shibata, 1996). Briefly, S. pombe wild-type (h−972) cells were arrested at S phase by hydroxyurea and permeabilized with Triton X-100 (Masuda et al., 1990, 1992). Sixty-microliter aliquots (106 cells per aliquot) were incubated for 5 min in 20 μl of fractions purified from HSEs in the presence of 1 mM MgATP at room temperature, then washed, and incubated for 10 min in 30 μl of 12.5 μM porcine tubulin, 5 μM taxol, and 3 mM GTP. Microtubules and SPBs were observed by immunofluorescent staining with a mouse monoclonal anti–α-tubulin antibody (B-5-1-2; Sigma, St. Louis, MO) and an affinity-purified rabbit anti–S. pombe γ-tubulin antibody (anti-Cγtb; Masuda and Shibata, 1996). In some experiments, ATP regeneration system (0.2 mg/ml creatine kinase, 7.5 mM phosphocreatine) and MgATP were added to the purified fraction for SPB activation.
Purification of an SPB Activator
An SPB activator was purified from HSEs by a series of conventional chromatographic procedures; the activity of the activator in the fractions was assessed using an in vitro SPB activation assay. HSEs were first fractionated on HiLoad Superdex 200-pg (Amersham Pharmacia Biotech Ltd., Uppsala, Sweden) gel-filtration columns in XB/EB (II) buffer (10 mM HEPES, 70 mM KCl, 5.9 mM MgCl2, 1 mM EGTA, 24 mM β-glycerophosphate, 35 mM sucrose, 0.1 mM trolox, 1 mM DTT, pH 7.6) supplemented with protease inhibitors. The fraction corresponding to 160–220 kDa was next subjected to ammonium sulfate precipitation. The major portion of the activity was found in the 20–45% ammonium sulfate cut, which was resuspended in XB/EB (III) buffer (10 mM HEPES, 5.9 mM MgCl2, 1 mM EGTA, 35 mM sucrose, 0.1 mM trolox, 1 mM DTT, pH 7.6) supplemented with protease inhibitors, and applied to a LiChrospher1000TMAE (Merck, Darmstadt, Germany) anion-exchange column and eluted stepwise with KCl. The activity was found in the fractions eluted at 0–150 mM KCl. The fractions were combined, dialyzed against 0.8 M (NH4)2SO4 in XB/EB (III) buffer supplemented with protease inhibitors, applied on a Phenyl Superose HR5/5 (Amersham Pharmacia Biotech Ltd.) hydrophobic interaction column, and eluted decreasing stepwise with (NH4)2SO4. The activity was found in the fractions eluted at 0.32–0.48 M (NH4)2SO4. The active fractions were combined, dialyzed against XB/EB (III) buffer supplemented with protease inhibitors, and purified on a second LiChrospher1000TMAE anion-exchange column with KCl gradient elution.
The 85-kDa polypeptide transferred onto polyvinylidene difluoride membrane (Immobilon-PSQ; Millipore, Bedford, MA) was sent to John Leszyk in the W.M. Keck Protein Chemistry Facility at the Worcester Foundation for Biomedical Research (Shrewsbury, MA) for amino acid sequence analysis. The sequence of two tryptic peptides of the protein was obtained: TDIDAAIETYNLLSEK and GAFIDQSQSLNIHVAEPNYGK. A 1.6-kb cDNA was amplified by PCR from a Xenopus cDNA library, kindly provided by Hiroshi Nojima (Osaka University), using degenerated primers designed from the tryptic peptides and was cloned into pUC119 (Takara, Japan) (pXRL522). The 1.6-kb fragment was sequenced.
Protein Expression and Purification
To obtain recombinant 6× histidine fusion mouse R1 protein, the BAC-TO-BAC baculovirus expression system (GIBCO BRL, Rockville, MD) was used. The full-length cDNA encoding murine R1 was amplified by PCR from a mouse cDNA library kindly provided by Hiroshi Miyazawa and Fumio Hanaoka (RIKEN) and was cloned into pFASTBACHT vector, which has a baculovirus-specific polyhedrin promoter for expression of proteins in insect cells. The recombinant plasmid was transformed into DH10BAC E. coli competent cells that contain a baculovirus shuttle vector (bacmid) with a mini-attTn7 target site and the helper plasmid. The mini-Tn7 element on the pFASTBAC donor plasmid transposed to the mini-attTn7 target site on the bacmid in the presence of transposition proteins provided by the helper plasmid. High-molecular-weight mini-prep DNA was prepared from selected E. coli colonies containing the recombinant bacmid. Sf9 insect cells were maintained in Sf900II SFM (GIBCO BRL) containing 5% fetal bovine serum, 50 U/ml penicillin, and 50 μg/ml streptomycin as monolayer cultures in plastic plates. To obtain the recombinant baculovirus, Sf9 cells were transfected with the recombinant bacmid using CELLFECTIN reagent (GIBCO BRL). Viruses (rBVMR1–3) were harvested from cell culture medium at 72 h after transfection and used for further amplification. To express the recombinant protein (His-R1), confluent Sf9 cells on four 150-mm plates were infected with rBVMR1-3 at a multiplicity of infection of 10. Cells were collected from the plates after 4 or 5 days after infection, washed once with PBS, frozen in liquid nitrogen, and stored at −80°C until needed. To purify His-R1 protein, frozen cells were thawed on ice and suspended in 30 ml of binding buffer (20 mM Tris-HCl, pH 7.9, 500 mM NaCl, 5 mM imidazole) containing 1% Triton X-100, and protease inhibitors. The cell suspension was briefly sonicated and centrifuged at 40,000 × g for 30 min. Supernatant was applied onto a 2.5-ml column of ProBond metal affinity resin (Invitrogen, Carlsbad, CA) equilibrated with binding buffer containing protease inhibitors. The column was washed with wash buffer (20 mM Tris-HCl, pH 7.9, 500 mM NaCl, 60 mM imidazole) containing protease inhibitors and bound proteins were eluted with an imidazole gradient. A fraction containing His-R1 protein was applied on a microspin G-25 column (Amersham Pharmacia Biotech) equilibrated with XB/EB containing protease inhibitors, and the eluate was used for the SPB activation assay.
Recombinant GST-mouse R1 fusion protein (GST-R1) was expressed in the bacteria system. The full-length mouse R1 cDNA was cloned into pGEX-4T (Amersham Pharmacia Biotech) and transformed into E. coli BL21(DE3)pLysS cells. To express the soluble fusion protein, cells were grown at 15°C with a minimal concentration (50 μM) of isopropylthio-β-d-galactoside (IPTG) as described (Davis et al., 1994). GST was expressed in BL21(DE3)pLysS using pGEX-4T. GST-R1 and GST were purified using Glutathione Sepharose 4B (Amersham Pharmacia Biotech) and dialyzed against XB/EB.
Xenopus γ-tubulin was expressed in E. coli BL21(DE3)pLysS as a histidine-tagged protein using a pRSET vector (Invitrogen), and purified using Probond metal affinity resin. The Xenopus γ-tubulin cDNA was kindly provided by Yuko Kiyosue and Nobuyuki Shiina (CREST, JST).
Peptides and Antibodies
Peptides used for antibody production: RNR-N corresponding to the N-terminus of mouse or human R1 (Table 1) and MRSS ((C)TENSFTLDADF) corresponding to the C terminus of mouse R2 with an N-terminal cysteine, or for inhibition of SPB activation and sperm aster formation: RNR-N, PM1, PM1B, X384, M572, RNR-C, and M783, were custom-synthesized (TANA Laboratories, L. C., Houston, TX) (Table 1).
Table 1.
Oligopeptides derived from R1 and comparison of the sequences with R1 from other species
| Species | Amino acid no. | Peptide name | Sequences |
|---|---|---|---|
| Mouse | 5–17 | RNR-N | KRDGRQERVMFDK(C) |
| Human | 5–17 | RNR-N | KRDGRQERVMFDK(C) |
| S. pombe | 5–17 | KRDGRQEKVAFDK | |
| Mouse | 572–583 | M572 | NVAPTDLWDWKP |
| Human | 572–583 | NVTPTDLWDWKV | |
| S. pombe | 571–582 | PM1 | NVNPTDLWDWAE(C) |
| 571–582 | PM1B | NVNPTDLWDWAE | |
| Xenopusa | X384 | NVTPTDLWDWTA | |
| Mouse | 754–772 | KEKLKDKEKALKEEEEKER | |
| Human | 754–772 | RNR-C | (C)KEKLKDKEKVSKEEEEKER |
| S. pombe | 753–771 | PVALRARNEESNEENKKPV | |
| Mouse | 783–792 | M783 | NREECLMCGS |
| Human | 783–792 | NRDECLMCGS | |
| S. pombe | 802–811 | NPEACEMCSA |
The amino acid sequence of Xenopus R1 was deduced from the nucleotide sequence of a PCR fragment that lacks amino- and carboxyl-terminal portions of R1 (see MATERIALS AND METHODS).
Antigens for use in the production of polyclonal anti-R1 and anti-R2 antibodies were prepared as follows: The insert of pXRL522 encoding a Xenopus R1 fragment XRL522 was recloned into pRSET vector (pRXRL522), and a mouse cDNA encoding the carboxyl-half of R1, CH-MR (aa 401–792), was amplified by PCR and cloned into pRSET (pRCHMR). The fragments were expressed in BL21(DE3)pLysS. The insoluble fraction of bacterial lysate were washed with 4 M urea, solubilized in 6 M urea, and mixed with ProBond Ni resin (Invitrogen, Carlsbad, CA). The bound proteins were eluted with 250 mM imidazole and separated on preparative SDS-PAGE. The recombinant proteins were extracted from the gel slice, dialyzed against PBS, and used as antigens. RNR-N and MRSS peptides were coupled to maleimide-activated keyhole limpet hemocyanin (Pierce, Rockford, IL) and used as antigens.
The antibodies were first purified on rProteinA Sepharose FF columns (Amersham Pharmacia Biotech). Antibodies against XRL522 and CH-MR were then affinity-purified using XRL522 and CH-MR fragments, respectively, coupled to CN-Br–activated Sepharose 4B. Antibodies against RNR-N and MRSS were affinity-purified using RNR-N and MRSS peptides, respectively, coupled to 2-fluoro-1-methylpyridium tolune-4-sulfonate cellulofine (Seikagaku Co., Tokyo, Japan).
Antibodies against the C-terminal fragment (aa323-446) of S. pombe γ-tubulin (anti-Cγtb) and against the C-terminal peptides of Xenopus γ-tubulin (anti-XLG) were prepared as described (Masuda and Shibata, 1996). A mouse monoclonal antibody against human RNR R1 (KM1466) was kindly provided by Kyowa Hakkou Co. (Cellbank, Tokyo Institute, Tokyo, Japan). An autoimmune serum against pericentrin (5051) was kindly provided by Zacheus Cande (University of California, Berkeley). Mouse monoclonal antibodies against α-tubulin (B-5-1-2; Sigma), γ-tubulin (GTU-88; Sigma), and actin (N350; Amersham) were obtained commercially.
Sperm Aster Formation Assay
Ten microliters of HSE was diluted with 20 μl of XB containing 60 mM HEPES (pH 7.7), 1 mM MgATP, and 1.5 mM DTT (XBHMD), with or without 0.5–1.5 mM synthetic peptides, and incubated on ice for 15 min. One microliter of 35 mM GTP, 1 μl of 100 μM tetra-methylrhodamine-labeled tubulin (Rd-tubulin), and 3 μl of Xenopus demembranated sperm heads (1500 nuclei/μl) were added to the extract and incubated for 15 min at room temperature. The extract was diluted with 1 ml BRB80 (80 mM PIPES, pH 6.8, 1 mM MgCl2, 1 mM EGTA) containing 0.5% Triton X-100 and 30% glycerol, and centrifuged on polylysine-coated coverslips through 2 ml of BRB80 containing 40% glycerol at 7000 rpm for 30 min. The coverslips with sperm nuclei attached were placed on slides with a drop of BRB80 containing 40% glycerol and 1 μg/ml DAPI. Microtubule asters and nuclei were observed under a fluorescent microscope. Rd-tubulin was prepared according to Hyman et al. (1991). The labeling stoichiometry was 0.6 per tubulin dimer. Xenopus demembranated sperm heads were kindly provided by Koji Okuhara (Institute of Molecular Cellular Bioscience, University of Tokyo).
For immunofluorescence of sperm centrioles with anti–γ-tubulin (anti-XLG) and anti–α-tubulin (B-5-1-2), 6 μl of HSE was diluted with 24 μl of XBHMD with or without 1.9 mM X384, and incubated on ice for 15 min. One microliter of 35 mM GTP, 3 μl of sperm heads, and 0.35 μl of 10 mM nocodazole were added to the extract and incubated for 15 min. The extract was centrifuged on polylysine-coated coverslips as described above. The sperm centrosomes were fixed by placing the coverslips in cold methanol for 10 min. The coverslips were washed in PBS and incubated for 30 min in PBS containing 3% BSA. They were then incubated for 1.5 h with the primary antibodies diluted in PBS containing 1% BSA, washed with Tw/PBS (0.02% Tween 20 in PBS), incubated for 1.5 h with the secondary antibodies, Cy3-anti-rabbit IgG and FITC-anti-mouse IgG (Jackson ImmunoResearch Laboratory, West Grove, PA), and washed with Tw/PBS containing 0.1 μg/ml DAPI.
To examine the localization of R1 on the sperm centrioles, 10 μl of HSE was diluted with 20 μl of XB containing 1 mM MgATP and 1.5 mM DTT, mixed with 1 μl of 35 mM GTP, 3 μl of sperm heads, and 0.75 μl of Rd-tubulin, and incubated for 15 min at room temperature. The mixture was centrifuged on polylysine-coated coverslips, and the sperm centrosomes attached were fixed with 2.6% paraformaldehyde and 0.1% glutaraldehyde in BRB80 containing 40% glycerol for 10 min. The coverslips were processed as described above using anti-XRL522 as the primary antibody and FITC-anti-rabbit IgG as the secondary antibody.
Immunofluorescent Staining of Animal Cells
Chinese hamster ovary (CHO-K1) cells were cultured in F-12 medium (GIBCO BRL, Rockville, MD) at 37°C, and Xenopus A6 cells were cultured in 50% L-15 medium (GIBCO BRL) at 24°C. The media contained 10% fetal bovine serum and antibiotics. CHO and A6 cells were grown on glass coverslips to 70–80% confluence. Some cultures were treated with 33 μM nocodazole for 4 h to depolymerize microtubules before use. The cells on the coverslips were washed with PHEM solution (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, pH 6.9), preextracted with 0.5% Triton X-100 in PHEM for 1 min, fixed in −20°C cold methanol for 5 min, and incubated for 30 min in block solution containing 3% BSA. Coverslips were then processed for immunofluorescence as described above.
Centrosome Isolation
Centrosomes were isolated from CHO cells according to Bornens
et al. (1987) with slight modifications. CHO cells were
grown in five dishes (100 mm in diameter) to 7–8 ×
106 cells/dish (70–80% of confluent) at 37°C
and incubated for 2 h in fresh medium containing 10 μg/ml
nocodazole and 5 μg/ml cytochalasin B. Cells were lysed in isolation
buffer (1 mM Tris-HCl, 1 mM EGTA, pH 8.0) containing 0.5% NP40, 0.1%
2-mercaptoethanol, and protease inhibitors. After addition of
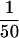 volume PE solution (500 mM PIPES, 1 mM EGTA, pH 7.2), the
cell lysate was centrifuged at 2300 × g for 5 min. The
supernatant (25 ml) was layered onto a 40/60% sucrose cushion (3.5
ml/3.5 ml) and centrifuged at 30,000 × g for 60 min in
a swinging bucket rotor. Fractions were collected from the bottom 4.5
ml of the centrifuged solution, dialyzed against PME (20 mM PIPES, pH
6.8, 1 mM EGTA, 0.5 mM MgCl2), and freeze-dried.
The proteins were dissolved in SDS-PAGE buffer, separated on SDS-PAGE,
and transferred to nitrocellulose membrane. The membrane was probed
with anti–γ-tubulin (GTU88) and anti-RNR R1 (KM1466) antibodies, and
visualized by ECL (Amersham Pharmacia Biotech Ltd.).
volume PE solution (500 mM PIPES, 1 mM EGTA, pH 7.2), the
cell lysate was centrifuged at 2300 × g for 5 min. The
supernatant (25 ml) was layered onto a 40/60% sucrose cushion (3.5
ml/3.5 ml) and centrifuged at 30,000 × g for 60 min in
a swinging bucket rotor. Fractions were collected from the bottom 4.5
ml of the centrifuged solution, dialyzed against PME (20 mM PIPES, pH
6.8, 1 mM EGTA, 0.5 mM MgCl2), and freeze-dried.
The proteins were dissolved in SDS-PAGE buffer, separated on SDS-PAGE,
and transferred to nitrocellulose membrane. The membrane was probed
with anti–γ-tubulin (GTU88) and anti-RNR R1 (KM1466) antibodies, and
visualized by ECL (Amersham Pharmacia Biotech Ltd.).
GST-Pulldown Assay
GST-R1 (37 μg; 1.7 μM at the final concentration) or GST (33 μg; 6 μM at the final concentration) was dissolved in 160 μl of XB/EB containing the energy mixture and protease inhibitors, mixed with 40 μl of HSE in the presence or absence of recombinant S. pombe γ-tubulin, incubated at room temperature for 20 min, and centrifuged at 15,000 rpm for 5 min. Forty microliters of 50% Glutathione Sepharose 4B slurry in XB/EB was added to each supernatant and incubated at 4°C for 1 h with gentle agitation; the resin was then pelleted by centrifugation at 3000 rpm for 2 min. The pellet was washed five times with 1 ml XB/EB containing 150 mM KCl, 1 mM MgATP, and 1% Triton X-100, resuspended in 100 μl of SDS-PAGE buffer, and incubated at 75°C for 5 min, and the resin was repelleted by centrifugation at 3000 rpm. The supernatants were subject to SDS-PAGE and transferred onto polyvinylidene difluoride membrane. The membrane was probed with anti–α-tubulin (B-5-1-2), anti–γ-tubulin (GTU88), antiactin (N350), and anti-R2 (anti-MRSS) antibodies, and visualized by ECL. Quantitative analysis of the proteins bound was performed according to manufacture's instruction using brain tubulin, histidine-tagged Xenopus γ-tubulin, and histidine-tagged mouse R2 as standards. S. pombe γ-tubulin was prepared as described (Masuda and Shibata, 1996).
To examine direct interactions of R1 with γ-tubulin and α/β-tubulin, GST-R1 or GST was incubated with S. pombe γ-tubulin or porcine α/β-tubulin in 200 μl of XB/EB containing 10 mg/ml BSA, the energy mixture, protease inhibitors, 2 mM DTT, and 1 mM GTP, and processed as described above. The dissociation constant (Kd) of the interaction was calculated using the data from quantitative immunoblot analysis.
RESULTS
Purification of an SPB Activator from Xenopus Egg Mitotic Extracts
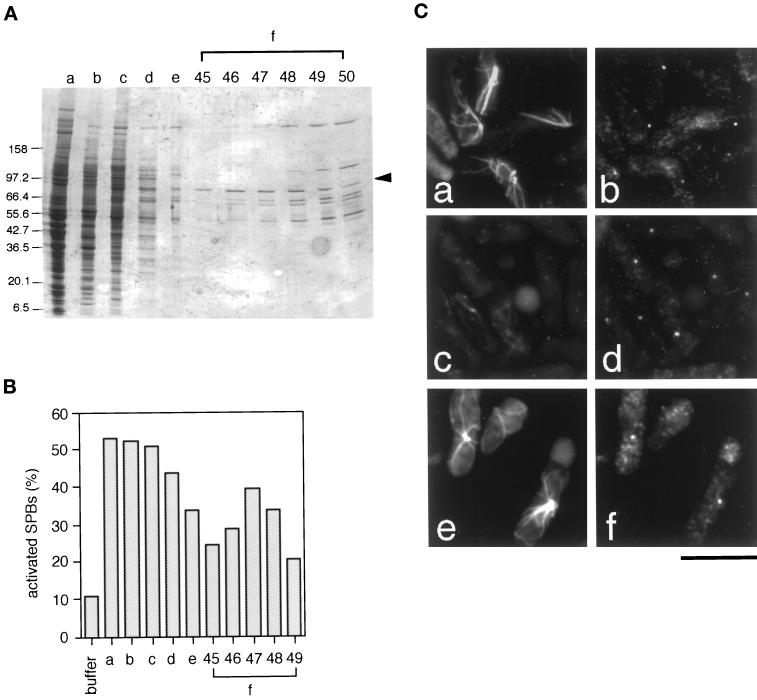
We previously reported that HSEs prepared from Xenopus unfertilized eggs contain an activator of the S. pombe SPB (Masuda et al., 1992; Masuda and Shibata, 1996). In this study we report the purification and characterization of this activator. To monitor the presence of the activator during purification, we used an in vitro SPB activation assay developed previously (Masuda et al., 1992; see Materials and Methods). We first fractionated HSEs on gel-filtration chromatography and obtained a major activity peak in the fraction corresponding to 160–220 kDa; we refer to this fraction as “the 200K fraction” (Figure 1A, lane b; Figure 1B, lane b). Proteins in the 200K fraction were then fractionated sequentially by ammonium sulfate precipitation (Figure 1A, lane c; Figure 1B, lane c), anion-exchange chromatography (Figure 1A, lane d; Figure 1B, lane d), hydrophobic interaction chromatography (Figure 1A, lane e; Figure 1B, lane e), and a second anion-exchange column (Figure 1A, lanes f; Figure 1B, lanes f). The activity from the second anion-exchange column eluted at 50–90 mM KCl (Figure 1B, lanes f; Figure 1C); we refer to this highly purified fraction as “fraction f.” ATP was required for SPB activation by fraction f (our unpublished data). The elution pattern of an 85-kDa protein (Figure 1A, arrowhead) showed strong correlation with SPB activation activity (Figure 1B, lanes f), whose peak was in fraction 47 (Figure 1B, lane f-47); a few other proteins were also seen in the active fractions, but their peaks were present in fractions that eluted at higher KCl concentrations (Figure 1A, lane f-50 or after).
Figure 1.
Purification of an SPB activator from Xenopus egg mitotic extracts. (A) Aliquots from the active fractions of each purification stage were separated on a 5–20% gradient SDS-polyacrylamide gel and stained with Coomassie Blue. (lane a) High-speed extracts (HSEs); (lane b) the 200K fraction from a HiLoad Superdex 200-pg column; (lane c) 20–45% ammonium sulfate cut; (lane d) the 150 mM KCl fraction from a LiChrospher1000TMAE column; (lane e) 0.32–0.48 M (NH4)2SO4 fraction from a Phenyl Superose column; and (lane f-45–49) fractions 45–49 (50–90 mM KCl) from the second anion-exchange column (LiChrospher1000TMAE). Arrowhead, the 85-kDa protein, which was subjected to amino acid sequence analysis. (B) In vitro assay of selected aliquots from each purification stage. Coordinates of the columns are as in (A). Vertical axis, the percentage of activated SPBs (N = 200). About 10% of SPBs nucleated microtubules without activation (buffer). (C) The SPB activation assay. S. pombe interphase SPBs were incubated with the 200K fraction (a and b); XB/EB (II) buffer (c and d); and the fraction 47 from the second anion-exchange column (e and f). Microtubules (a, c, and e) and SPBs (b, d, and f) were visualized by double-immunofluorescence with anti–α-tubulin (B-5-1-2) and anti–γ-tubulin (anti-Cγtb) antibodies, respectively. Bar, 10 μm.
Identification of the 85-kDa Protein as Ribonucleotide Reductase Large Subunit R1
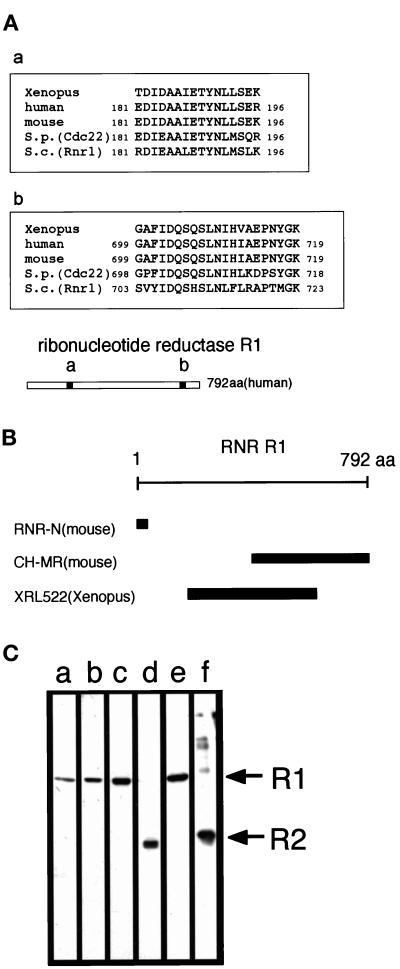
We subjected the 85-kDa protein to amino acid microsequencing analysis. Sequences of two tryptic peptides were determined, and a database search revealed that they were almost identical to sequences of ribonucleotide reductase (RNR) R1 proteins reported from several other organisms (Figure 2A). Because the Xenopus homologue of R1 has not yet been cloned, we amplified a 1.6-kb cDNA fragment by PCR from a Xenopus cDNA library using degenerate primers designed from DIDAAIE (a part of region a, Figure 2A) and HVAEPNYGK (a part of region b, Figure 2A). The amino acid sequence predicted from this cDNA fragment showed 88 and 72% identity with the mouse and S. pombe R1 equivalent, respectively (Caras et al., 1985; Fernandez Sarabia et al., 1993); nucleotide sequence data have been submitted to the GenBank database under accession number AF168794. We raised three polyclonal antibodies against R1 by immunizing rabbits with two fragments from mouse R1 (RNR-N and CH-MR) and with one fragment from Xenopus R1 (XRL522; Figure 2B). All three antibodies, which were affinity-purified with antigens, recognized the 85-kDa protein in the egg extract (Figure 2C, lanes a-c).
Figure 2.
Identification of an SPB activator as RNR R1. (A) Sequences of tryptic peptides from the isolated 85-kDa protein and their alignment with RNR R1 sequences from human, mouse, S. pombe (Cdc22), and S. cerevisiae (Rnr1) (EMBL accession no. X59543, K02927, X65116, and U18813, respectively) (a and b). The location of the corresponding peptides in the human R1 is shown (regions a and b in the bar). (B) Antigens used in the production of the anti-R1 polyclonal antibodies used in this study (See text for details). (C) Anti-R1 antibodies recognize the 85-kDa protein in Xenopus egg mitotic extracts. (lanes a–c) The 200K fraction of egg mitotic extracts was loaded on 10% SDS-PAGE, transferred onto nitrocellulosemembrane, and blotted with affinity-purified anti–RNR-N (a), anti–CH-MR (b), and anti-XRL522 (c) antibodies. (lanes d and f) The anti-R2 antibody (anti-MRSS) recognizes recombinant mouse R2 (f) and a 45-kDa polypeptide in the 200K fraction, a putative Xenopus homologue of RNR R2 (d). (lane e) The SPB activator purified from Xenopus egg mitotic extracts (fraction f from the second anion-exchange column, Figure 1A, lanes f) was probed with a mixture of anti-R1 (anti-XRL522) and anti-R2 (anti-MRSS) antibodies.
Recombinant R1 Activates Interphase SPBs
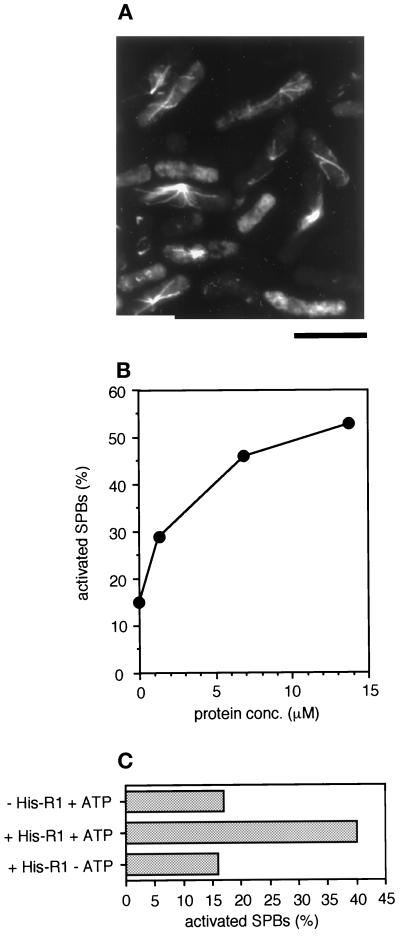
To confirm that R1 has the ability to activate SPBs, we expressed 6X histidine fusion mouse R1 (His-R1) protein in baculovirus-infected Sf9 cells and examined whether it could activate interphase SPBs. Figure 3A shows that His-R1 protein promotes microtubule assembly from the interphase SPBs. We found that His-R1 protein must be freshly prepared from Sf9 cells just before performing an SPB activation assay to be fully effective in activating SPBs. The efficiency of SPB activation was dependent on His-R1 protein concentration (Figure 3B). We obtained >50% activation, which is comparable to the activation level achieved by a Xenopus egg HSE, with recombinant His-R1 protein (Figure 3B). We also obtained SPB activation using recombinant His-R1 expressed in E. coli, but less effectively (our unpublished data). These results indicate that R1 is a protein responsible for SPB activation and that R2, another essential subunit of RNR, is not required for SPB activation. This is consistent with the result that the active fraction (fraction f) purified from Xenopus egg extracts contained no R2 protein, as judged by immunoblot analysis with anti-R2 antibody (Figure 2C, lane e). ATP was required for the SPB activation by recombinant His-R1 protein (Figure 3C), as shown by fraction f or by Xenopus egg mitotic extracts (Masuda et al., 1992). R1 needs binding of ATP to the activity site for RNR function (Brown and Reichard, 1969). Our result suggests that ATP binding to R1 is also essential for SPB activator function.
Figure 3.
SPB activation by recombinant His-mouse R1 fusion. (A) The SPB activation assay. Permeabilized S. pombe interphase cells were incubated with 59 μM recombinant histidine-tagged mouse R1 (His-R1). Polymerized microtubules were visualized by immunofluorescence with anti–α-tubulin (B-5-1-2). Bar, 10 μm. (B) Protein-concentration dependency of SPB activation by recombinant His-R1 protein. Horizontal axis, the protein concentration of His-R1 during incubation with permeabilized S. pombe interphase cells (N = 300). (C) ATP dependency of SPB activation by recombinant His-R1 protein. Permeabilized S. pombe interphase cells were incubated with a control buffer containing 1 mM Mg-ATP and ATP regeneration system (−His-R1 + ATP) or with 30 μM His-R1 protein in the presence (+His-R1 + ATP) or absence (+His-R1 − ATP) of 1 mM Mg-ATP and ATP regeneration system. Horizontal axis, the percentage of activated SPBs (N = 300).
Native or recombinant R1 is known to be a mixture of monomer, dimer, and tetramer in solution (Thelander et al., 1980; Ingemarson and Thelander, 1996; Chabes et al., 1999). Recombinant His-R1 fraction expressed in insect cells may also contain a variety of R1 composites. The active form of R1 as an SPB activator is probably a dimeric R1, because an SPB activator in Xenopus egg extracts has molecular sizes of 160–220 kDa (200K fraction) on gel-filtration chromatography, corresponding to the size of an R1 dimer.
Anti-R1 Antibodies and PM1 Peptides Inhibit SPB Activation
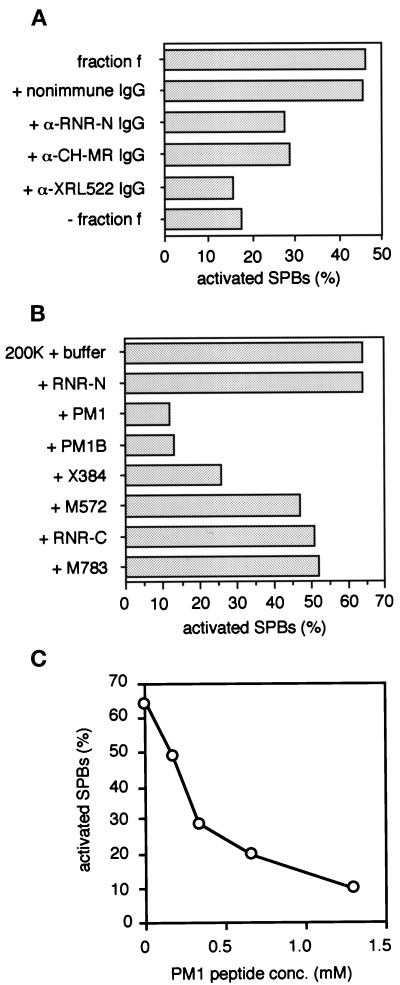
Other lines of evidence further support the idea that R1 possesses the ability to activate SPBs. We examined the effects of anti-R1 antibodies on SPB activation. Preincubation of fraction f with affinity-purified anti-R1 antibodies (anti–RNR-N, anti–CH-MR, and anti-XRL522) inhibited SPB activation, whereas nonimmune IgG had no effect (Figure 4A). In particular, the anti-XRL522 antibody completely suppressed activation. These antibodies should inhibit SPB activation by blocking R1 function in fraction f, because they did not react with R1 in crude extracts prepared from S. pombe cells (our unpublished data).
Figure 4.
Effects of Anti-R1 antibodies and R1 partial peptides on SPB activation. (A) Anti-R1 antibodies inhibit SPB activation. Affinity purified polyclonal antibodies against R1—anti–RNR-N, anti–CH-MR, and anti-XRL522—and nonimmune rabbit IgG were added separately to the SPB activator purified from Xenopus egg mitotic extracts (Figure 1A, lanes f) to a final concentration of 50–140 μg/ml, incubated on ice for 2 h, and then subjected to the SPB activation assay. Horizontal axis, the percentage of activated SPBs (N = 400). (B) Effects of R1 partial peptide addition to the 200K fraction of Xenopus egg mitotic extracts. The 200K fraction was mixed with each of the peptides, RNR-N, PM1, PM1B, X384, M572, RNR-C, or M783, at 1.2 mM final concentration andsubjected to the SPB activation assay. Horizontal axis, the percentage of activated SPBs (N = 300). (C) The PM1 peptide suppresses SPB activation by the 200K fraction in a concentration-dependent manner. The 200K fraction was mixed with the PM1 peptide at various concentrations (0.17–1.3 mM) and subjected to the SPB activation assay. Horizontal axis, the peptide concentration (mM); vertical axis, the percentage of activated SPBs (N = 100).
We also examined the effects of oligopeptides derived from R1 on SPB activation. We first tested five oligopeptides: RNR-N, which corresponds to amino acid 5 to 17 of mouse or human R1 with a C-terminal cysteine; PM1 or PM1B, which corresponds to amino acid 571 to 582 of S. pombe R1 (cdc22) with (PM1) or without (PM1B) a C-terminal cysteine; RNR-C, which corresponds to amino acid 754 to 772 of human R1 with a N-terminal cysteine; and M783, which corresponds to amino acid 783 to 792 of mouse R1 (Table 1). By analogy to the E. coli RNR R1 crystal structure (Uhlin and Eklund, 1994), these five oligopeptides are likely located on distinct regions of the surface of the R1 dimer, and therefore, may participate in association of R1 with R1 interacting factors (Uhlin and Eklund, 1994; Eriksson et al., 1997). Addition of the RNR-C and M783 oligopeptides to the 200K fraction of the Xenopus activator had only a slight inhibitory effect on SPB activation, and the RNR-N oligopeptide had no measurable effect (Figure 4B). Addition of the PM1 oligopeptide to the 200K fraction, on the other hand, dramatically inhibited SPB activation in a dose-dependent manner (Figure 4C). These results suggest that the PM1 oligopeptide competitively disrupts interaction between the PM1 segment of R1 and some unknown factor(s) and that the interaction is critical for SPB activation. We also tested X384 and M572 peptides, which correspond to the PM1 segment of Xenopus and mouse R1, respectively. These peptides were different from the PM1 or PM1B peptide in degree of effect on SPB activation. Addition of X384 to the 200K fraction moderately inhibited the SPB activation, and M572 slightly inhibited (Figure 4B).
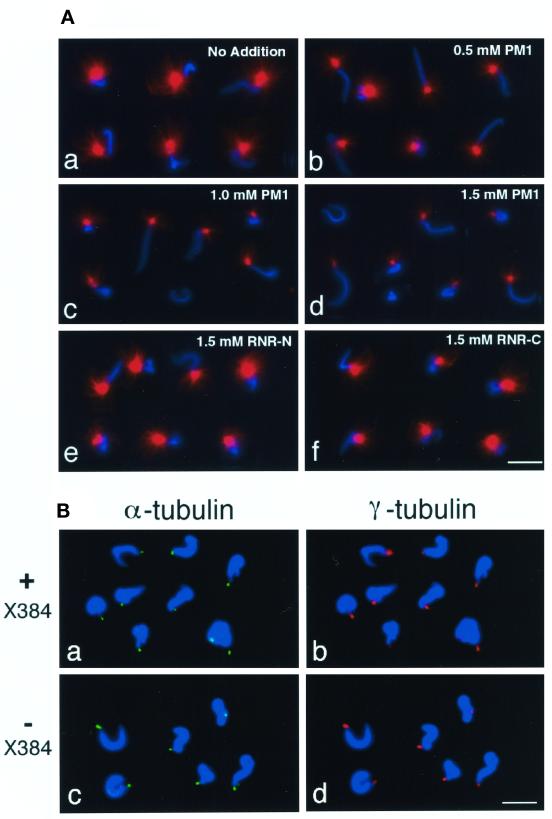
PM1 Peptides also Affect Sperm Aster Formation
To determine whether R1 also affects the extent of microtubule nucleation by the animal centrosome, we used an in vitro aster formation assay, which exploits the ability of Xenopus egg extracts to convert the centrioles of demembranated sperm heads into mature centrosomes that nucleate microtubule asters. The conversion seems to depend on accumulation of pericentriolar material, including γ-tubulin complexes, from the egg extract (Felix et al., 1994; Stearns and Kirschner, 1994). We examined the effects of the R1 partial peptides PM1, RNR-N, and RNR-C on aster formation (Figure 5A). We first incubated mitotic extracts with each of the peptides in the presence of ATP and then added rhodamine-labeled tubulin and demembranated sperm heads. Microtubule asters (red in Figure 5A) were observed at one end of sperm chromatin stained with DAPI (blue in Figure 5A). Aster formation on the reconstituted centrosome was effectively inhibited by the PM1 peptide in a concentration-dependent manner (Figure 5A, a-d). X384 and M572 peptides, which correspond to the PM1 segment of Xenopus and mouse R1, respectively, were also effective for inhibition of aster formation (our unpublished data). The RNR-C peptide had little effect on aster formation and RNR-N had no apparent effect (Figure 5A, e and f). These results suggest that R1 is also involved in formation of active centrosomes in Xenopus egg extracts.
Figure 5.
Effects of R1 partial peptides on sperm aster formation and γ-tubulin accumulation at sperm centrioles. (A) PM1 peptide inhibits aster formation at centrosomes reconstituted in Xenopus egg mitotic extracts. Demembranated Xenopus sperm heads were incubated in a Xenopus egg mitotic extract (HSE) containing rhodamine-labeled tubulin in the presence of PM1, RNR-N, or RNR-C peptides. (a) No addition; (b) 0.5 mM PM1; (c) 1.0 mM PM1; (d) 1.5 mM PM1; (e) 1.5 mM RNR-N; and (f) 1.5 mM RNR-C. Representative images of microtubule asters (red) and sperm chromatin stained with DAPI (blue) are shown. Several images were combined digitally to make each panel. (B) γ-Tubulin is accumulated at sperm centrioles either in the presence or absence of X384. Demembranated Xenopus sperm heads were incubated in a Xenopus egg mitotic extract (HSE) containing nocodazole in the presence (a and b) or absence (c and d) of 1.9 mM X384. Centrioles were stained with anti–α-tubulin (B-5-1-2; green in a and c), γ-tubulin recruited from HSE on the centrioles was detected with anti-Xenopus γ-tubulin (red in b and d), and sperm chromatin was stained with DAPI (blue). Bar, 20 μm.
We next examined whether the inhibitory peptides affect recruitment of γ-tubulin complexes to sperm centrioles. In these experiments, we searched for conditions where microtubule assembly at sperm centrioles is significantly suppressed by addition of the X384 peptide: under these conditions, >75% of centrioles had no detectable microtubules in the presence of X384, whereas > 90% of centrioles were competent for nucleating microtubules in the absence of the peptide (our unpublished data). For comparison of the amount of γ-tubulin by immunofluorescence, microtubule formation at sperm centrioles was suppressed by a microtubule depolymerizing agent nocodazole, because the presence of microtubules nucleated tended to cover centrosomal antigens and to reduce or remove the reactivity of the antibodies. Addition of nocodazole to Xenopus egg extracts does not inhibit accumulation of pericentriolar material including γ-tubulin at centrioles or affect the microtubule-nucleating ability of the centrioles (Felix et al., 1994; Stearns and Kirschner, 1994). Sperm centrioles and γ-tubulin were recognized with anti–α-tubulin (green in Figure 5B, a and c) and anti-Xenopus γ-tubulin (red in Figure 5B, b and d) antibodies, respectively. γ-Tubulin was accumulated at centrioles either in the presence (Figure 5B, a and b) or absence (Figure 5B, c and d) of the peptide. This suggests that recruitment of γ-tubulin complex from the extract to the sperm centriole is not sufficient for microtubule assembly and that R1 may play a role in microtubule assembly from γ-tubulin complexes rather than γ-tubulin recruitment.
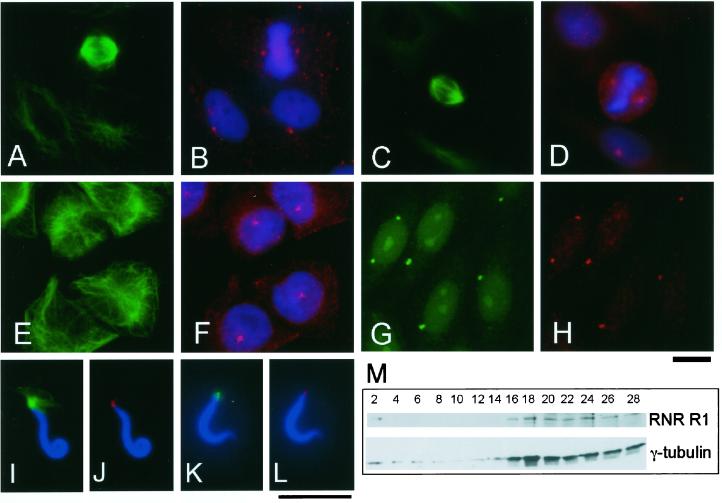
R1 Is a Centrosomal Component in Animal Cells
Because R1 is able to activate microtubule nucleation on the centrosome, we examined whether R1 localized to the centrosome in Xenopus A6 (Figure 6,A and B), Chinese hamster ovary (CHO; Figure 6, C–F), HeLa, and mouse 3Y1 (our unpublished data) cells by indirect immunofluorescence with polyclonal antibodies against R1 (red in Figure 6, B, D, and F) and against α-tubulin (green in Figure 6, A, C, and E); chromosomes were stained with DAPI (blue in Figure 6, B, D, and F). In mitotic cells, the spindle poles were stained by anti-R1 antibodies (Figure 6, A–D). In interphase cells, most R1 showed a diffuse cytoplasmic distribution, as was previously shown by Engstrom et al. (1984), but we found that part of R1 was highly concentrated in small regions centering on microtubule arrays (Figure 6, A, B, E, and F). To ascertain that the regions where R1 was highly concentrated corresponded to the centrosome, CHO cells were incubated in nocodazole and double-stained with an anti-R1 antibody (anti–RNR-N; red in Figure 6H) and antibodies that recognize the centrosome: anti-pericentrin (Doxsey et al., 1994; green in Figure 6G) or anti–γ-tubulin (our unpublished data). R1 colocalized with pericentrin or γ-tubulin at the centrosome, showing that localization of R1 on the centrosome does not depend on the presence of microtubules. We also found by immunofluorescence with anti-XRL522 that R1 localized to the centrosome reconstituted in Xenopus egg extract (Figure 6, I and J) and at sperm centrioles (Figure 6, K and L).
Figure 6.
R1 is a centrosomal component in animal cells. (A–F) R1 localizes to the centrosome in animal cells. Xenopus A6 (A and B) and CHO cells (C–F) were triple-stained with anti–α-tubulin (B-5-1-2; green in A, C, and E), DAPI (blue in B, D, and F), and anti-R1 antibodies: anti–CH-MR (red in B) or anti-XRL522 (red in D and F). (G and H) R1 is an integral component of the centrosome. CHO cells incubated with nocodazole for 4 h before fixation were double-stained with anti-pericentrin (green in G) and anti–RNR-N (red in H). Bar, 10 μm. (I–L) R1 localizes to the sperm centrosome. Demembranated Xenopus sperm heads were incubated in a Xenopus egg mitotic extract (HSE) containing Rd-tubulin (I and J) or in a control buffer (K and L). (I and J) The sperm heads were stained with anti-XRL522 (red in J) and DAPI (blue). In (I), a microtubule aster of Rd-tubulin is shown in green. (K and L) The sperm heads were stained with anti–α-tubulin (B-5-1-2; green in K), anti-XRL522 (red in L), and DAPI (blue). Bar, 10 μm. (M) Centrosomes isolated from CHO cells contain R1. The centrosomes from CHO cells were fractionated by sucrose density gradient centrifugation, and the resulting fractions were processed for immunoblot analysis with anti–γ-tubulin (GTU88) and anti-human R1 (KM1466) antibodies. R1 cosediments with the fractions that are recognized by the γ-tubulin antibody.
Furthermore, we found that centrosomes isolated from cultured mammalian cells contained R1. Centrosomes were isolated from CHO cells by discontinuous sucrose density gradient centrifugation. Proteins in fractions from the centrifugation were separated on SDS-PAGE and probed with antihuman R1 (KM1466) and anti–γ-tubulin antibodies (Figure 6M). The sedimentation profile of the centrosomes was determined using the γ-tubulin antibody, and R1 was found to cosediment with the centrosomes (Figure 6M). Taken together, our results indicate that R1 is an integral component of the centrosome in animal cells.
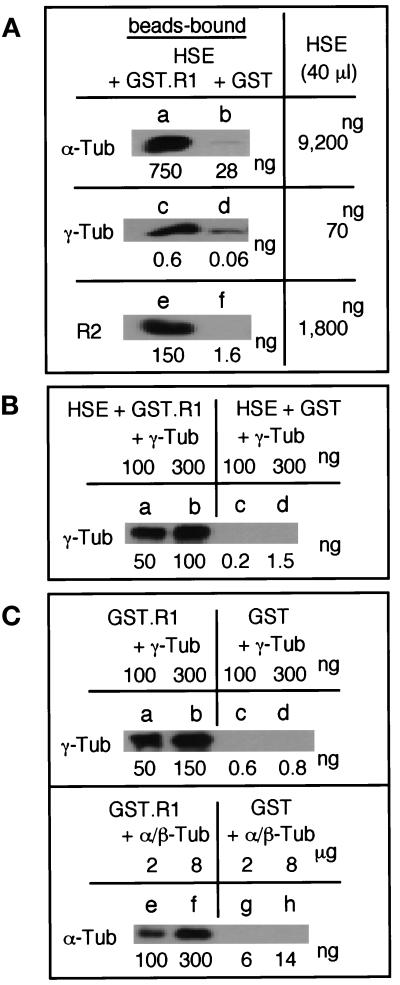
R1 Interacts with γ-Tubulin and α/β-Tubulin In Vitro
Cosedimentation of R1 with centrosomes (Figure 6M)
implies that it interacts with centrosomal component(s). The activator
in Xenopus egg mitotic extracts appears to interact directly
with γ-tubulin located on the SPBs (Masuda and Shibata, 1996). We
investigated the ability of R1 to interact with various proteins in
Xenopus egg extracts using a GST-pulldown experiment with
recombinant GST-mouse R1 fusion protein. Purified GST-R1 fusion protein
or GST was incubated in a mitotic extract (HSE) and recovered with
Glutathione-Sepharose beads. The proteins bound to the beads were
separated on SDS-PAGE and probed with antibodies against R2,
γ-tubulin, α-tubulin, and actin. R2, α-tubulin, and γ-tubulin
in HSE interacted with GST-R1 fusion protein (Figure
7A), whereas actin did not (our
unpublished data). Quantitative immunoblot analysis shows
that HSE contains 1.8 μg R2, 9.2 μg α-tubulin, 70 ng γ-tubulin,
and 4 μg R1 per 40 μl HSE. About 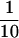 of R2,
of R2, 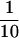 of
α-tubulin, and
of
α-tubulin, and 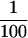 of γ-tubulin present in the extract
were pulled down with GST-R1 when 37 μg of GST-R1 was incubated with
40 μl of HSE. To confirm that γ-tubulin and α/β-tubulin have
the ability to bind to R1, recombinant S. pombe γ-tubulin
or porcine α/β-tubulin was incubated with GST-R1 or GST (Figure 7,
B and C). The recombinant γ-tubulin effectively bound to GST-R1
(Kd = ∼1.7 ×
10−6 M), but not to GST either in the presence
or absence of HSE. Porcine α/β-tubulin weakly bound to GST-R1
(Kd = ∼3.8 ×
10−5 M). These results suggest that γ-tubulin
itself has R1 binding activity, but the activity of most γ-tubulin is
blocked in the extract.
of γ-tubulin present in the extract
were pulled down with GST-R1 when 37 μg of GST-R1 was incubated with
40 μl of HSE. To confirm that γ-tubulin and α/β-tubulin have
the ability to bind to R1, recombinant S. pombe γ-tubulin
or porcine α/β-tubulin was incubated with GST-R1 or GST (Figure 7,
B and C). The recombinant γ-tubulin effectively bound to GST-R1
(Kd = ∼1.7 ×
10−6 M), but not to GST either in the presence
or absence of HSE. Porcine α/β-tubulin weakly bound to GST-R1
(Kd = ∼3.8 ×
10−5 M). These results suggest that γ-tubulin
itself has R1 binding activity, but the activity of most γ-tubulin is
blocked in the extract.
Figure 7.
R1 interacts with γ- and α/β-tubulin and R2 in vitro. (A) GST-R1 interacts with γ- and α-tubulin and R2 in Xenopus egg mitotic extracts. GST-R1 (37 μg; a, c, and e) or GST (33 μg; b, d, and f) was incubated with 40 μl of a Xenopus egg mitotic extract (HSE) and recovered with Glutathione-Sepharose beads. Total amounts of α- and γ-tubulin and R2 in the beads-bound fractions and HSE were determined by quantitative immunoblotting with anti–α-tubulin (B-5-1-2), anti–γ-tubulin (GTU88), and anti-R2 (anti-MRSS) antibodies using purified proteins as standards. (B) Exogenous γ-tubulin interacts with GST-R1 in Xenopus egg mitotic extracts. GST-R1 (a and b) or GST (c and d) was incubated with HSE containing 100 ng (a and c) or 300 ng (b and d) of S. pombe γ-tubulin and processed as in (A). (C) γ-Tubulin and α/β-tubulin directly bind to GST-R1. GST-R1 (a, b, e, and f) or GST (c, d, g, and h) was incubated with 100 ng (a and c) or 300 ng (b and d) of S. pombe γ-tubulin or with 2 μg (e and g) or 8 μg (f and h) of porcine α/β-tubulin and processed as in (A).
DISCUSSION
R1 Acts as a Universal Activator of the MTOCs
In this article, we have identified an 85-kDa SPB activator purified from Xenopus egg mitotic extracts as the R1 subunit of RNR. R1 acts as an activator of both the SPB and the centrosome; the yeast SPBs and the animal centrosomes are structurally different from each other but are functionally equivalent as the major MTOCs that act as the spindle poles during mitosis. The ubiquitous distribution of γ-tubulin, a protein that plays a key role in microtubule nucleation, on SPBs and centrosomes suggests that the fundamental mechanisms for microtubule nucleation are common between SPBs and centrosomes. Horio and Oakley (1994) have reported that human γ-tubulin can substitute for the fission yeast counterpart, suggesting that molecular machines and mechanisms for microtubule nucleation are evolutionarily conserved. In fact, homologues of Spc98p and Spc97p, the Saccharomyces cerevisiae SPB components that interact with γ-tubulin (Geissler et al., 1996; Knop et al., 1997) are found in Xenopus and mammalian γ-tubulin complexes and are involved in microtubule nucleation on the centrosome (Martin et al., 1998; Murphy et al., 1998; Tassin et al., 1998). Spc98p and Spc97p homologues are also found in S. pombe (SPBC428.20C/Alp6 and Alp4; Vardy and Toda, 2000). Here we have shown that Xenopus and mouse R1 proteins are able to activate the S. pombe SPB. Consistently with this observation, R1 is extremely well conserved in eukaryotes: S. pombe and human R1 share 66% amino acid sequence identity, and the sequence similarities are distributed along the whole polypeptide chain (Parker et al., 1991; Fernandez Sarabia et al., 1993). Our results, then, suggest that the microtubule-nucleating activity of the SPB and the centrosome is regulated through a similar mechanism mediated by a common component, R1.
We suggest that R1 plays a role in activation of multiprotein complexes containing γ-tubulin (γ-tubulin complexes), which has been proposed to act as seeds or templates of microtubule assembly (Oakley, 1992; Zheng et al., 1995; Erickson and Stoffler, 1996), to nucleate microtubules on the centrosome/SPB. In S. pombe, γ-tubulin complexes already exist at the interphase SPB, and the amount of γ-tubulin does not change significantly at the onset of mitosis when the SPB becomes active for microtubule nucleation (Horio et al.,1991; Masuda et al., 1992; Ding et al., 1997). This means that γ-tubulin complexes on the SPB must be activated for nucleating microtubules as the cell enters mitosis. Our results suggest that R1 is involved in this activation of γ-tubulin complexes on the SPB. The activation could simply be due to a structural alteration of the complex by binding of R1 or by a posttranslational modification of the component(s). Alternatively, it could be due to a promotion of microtubule-nucleation that is directly induced by R1 at the complex when microtubules assemble. We found that γ-tubulin has the ability to bind to GST-R1 with a moderate affinity (Kd = ∼1.7 × 10−6 M) in vitro (Figure 7, B and C). On the other hand, only 1% of γ-tubulin in Xenopus egg mitotic extracts bound to exogenous GST-R1 (Figure 7A). In the Xenopus egg cytoplasm, a large part of γ-tubulin exists as a component of γ-tubulin ring complex (γTuRC; Zheng et al., 1995). R1, which is eluted at 160–220 kDa on gel-filtration column chromatography, is not likely a component of γTuRC because γTuRC has 2000 kDa of molecular mass and is eluted at 25S (Zheng et al., 1995). Considering these results together with our previous observation that the activator in egg mitotic extracts activates SPBs through direct interaction with γ-tubulin present on the SPBs (Masuda and Shibata, 1996), we propose that interaction of R1 with γ-tubulin is locally restricted on the SPB so that the activation of the γ-tubulin complex occurs only at the SPB. The ability of R1 to bind α/β-tubulin suggests that R1 may increase local concentrations of α/β-tubulin at the γ-tubulin complex or more directly promote interaction of the γ-tubulin complex and α/β-tubulin on the SPB.
Unlike the S. pombe SPB, the centrosome in animal cells seems to be activated through more than one mechanism. The amount of γ-tubulin on the centrosome increases at the onset of mitosis (Dictenberg et al., 1998; Khodjakov and Rieder, 1999), suggesting that recruitment of γ-tubulin complexes from the cytoplasm increases the number of nucleation sites. However, the recruitment of γ-tubulin complexes may be insufficient for microtubule assembly. The amount of γ-tubulin per centrosome in CHO cells increases 2.5-fold, progressively from G1 through G2 (Dictenberg et al., 1998), whereas the microtubule nucleating activity per centrosome does not change during this period (Kuriyama and Borisy, 1981). Moreover, we found that R1 partial peptides inhibit microtubule assembly at the sperm centrosome in Xenopus egg extracts without affecting the accumulation of γ-tubulin on the centrosome (Figure 5B, a–d). We suppose that as found in S. pombe SPB activation, γ-tubulin complexes on the centrosome could be activated for nucleating microtubules and R1 may play a role in the activation. This hypothesis is supported by our finding that R1 localizes at the centrosome in animal cells (Figure 6).
Potential Roles of a Bifunctional R1 Protein in Cell Cycle Progression
Our results suggest that R1 is a bifunctional protein that is essential for two distinct cellular events: as a subunit of RNR, R1 is required for DNA synthesis, and as an activator of MTOCs, R1 is required for spindle formation. Interestingly, certain cold-sensitive and deletion mutants of RNR4, the second RNR R2 gene in S. cerevisiae (Huang and Elledge, 1997; Wang et al., 1997), are resistant to the microtubule-depolymerizing drug benomyl (Wang et al., 1997). Resistance to benomyl is also achieved by treating wild-type cells with hydroxyurea; hydroxyurea binds to the R2 subunit and inhibits RNR activity (Wang et al., 1997). These results suggest a link between RNR activity and microtubule organization.
In S. pombe, R1 and R2 are encoded by the essential genes cdc22+ and suc22+, respectively (Fernandez Sarabia et al., 1993). Only two cdc22 mutant alleles (cdc22-M45 and C11) have been reported, both of which show S phase arrest at the restrictive temperature (Nasmyth and Nurse, 1981). Nevertheless, a recent report on Liz1p has provided an implication for a possible G2/M function for RNR (Moynihan and Enoch, 1999): in the absence of Liz1p function, inactivation of RNR, by either hydroxyurea or the cdc22-M45 mutation, causes an aberrant mitosis, resulting in chromosome missegregation and late mitotic arrest.
If R1 is a bifunctional protein as we suggest, its functions must be regulated during the cell cycle such that it enables DNA synthesis in S phase and activation of microtubule nucleation in M phase. For RNR activity, an R1 homodimer must form a tetrameric complex with an R2 homodimer, whereas for activation of microtubule nucleation, the R1 dimer does not interact with R2, suggesting that it may induce MTOC activation only when it is dissociated from R2. The relative amount of R1 to R2 may influence R1–R2 complex formation, and as a result, switch on/off R1 function as an MTOC activator. In support of this idea, although the amount of R1 is relatively constant throughout the cell cycle, the amount of R2 oscillates, reaching a maximum in S phase and correlating to peak RNR activity (Eriksson et al., 1984; Engstrom et al., 1985). Simple dissociation of R1 from R2, however, may not be sufficient for MTOC activation. Importantly, native R1 purified from Xenopus eggs was much more efficient than recombinant mouse R1 in activating SPBs: to activate SPBs to the same extent as native Xenopus R1, more than five times higher concentrations of recombinant mouse R1 were required (our unpublished data). We speculate that phosphorylation or some other modification of R1 occurring downstream of CDK1 protein kinase activates the MTOC activator function of R1, because the key mitotic kinase CDK1 indirectly enhances microtubule nucleation at SPBs (Masuda et al., 1992) and centrosomes (Buendia et al., 1992; Ohta et al., 1993).
We propose that R1, because of its bifunctionality, may play an active role in preventing mitosis in the presence of DNA damage or replication blocks. RNR activity is known to increase in response to those events. In S. cerevisiae, DNA replication/damage checkpoint proteins, Mec1p and Rad53p, are involved in the regulation of dNTP levels: they seem to be required to induce RNR genes (Huang et al., 1998) and relieve inhibition of RNR activity caused by binding of Sml1p to Rnr1p (S. cerevisiae RNR R1; Zhao et al., 1998; Chabes et al., 1999). In human cells, DNA damage induces expression of p53R2, a RNR R2 homolog, in a p53-dependent manner. Induction of p53R2 and the resulting RNR activity seem to be required for DNA repair and G2/M arrest (Tanaka et al., 2000). We speculate that activation of R1 function as a subunit of RNR suppresses R1 function as the MTOC activator, thereby preventing the spindle pole activation and subsequent spindle formation.
Loss of centrosome function in response to DNA defects has been observed in Drosophila syncytial divisions in early development. DNA replication block or DNA damage induces mitosis-specific centrosome inactivation involving dissociation of γ-tubulin complex components and results in spindle defects when the replication/damage checkpoint fails (Sibon et al., 2000). This observation suggests the presence of a mechanism that monitors DNA defects and directly signals to the centrosome.
The centrosome possibly functions as a cellular control center where essential processes are monitored and signals for progress through or arrest the cell cycle are produced, and R1 may be involved in this process. Further studies on regulation of the R1 dual functions will provide new insights into the cell cycle control mechanism involving the centrosome.
ACKNOWLEDGMENTS
We thank Hiroshi Miyazawa, Fumio Hanaoka (RIKEN), and Hiroshi Nojima (Osaka University) for cDNA libraries, Zac Cande (University of California, Berkeley), Nobuo Hanai and Akiko Furuya (Kyowa Hakkou Co.) for antibodies, Yuko Kiyosue and Nobuyuki Shiina (JST) for plasmids, Tomohiko Itoh (Nagoya University) for tubulin pellets, Koji Okuhara (University of Tokyo) for Xenopus sperm, Sumiko Gomi (RIKEN) for technical assistance on baculovirus expression system, and Yasue Ichikawa (Biodesign DNA sequencing facility, RIKEN) for DNA sequence analysis. Rabbit antisera were generated at the Department of Research Fundamental Technology, Division of Laboratory Animal Research, RIKEN. We also thank Takashi Toda (Imperial Cancer Research Fund) for communicating information before publication, and David Alexander (Kyoto University), Zac Cande, Satoru Uzawa (University of California, Berkeley), and members of our laboratories for critical reading of the manuscript. This work was supported by Special Postdoctoral Researchers Program of RIKEN to S.T., grants from Japan Science and Technology Corporation to H.M. and Y.H., a grant from Human Frontier Science Program to H.M., and Biodesign Research Program of RIKEN.
Abbreviations used:
- MTOC
microtubule-organizing center
- SPB
spindle pole body
- RNR
ribonucleotide reductase
REFERENCES
- Bailly E, Doree M, Nurse P, Bornens M. p34cdc2 is located in both nucleus and cytoplasm; part is centrosomally associated at G2/M and enters vesicles at anaphase. EMBO J. 1989;8:3985–3995. doi: 10.1002/j.1460-2075.1989.tb08581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly E, Pines J, Hunter T, Bornens M. Cytoplasmic accumulation of cyclin B1 in human cells: association with a detergent-resistant compartment and with the centrosome. J Cell Sci. 1992;101:529–545. doi: 10.1242/jcs.101.3.529. [DOI] [PubMed] [Google Scholar]
- Bornens M, Paintrand M, Berges J, Marty MC, Karsenti E. Structural and chemical characterization of isolated centrosomes. Cell Motil Cytoskelet. 1987;8:238–49. doi: 10.1002/cm.970080305. [DOI] [PubMed] [Google Scholar]
- Brinkley BR, Goepfert TM. Supernumerary centrosomes and cancer: Boveri's hypothesis resurrected. Cell Motil Cytoskelet. 1998;41:281–288. doi: 10.1002/(SICI)1097-0169(1998)41:4<281::AID-CM1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Brown CR, Doxsey SJ, White E, Welch WJ. Both viral (adenovirus E1B) and cellular (hsp 70, p53) components interact with centrosomes. J Cell Physiol. 1994;160:47–60. doi: 10.1002/jcp.1041600107. [DOI] [PubMed] [Google Scholar]
- Brown NC, Reichard P. Role of effector binding in allosteric control of ribonucleotide diphosphate reductase. J Mol Biol. 1969;46:39–55. doi: 10.1016/0022-2836(69)90056-4. [DOI] [PubMed] [Google Scholar]
- Buendia B, Draetta G, Karsenti E. Regulation of the microtubule nucleating activity of centrosomes in Xenopus egg extracts: role of cyclin A-associated protein kinase. J Cell Biol. 1992;116:1431–1442. doi: 10.1083/jcb.116.6.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caras IW, Levinson BB, Fabry M, Williams SR, Martin DW., Jr Cloned mouse ribonucleotide reductase subunit M1 cDNA reveals amino acid sequence homology with Escherichia coli and herpesvirus ribonucleotide reductases. J Biol Chem, 1985;260:7015–7022. [PubMed] [Google Scholar]
- Chabes A, Domkin V, Thelander L. Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J Biol Chem. 1999;274:36679–36683. doi: 10.1074/jbc.274.51.36679. [DOI] [PubMed] [Google Scholar]
- Davis R, Thelander M, Mann GJ, Behravan G, Soucy F, Beaulieu P, Lavallee P, Graslund A, Thelander L. Purification, characterization, and localization of subunit interaction area of recombinant mouse ribonucleotide reductase R1 subunit. J Biol Chem. 1994;269:23171–2376. [PubMed] [Google Scholar]
- Dictenberg JB, Zimmerman W, Sparks CA, Young A, Vidair C, Zheng Y, Carrington W, Fay FS, Doxsey SJ. Pericentrin, and γ-tubulin. form a protein complex and are organized into a novel lattice at the centrosome. J Cell Biol. 1998;141:163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R, West RR, Morphew DM, Oakley BR, McIntosh JR. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol Biol Cell. 1997;8:1461–1479. doi: 10.1091/mbc.8.8.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey SJ, Stein P, Evans L, Calarco PD, Kirschner M. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell. 1994;76:639–650. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- Elledge SJ, Zhou Z, Allen JB, Navas TA. DNA damage and cell cycle regulation of ribonucleotide reductase. BioEssays. 1993;15:333–339. doi: 10.1002/bies.950150507. [DOI] [PubMed] [Google Scholar]
- Engstrom Y, Rozell B, Hansson S, Thelander L. Localization of ribonucleotide reductase in mammalian cells. EMBO J. 1984;3:863–867. doi: 10.1002/j.1460-2075.1984.tb01897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom Y, Eriksson S, Jildevik I, Skog S, Thelander L, Tribukait B. Cell cycle-dependent expression of mammalian ribonucleotide reductase. Differential regulation of the two subunits. J Biol Chem. 1985;260:9114–9116. [PubMed] [Google Scholar]
- Erickson HP, Stoffler D. Protofilaments and rings, two conformations of the tubulin family conserved from bacterial FtsZ to α/β and γ-tubulin. J Cell Biol. 1996;135:5–8. doi: 10.1083/jcb.135.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S, Graslund A, Skog S, Thelander L, Tribukait B. Cell-cycle dependent regulation of mammalian ribonucleotide reductase. The S phase-correlated increase in subunit M2 is regulated by de novo protein synthesis. J Biol Chem. 1984;259:11695–11700. [PubMed] [Google Scholar]
- Eriksson M, Uhlin U, Ramaswamy S, Ekberg M, Regnstrom K, Sjoberg BM, Eklund H. Binding of allosteric effectors to ribonucleotide reductase protein R1: reduction of active-site cysteines promotes substrate binding. Structure. 1997;5:1077–1092. doi: 10.1016/s0969-2126(97)00259-1. [DOI] [PubMed] [Google Scholar]
- Fava F, Raynaud-Messina B, Leung-Tack J, Mazzolini L, Li M, Guillemot JC, Cachot D, Tollon Y, Ferrara P, Wright M. Human 76p: a new member of the γ-tubulin-associated protein family. J Cell Biol. 1999;147:857–868. doi: 10.1083/jcb.147.4.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix M-A, Antony C, Wright M, Maro B. Centrosome assembly in vitro: role of γ-tubulin recruitment in Xenopus sperm aster formation. J Cell Biol. 1994;124:19–31. doi: 10.1083/jcb.124.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez Sarabia M-J, McInerny C, Harris P, Gordon C, Fantes P. The cell cycle genes cdc22+ and suc22+ of the fission yeast Schizosaccharomyces pombe encode the large and small subunits of ribonucleotide reductase. Mol Gen Genet. 1993;238:241–251. doi: 10.1007/BF00279553. [DOI] [PubMed] [Google Scholar]
- Gallant P, Nigg EA. Cyclin B2 undergoes cell cycle-dependent nuclear translocation and, when expressed as a non-destructible mutant, causes mitotic arrest in HeLa cells. J Cell Biol. 1992;117:213–224. doi: 10.1083/jcb.117.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler SG, Pereira G, Spang A, Knop M, Soues S, Kilmartin J, Schiebel E. The spindle pole body component Spc98p interacts with the γ-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. EMBO J. 1996;15:3899–3911. [PMC free article] [PubMed] [Google Scholar]
- Hagan I, Hyams JS. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Hagan IM, Gull K, Glover DM. Poles apart? Spindle pole bodies and centrosomes differ in ultrastructure yet their function and regulation are conserved. In: Endow SA, Glover DM, editors. Dynamics of Cell Division. Oxford, United Kingdom: Oxford University Press; 1998. pp. 57–96. [Google Scholar]
- Horio T, Uzawa S, Jung MK, Oakley BR, Tanaka K, Yanagida M. The fission yeast γ-tubulin is essential for mitosis and is localized at microtubule organizing centers. J Cell Sci. 1991;99:693–700. doi: 10.1242/jcs.99.4.693. [DOI] [PubMed] [Google Scholar]
- Horio T, Oakley BR. Human γ-tubulin functions in fission yeast. J Cell Biol. 1994;126:1465–1473. doi: 10.1083/jcb.126.6.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Elledge SJ. Identification of RNR4, encoding a second essential small subunit of ribonucleotide reductase in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6105–6113. doi: 10.1128/mcb.17.10.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Zhou Z, Elledge SJ. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 1998;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- Hyman A, Drechsel D, Kellogg D, Salser S, Sawin K, Steffen P, Wordeman L, Mitchison T. Preparation of modified tubulins. Methods Enzymol. 1991;196:478–485. doi: 10.1016/0076-6879(91)96041-o. [DOI] [PubMed] [Google Scholar]
- Ingemarson R, Thelander L. A kinetic study on the influence of nucleoside triphosphate effectors on subunit interaction in mouse ribonucleotide reductase. Biochemistry. 1996;35:8603–8609. doi: 10.1021/bi960184n. [DOI] [PubMed] [Google Scholar]
- Joshi H, Palacios MJ, McNamara L, Cleaveland DW. γ-Tubulin is a centrosomal protein required for cell cycle-dependent microtubule nucleation. Nature. 1992;356:80–82. doi: 10.1038/356080a0. [DOI] [PubMed] [Google Scholar]
- Kalt A, Schliwa M. Molecular components of the centrosome. Trends Cell Biol. 1993;3:118–128. doi: 10.1016/0962-8924(93)90174-y. [DOI] [PubMed] [Google Scholar]
- Kellogg DR, Moritz M, Alberts BM. The centrosome and cellular organization. Annu Rev Biochem. 1994;63:639–674. doi: 10.1146/annurev.bi.63.070194.003231. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL. The sudden recruitment of γ-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J Cell Biol. 1999;146:585–596. doi: 10.1083/jcb.146.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, Schiebel E. The spindle pole body component Spc97p interacts with the gamma-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 1997;16:1550–1564. doi: 10.1093/emboj/16.7.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama R, Borisy GG. Microtubule-nucleating activity of centrosomes in Chinese hamster ovary cells is independent of the centriole cycle but coupled to the mitotic cycle. J Cell Biol. 1981;91:822–826. doi: 10.1083/jcb.91.3.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Codina G, Glover DM. Cyclins A and B associate with chromatin and the polar regions of spindles, respectively, and do not undergo complete degradation at anaphase in syncytial Drosophila embryos. J Cell Biol. 1992;116:967–976. doi: 10.1083/jcb.116.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall LG, Jeng RL, Mulholland J, Stearns T. Analysis of Tub4p, a yeast γ-tubulin-like protein: implications for microtubule-organizing center function. J Cell Biol. 1996;134:443–454. doi: 10.1083/jcb.134.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin OC, Gunawardane RN, Iwamatsu A, Zheng Y. Xgrip109: a γ tubulin-associated protein with an essential role in γ tubulin ring complex (γTuRC) assembly and centrosome function. J Cell Biol. 1998;141:675–687. doi: 10.1083/jcb.141.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H, Hirano T, Yanagida M, Cande WZ. In vitro reactivation of spindle elongation in fission yeast nuc2 mutant cells. J Cell Biol. 1990;110:417–425. doi: 10.1083/jcb.110.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H, Sevik M, Cande WZ. In vitro microtubule-nucleating activity of spindle pole bodies in fission yeast Schizosaccharomyces pombe: cell cycle-dependent activation in Xenopus cell-free extracts. J Cell Biol. 1992;117:1055–1066. doi: 10.1083/jcb.117.5.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H, Shibata T. Role of γ-tubulin in mitosis- specific microtubule nucleation from the Schizosaccharomyces pombe spindle pole body. J Cell Sci. 1996;109:165–177. doi: 10.1242/jcs.109.1.165. [DOI] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Sedat JW, Alberts B, Agard DA. Microtubule nucleation by γ-tubulin-containing rings in the centrosome. Nature. 1995;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- Moritz M, Zheng Y, Alberts BM, Oemega K. Recruitment of the γ-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. J Cell Biol. 1998;142:775–786. doi: 10.1083/jcb.142.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moudjou M, Bordes N, Paintrand M, Bornens M. γ-Tubulin in mammalian cells: the centrosomal and the cytosolic forms. J Cell Sci. 1996;109:875–887. doi: 10.1242/jcs.109.4.875. [DOI] [PubMed] [Google Scholar]
- Moynihan EB, Enoch T. Liz1p, a novel fission yeast membrane protein, is required for normal cell division when ribonucleotide reductase is inhibited. Mol Biol Cell. 1999;10:245–257. doi: 10.1091/mbc.10.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SM, Urbani L, Stearns T. The mammalian γ-tubulin complex contains homologues of the yeast spindle pole body components Spc97p and Spc98p. J Cell Biol. 1998;141:663–674. doi: 10.1083/jcb.141.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Nakamura M, Masuda H, Horii J, Kuma K, Yokoyama N, Ohba T, Nishitani H, Miyata T, Tanaka M, Nishimoto T. When overexpressed, a novel centrosomal protein, RanBPM, causes ectopic microtubule nucleation similar to γ-tubulin. J Cell Biol. 1998;143:1041–1052. doi: 10.1083/jcb.143.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Nurse P. Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1981;182:119–124. doi: 10.1007/BF00422777. [DOI] [PubMed] [Google Scholar]
- Oakley BR. γ-tubulin. The microtubule organizer? Trends Cell Biol. 1992;2:1–5. doi: 10.1016/0962-8924(92)90125-7. [DOI] [PubMed] [Google Scholar]
- Oakley BR, Oakley CE, Yoon Y, Jung MK. γ-Tubulin is a component of the spindle pole body that is essential for microtubule function on Aspergillus nidulans. Cell. 1990;61:1289–1301. doi: 10.1016/0092-8674(90)90693-9. [DOI] [PubMed] [Google Scholar]
- Ohta K, Shiina N, Okumura E, Hisanaga S, Kishimoto T, Endo S, Gotoh Y, Nishida E, Sakai H. Microtubule nucleating activity of centrosomes in cell-free extracts from Xenopus eggs: involvement of phosphorylation and accumulation of pericentriolar material. J Cell Sci. 1993;104:125–137. doi: 10.1242/jcs.104.1.125. [DOI] [PubMed] [Google Scholar]
- Parker NJ, Begley CG, Fox RM. Human M1 subunit of ribonucleotide reductase: cDNA sequence and expression in stimulated lymphocytes. Nucleic Acids Res. 1991;19:3741. doi: 10.1093/nar/19.13.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. Nuclear transport protein does double duty in mitosis. Science. 1999;284:1260–1261. doi: 10.1126/science.284.5418.1260. [DOI] [PubMed] [Google Scholar]
- Pereira G, Schiebel E. Centrosome-microtubule nucleation. J Cell Sci. 1997;110:295–300. doi: 10.1242/jcs.110.3.295. [DOI] [PubMed] [Google Scholar]
- Pereira G, Knop M, Schiebel E. Spc98p directs the yeast γ-tubulin complex into the nucleus and is subject to cell-cycle-dependent phosphorylation on the nuclear side of the spindle pole body. Mol Biol Cell. 1998;9:775–793. doi: 10.1091/mbc.9.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annu Rev Biochem. 1988;57:349–374. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- Reichard P. From RNA to DNA, why so many ribonucleotide reductases? Science. 1993;260:1773–1777. doi: 10.1126/science.8511586. [DOI] [PubMed] [Google Scholar]
- Reinsch S, Gonczy P. Mechanisms of nuclear positioning. J Cell Sci. 1998;111:2283–2295. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- Sibon OCM, Kelkar A, Lemstra W, Theurkauf WE. DNA-replication/DNA-damage-dependent centrosome inactivation in Drosophila embryos. Nature Cell Biol. 2000;2:90–95. doi: 10.1038/35000041. [DOI] [PubMed] [Google Scholar]
- Snyder JA, McIntosh JR. Initiation and growth of microtubules from mitotic centers in lysed mammalian cells. J Cell Biol. 1975;67:744–760. doi: 10.1083/jcb.67.3.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel SG, Snyder M. A high divergent γ-tubulin gene is essential for cell growth and proper microtubule organization in Saccharomyces cerevisiae. J Cell Biol. 1995;131:1775–1788. doi: 10.1083/jcb.131.6.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Geissler S, Grein K, Schiebel E. γ-Tubulin-like Tub4p of Saccharomyces cerevisiae is associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J Cell Biol. 1996;134:429–441. doi: 10.1083/jcb.134.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Kirschner M. In vitro reconstitution of centrosome assembly and function: the central role of γ-tubulin. Cell. 1994;76:623–637. doi: 10.1016/0092-8674(94)90503-7. [DOI] [PubMed] [Google Scholar]
- Sunkel CE, Gomes R, Sampaio P, Perdigao J, Gonzales C. γ-tubulin is required for the structure and function of the microtubule organizing center in Drosophila neuroblasts. EMBO J. 1995;14:28–36. doi: 10.1002/j.1460-2075.1995.tb06972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- Tassin A-M, Celati C, Moudiou M, Bornens M. Characterization of the human homologue of the yeast Spc98p and its association with γ-tubulin. J Cell Biol. 1998;255:7426–7432. doi: 10.1083/jcb.141.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelander L, Reichard P. Reduction of ribonucleotides. Annu Rev Biochem. 1979;48:133–158. doi: 10.1146/annurev.bi.48.070179.001025. [DOI] [PubMed] [Google Scholar]
- Thelander L, Eriksson S, Akerman M. Ribonucleotide reductase from calf thymus. J Biol Chem. 1980;255:7426–7432. [PubMed] [Google Scholar]
- Thelander M, Graslund A, Thelander L. Subunit M2 of mammalian ribonucleotide reductase. J Biol Chem. 1985;260:2737–2741. [PubMed] [Google Scholar]
- Tugendreich S, Tomkiel J, Earnshaw W, Hieter P. CDC27Hs colocalizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell. 1995;81:261–268. doi: 10.1016/0092-8674(95)90336-4. [DOI] [PubMed] [Google Scholar]
- Uhlin U, Eklund H. Structure of ribonucleotide reductase protein R1. Nature. 1994;370:533–539. doi: 10.1038/370533a0. [DOI] [PubMed] [Google Scholar]
- Vardy, L., and Toda, T. (2000). The fission yeast γ-tubulin complex is required in G1 phase and is a component of the spindle-assembly checkpoint. EMBO J. (in press). [DOI] [PMC free article] [PubMed]
- Vernos I, Karsenti E. Motors involved in spindle assembly and chromosome segregation. Curr Opin Cell Biol. 1996;8:4–9. doi: 10.1016/s0955-0674(96)80041-x. [DOI] [PubMed] [Google Scholar]
- Vogel JM, Stearns T, Rieder CL, Palazzo RE. Centrosomes isolated from Spisula solidissima oocytes contain rings and an unusual stoichiometric ratio of α/β tubulin. J Cell Biol. 1997;137:193–202. doi: 10.1083/jcb.137.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PJ, Chabes A, Casagrande R, Tian XC, Thelander L, Huffaker TC. Rnr4p, a novel ribonucleotide reductase small-subunit protein. Mol Cell Biol. 1997;17:6114–6121. doi: 10.1128/mcb.17.10.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC, Salmon ED. Pathways of spindle assembly. Curr Opin Cell Biol. 1997;9:37–43. doi: 10.1016/s0955-0674(97)80149-4. [DOI] [PubMed] [Google Scholar]
- Wigley WC, Fabunmi RP, Lee MG, Marino CR, Mueallem S, DeMartino GN, Thomas PJ. Dynamic association of proteasomal machinery with the centrosome. J Cell Biol. 1999;145:481–490. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wong ML, Alberts B, Michison T. Nucleation of microtubule assembly by γ-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]