Abstract
Bovine leukemia virus (BLV) is silent in most cells detectable in vivo, and the repression of its expression allows BLV to evade the host's immune response. In this study, we examined whether CpG methylation of DNA might be involved in the regulation of the expression of BLV in vivo. To investigate the effects of CpG methylation on the activity of the long terminal repeat (LTR) of BLV, we measured the transactivation activity of this region after treatment with the CpG methyltransferase SssI by using a luciferase reporter system. The activity of methylated LTR was significantly lower than that of nonmethylated LTR. Therefore, we examined the extent of CpG methylation of the U3 region and part of the R region of the LTR in BLV-infected cattle and in experimentally BLV-infected sheep at various clinical stages by the bisulfite genomic sequencing method. We detected no or minimal CpG methylation at all stages examined in cattle and sheep, and our results indicate that CpG methylation probably does not participate in the silencing of BLV in vivo.
Bovine leukemia virus (BLV), which is closely related to human T-cell leukemia virus types 1 and 2 (HTLV-1 and HTLV-2), is the etiologic agent of enzootic bovine leukosis (EBL), a disease characterized by a very extended course that often involves persistent lymphocytosis (PL) and culminates in B-cell lymphosarcoma (2). Sheep that are experimentally inoculated with BLV develop B-cell tumors at a higher frequency and with a shorter latency period than naturally infected cattle (1, 8). BLV encodes at least two apparently unique regulatory proteins, Tax and Rex. Tax acts on a triplicate 21-bp motif, known as the Tax-responsive element, in the U3 region of the long terminal repeat (LTR), and it stimulates transactivation of the viral genome (6, 37). Rex interacts with a sequence known as the Rex-responsive element in the R region of the 3′ LTR of BLV mRNA and enhances the cytoplasmic accumulation of singly or nonspliced transcripts that encode structural proteins (Gag, Pol, and Env proteins) (7, 25).
After infection of cattle, the expression of BLV is blocked at the transcriptional level during the latency period (14, 17), and such repression appears to be very important in the escape of BLV from the host's immunosurveillance system. Such silencing is also observed in sheep infected with BLV (17, 25). By contrast, the synthesis of viral RNA is induced within 1 h when peripheral blood lymphocytes (PBL), purified from the blood of infected animals, are placed in culture, even in the presence of cycloheximide, an inhibitor of protein synthesis (17, 25).
The mechanism responsible for the BLV silencing is unknown. It has been reported previously that BLV-infected cattle retain a full-length proviral genome throughout the course of the disease, suggesting that BLV silencing is not associated with genetic deletion(s) in the BLV proviral genome (32). However, recent findings indicate that a viral enhancer sequence appears to be suitable for both the activation and the repression of viral expression (18). Rex might decrease the expression of viral proteins by suppressing the expression of tax and rex RNA (7, 25). However, the continuous expression of viral transcripts can be observed in many types of cultured cells that have been stably transfected with an infectious molecular clone of BLV (11), indicating that Rex alone is insufficient for the repression of viral expression.
In mammals, 60 to 90% of cytosine residues in all CpG dinucleotides are methylated and many of the nonmethylated CpG sites are found in so-called “CpG islands,” which usually include functional promoters. The CpG methylation is correlated with the repression of gene expression, but the mechanism responsible for this phenomenon remains unknown. Recent studies have provided insight into the mechanism of transcriptional repression by methyl-CpG-binding proteins (12, 22). For example, MeCP2, which is a methyl-CpG-binding protein, interacts with the transcriptional corepressor Sin3, which is a histone deacetylase, and this observation suggests that the repression of gene expression by CpG methylation might involve chromatin modification (12, 21). The extent of methylation of many proviral genomes has also been examined in studies with methylation-sensitive restriction enzymes that can discriminate between the presence and the absence of methylation of cytosine residues at their recognition sites. Such studies demonstrated that proviral DNA is methylated in the PBL from BLV-infected cattle and HTLV-1-infected patients (5, 13, 15, 23). In MT4 cells transformed with HTLV-1, which express a low level of viral RNA, the proviral genome is heavily methylated, and the treatment of such cells with 5-azacytidine, a demethylating agent, results in an increase in viral expression (30). In vitro methylation of the HTLV-1 LTR-chloramphenicol acetyltransferase construct or an LTR-luciferase construct resulted in significant inhibition of the transcription of the genes in transfected cells (3, 31). These findings suggest that methylation of the proviral genome might serve to prevent viral expression. On the other hand, differences in the extents of methylation have been noted among various regions of the proviral genome in PBL of BLV-infected cattle and HTLV-1-infected patients (15, 23). However, studies with restriction enzymes lack overall sensitivity because of their dependence on restriction sites and, hence, the pattern of CpG methylation within an integrated proviral LTR remains to be clarified. A more sensitive method for the detection of DNA methylation, known as bisulfite genomic sequencing, was developed (4). Using this method, Koiwa et al. demonstrated the hypermethylation of the 5′ LTR and the accompanying hypomethylation of the 3′ LTR both in latently HTLV-1-infected cell lines and in HTLV-1-induced-adult T-cell leukemia cells that harbored a complete provirus (16). These findings indicate that it is necessary to investigate the methylation status of the proviral genome of BLV, and that of the LTR in particular, by a more sensitive method than restriction enzyme digestion. In this study, we examined the possible role of CpG methylation of the BLV LTR in the silencing of BLV by using a reporter gene and an LTR that had been methylated in vitro. We also examined the levels of CpG methylation of parts of the LTR in BLV-positive cattle and experimentally BLV-infected sheep by bisulfite genomic sequencing.
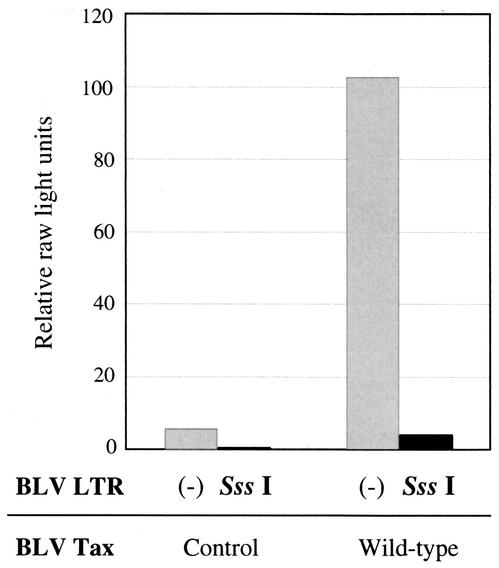
We investigated the effects of CpG methylation of the BLV LTR on the transactivation activity of the LTR by using a luciferase assay. A reporter plasmid, pGV-BLTR, in which the entire LTR was linked to the upstream region of a firefly luciferase gene (33) was methylated with CpG methyltransferase SssI (New England Biolabs, Beverly, Mass.). Human embryonic kidney 293T cells (3 × 105 cells/well in 6-well culture plates) were transfected with 1 μg of SssI-treated or untreated pGV-BLTR, 1 μg of the control plasmid pME18Neo or the Tax-expressing plasmid pME18Neo that encoded wild-type BLV Tax (33), and 0.3 μg of the reference plasmid pRL-SV40, which encoded Renilla luciferase (Promega, Madison, Wis.) as described previously (33). Sixty hours after transfection, cells were harvested and the activities of firefly luciferase and Renilla luciferase were measured in cell lysates (Fig. 1). We observed Tax-driven transactivation of the LTR when cells were cotransfected with the Tax-expressing plasmid and pGV-BLTR that had not been treated with SssI. The transactivation activity of Tax was markedly decreased when untreated pGV-BLTR was replaced by methylated pGV-BLTR (the percentage of the inhibition of transcription by SssI methylation was 96.0%). We observed a similar phenomenon when SssI-methylated pGV-BLTR was used together with the control plasmid to transfect cells (the percentage of the inhibition of transcription by SssI methylation was 92.8%). Similar results were also obtained when we used bovine lymph node 23CLN cells for the luciferase assay (data not shown). These results demonstrated that CpG methylation of the BLV LTR repressed the activity of the LTR, irrespective of the presence or absence of Tax.
FIG. 1.
Inhibition of the activity of BLV LTR by CpG methyltransferase SssI. Human embryonic kidney 293T cells were transfected with pME18Neo that encoded the wild-type Tax protein or with the control plasmid pME18Neo, pGV-BLTR that had been treated (SssI) or not treated (-) with SssI, and the reference plasmid pRL-SV40. At 60 h after transfection, cells were recovered and the activities of firefly luciferase and Renilla luciferase were measured in cell lysates. For each sample, relative raw light units were calculated by dividing the activity of firefly luciferase by that of Renilla luciferase.
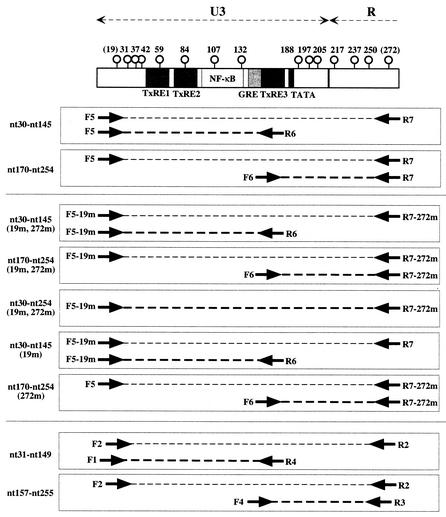
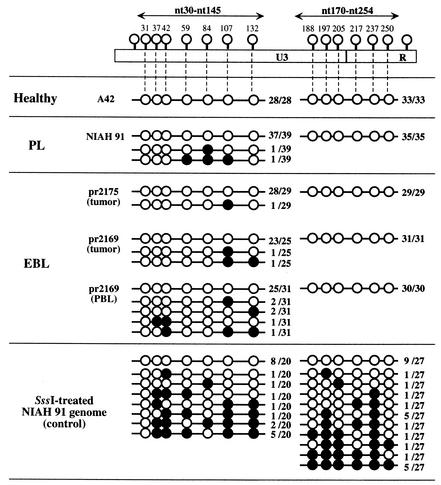
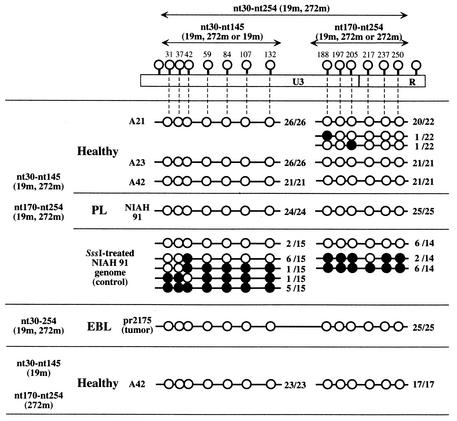
The results described above suggested that CpG methylation might regulate the expression of BLV in vivo. Therefore, we attempted to characterize the extent of methylation of the U3 region and part of the R region, which are the major regulatory regions of the proviral LTR that controls the expression of BLV in BLV-positive cattle, by the bisulfite genomic sequencing method (4), with slight modifications. We prepared genomic DNA from PBL of a BLV-infected but clinically healthy cow (A42) and a cow with PL (NIAH 91) and from the tumor tissues and PBL of cows with EBL (pr2169 and pr2175) by standard methods, as described elsewhere (10). We denatured 3-μg aliquots of DNA in 30 μl of 0.25 M sodium hydroxide for 10 min at 37°C, added 460 μl of 3 M sodium bisulfite (pH 5.0) and 10 μl of 25 mM hydroquinone, and then incubated each mixture for 15 h at 55°C. The bisulfite-modified DNA was purified with a Wizard DNA Clean-Up System (Promega), denatured in 0.25 M sodium hydroxide, precipitated, and resuspended in 50 μl of distilled water. Aliquots (1 μl) of bisulfite-modified DNA were amplified by PCR with primers F5 and R7 (as shown in Table 1 and Fig. 2) and EX Taq DNA polymerase (Ohtsu, Shiga, Japan). Amplification was achieved by 30 cycles of incubation at 94°C for 30 s, at 60°C for 30 s, and at 72°C for 30 s. Aliquots (1 μl) of the products of the first PCR were also amplified by seminested PCR by using the same conditions as those of the first PCR, with primers F5 and R6 (nucleotides [nt] 30 to 145) and primers F6 and R7 (nt 170 to 254) (Table 1 and Fig. 2). The products of PCR were blunted with T4 DNA polymerase and introduced into the EcoRV site of pBluescript II SK(+) (Stratagene, La Jolla, Calif.). The cloned DNAs were then sequenced as described previously (33) (Fig. 3). We obtained no or very few clones derived from methylated DNA from either symptomatic or asymptomatic cattle. Furthermore, in clones derived from methylated DNA, only a few CpG sites had been methylated. In this experiment, we used primers that corresponded to an LTR sequence in which two CpG sites (nt 19 and nt 272) were not methylated, and such primers might be inadequate for the amplification of LTR with significant methylation. Therefore, we next examined the methylation statuses of proviral LTRs from BLV-infected but healthy cows (A21, A23, and A42), a cow with PL (NIAH 91), and a cow with EBL (pr2175) by using primers F5-19m and R7-272m that corresponded to LTR sequences in which positions 19 and 272, respectively, were methylated (Table 1 and Fig. 2). As in the experiments described above, hardly any of the sequenced clones were clones of methylated DNA (Fig. 4). We also examined the methylation status of the SssI-treated proviral genome from cattle with PL, after preparation of genomic DNA, as a positive control (Fig. 3 and 4). The relative number of clones of methylated DNA amplified from the SssI-treated genome from cattle with PL was significantly higher than that of methylated DNA amplified from the untreated genome; approximately 50% of sequenced clones were clones of DNA that had been methylated at many CpG sites, indicating that the deduced hypomethylation of the source DNA of most of the LTR clones described above was not due to methodological problems.
TABLE 1.
Oligonucleotide primers used for bisulfite genomic sequencing analysis
| Primer | Sequence | Coordi- natesa (nt) |
|---|---|---|
| F1 | 5′-TATATAAAAAATCATACCAACCTA | 1-24 |
| F2 | 5′-AAAAATCATACCAACCTAAAAACC | 7-30 |
| F4 | 5′-AAAAATCCACACCCTAAACTACTA | 133-156 |
| F5 | 5′-TATATAAAAAATCATACCAACCTAAAAAC | 1-29 |
| F5-19m | 5′-TATATAAAAAATCATACCGACCTAAAAAC | 1-29 |
| F6 | 5′-CACACCCTAAACTACTAACCTCACCTACTA | 140-169 |
| F7 | 5′-ATTACAACTACTAAAAAATAAATAACTCTCC | 8130-8160 |
| F8 | 5′-CAAACTACTAATAATTAACAATATCTTTTAAA | |
| F9 | 5′-AATAATAATAACACAATATAATAACTATAC | |
| R2 | 5′-TAGAAGGTTTTGGGAGTAAGAGAGT | 282-258 |
| R3 | 5′-GAAGGTTTTGGGAGTAAGAGAGTTT | 280-256 |
| R4 | 5′-ATTTATTAGTAGGTGAGGTTAGTAGT | 175-150 |
| R6 | 5′-TAATTTATTAGTAGGTGAGGTTAGTAGTTTA | 177-146 |
| R7 | 5′-GATTAGAAGGTTTTGGGAGTAAGAGAGTTTA | 285-255 |
| R7-272m | 5′-GATTAGAAGGTTTCGGGAGTAAGAGAGTTTA | 285-255 |
FIG. 2.
Schematic representation of the U3 region and part of the R region of the BLV LTR and regions amplified by PCR for bisulfite genomic sequencing analysis. The upper panel shows the location of cis-acting elements in the U3 region. Open circles and numbers indicate cytosine residues in CpG dinucleotides and their positions relative to the beginning of the genome (29), respectively. Dotted lines indicate regions amplified by PCR. Primer pairs used for PCR are also indicated. TxRE, Tax-responsive element; GRE, glucocorticoid-responsive element.
FIG. 3.
CpG methylation of the U3 region and part of the R region of the LTR of BLV provirus in BLV-infected cattle at various stages of infection. The methylation status that gives rise to the various clones is indicated for each sample. Methylated and unmethylated sites are shown as filled circles and open circles, respectively. The numbers of methylated and unmethylated sequences that were cloned relative to the total number of clones examined are given to the right of the circles. Arrows indicate regions that were amplified, cloned, and sequenced in this experiment.
FIG. 4.
Levels of CpG methylation of the U3 region and part of the R region of the LTR of BLV provirus in BLV-infected cattle at various stages of infection, as assessed with primers that corresponded to the sequence of proviral LTR that was methylated at positions 19 and 252. The methylation status of each sample is presented in terms of the clones analyzed. Methylated and unmethylated sites are shown as filled circles and open circles, respectively. The numbers of clones of methylated and unmethylated DNA relative to the total number of clones examined are given to the right of the circles. The arrows represent the regions that were amplified, cloned, and sequenced in this experiment.
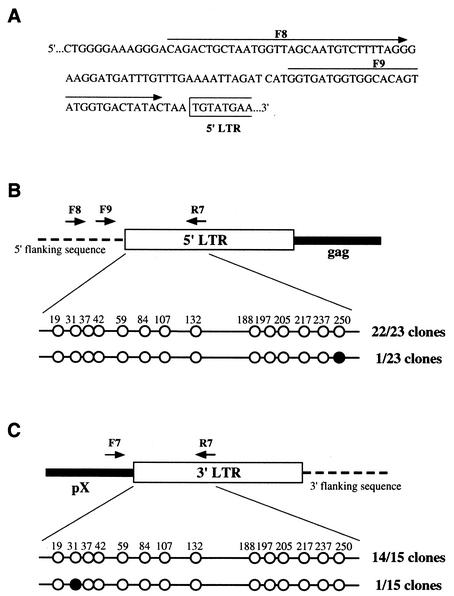
In the experiments described above, we characterized the extent of CpG methylation in both the 5′ LTR and the 3′ LTR at the same time. In the proviral genome of HTLV-1, however, the 5′ LTR is hypermethylated while the 3′ LTR is hypomethylated (16). Therefore, we next tried to examine the methylation status of the two LTRs independently. We first cloned the 5′ flanking region of the proviral genome by inverse PCR to determine the nucleotide sequence of the region. Genomic DNA from the tumor tissue of a cow with EBL (pr2169) was digested with BamHI, BclI, BglII, and BstYI, and the fragments were self-ligated. We then amplified the self-ligated DNA by PCR with primers 5′-GAGCTCTCTTGCTCCCGAGAC-3′ (nt 256 to 276) and 5′-GTCGACGTCTCTGTCTGGTTTACG-3′ (nt 64 to 41) and EX Taq DNA polymerase. Amplification was achieved by 35 cycles of incubation at 94°C for 30 s, at 60°C for 30 s, and at 72°C for 2 min. The PCR products were subcloned into plasmid pGEM-T (Promega), and their sequences were determined (Fig. 5A). The resultant 5′ flanking sequences were used to design two sense primers, F8 and F9. We also designed a sense primer, F7, on the basis of the sequence of the pX region of BLV for amplification of the bisulfite-modified 3′ LTR. We then characterized the CpG methylation of the U3 region and part of the R region of the 5′ LTR by using primers F8 and R7 for the first PCR and primers F9 and R7 for the second, seminested PCR and that of the 3′ LTR by using primers F7 and R7, as described above (Fig. 5B and C). We found that only 1 clone among 23 clones of the 5′ LTR and 1 clone among 15 clones of the 3′ LTR were methylated and that each of these clones had only one site of CpG methylation. Thus, our results demonstrated that, in the host cattle tested, the transcription-regulating region of proviral BLV was barely methylated, irrespective of the clinical stage of infection. Therefore, CpG methylation appears not to be associated with the latency of BLV.
FIG. 5.
Levels of CpG methylation of the U3 region and part of the R region of the LTR of BLV in tumor cells from a cow with EBL (pr2169). (A) Nucleotide sequence of the 5′ flanking region of the provirus in pr2169 tumor cells. Arrows indicate the positions of primers F8 and F9, which were used for bisulfite genomic sequencing analysis. (B) Results of 5′-LTR-selective bisulfite genomic sequencing analysis. Bisulfite-treated DNA was amplified with primers F8 and R7 for the first PCR and primers F9 and R7 for the second PCR. Methylated and unmethylated CpG sites are shown as filled and open circles, respectively. The numbers of methylated and unmethylated sequences relative to the total number of clones examined are indicated to the right of the circles. (C) Results of 3′-LTR-selective bisulfite genomic sequencing analysis. Bisulfite-modified DNA was amplified with primers F7 and R7. Methylated CpG sites are shown as filled circles, and unmethylated CpG sites are shown as open circles. The numbers of methylated and unmethylated sequences relative to the total number of clones examined are given to the right of the circles.
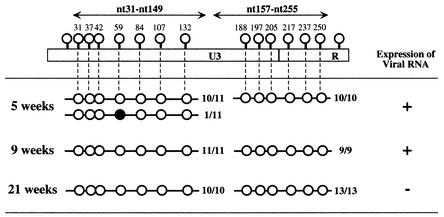
Sheep can easily be infected with BLV under experimental conditions, and we next investigated whether the methylation status in the U3 region and part of the R region of the BLV LTR in BLV-infected sheep might change with the level of expression of viral RNA. We inoculated sheep G279 with an infectious molecular clone of BLV (35), and then we collected blood 5, 9, and 21 weeks after inoculation. The methylation statuses of regions from nt 31to 149 and from nt 157 to 255 were examined by using the primers shown in Fig. 2 (Fig. 6). The expression of BLV RNA in vivo was also checked by RT-PCR as described previously (34). Viral RNA was clearly detectable 5 and 9 weeks after infection, but the level of RNA was significantly lower 21 weeks after infection. However, we detected no differences in the relative numbers of methylated clones from the various time points, and moreover, almost all clones examined were derived from unmethylated DNA. This result confirmed that in BLV-infected sheep, as well as in BLV-infected cattle, BLV expression is not correlated with the methylation of the U3 region and part of the R region of the proviral genome.
FIG. 6.
Levels of CpG methylation of the U3 region and part of the R region of the BLV LTR in PBL from a sheep, G279, that had been experimentally infected with BLV. Five, 9, and 21 weeks after the injection of an infectious BLV clone (pBLV-IF), genomic DNA was isolated from PBL and subjected to bisulfite genomic sequencing analysis. The methylation statuses of sources of clones for each time point are presented. Methylated and unmethylated sites are shown as filled circles and open circles, respectively. The numbers of methylated and unmethylated sequences relative to the total number of clones examined are given to the right of the circles. Arrows indicate the regions that were amplified, cloned, and sequenced in this experiment. The expression of viral RNA in PBL prepared from the sheep at each time point is also indicated, as follows: +, detected; −, not detected.
In the present study, we evaluated the possible involvement of CpG methylation of the BLV LTR in the regulation of viral expression. We characterized the extent of CpG methylation of the U3 region and part of the R region of the LTR, which includes most of the elements that regulate the expression of BLV, in BLV-infected cattle and in experimentally infected sheep at various stages by the bisulfite genomic sequencing method. Our observations indicated that most of the CpG sites in the U3 region and part of the R region of the proviral BLV genome were not methylated in BLV-positive animals. Nonetheless, we also showed that the transactivation activity of LTR was reduced after treatment with the CpG methyltransferase SssI. These results raise the possibility that CpG methylation is required for silencing induction but not for maintenance of BLV silencing. However, in our reporter assay, the reporter plasmid pGV-BLTR was not integrated into the cellular genome in transfected cells, an observation that indicated that the repression of BLV expression by CpG methylation in vitro is not necessarily reflected by events in PBL that harbor an integrated proviral genome. Therefore, we conclude that CpG methylation does not contribute to BLV silencing.
In contrast to our results for BLV, the 5′ LTR in HTLV-1 is hypermethylated and the 3′ LTR is hypomethylated in both latently infected cell lines and in adult T-cell leukemia cells with a complete provirus and, moreover, the expression of proviral genes is activated by an inhibitor of DNA methyltransferases in latently infected cell lines (16, 30). These findings indicate that hypermethylation of the 5′ LTR is involved in the silencing of HTLV-1 gene expression. Thus, it appears that HTLV-1 and BLV repress viral expression by different mechanisms in vivo. It remains to be determined why two such closely related viruses have developed different silencing mechanisms, but the difference might be related to the fact that the target cells of the viruses are different: the target cells of BLV and HTLV-1 are T cells and B cells, respectively. Other reasons, such as differences in the host species, also might be involved.
The expression of BLV is induced within 1 h when PBL purified from the blood of infected animals are cultured at 37°C (17, 25), and moreover, levels of BLV RNA increase within a few hours when whole blood from BLV-infected cattle is incubated at 37°C (S. Tajima, unpublished data). Therefore, it is possible that CpG demethylation would not be appropriate for rapid induction of the synthesis of viral RNA. What regulates the expression of BLV in vivo? It has been proposed that certain factors present in the plasma of BLV-infected animals might silence viral expression (9, 36, 38), and interleukin-10 reduces the level of expression of BLV mRNA in lymphocytes from BLV-infected cattle after culture in vitro (26-28). A recent study also suggests that the acetylation and deacetylation of histones and/or transcription factors might be involved in the regulation of the expression of BLV (19). Furthermore, some reports have showed the presence of a methylation-independent mechanism for the silencing of retroviral expression (20, 24). Mizutani et al. demonstrated the possibility that factors that participate in chromatin remodeling but not in DNA methylation might play a critical role in retrovirus silencing (20). Indeed, the inhibition of histone deacetylases stimulates BLV expression in vivo, indicating that the deacetylation of histones and/or certain transcription factors might be important for the repression of BLV expression (19). Detailed analysis of the acetylation, methylation, and phosphorylation of histones and transcription factors that interact with the BLV LTR in vivo is necessary to understand the mechanism of BLV silencing.
Acknowledgments
We thank K. Okada (Faculty of Agriculture, Iwate University, Morioka, Iwate, Japan) for kindly providing blood samples from sheep, H. Sentsui (National Institute of Animal Health, Tsukuba, Ibaraki, Japan) for kindly providing blood samples from cattle, and T. Watanabe and T. Ishida (University of Tokyo, Tokyo, Japan) for helpful discussions.
This study was supported by a grant-in-aid for young scientists (B) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, by Grants-in-Aid for Scientific Research (A and B) from the Japan Society for the Promotion of Science (JSPS), by a grant for a special postdoctoral researcher, and by special grants from RIKEN for the promotion of research.
REFERENCES
- 1.Aida, Y., M. Miyasaka, K. Okada, M. Onuma, S. Kogure, M. Suzuki, P. Minoprio, D. Levy, and Y. Ikawa. 1989. Further phenotypic characterization of target cells for bovine leukemia virus experimental infection in sheep. Am. J. Vet. Res. 50:1946-1951. [PubMed] [Google Scholar]
- 2.Burny, A., Y. Cleuter, R. Kettmann, M. Mammerickx, G. Marbaix, D. Portetelle, A. van den Broeke, L. Willems, and R. Thomas. 1988. Bovine leukaemia: facts and hypotheses derived from the study of an infectious cancer. Vet. Microbiol. 17:197-218. [DOI] [PubMed] [Google Scholar]
- 3.Cassens, S., U. Ulrich, P. Beimling, and D. Simon. 1994. Inhibition of human T cell leukaemia virus type I long terminal repeat expression by DNA methylation: implications for latency. J. Gen. Virol. 75:3255-3259. [DOI] [PubMed] [Google Scholar]
- 4.Clark, S. J., J. Harrison, C. L. Paul, and M. Frommer. 1994. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22:2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke, M. F., C. D. Trainor, D. L. Mann, R. C. Gallo, and M. S. Reitz. 1984. Methylation of human T-cell leukemia virus proviral DNA and viral RNA expression in short- and long-term cultures of infected cells. Virology 135:97-104. [DOI] [PubMed] [Google Scholar]
- 6.Derse, D. 1987. Bovine leukemia virus transcription is controlled by a virus-encoded trans-acting factor and by cis-acting response elements. J. Virol. 61:2462-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derse, D. 1988. trans-acting regulation of bovine leukemia virus mRNA processing. J. Virol. 62:1115-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Djilali, S., A. L. Parodi, D. Levy, and G. L. Cockerell. 1987. Development of leukemia and lymphosarcoma induced by bovine leukemia virus in sheep: a hematopathological study. Leukemia 1:777-781. [PubMed] [Google Scholar]
- 9.Gupta, P., and J. F. Ferrer. 1982. Expression of bovine leukemia virus genome is blocked by a nonimmunoglobulin protein in plasma from infected cattle. Science 215:405-407. [DOI] [PubMed] [Google Scholar]
- 10.Hughes, S. H., P. R. Shank, D. H. Spector, H. J. Kung, J. M. Bishop, H. E. Varmus, P. K. Vogt, and M. L. Breitman. 1978. Proviruses of avian sarcoma virus are terminally redundant, co-extensive with unintegrated linear DNA and integrated at many sites. Cell 15:1397-1410. [DOI] [PubMed] [Google Scholar]
- 11.Inabe, K., K. Ikuta, and Y. Aida. 1998. Transmission and propagation in cell culture of virus produced by cells transfected with an infectious molecular clone of bovine leukemia virus. Virology 245:53-64. [DOI] [PubMed] [Google Scholar]
- 12.Jones, P. A., and D. Takai. 2001. The role of DNA methylation in mammalian epigenetics. Science 293:1068-1070. [DOI] [PubMed] [Google Scholar]
- 13.Kashmiri, S. V., R. Mehdi, P. Gupta, and J. F. Ferrer. 1985. Methylation and expression of bovine leukemia proviral DNA. Biochem. Biophys. Res. Commun. 129:126-133. [DOI] [PubMed] [Google Scholar]
- 14.Kettmann, R., J. Deschamps, Y. Cleuter, D. Couez, A. Burny, and G. Marbaix. 1982. Leukemogenesis by bovine leukemia virus: proviral DNA integration and lack of RNA expression of viral long terminal repeat and 3′ proximate cellular sequences. Proc. Natl. Acad. Sci. USA 79:2465-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitamura, T., M. Takano, H. Hoshino, K. Shimotohno, M. Shimoyama, M. Miwa, F. Takaku, and T. Sugimura. 1985. Methylation pattern of human T-cell leukemia virus in vivo and in vitro: pX and LTR regions are hypomethylated in vivo. Int. J. Cancer. 35:629-635. [DOI] [PubMed] [Google Scholar]
- 16.Koiwa, T., A. Hamano-Usami, T. Ishida, A. Okayama, K. Yamaguchi, S. Kamihira, and T. Watanabe. 2002. 5′-long terminal repeat-selective CpG methylation of latent human T-cell leukemia virus type 1 provirus in vitro and in vivo. J. Virol. 76:9389-9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagarias, D. M., and K. Radke. 1989. Transcriptional activation of bovine leukemia virus in blood cells from experimentally infected, asymptomatic sheep with latent infections. J. Virol. 63:2099-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merezak, C., C. Pierreux, E. Adam, F. Lemaigre, G. G. Rousseau, C. Calomme, C. Van Lint, D. Christophe, P. Kerkhofs, A. Burny, R. Kettmann, and L. Willems. 2001. Suboptimal enhancer sequences are required for efficient bovine leukemia virus propagation in vivo: implications for viral latency. J. Virol. 75:6977-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merezak, C., M. Reichert, C. Van Lint, P. Kerkhofs, D. Portetelle, L. Willems, and R. Kettmann. 2002. Inhibition of histone deacetylases induces bovine leukemia virus expression in vitro and in vivo. J. Virol. 76:5034-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizutani, T., T. Ito, M. Nishina, N. Yamamichi, A. Watanabe, and H. Iba. 2002. Maintenance of integrated proviral gene expression requires Brm, a catalytic subunit of SWI/SNF complex. J. Biol. Chem. 277:15859-15864. [DOI] [PubMed] [Google Scholar]
- 21.Nan, X., H. H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Eisenman, and A. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386-389. [DOI] [PubMed] [Google Scholar]
- 22.Ng, H. H., and A. Bird. 1999. DNA methylation and chromatin modification. Curr. Opin. Genet. Dev. 9:158-163. [DOI] [PubMed] [Google Scholar]
- 23.Ogawa, Y., N. Sagata, J. Tsuzuku-Kawamura, M. Onuma, H. Izawa, and Y. Ikawa. 1985. Methylation pattern of the bovine leukemia provirus genome in bovine leukemic cells. Jpn. J. Cancer Res. 76:5-8. [PubMed] [Google Scholar]
- 24.Pannell, D., C. S. Osborne, S. Yao, T. Sukonnik, P. Pasceri, A. Karaiskakis, M. Okano, E. Li, H. D. Lipshitz, and J. Ellis. 2000. Retrovirus vector silencing is de novo methylase independent and marked by a repressive histone code. EMBO J. 19:5884-5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Powers, M. A., and K. Radke. 1992. Activation of bovine leukemia virus transcription in lymphocytes from infected sheep: rapid transition through early to late gene expression. J. Virol. 66:4769-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pyeon, D., K. L. O'Reilly, and G. A. Splitter. 1996. Increased interleukin-10 mRNA expression in tumor-bearing or persistently lymphocytotic animals infected with bovine leukemia virus. J. Virol. 70:5706-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pyeon, D., and G. A. Splitter. 1999. Regulation of bovine leukemia virus tax and pol mRNA levels by interleukin-2 and -10. J. Virol. 73:8427-8434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pyeon, D., F. J. Diaz, and G. A. Splitter. 2000. Prostaglandin E(2) increases bovine leukemia virus tax and pol mRNA levels via cyclooxygenase 2: regulation by interleukin-2, interleukin-10, and bovine leukemia virus. J. Virol. 74:5740-5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sagata, N., T. Yasunaga, J. Tsuzuku-Kawamura, K. Ohishi, Y. Ogawa, and Y. Ikawa. 1985. Complete nucleotide sequence of the genome of bovine leukemia virus: its evolutionary relationship to other retroviruses. Proc. Natl. Acad. Sci. USA 82:677-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saggioro, D., M. Panozzo, and L. Chieco-Bianchi. 1990. Human T-lymphotropic virus type I transcriptional regulation by methylation. Cancer Res. 50:4968-4973. [PubMed] [Google Scholar]
- 31.Saggioro, D., M. Forino, and L. Chieco-Bianchi. 1991. Transcriptional block of HTLV-I LTR by sequence-specific methylation. Virology 182:68-75. [DOI] [PubMed] [Google Scholar]
- 32.Tajima, S., Y. Ikawa, and Y. Aida. 1998. Complete bovine leukemia virus (BLV) provirus is conserved in BLV-infected cattle throughout the course of B-cell lymphosarcoma development. J. Virol. 72:7569-7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tajima, S., and Y. Aida. 2000. The region between amino acids 245 and 265 of the bovine leukemia virus (BLV) Tax protein restricts transactivation not only via the BLV enhancer but also via other retrovirus enhancers. J. Virol. 74:10939-10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tajima, S., and Y. Aida. 2002. Mutant Tax protein from bovine leukemia virus with enhanced ability to activate the expression of c-fos. J. Virol. 76:2557-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tajima, S., M. Takahashi, S. Takeshima, S. Konnai, S. A. Yin, S. Watarai, Y. Tanaka, M. Onuma, K. Okada, and Y. Aida. 2003. A mutant form of the Tax protein of bovine leukemia virus (BLV), with enhanced transactivation activity, increases the expression and propagation of BLV in vitro but not in vivo. J. Virol. 77:1894-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsukiyama, K., M. Onuma, and H. Izawa. 1987. Effect of platelet-derived factor on expression of bovine leukemia virus genome. Arch. Virol. 96:89-96. [DOI] [PubMed] [Google Scholar]
- 37.Willems, L., A. Gegonne, G. Chen, A. Burny, R. Kettmann, and J. Ghysdael. 1987. The bovine leukemia virus p34 is a transactivator protein. EMBO J. 6:3385-3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zandomeni, R. O., E. Esteban, M. Carrera-Zandomeni, and J. F. Ferrer. 1994. Host soluble factors with blocking and stimulating activity on the expression of the bovine leukemia virus. J. Infect. Dis. 170:787-794. [DOI] [PubMed] [Google Scholar]