Abstract
Persistence of human immunodeficiency virus type 1 (HIV-1) constitutes a major obstacle in the control of HIV-1 infection. Here we investigated whether the CpG methylation of the HIV-1 promoter can directly influence the expression of the HIV-1 genome and thereby contribute to the persistence and latency of HIV-1. The levels of CpG methylation in the promoter of HIV-1 were studied after bisulfite-induced modification of DNA in five Jurkat clonal cell lines transduced by an HIV-1 long terminal repeat (LTR)-driven retroviral vector and expressing enhanced green fluorescent protein (GFP) and in primary resting memory T cells challenged with HIV-1 or with an HIV-1-derived retroviral vector. Basal HIV-1 promoter activities were low or undetectable in three tested HIV-1 LTR-GFP clones, one of which encoded the Tat protein, and they reached medium or high levels in two other clones. The CpG dinucleotide that occurred in a latently infected clonal cell line 240 nucleotides upstream of the transcription start remained methylated after reactivation of HIV-1 transcription with 10 nM phorbol-12-myristate-13-acetate. In two clones showing a medium promoter activity and in resting memory T cells, the HIV-1 LTR was generally not methylated. Our results show that the methylation profiles of the HIV-1 LTR, including those present in latently infected cells, are low and do not correlate with the transcriptional activity. We suggest that, in a noncloned cellular population in which the HIV-1 proviruses are randomly integrated in the human genome, HIV-1 latency is imperfectly controlled by CpG methylation and is inherently accompanied by residual replication.
Persistence of human immunodeficiency virus type 1 (HIV-1) constitutes a major obstacle in the control of HIV-1 infection. Although highly active antiretroviral therapy reduces HIV-1 plasma viremia to undetectable levels in a substantial proportion of treated patients, replication-competent HIV persists in resting memory CD4+ T cells (12, 19, 47) and in other anatomical or cellular compartments inaccessible to current HIV-1 therapy (8). In resting memory CD4+ T cells, HIV-1 persists in a latent form (7, 11, 12, 18) or replicates presumably at a reduced residual level (2, 21, 42, 51, 52). The expression of HIV-1 from latently infected resting CD4+ T cells can be induced in vitro with a variety of agents, such as phytohemagglutinin (PHA) and anti-CD3/CD28 antibody (12, 19, 20, 37, 47, 49, 50), or with different cytokines, including interleukin-2 (IL-2), tumor necrosis factor (TNF-α), and IL-6 (9, 30, 46). Intermittent administration of IL-2 to HIV-infected individuals along with antiretroviral therapy significantly decreases the pool of infected resting CD4+ T cells, presumably due to the killing of reactivated latent HIV (10).
Here, we investigate whether the CpG methylation of the HIV-1 promoter can directly influence the expression of the HIV-1 genome and thereby contribute to the persistence and latency of HIV-1. A clear regulatory role of CpG methylation in enhancer/promoter sequences was demonstrated by using in vitro-methylated provirus reporters in both human T-cell lymphotropic virus type 1 (38, 39) and simple retroviruses (24-26). CpG methylation of the HIV-1 long terminal repeat (LTR) was first reported in stably transfected fibroblasts (5). At the functional level, CpG methylation of the HIV-1 promoter inhibits transcription of in vitro-methylated plasmids transfected into cells (3, 23, 40), and it was suggested as a mechanism to maintain HIV-1 latency in long-term infected U937 cells (44). Demethylation of the HIV-1 promoter occurred in the presence of a DNA methylation inhibitor, 5-azacytidine, concomitantly with activation of the latent provirus in ACH-2 cells (35). Two mechanisms have been proposed to explain how methylation attenuates expression of the HIV-1 genome. First, the methyl-CpG-binding protein 1 complex and methyl-CpG-binding protein 2 may bind to methylated Sp1 sites and inhibit the binding of Sp1 proteins (28, 41). Second, transcription factors NF-κB (4) and USF (16) loose their affinity for the methylated HIV-1 LTR cognate motifs. On the other hand, the site of HIV-1 integration in the human genome, but not of DNA methylation and histone acetylation, was proposed to determine the basal transcriptional activity and response to Tat transactivation in a study of Jurkat clonal cell lines (29).
To facilitate the investigation of HIV-1 persistence and latency, we developed an in vitro model of infection of HIV-1 in primary resting memory T cells (22). Viral latency (defined by the absence of nonspliced HIV-1 RNA and the absence of viral particles) was established in approximately 40% of infected resting T cells and could be reactivated by PHA treatment. We studied the CpG methylation of the HIV-1 promoter in this cell system and in Jurkat clonal cell lines, each containing a single integration of the HIV-1 vector expressing the green fluorescent protein (GFP) reporter gene at different levels under the control of the HIV-1 promoter (29). Previous studies of methylation of the HIV promoter were based on the resistance of the occasionally occurring restriction endonuclease target sequences containing methylated CpG dinucleotides in the promoter sequence. We sequenced genomic DNA (13, 36) that had been modified by bisulfite treatment and observed no correlation between the transcriptional activity of the HIV-1 promoter and the level of CpG methylation.
MATERIALS AND METHODS
Clonal cell lines.
We prepared and cultured Jurkat clonal cell lines as described previously (29). Each contained a single integration of the HIV-1 vector expressing the GFP reporter gene at different levels under the control of the HIV-1 promoter. Clonal cell line 82 harboring latent HIV-1 LTR-Tat-internal ribosome entry site (IRES)-GFP provirus was treated with 10 nM phorbol-12-myristate-13-acetate (PMA) to reactivate production of GFP. Two days later, the GFP-positive and -negative cells were separated by cell sorting with a FACSVantage cell sorter (Becton Dickinson).
Preparation of HIV-1-infected resting memory T cells derived from PBMC.
Peripheral blood mononuclear cells (PBMC) from healthy donors were separated on Ficoll-Hypaque gradients. Aliquots of 2 × 108 PBMC depleted of monocytes by adherence to a plastic culture flask were activated with 2 μg of PHA-P (Difco, Franklin Lakes, N.J.)/ml in RPMI 1640 supplemented with 200 U of recombinant IL-2 (Chiron, Seresne, France)/ml, 15% fetal calf serum, and antibiotics for 3 days. After disintegration of cell clumps 3 days later, 2 × 107 peripheral blood lymphocytes per ml were treated for 1 h with the anti-CD8 antibody at the saturating concentration at 4°C. The cell suspension was incubated with magnetic beads coated with goat anti-mouse antibody (Miltenyi Biotech, Bergisch Gladbach, Germany), and the positively labeled cells were removed as recommended by the manufacturer. CD4+ T cells were then infected with HIV-1 NL4-3 (1) at a multiplicity of infection of 0.01 or 0.1 tissue culture infectious doses/cell 10 days after PHA activation. Alternatively, CD4+ T cells were transduced by an HIV-1-derived vector (HDV) (kindly provided by D. R. Littman, Skirball Institute of Biomolecular Medicine, New York, N.Y.), prepared, and used as described by Unutmaz et al. (46). HDV pseudotyped by vesicular stomatitis virus glycoprotein G typically had a titer of 2 × 106 HeLa tissue culture infectious doses/ml. Peripheral blood lymphocytes were cultured at a concentration of 106 cells/ml in the presence of IL-2 and fed and analyzed every 3 or 4 days until the expression of activation markers CD25 and HLA-DR had diminished to below 15 and 30% of maximum levels, respectively. Approximately 3 weeks after PHA activation, residual activated T cells were removed from the cell culture by incubation with monoclonal antibody against CD25, CD69, and HLA-DR and magnetic bead separation. Typically, 5 × 106 to 10 × 106 cells were recovered after immunodepletion. In some experiments, the resting T cells generated in vitro were reactivated by 2 μg of PHA/ml.
Flow cytometry analysis.
Lymphocytes were identified by light scattering and expression of lymphocyte markers.
Bisulfite cytosine methylation analysis.
The samples of total genomic DNA (700 ng) isolated with the QIAamp DNA blood mini kit (Qiagen, Inc.) and digested overnight with EcoRI in a volume of 20 μl were prepared for methylation analysis as described by Olek et al. (36). Briefly, the digested DNA, which had been boiled for 5 min, quickly chilled on ice, and incubated in 0.3 M NaOH at 50°C for 15 min, was mixed with 2.5 volumes of 2% low-melting-point agarose (SeaPlaque agarose; FMC). Ten-microliter volumes of agarose-DNA mixture were pipetted into 750 μl of chilled mineral oil to form agarose beads. Aliquots of 1 ml of 2.5 M sodium metabisulfite (Sigma) and 125 mM hydroquinone (Sigma) (pH 5.0) were then added to mineral oil containing up to 5 agarose beads. The reaction mixtures were kept on ice for 30 min and then incubated in the dark at 50°C for 3.5 h. The agarose beads were equilibrated 4 times (for 15 min each time) with 1 ml of Tris-EDTA (pH 8.0) and DNA desulfonated in 500 μl of 0.2 M NaOH (2 times, for 15 min each time) at room temperature. Finally, the beads were washed with 1 ml of Tris-EDTA (pH 8.0) (3 times, for 10 min each time) and kept at 4°C.
Bisulfite-treated DNA in melted agarose was amplified by PCR in a 50-μl reaction mixture containing 50 mM Tris-HCl (pH 9.2), 1.75 mM MgCl2, 350 μM concentrations of each deoxynucleoside triphosphate, and 45 pmol each of MB (5′-TGGTAGAATTATATATTAGGGTTAGGGATT-3′, nucleotides [nt] 81 to 110, sense) and ME (5′-ATATTTATCTACAACCTTCTAATATCTCTA-3′, nt 941 to 970, antisense) primers complementary to the HIV-1 LTR and Gag-coding region (Fig. 1), with 1 U of Taq polymerase. The amplification products were subjected to a second round of PCR with nested primers MC (5′-AGAGTAAGTAGAAGAGGTTAAATAAGGAGA-3′, nt 161 to 190, sense) and MD (5′-AACCAAAATTAACTACAAATCATTCTAACT-3′, nt 908 to 937, antisense). In some experiments, the primer MD was replaced by the primer MF (5′-AAATCTAAAAAATCTCTAATTACCAAAATC-3′, nt 577 to 606, antisense) or the primer MR (5′-ACTCCCAAACTCAAATCTAATCTAACCAAA-3′, nt 461 to 490, antisense). The sense primers contained T and the antisense primers contained A instead of C in positions complementary to nonmethylable C (i.e., C out of CpG dinucleotides). PCR was performed with about 50 ng of genomic DNA (out of 140 ng of DNA per 1 agarose bead) for 40 cycles at 95°C for 45 s, 58°C for 30 s, and 72°C for 30 s. Amplification products were cloned in the pGEM-Easy vector system (Promega, Madison, Wis.) and sequenced.
FIG. 1.
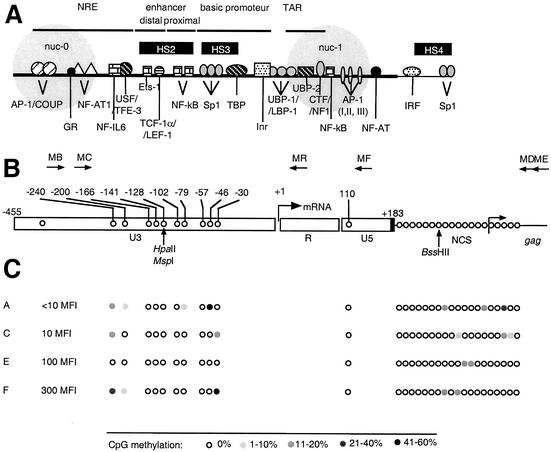
CpG methylation pattern of the LTR region in LTR-GFP clonal cell lines expressing different levels of GFP. (A) Transcription elements in the HIV-1 LTR. NRE, negative regulatory element; nuc-0 and nuc-1, nucleosomes. Black boxes, HS, DNase I-hypersensitive sites. (B) Positions and orientations of PCR primers (MB, MC, MD, ME, MF, and MR) used to amplify bisulfite-treated HIV-1 DNA. The arrow at the U3-R junction denotes the start site of transcription (nucleotide +1). Positions of CpG dinucleotides within the HIV-1 LTR, the adjacent leader region, and the coding region of Gag are shown by open circles. Positions are noted with respect to nucleotide +1. NCS, noncoding sequence. Positions of restriction endonucleases sensitive to CpG methylation are shown by vertical arrows. (C) Positions of CpG dinucleotides within the HIV-1 LTR are shown by open circles and noted with respect to nucleotide +1. Open circles, nonmethylated CpG residues; closed circles, methylated CpG residues, grey scale indicates the percentage of CpG methylation. The MFIs of clonal cell lines A, C, E, and F are shown. Low basal activity is indicated by an MFI of <0.15, and a high basal activity is indicated by an MFI of >100.
Quantitative competitive PCR and RT-PCR.
Plasmid pSPLI-II, obtained from M. Giacca, was used as a competitor for the competitive determination of the copy number of HIV-1 proviral DNA and of the reference single-copy β-globin gene in DNA samples. The in vitro transcription products from plasmid pSPLI-II were used to quantify HIV-1 viral mRNA and cellular β-actin mRNA in cellular RNA samples by reverse transcription-PCR (RT-PCR) (14, 15). Competitive quantitative PCR and RT-PCR were performed and analyzed as described previously (15).
RESULTS
Methylation level of the HIV-1 promoter does not correlate with transcriptional suppression.
First, we investigated the methylation profiles of the HIV-1 LTR in four clonal Jurkat cell lines, each containing a single integration of the HIV-1 LTR-GFP vector and differing in expression levels of the GFP reporter gene (Fig. 1). CpG methylation levels were low in these clones, which expressed the HIV-1 genome at basal levels in the absence of the protein Tat. In two of these clones, one expressing a low level of GFP (mean fluorescence intensity [MFI] of <10, clone A) and the other expressing a high level of GFP (300 MFI, clone F), the methylation profiles showed distinct patterns. Cytosine at nt −46 was methylated in 56% of the tested sequences in clone A, whereas cytosines at nt −30 (50%) and −240 (36%) were methylated in clone F. No changes of the methylation profile were found in the target sequences of HpaII, MspI, and BssHII restriction endonucleases previously used for the detection of CpG methylation (29) (Fig. 1). These results show that the CpG methylation levels and patterns of the HIV-1 LTR do not correlate with the transcriptional activity of the HIV-1 promoter.
Methylation level of the HIV-1 promoter in latently infected cell clones is low and shows no change after reactivation of the transcriptional activity with PMA.
We next examined the methylation profile of the HIV-1 LTR in a Jurkat clonal cell line latently infected with the LTR-Tat-IRES-GFP HIV-1-derived vector (29). Both the Tat and GFP open reading frames were transcribed by the HIV promoter into a single transcript in this clone. We expected that the HIV-1 LTR would be hypermethylated in these latently infected cells. However, the bisulfite analysis showed that the methylation level of CpG dinucleotides in the latent HIV-1 promoter was low and was not significantly changed after reactivation by 10 nM PMA, which increased GFP expression more than 70-fold (Fig. 2). Low methylation profiles were observed in both the U3 and U5 regions, in the noncoding sequence, and in the 5′ terminus of the Gag-coding sequence. Thus, the transcriptional activity of the HIV-1 promoter in this latently infected cell clone does not correlate with LTR methylation.
FIG. 2.
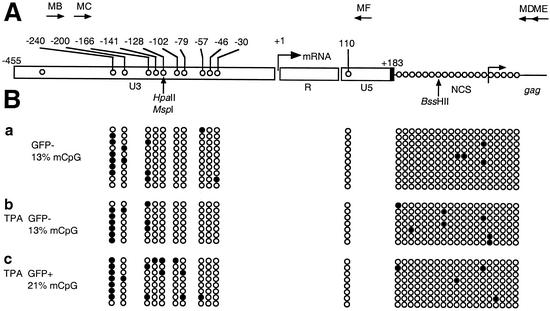
CpG methylation pattern of the LTR region in a Jurkat clonal cell line latently infected with an LTR-Tat-IRES-GFP HIV-1-derived vector before and after reactivation of HIV-1 with 10 nM PMA. (A) Positions of CpG in the HIV-1 LTR. The arrow at the U3-R junction denotes the start site of transcription (nucleotide +1). (B) Bisulfite sequencing of HIV-1 promoters from top to bottom. (a) Nonstimulated, latently infected cells; (b) 10 nM PMA-stimulated cells, GFP-negative cell sorting; (c) 10 nM PMA-stimulated cells, GFP-positive cell sorting. Open circles, nonmethylated CpG residues; closed circles, methylated CpG residues. The total percentages of methylated CpG (mCpG) for nonstimulated and PMA-stimulated cells (GFP negative and GFP positive) are indicated in panel B.
A strikingly high frequency of methylation (6 of 10 tested sequences) was observed at the CpG dinucleotide localized 240 nt upstream of the transcription start, in the negative regulatory element of the HIV-1 LTR. The high level of methylation at this position remained stable (7 of 8 tested sequences) after reactivation of HIV-1 latency by 10 nM PMA (Fig. 2). Therefore, methylation of dinucleotide −240 does not correlate with transcriptional silencing of this latently infected cell clone.
HIV-1 expression in infected primary resting memory T cells.
After the methylation profiles of the HIV-1 LTR in clonal Jurkat cell lines were analyzed, CpG methylation in primary resting memory T cells infected in vitro was investigated (22). Previously, it was shown that the virus production is reduced to about 5% in this cell system and that approximately half of the cell population is latently infected (22).
To obtain resting memory CD4+ T cells containing the HIV-1 provirus, PHA-activated PBMC were challenged with T-cell-tropic HIV-1 NL4-3 clone (Fig. 3A), and residual CD25+, CD69+, and HLA-DR+ cells were immunodepleted after a 3-week cultivation period. A high proportion (27%) of purified resting memory T cells expressed intracellular p24gag after depletion of residually activated cells, although they expressed only a residual amount of CD25.
FIG. 3.
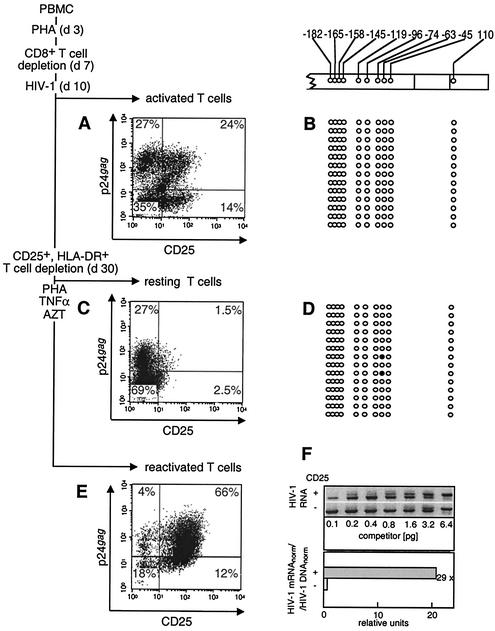
Expression of the HIV-1 genome and CpG methylation pattern in the LTR region in primary resting memory T cells infected in vitro. (A and B) PHA-activated PBMC infected with HIV-1 10 days after activation (d10). (C and D) The same culture that developed the resting memory phenotype 20 days later (d30). The depletion of CD25- and HLA-DR-positive cells was performed 30 days after PBMC activation. (E) Reactivation of HIV-1 production in resting memory T cells after their reactivation with PHA and TNF-α. (A, C, and E) The immunofluorescence of the p24gag- and CD25-labeled cells was determined in activated (A), resting (C), and reactivated (E) cells. Numbers at the corner of each quadrant are percentages of positive cells. The CpG methylation patterns of the LTR region in primary PHA-activated (B) and resting memory T (D) cells are shown. Open circles, nonmethylated CpG residues; closed circles, methylated CpG residues. (F) Quantitation of the transcriptional activity of the HIV-1 promoter in activated (CD25+) and resting (CD25−) T cells. (F, upper panel) Quantitative competitive RT-PCR with primers that recognize HIV-1 unspliced RNA, with indicated amounts of competitor RNA (14, 15). (F, lower panel) The bars show the relative quantity of HIV-1 transcripts normalized for the β-actin mRNA in the same sample (HIV-1 mRNAnorm) and standardized to the relative quantity of HIV-1 proviruses normalized for the β-globin DNA (HIV-1 DNAnorm). The overall ratio of HIV-1 mRNAnorm to HIV-1 DNAnorm in activated and resting T cells (29 in the example shown) indicates reduction of the transcriptional activity in resting memory T cells compared to activated cells.
To assess the transcriptional activity of the HIV-1 provirus in these cells, we used quantitative competitive PCR and RT-PCR with a single multicompetitor molecule permitting the determination of the copy number of HIV-1 proviral DNA and of the reference single-copy β-globin gene in DNA samples as well as quantification of HIV-1 viral mRNA and of cellular β-actin mRNA in cellular RNA samples (15) (Fig. 3F). This procedure permits the exact quantification of the relative abundance of different DNA and RNA species. We found that expression of HIV-1 dropped 29 times during the return of activated HIV-challenged cells to the quiescent phase, showing that the transcriptional activity of the HIV-1 promoter was reduced to a low basal level.
The population of resting memory T cells was reactivated for 3 days by PHA and TNF-α in the presence of 10 μM azidothymidine (AZT) to determine the proportion of latently infected cells (Fig. 3E). AZT limits the secondary cycles of HIV-1 infection and permits analysis of the cell population infected before reactivation. In this population (Fig. 3C and E), 4% of an original 27% of p24+ CD25− resting T cells (15% of the total), and 18% of an original 69% of p24− CD25− resting T cells (26% of the total) remained in the resting state after reactivation. This suggests that PHA and TNF-α reactivated similar proportions of both p24+ (85%) and p24− (74%) resting memory T cells. The cell viability was reduced by about 50% at 3 days after PHA and TNF-α reactivation (22). Whereas 28.5% of resting memory T cells expressed HIV-1 (Fig. 3C), virus production was detected in 70% of reactivated cells (Fig. 3E). Equal proportions of reactivated T cells within the p24+ and p24− subsets (Fig. 3C and E) and the high cytotoxicity of HIV-1 for reactivated T cells (22) suggest that the increased number of virus-producing cells following reactivation is not due to the outgrowth of cells that were already p24 positive but due to their recruitment from the pool of p24− CD25− cells. A twofold increase in the number of p24+ T cells was observed also after nonmitogenic stimulation of resting memory T cells by 10 nM PMA (data not shown). Taken together, these results suggest that about half of the population of infected resting memory T cells harbored latent HIV-1 proviruses.
Low CpG methylation of HIV-1 LTR in HIV-1-infected resting memory T cells.
We next determined the CpG methylation in activated and resting T cells (Fig. 3B and D). Only very low levels and no distinct patterns of CpG methylation were observed in all populations, including resting memory T cells. A stable methylation profile of the HIV LTR despite the 29-fold reduction of HIV expression during the return of activated HIV-infected cells to the quiescent phase shows that the transcriptional suppression does not correlate with proviral DNA methylation. Furthermore, because the proportions of cells harboring latent and residually replicating proviruses in the population of resting memory T cells were equal, about half of the tested methylation-free sequences originated from latently infected cells. This suggests that the HIV-1 LTR is also hypomethylated in latently infected primary resting memory T cells.
Hypomethylation of HIV-1 LTR in primary resting memory T cells transduced by HDV.
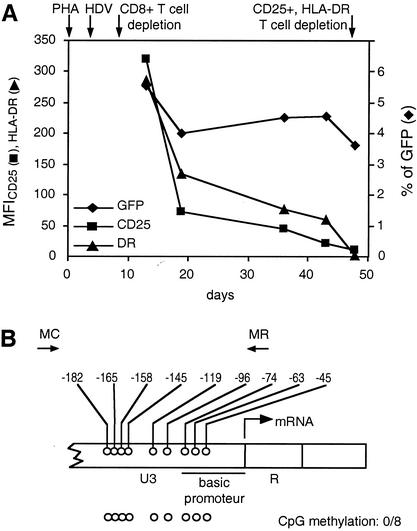
To measure the CpG methylation in resting T lymphocytes without confounding the effects of repeated infection cycles and the death of infected cells, we challenged PBMC with HDV (46) 3 days after PHA activation and determined the methylation profiles of the HIV-1 LTR 48 days later, after the return of T cells into the quiescent phase and the depletion of residual CD25+ and HLA-DR+ T cells (22) (Fig. 4). HDV is a replication incompetent and noncytopathic vector with deletions of the vif, vpr, vpu, env, and nef genes and pseudotyped with the vesicular stomatitis virus glycoprotein G. Expression of HDV can be monitored by the fluorescence of the GFP inserted in the open reading frame of the nef gene. HDV can challenge primary T lymphocytes but is unable to form infectious progeny. We found that the percentage of GFP-positive cells decreased by about 30% during the return of activated T cells to the quiescent state while the MFI of CD25 and HLA-DR diminished by more than 95% during the same period (22) (Fig. 4A). Reactivation of the HDV-challenged resting memory T cells for 2 days with PHA resulted in about a twofold increase of the percentage of GFP-positive cells (data not shown). This suggests that about 50% of transduced resting memory T cells harbored latent HDV proviruses. No CpG methylation was observed in the promoter region of HDV in resting memory T cells (Fig. 4B).
FIG. 4.
Expression of the HDV, cell-surface activation markers, and CpG methylation pattern in the LTR region in primary resting memory T cells challenged in vitro. (A) The PHA-activated PBMC were transduced with HDV, and after the CD8+-T-cell depletion, the percentage of GFP+ cells (⧫) and the MFI for CD25 (▪) and HLA-DR (▴) were monitored for up to 50 days after activation. The time schedule of cell culture and HDV challenge are indicated by arrows. (B) CpG methylation pattern of the LTR region in resting CD4+ T cells at day 50 after PHA activation. Positions and orientations of PCR primers MC and MR used to amplify bisulfite-treated HDV DNA are shown by arrows. The arrow at the U3-R junction denotes the start site of transcription (nucleotide +1). Open circles, nonmethylated CpG residues in 8 tested promoter sequences.
DISCUSSION
In this study, we found that the CpG methylation levels were generally low in the HIV-1 LTR of Jurkat clonal cell lines transduced by the HIV-1-derived retroviral vector and expressing different levels of GFP and in primary resting memory T cells infected in vitro with HIV-1 and HDV. We also found that these levels did not change after reactivation of latent HIV-1. We reexamined hypomethylation in some clones, which were monitored previously by cleavage with CpG-methylation-sensitive restriction endonucleases (29), by using the more-sensitive technique of bisulfite genomic sequencing. Despite the low methylation levels, the LTR sequences in three of five clonal cell lines showed distinct methylation patterns. In one clone that expressed no GFP, a methylated CpG dinucleotide was localized 240 and 46 nt upstream of the transcription start. In another, which had high basal expression of GFP, two methylated CpG dinucleotides at nt −240 and −30 were detected. Our results show that the methylation profiles of the HIV-1 LTR, including those present in latently infected cells, are low and do not correlate with the transcriptional activity of the HIV-1 promoter. They suggest that the chromatin structure surrounding the integration site may influence the methylation of the HIV-1 LTR. Methylation of other retroviruses is influenced by the local DNA environment at the integration site (24, 27, 48).
In addition to highly homogeneous Jurkat clonal cell lines transduced by an HIV-1 LTR-driven retroviral vector, we studied the methylation profile of the HIV-1 promoter in a more-complex but biologically more-relevant system of primary resting memory T lymphocytes that contain about 50% of the HIV-1 provirus in a latent state. To this end, it was shown previously that the titers of provirus-expressing primary resting memory T cells determined by RT-PCR and coculture with C8166 indicator cells represent about 50% of the titers of provirus-harboring cells determined by PCR (22). Consistently with this observation, the population of HIV-1-expressing cells more than doubled after reactivation of resting T cells by PHA and TNF-α in the presence of 10 μM AZT. The hypomethylation of HIV-1 LTRs in the population of primary resting memory T cells suggests that the latent state of HIV-1 or HDV in this culture does not correlate with the CpG methylation.
Previous studies, in which the HIV-1 LTR methylated in vitro was introduced into target cells by transfection, proposed that the HIV-1 promoter was inactivated by CpG methylation (3, 23, 40). Scarce methylation sites detected in the present study in two clonal cell lines infected with the retroviral vector did not result in gene silencing. Thus, two methylated CpGs present 240 and 30 nt upstream of the transcription start were not sufficient to abolish the high basal expression of the HIV-1 promoter in clone F. Similarly, the persistence of 5-methylcytosine (nt −240) after reactivation of a latently infected cell clone indicates that methylation at this position is not responsible for HIV-1 latency.
Several mechanisms can explain the hypomethylation of the HIV-1 promoter. By analogy with other viruses which establish latency, the time elapsed from the infection of activated primary T cells to the development of the resting memory T-cell phenotype might be insufficient for the methylation of the HIV-1 promoter. The integrated viral genome of Moloney murine leukemia virus in undifferentiated teratocarcinoma cells is methylated within 15 days after infection (34), and methylation of the Wp latent promoter in B-cell cultures infected with Epstein-Barr virus occurs between 7 and 21 days after infection (45). Therefore, the 20 days from HIV-1 integration to development of the resting memory cell phenotype (Fig. 3) should be just sufficient for promoter methylation. However, this argument does not apply to hypomethylation in clonal cell lines kept in culture for many cell cycles and for primary T cells challenged by HDV. Hypomethylation of the HIV-1 LTR in latently infected cells or in cells expressing HIV-1 at low basal levels suggests that HIV-1 developed a mechanism to avoid methylation de novo or to increase the demethylation rate. HIV-1 infection upregulates DNA methyltransferases and results in de novo methylation of some cellular gene promoters, such as gamma interferon and p16INK4A, and subsequent downregulation of respective protein products (17, 31-33). Thus, a negative methylation control may play an important role in virus replication strategy. Extremely low frequencies of CpG dinucleotides found in HIV-1 LTR (43) and the chromatin structure around the HIV-1 promoter could be involved in such a mechanism.
Methylation of foreign DNA has been recognized as a potential cellular defense mechanism to inactivate transcription of foreign sequences, transposons, and retroviruses (48). Methylation of viral DNA may be an essential part of the retroviral life cycle in vivo and may explain the persistence of virus-infected cells despite the immune surveillance (6). Methylation of proviral DNA, which retains the ability to be reactivated, allows the virus to survive by escaping the immune surveillance. A stringent control of viral latency by methylation could be fundamentally important for the development and maintenance of cell transformation as shown for human T-cell lymphotropic virus type 1 (38, 39) and simple retroviruses (24-26).
Recent investigations of clonal cell lines transduced by the HIV-1 LTR-GFP construct demonstrated that the chromatin environment, not CpG methylation, influences the basal HIV-1 gene expression and latency (29). We hypothesize that the absence of the CpG methylation mechanism causes this control mechanism of basal HIV-1 gene expression to be leaky at many integration sites. Indeed, the basal transcriptional activity in as much as 20% of HIV-1 LTR-GFP clonal cell lines cultured for many generations was moderate to high (29). Similarly, besides latently infected cells, the high proportion of resting memory T cells infected in vitro produced a residual quantity of the virus (22).
In conclusion, imperfect control of HIV-1 latency in the absence of CpG methylation could explain the persistent residual replication of HIV-1 in infected individuals (2, 21, 42, 51, 52). The imperfect control of latency could be a part of HIV-1 replication strategy permitted by ample mechanisms by which HIV-1 evades immune mechanisms and can escape the immunologic surveillance even when it residually replicates.
Acknowledgments
This work was supported by INSERM and ANRS (grants to I.H.), by a Public Health Service grant from the NIH (to E.V.) and by the Grant Agency of the Czech Republic (grant no. 204/01/0632 to J.H.). M.P. was supported by the Sidaction-ACS and by the Fondation pour la Recherche Medicale (FRM). A.B. was the recipient of predoctoral fellowships from the Ministère de la Recherche et de la Technologie (MRT).
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appay, V., P. Hansasuta, J. Sutton, R. D. Schrier, J. K. Wong, M. Furtado, D. V. Havlir, S. M. Wolinsky, A. J. McMichael, D. D. Richman, S. L. Rowland-Jones, and C. A. Spina. 2002. Persistent HIV-1-specific cellular responses despite prolonged therapeutic viral suppression. AIDS 16:161-170. [DOI] [PubMed] [Google Scholar]
- 3.Bednarik, D. P., J. A. Cook, and P. M. Pitha. 1990. Inactivation of the HIV LTR by DNA CpG methylation: evidence for a role in latency. EMBO J. 9:1157-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bednarik, D. P., C. Duckett, S. U. Kim, V. L. Perez, K. Griffis, P. C. Guenthner, and T. M. Folks. 1991. DNA CpG methylation inhibits binding of NF-kappa B proteins to the HIV-1 long terminal repeat cognate DNA motifs. New Biol. 3:969-976. [PubMed] [Google Scholar]
- 5.Bednarik, D. P., J. D. Mosca, and N. B. Raj. 1987. Methylation as a modulator of expression of human immunodeficiency virus. J. Virol. 61:1253-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bird, A. P., and A. P. Wolffe. 1999. Methylation-induced repression-belts, braces, and chromatin. Cell 99:451-454. [DOI] [PubMed] [Google Scholar]
- 7.Chun, T. W., L. Carruth, D. Finzi, X. Shen, J. A. DiGiuseppe, H. Taylor, M. Hermankova, K. Chadwick, J. Margolick, T. C. Quinn, Y. H. Kuo, R. Brookmeyer, M. A. Zeiger, P. Barditch-Crovo, and R. F. Siliciano. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183-188. [DOI] [PubMed] [Google Scholar]
- 8.Chun, T. W., R. T. Davey, Jr., M. Ostrowski, J. Shawn Justement, D. Engel, J. I. Mullins, and A. S. Fauci. 2000. Relationship between pre-existing viral reservoirs and the re-emergence of plasma viremia after discontinuation of highly active anti-retroviral therapy. Nat. Med. 6:757-761. [DOI] [PubMed] [Google Scholar]
- 9.Chun, T. W., D. Engel, S. B. Mizell, L. A. Ehler, and A. S. Fauci. 1998. Induction of HIV-1 replication in latently infected CD4+ T cells using a combination of cytokines. J. Exp. Med. 188:83-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chun, T. W., D. Engel, S. B. Mizell, C. W. Hallahan, M. Fischette, S. Park, R. T. Davey, Jr., M. Dybul, J. A. Kovacs, J. A. Metcalf, J. M. Mican, M. M. Berrey, L. Corey, H. C. Lane, and A. S. Fauci. 1999. Effect of interleukin-2 on the pool of latently infected, resting CD4+ T cells in HIV-1-infected patients receiving highly active anti-retroviral therapy. Nat. Med. 5:651-655. [DOI] [PubMed] [Google Scholar]
- 11.Chun, T. W., D. Finzi, J. Margolick, K. Chadwick, D. Schwartz, and R. F. Siliciano. 1995. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat. Med. 1:1284-1290. [DOI] [PubMed] [Google Scholar]
- 12.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark, S. J., J. Harrison, C. L. Paul, and M. Frommer. 1994. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22:2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Comar, M., G. Marzio, P. D'Agaro, and M. Giacca. 1996. Quantitative dynamics of HIV type 1 expression. AIDS Res. Hum. Retrovir. 12:117-126. [DOI] [PubMed] [Google Scholar]
- 15.Comar, M., C. Simonelli, S. Zanussi, P. Paoli, E. Vaccher, U. Tirelli, and M. Giacca. 1997. Dynamics of HIV-1 mRNA expression in patients with long-term nonprogressive HIV-1 infection. J. Clin. Investig. 100:893-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.di Fagagna, F. D., G. Marzio, M. I. Gutierrez, L. Y. Kang, A. Falaschi, and M. Giacca. 1995. Molecular and functional interactions of transcription factor USF with the long terminal repeat of human immunodeficiency virus type 1. J. Virol. 69:2765-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang, J. Y., J. A. Mikovits, R. Bagni, C. L. Petrow-Sadowski, and F. W. Ruscetti. 2001. Infection of lymphoid cells by integration-defective human immunodeficiency virus type 1 increases de novo methylation. J. Virol. 75:9753-9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosenberg, B. Walker, S. Gange, J. Gallant, and R. F. Siliciano. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512-517. [DOI] [PubMed] [Google Scholar]
- 19.Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. Gallant, M. Markowitz, D. D. Ho, D. D. Richman, and R. F. Siliciano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295-1300. [DOI] [PubMed] [Google Scholar]
- 20.Folks, T., J. Kelly, S. Benn, A. Kinter, J. Justement, J. Gold, R. Redfield, K. W. Sell, and A. S. Fauci. 1986. Susceptibility of normal human lymphocytes to infection with HTLV-III/LAV. J. Immunol. 136:4049-4053. [PubMed] [Google Scholar]
- 21.Furtado, M. R., D. S. Callaway, J. P. Phair, K. J. Kunstman, J. L. Stanton, C. A. Macken, A. S. Perelson, and S. M. Wolinsky. 1999. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N. Engl. J. Med. 340:1614-1622. [DOI] [PubMed] [Google Scholar]
- 22.Gondois-Rey, F., A. Biancotto, M. Pion, A. L. Chenine, P. Gluschankof, V. Horejsi, C. Tamalet, R. Vigne, and I. Hirsch. 2001. Production of HIV-1 by resting memory T lymphocytes. AIDS 15:1931-1940. [DOI] [PubMed] [Google Scholar]
- 23.Gutekunst, K. A., F. Kashanchi, J. N. Brady, and D. P. Bednarik. 1993. Transcription of the HIV-1 LTR is regulated by the density of DNA CpG methylation. J. Acquir. Immune Defic. Syndr. 6:541-549. [PubMed] [Google Scholar]
- 24.Harbers, K., A. Schnieke, H. Stuhlmann, D. Jahner, and R. Jaenisch. 1981. DNA methylation and gene expression: endogenous retroviral genome becomes infectious after molecular cloning. Proc. Natl. Acad. Sci. USA 78:7609-7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hejnar, J., J. Plachy, J. Geryk, O. Machon, K. Trejbalova, R. V. Guntaka, and J. Svoboda. 1999. Inhibition of the rous sarcoma virus long terminal repeat-driven transcription by in vitro methylation: different sensitivity in permissive chicken cells versus mammalian cells. Virology 255:171-181. [DOI] [PubMed] [Google Scholar]
- 26.Hoeben, R. C., A. A. Migchielsen, R. C. van der Jagt, H. van Ormondt, and A. J. van der Eb. 1991. Inactivation of the Moloney murine leukemia virus long terminal repeat in murine fibroblast cell lines is associated with methylation and dependent on its chromosomal position. J. Virol. 65:904-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jahner, D., H. Stuhlmann, C. L. Stewart, K. Harbers, J. Lohler, I. Simon, and R. Jaenisch. 1982. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature 298:623-628. [DOI] [PubMed] [Google Scholar]
- 28.Joel, P., W. Shao, and K. Pratt. 1993. A nuclear protein with enhanced binding to methylated Sp1 sites in the AIDS virus promoter. Nucleic Acids Res. 21:5786-5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jordan, A., P. Defechereux, and E. Verdin. 2001. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 20:1726-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kutsch, O., E. N. Benveniste, G. M. Shaw, and D. N. Levy. 2002. Direct and quantitative single-cell analysis of human immunodeficiency virus type 1 reactivation from latency. J. Virol. 76:8776-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikovits, J. A., N. C. Lohrey, R. Schulof, J. Courtless, and F. W. Ruscetti. 1992. Activation of infectious virus from latent human immunodeficiency virus infection of monocytes in vivo. J. Clin. Investig. 90:1486-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mikovits, J. A., Raziuddin, M. Gonda, M. Ruta, N. C. Lohrey, H. F. Kung, and F. W. Ruscetti. 1990. Negative regulation of human immune deficiency virus replication in monocytes. Distinctions between restricted and latent expression in THP-1 cells. J. Exp. Med. 171:1705-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mikovits, J. A., H. A. Young, P. Vertino, J. P. Issa, P. M. Pitha, S. Turcoski-Corrales, D. D. Taub, C. L. Petrow, S. B. Baylin, and F. W. Ruscetti. 1998. Infection with human immunodeficiency virus type 1 upregulates DNA methyltransferase, resulting in de novo methylation of the gamma interferon (IFN-γ) promoter and subsequent downregulation of IFN-γ production. Mol. Cell. Biol. 18:5166-5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niwa, O., Y. Yokota, H. Ishida, and T. Sugahara. 1983. Independent mechanisms involved in suppression of the Moloney leukemia virus genome during differentiation of murine teratocarcinoma cells. Cell 32:1105-1113. [DOI] [PubMed] [Google Scholar]
- 35.O'Brien, M. C., T. Ueno, N. Jahan, M. Zajac-Kaye, and H. Mitsuya. 1995. HIV-1 expression induced by anti-cancer agents in latently HIV-1-infected ACH2 cells. Biochem. Biophys. Res. Commun. 207:903-909. [DOI] [PubMed] [Google Scholar]
- 36.Olek, A., J. Oswald, and J. Walter. 1996. A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res. 24:5064-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pantaleo, G., and A. S. Fauci. 1996. Immunopathogenesis of HIV infection. Annu. Rev. Microbiol. 50:825-854. [DOI] [PubMed] [Google Scholar]
- 38.Saggioro, D., M. Forino, and L. Chieco-Bianchi. 1991. Transcriptional block of HTLV-I LTR by sequence-specific methylation. Virology 182:68-75. [DOI] [PubMed] [Google Scholar]
- 39.Saggioro, D., M. Panozzo, and L. Chieco-Bianchi. 1990. Human T-lymphotropic virus type I transcriptional regulation by methylation. Cancer Res. 50:4968-4973. [PubMed] [Google Scholar]
- 40.Schulze-Forster, K., F. Gotz, H. Wagner, H. Kroger, and D. Simon. 1990. Transcription of HIV1 is inhibited by DNA methylation. Biochem. Biophys. Res. Commun. 168:141-147. [DOI] [PubMed] [Google Scholar]
- 41.Shao, W. 1997. Characterization of HMBP-2, a DNA-binding protein that binds to HIV-1 LTR when only one of the three Sp1 sites is methylated. J. Biomed. Sci. 4:39-46. [DOI] [PubMed] [Google Scholar]
- 42.Sharkey, M. E., I. Teo, T. Greenough, N. Sharova, K. Luzuriaga, J. L. Sullivan, R. P. Bucy, L. G. Kostrikis, A. Haase, C. Veryard, R. E. Davaro, S. H. Cheeseman, J. S. Daly, C. Bova, R. T. Ellison III, B. Mady, K. K. Lai, G. Moyle, M. Nelson, B. Gazzard, S. Shaunak, and M. Stevenson. 2000. Persistence of episomal HIV-1 infection intermediates in patients on highly active anti-retroviral therapy. Nat. Med. 6:76-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shpaer, E. G., and J. I. Mullins. 1990. Selection against CpG dinucleotides in lentiviral genes: a possible role of methylation in regulation of viral expression. Nucleic Acids Res. 18:5793-5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh, M. K., and C. D. Pauza. 1992. Extrachromosomal human immunodeficiency virus type 1 sequences are methylated in latently infected U937 cells. Virology 188:451-458. [DOI] [PubMed] [Google Scholar]
- 45.Tierney, R. J., H. E. Kirby, J. K. Nagra, J. Desmond, A. I. Bell, and A. B. Rickinson. 2000. Methylation of transcription factor binding sites in the Epstein-Barr virus latent cycle promoter Wp coincides with promoter down-regulation during virus-induced B-cell transformation. J. Virol. 74:10468-10479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Unutmaz, D., V. N. KewalRamani, S. Marmon, and D. R. Littman. 1999. Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. J. Exp. Med. 189:1735-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong, J. K., M. Hezareh, H. F. Gunthard, D. V. Havlir, C. C. Ignacio, C. A. Spina, and D. D. Richman. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291-1295. [DOI] [PubMed] [Google Scholar]
- 48.Yoder, J. A., C. P. Walsh, and T. H. Bestor. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335-340. [DOI] [PubMed] [Google Scholar]
- 49.Zack, J. A., A. J. Cann, J. P. Lugo, and I. S. Chen. 1988. HIV-1 production from infected peripheral blood T cells after HTLV-I induced mitogenic stimulation. Science 240:1026-1029. [DOI] [PubMed] [Google Scholar]
- 50.Zagury, D., J. Bernard, R. Leonard, R. Cheynier, M. Feldman, P. S. Sarin, and R. C. Gallo. 1986. Long-term cultures of HTLV-III-infected T cells: a model of cytopathology of T-cell depletion in AIDS. Science 231:850-853. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, L., B. Ramratnam, K. Tenner-Racz, Y. He, M. Vesanen, S. Lewin, A. Talal, P. Racz, A. S. Perelson, B. T. Korber, M. Markowitz, and D. D. Ho. 1999. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N. Engl. J. Med. 340:1605-1613. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, Z., T. Schuler, M. Zupancic, S. Wietgrefe, K. A. Staskus, K. A. Reimann, T. A. Reinhart, M. Rogan, W. Cavert, C. J. Miller, R. S. Veazey, D. Notermans, S. Little, S. A. Danner, D. D. Richman, D. Havlir, J. Wong, H. L. Jordan, T. W. Schacker, P. Racz, K. Tenner-Racz, N. L. Letvin, S. Wolinsky, and A. T. Haase. 1999. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science 286:1353-1357. [DOI] [PubMed] [Google Scholar]