Abstract
The lytic cycle-associated lytic latent membrane protein-1 (lyLMP-1) of Epstein-Barr virus (EBV) is an amino-terminally truncated form of the oncogenic LMP-1. Although lyLMP-1 shares none of LMP-1's transforming and signal transducing activities, we recently reported that lyLMP-1 can negatively regulate LMP-1-stimulated NF-κB activation. The lyLMP-1 protein encoded by the B95-8 strain of EBV initiates from methionine 129 (Met129) of the LMP-1 open reading frame (ORF). The recent report that Met129 in the B95-8 LMP-1 ORF is not conserved in the Akata strain of EBV prompted us to screen a panel of EBV-positive cell lines for conservation of Met129 and lyLMP-1 expression. We found that 15 out of 16 tumor-associated virus isolates sequenced encoded an ATT or ACC codon in place of ATG in the LMP-1 ORF at position 129, and tumor cell lines harboring isolates lacking an ATG at codon 129 did not express the lyLMP-1 protein. In contrast, we found that EBV DNA from 22 out of 37 healthy seropositive donors retained the Met129 codon. Finally, the lyLMP-1 initiator occurs variably within distinct EBV strains and its presence cannot be predicted by EBV strain identity. Thus, Met129 is not peculiar to the B95-8 strain of EBV, but rather can be found in the background of several evolutionarily distinct EBV strains. Its absence from EBV isolates from tumors raises the possibility of selective pressure on Met129 in EBV-dependent tumors.
Epstein-Barr virus (EBV) is a human herpesvirus well known for its causative role in infectious mononucleosis (IM). EBV is associated with a number of malignancies, including endemic Burkitt's lymphoma (BL) (20), nasopharyngeal carcinoma (NPC) (10), and Hodgkin's disease (40). Immunocompromised individuals, such as transplant recipients and AIDS patients, are at high risk for development of EBV-dependent lymphoproliferative diseases and malignancies.
EBV is a latent virus, persists for the lifetime of the host, and rarely enters the lytic phase of its life cycle. Viral gene expression in latently infected B cells is restricted to ∼10 out of 100 open reading frames (ORFs) (reviewed in reference 46). Of the 10 expressed viral proteins, 5 are required for B-cell immortalization by EBV in vitro (reviewed in reference 42). Studies of transformed cells in vitro indicate that approximately 1 in 103 to 1 in 106 latently infected B cells per generation spontaneously enters the lytic cycle, during which virion production and release are accompanied by cell death (45, 54). Very little is known about EBV reactivation in vivo. Lytic cycle entry in EBV-immortalized B cells can be stimulated in vitro by superinfection with P3HR1 virus (5) or by manipulation of cell signaling pathways by chemical or immunoglobulin treatment (47, 53).
Our research is focused on the role of two related, yet differentially expressed, viral membrane proteins in EBV's life cycle. Latent membrane protein-1 (LMP-1) is expressed during the latent phase of EBV's life cycle, functions as EBV's transforming protein, and is indispensable for B-cell immortalization in vitro (42). LMP-1 is localized in patches in the plasma membrane where it is associated with the cytoskeleton (reviewed in reference 28). The large cytoplasmic carboxy terminus of LMP-1 is critical for immortalization of primary B cells by EBV, TRAF binding, and activation of cellular signaling pathways (reviewed in references 12 and 15).
A late lytic cycle promoter within the LMP-1 gene activates expression of a gene product whose translation is predicted to initiate at the 129th codon (ATG) of the B95-8 LMP-1 ORF (Fig. 1) (25). This truncated LMP-1 protein is called lyLMP-1 (also known as D1LMP-1, or trLMP-1) because of its association with EBV's lytic cycle (3, 13). The lyLMP-1 protein is associated with extracellular EBV virions and is detectable in EBV-infected cells within minutes after infection, independent of protein synthesis (13). Although lyLMP-1 encodes the carboxy-terminal signaling domain of LMP-1, it shares none of LMP-1's identified transforming or signaling activities, nor does it localize with LMP-1 or share its biochemical properties (3, 18, 26, 35, 38, 39, 51, 52). To date the only identified biological activity of lyLMP-1 is its ability to negatively regulate LMP-1 signaling (14). The recent report that LMP-1's activation of NF-κB prevents lytic cycle activation (1), together with our findings that lyLMP-1 negatively regulates LMP-1 signaling and is associated with extracellular EBV virions, strongly support a role for lyLMP-1 in progression of EBV's lytic cycle.
FIG. 1.
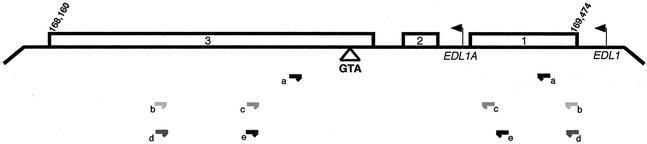
Schematic of the BNLF-1 region of EBV encoding the LMP-1 and lyLMP-1 ORFs. Leftward arrows indicate the position of the EDL1 (LMP-1) and EDL1A (lyLMP-1) promoters; open rectangles represent the three exons of LMP-1 and are numbered accordingly; the triangle below exon 3 shows the position of codon 129. Numbers (169,474 and 168,160) correspond to B95-8 genome coordinates. Horizontal arrows show positions of the PCR primers used and are summarized in Table 1.
The EBV prototype strain, B95-8, originating from a patient with IM, encodes a methionine codon (ATG) at position 129 in the LMP-1 ORF which is predicted to function as lyLMP-1's initiating methionine (2, 25). The lyLMP-1 promoter, transcript, and protein were initially characterized in B95-8 cells (16, 25). B95-8 cells are distinct from EBV-positive tumor cell lines in that they are an in vitro-immortalized lymphoblastoid cell line (not tumor derived) and are permissive for virus replication and release. B95-8 cells can be induced by treatment with phorbol 12-tetradecanoate 13-acetate (TPA) and butyrate to enter the lytic cycle with concomitant upregulation of lyLMP-1 mRNA and protein (3, 13, 25). Induced B95-8 cells produce and release infectious virus. Lytic cycle induction in EBV-positive Akata cells results in the upregulation of lyLMP-1 mRNA but, unlike induced B95-8 cells, the lyLMP-1 protein is undetectable (48). However, high levels of full-length LMP-1 and variable levels of LMP-1 degradation products, some of which migrate with an apparent molecular weight similar to that of lyLMP-1, are detected in induced Akata cells. Nucleotide sequence analysis of the Akata virus LMP-1 ORF reveals an ATT codon in the position of Met129 of the B95-8 strain, resulting in an amino acid change to isoleucine (48). This report raised the question that Met129, and therefore the lyLMP-1 ORF, is peculiar to the B95-8 laboratory-adapted strain of EBV, is found only in EBV strains found in permissive cell lines, or is not conserved in EBV isolates from tumors.
Here we report a series of experiments designed to test whether Met129 functions as the lyLMP-1 initiator and if Met129 is unique to the B95-8 strain of EBV. We found that Met129 does serve as the lyLMP-1 initiator and thus specifies the lyLMP-1 ORF. Further, we found that Met129 is not unique to the laboratory-adapted B95-8 strain of EBV and is not conserved in isolates from tumor cell lines or biopsies. Finally, Met129 occurs variably in distinct strains of EBV and, thus, its presence cannot be predicted by strain identity.
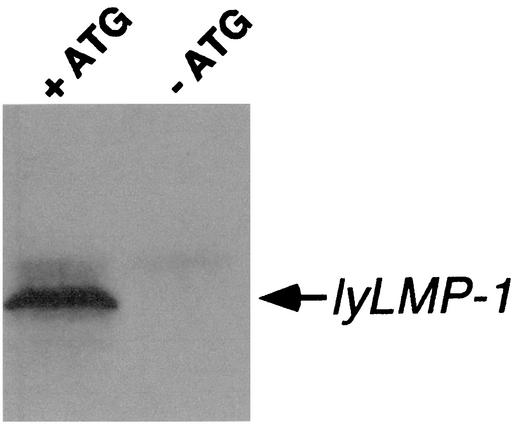
Met129 functions as the initiator for lyLMP-1 protein. To ensure that codon 129 encodes the initiating methionine for the lyLMP-1 protein, we translated lyLMP-1 in vitro using pcDNA3-based plasmids encoding the lyLMP-1 ORF with or without its initiating methionine. lyLMP-1 was transcribed and translated in a reticulocyte lysate in the presence of pancreatic microsomes (Promega) from a pcDNA3-based lyLMP-1 expression vector encoding the lyLMP-1 transcript (14). The ATG codon corresponding to Met129 of LMP-1 was mutated to GCG by site-directed mutagenesis. Translated products were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), analyzed by Western blotting using affinity-purified anti-LMP-1 antiserum, and visualized by ECL (Amersham) as described previously (14). We found that substitution of ATG at codon 129 with GCG, resulting in an amino acid change from methionine to alanine, inhibited translation of lyLMP-1 protein (Fig. 2). The essential role of this methionine residue in lyLMP-1 translation (Fig. 2) and the observation that Met129 is not encoded by Akata virus (48) prompted us to screen a panel of EBV isolates and EBV DNA from clinical specimens, with the goal of determining whether Met129 is restricted to the B95-8 strain of EBV.
FIG. 2.
Met129 is required for lyLMP-1 translation. The lyLMP-1 ORF was expressed in vitro from the T7 promoter in pcDNA3-based expression plasmids encoding Met129 (+ATG) or Ala129 (-ATG). Translated products were resolved by SDS-PAGE and analyzed by Western blotting using affinity-purified anti-LMP-1 rabbit antiserum recognizing LMP-1's carboxy terminus (14) followed by anti-rabbit-horseradish peroxidase-conjugated secondary antibody. lyLMP-1 was visualized by ECL (Amersham).
Tumor virus isolates do not encode Met129.
We screened a panel of EBV-positive tumor cell lines to determine if EBV strains other than B95-8 encode Met129. These EBV-positive tumor cell lines differ from the in vitro-immortalized marmoset lymphoblastoid cell line B95-8 in that they were derived from BL tumor biopsies. EBV DNA was isolated from tumor cell lines and the region encompassing the Met129 codon was amplified by PCR with primer set a (Fig. 1; Table 1). Sequence analysis was performed simultaneously on viral DNA isolated from B95-8 (2), Raji (19), and Akata (48) cells to confirm the high fidelity of the DNA polymerase (Bio-X-Act) used in the PCR amplification (Fig. 3).
TABLE 1.
PCR primers
| Source of amplified DNA (primer set) | Sequence | B95-8 coordinates |
|---|---|---|
| Tumor virus isolates | ||
| (a)Sense | 5′CCCTAGGCCTTGCTCTCC3′ | 169,404-169,387 |
| Antisense | 5′AATCGCCAGAAACAGGAGGAG3′ | 168,781-168,801 |
| EBV DNA from healthy donorsa | ||
| (b)Sense | 5′ATGGAACACGACCTTGAGAGG3 | 169,474-169,454 |
| Antisense | 5′ATCATTTCCAGCAGAGTCGCT3′ | 168,370-168,390 |
| (c)Sense | 5′AGACCTTCTCTGTCCACTTGG3′ | 169,253-169,233 |
| Antisense | 5′AGAATCATCGGTAGCTTGTTG3′ | 168,689-168,708 |
| EBV DNA from clinical specimens | ||
| (d)Sense | 5′TGGAACACGACCTTGAGAGG3′ | 169,473-169,455 |
| Antisense | 5′CATCATTTCCAGCAGAGTCG3′ | 168,369-168,388 |
| (e)Sense | 5′TCCACTTGGAGCCCTTTGTA3′ | 169,241-169,222 |
| Antisense | 5′ATGGCCAGAATCATCGGTAG3′ | 168,682-168,701 |
These primers have been described previously (32).
FIG. 3.
LMP-1 sequence flanking Met129 codon from tumor virus isolates. A region of the LMP-1 ORF was amplified by PCR from genomic DNA isolated from EBV-positive tumor cell lines (HH514, BL74, Jijoye, and Elijah) using primer set a (Fig. 1; Table 1). A portion of the sequence obtained from the PCR products is shown and corresponds to EBV coordinates 168,971 to 168,835 of B95-8 virus. LMP-1's 129th codon (lyLMP-1's initiating methionine) is shown in boldface, as are mutations of the ATG codon. Numbers (119 to 164) above the B95-8 sequence indicate the codon numbers of the LMP-1 ORF; shaded areas represent LMP-1's fourth and fifth transmembrane domains. Base substitutions and deletions (-) (relative to B95-8 reference strain) are shown and dots represent identity. HH514 cells are derived from the P3HR1 clone of the Jijoye BL cell line (41) and are permissive for virus replication; B95-8 cells are an EBV-positive marmoset lymphoblastoid cell line transformed in vitro by EBV (36). BL74, Jijoye, Elijah, Akata, Raji, and AG876 are virus isolates from BL cell lines; CAO, C15, and Xeno-2117 are virus isolates from NPC xenografts; NPC3, NPC6, NPC10, and NPC30 are virus isolates from primary NPC biopsies; NPC1510 is a phage clone isolated from NPC biopsy no. 122 (published sequences are cited in the figure). EBV DNA was isolated from 107 cells resuspended in TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA) at 4 × 107 cells/ml plus 10 volumes of lysis buffer (10 mM Tris [pH 8.0], 100 mM EDTA, 20 μg of pancreatic RNase/ml, 0.5% SDS) and incubated at 37°C for 1.5 h. Proteinase K was added to the lysate at a final concentration of 100 μg/ml and the mixture was incubated at 50°C for 4 h. The lysate was phenol extracted and DNA was precipitated and resuspended in TE buffer at 100 ng/μl. The region encompassing the Met129 codon was amplified by PCR with primer set a (Fig. 1; Table 1) using a DNA Engine PTC-200 (MJ Research) under the following conditions: 1 cycle of 94°C for 4 min; 35 cycles of 94°C for 15 s, 59.3°C for 30 s, 72°C for 30 s; 1 cycle of 72°C for 5 min. PCR-amplified products were resolved by agarose gel electrophoresis, excised from the gel, and purified using a QIAquick gel extraction kit (Qiagen). Purified products were sequenced by automated cycle sequencing using the antisense primer of primer set a (Fig. 1; Table 1).
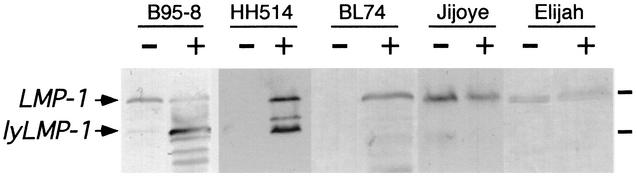
Sequence analysis revealed that EBV isolates from three of four tumor lines (BL74, Elijah, and Jijoye) did not encode the lyLMP-1 ORF (Fig. 3). Elijah and Jijoye virus encoded a G-to-T substitution in Met129, resulting in an amino acid change to isoleucine. BL74 virus encoded ACC in place of ATG at codon 129, resulting in an amino acid change from methionine to threonine. LMP-1 protein expression in these Met129-negative cell lines was assessed to determine if, as in Akata cells, a “lyLMP-1-like” LMP-1 proteolytic fragment was detectable after treatment with TPA and butyrate. We found that none of these cell lines expressed lyLMP-1, or LMP-1 cleavage products comigrating with lyLMP-1, following treatment with TPA and butyrate (Fig. 4), nor did they enter the lytic cycle as assessed by immunofluorescence staining with anti-gp350 antibody (gift of L. Hutt-Fletcher) (data not shown). The BL74 cell line expressed the early antigen complex but not late viral antigens following TPA and butyrate treatment, suggesting that it may enter an abortive lytic cycle as has been shown for Raji cells (19). EBV from HH514 BL cells was found to encode Met129 and to express lyLMP-1 protein following TPA and butyrate treatment (Fig. 3 and 4). Interestingly, HH514 cells, like B95-8 cells, are permissive for lytic replication and thus differ in this regard from the other tightly latent BL lines sequenced (53) (data not shown). Consistent with these results, a literature search revealed that LMP-1 sequences from additional tumor-associated EBV strains, either from tumor cell lines, NPC xenografts, or primary NPC biopsies, have a G-to- T substitution at codon 129 (Fig. 3) (reference 17 and references shown in Fig. 3) and thus do not encode the lyLMP-1 ORF.
FIG. 4.
LMP-1 expression in EBV-positive cell lines. Exponentially growing cells were maintained in 20 ng of TPA/ml and 3.5 mM butyrate for 72 h. Sonicated cell lysates were resolved by SDS-PAGE and analyzed by Western blotting using anti-LMP-1 antiserum. −, lysate of cells harvested prior to adding TPA and butyrate; +, lysate of cells treated with TPA and butyrate for 72 h. Each cell line is indicated above the Western blot. Upper and lower arrows show migration of LMP-1 and lyLMP-1 proteins, respectively. Molecular mass markers are shown to the right of the blot and are 68 and 43 kDa, respectively.
Met129 is found in EBV DNA from healthy individuals.
The association of specific LMP-1 sequences with disease has been extensively studied. Some studies reveal no relationship between specific EBV strains and disease (11, 27, 50), whereas others have shown that sequence variation in LMP-1 (compared to the B95-8 reference strain) is associated with more oncogenic phenotypes (21-24, 29-31, 44, 49). Thus, the ATG-to-ATT/ACC substitution in the LMP-1 gene in tumor-associated strains may not be representative of virus found in healthy individuals. Therefore, we determined if Met129 was conserved in EBV DNA from healthy seropositive donors. Genomic DNA was prepared from hyperplastic tonsils of 37 EBV-seropositive children from Switzerland and amplified using primer set b (Fig. 1; Table 1).
The region of the LMP-1 sequence corresponding to nucleotides 168,966 to 168,830 of B95-8 is shown in Fig. 5. Strikingly, unlike the sequence in tumor-associated virus strains, Met129 was conserved in EBV DNA from 22 of 37 (60%) healthy seropositive donors (Fig. 5). Thus, the lyLMP-1 ORF is not peculiar to laboratory-adapted B95-8 virus from B95-8 cells, but rather is well represented in vivo in viral DNA isolated from healthy donors. We observed a difference in the incidence of Met129 in tumor-associated isolates (1 of 16) compared to that in EBV DNA from healthy donors (22 of 37). These results suggest a negative correlation between the conservation of the lyLMP-1 ORF and the association of these EBV strains with human tumors. Alternatively, the Met129-positive healthy donors reported in Fig. 5 could be infected with B95-8-like strains, with the prevalence of Met129 simply reflecting the prevalence of B95-8-like EBV strains infecting this population.
FIG. 5.
LMP-1 sequence flanking Met129 codon from viral DNA from healthy seropositive donors. A region of the LMP-1 ORF was amplified by nested PCR from viral DNA isolated from tonsils using primer sets b and c (Fig. 1; Table 1). A portion of the sequence obtained from the PCR products is shown and corresponds to EBV coordinates 168,971 to 168,835 of B95-8 virus. Individual donors are designated by the numbers to the left of each sequence. All else is as described in the legend to Fig. 3. Total genomic DNA was prepared from hyperplastic tonsils of 37 EBV-seropositive children from Switzerland as described in the text, using a QIAamp blood kit following the manufacturer's instructions (Qiagen, Inc.) (4). PCRs using primer set b (Fig. 1; Table 1) were performed on the Perkin-Elmer Gene Amp system 9700 with the following conditions for the primary PCR: 1 cycle of 93°C for 10 min; 2 cycles of 94°C for 30 s, 58°C for 30 s, 72°C for 2 min; 3 cycles of 94°C for 30 s, 66°C for 30 s, 72°C for 2 min; 25 cycles of 94°C for 30 s, 64°C for 30 s, 72°C for 90 s; 1 cycle of 72°C for 10 min. The second round of PCR was performed using the following conditions: 1 cycle of 93°C for 10 min; 5 cycles of 94°C for 30 s, 63°C for 30 s, 72°C for 2 min; 25 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 90 s; 1 cycle of 72°C for 10 min. Cycle sequencing was performed using primer set c (Fig. 1; Table 1) with the Perkin-Elmer Gene Amp system 9700 and ABI Prism 310.
The presence of Met129 cannot be predicted by strain identity.
Naturally occurring EBV genotypes can be defined as strains based on evolutionarily stable patterns of sequence variation in the carboxy-terminal sequences of the LMP-1 gene (50). To determine if the occurrence of Met129 in vivo is associated with a particular strain of EBV (i.e., B95-8), we identified 23 clinical specimens (EBV-infected, healthy, and diseased clinical specimens), each infected with a single EBV strain as determined by PCR amplification, cloning, and sequencing of the LMP-1 gene carboxy terminus (50). All specimens were from different donors, with the exception of numbers 26 and 28, which came from the same donor. As detailed in the fourth column in Table 2 (sample source), numbered specimens originated from the following donors: eight from healthy donors, one from an individual with IM, five from human immunodeficiency virus-positive (HIV+) individuals without disease pathology, six from HIV+ individuals with hairy leukoplakia (HLP), two from individuals with AIDS lymphoma, and one from an individual with posttransplant lymphoproliferative disorder. We then sequenced the LMP-1 Met129 region of EBV LMP-1 DNA from each of these 23 clinical specimens containing single EBV strains. This approach ensured that our analysis and interpretation of the Met129 sequences would not be complicated by a mixed population of EBV strains in the genomic DNA used for amplification of Met129.
TABLE 2.
Incidence of Met129 in clinical specimens, tumor biopsies, and tumor cell linesa
| Specimen no.,b tumor line, or tumor biopsy | EBV strain | Met129 status | Sample source |
|---|---|---|---|
| 26 | B958a-B958 | Positive | Healthy individual PBMC |
| 10 | B958a-B958 | Positive | HIV+ PBMC |
| 19 | B958a-B958 | Positive | HIV+ normal tongue |
| 15 | B958a-B958 | Positive | HIV+ HLP |
| Jijoye BL | B958a-F2 | Negative | EBV+ BL cell line |
| 22 | B958b-F2 | Positive | Healthy individual saliva |
| 23 | B958b-F2 | Positive | Healthy individual saliva |
| 28 | 1-G | Positive | Healthy individual PBMC |
| Raji BL* | 1-G | Negative | EBV+ BL cell line |
| 29 | 2a-G | Positive | Healthy individual saliva |
| 7 | 2a-G | Positive | HIV+ normal tongue |
| 16 | 2a-G | Positive | HIV+ HLP |
| 17 | 2a-G | Negative | HIV+ lymphoma |
| 6 | 2a-F1 | Positive | HIV+ normal tongue |
| 2 | 2a-F1 | Negative | HIV+ HLP |
| 18 | 2a-F1 | Positive | HIV+ lymphoma |
| Akata* | 2a-F1 | Negative | Akata BL cell line |
| CAO* | 2a-F1 | Negative | NPC xenograft |
| Xeno-2117 | 2a-F1 | Negative | NPC xenograft |
| NPC10 | 2a-F1 | Negative | Primary NPC biopsy |
| NPC3* | 2a-F1 | Negative | Primary NPC biopsy |
| NPC6* | 2a-F1 | Negative | Primary NPC biopsy |
| NPC 1510* | 2b-F1 | Negative | Primary NPC biopsy |
| C15* | 2c-C | Negative | NPC xenograft |
| AG876* | 2a-C | Negative | EBV+ BL cell line |
| 20 | 3c-C | Positive | Healthy individual saliva |
| 21 | 3c-D | Negative | Healthy individual saliva |
| 24 | 3c-D | Negative | Healthy individual saliva |
| 8 | 3c-D | Positive | HIV+ normal tongue |
| 1 | 3c-D | Negative | HIV+ HLP |
| 11 | 4-D | Negative | IM saliva |
| 3 | 4-D | Negative | HIV+ HLP |
| 4 | 5-D | Negative | HIV+ HLP |
| NPC30* | 7-H | Negative | Primary NPC biopsy |
| 14 | 8-F3 | Positive | Posttransplant lymphoma |
Numbered clinical specimens harbor a single strain of EBV (50), as do shown BL cell lines and NPC biopsies. The strain identity of infecting EBV is shown with its Met129 status. The strain identities, based on C-terminal LMP-1 sequences (50), of several of the tumor isolates listed in Fig. 3 were derived from published sequences, and the status of Met129 for these isolates is included for comparison. *, references for tumor cell lines or biopsies are cited in Fig. 3.
Human subjects guidelines of each participating institution were followed in the conduct of this research with human tissues.
Total genomic DNA was prepared from clinical specimens as described by Walling et al. (50). Between 0.1 and 0.5 μg of DNA was amplified using primers from the carboxy terminus of the LMP-1 gene (50). PCR products were cloned, and at least 8 to 10 clones from each specimen were sequenced to determine the EBV strain(s) present in the tissue, as described by Walling et al. (50). Between 0.1 and 2.5 μg of DNA was amplified using a DNA Engine PTC-200 (MJ Research) by nested PCR using primer pairs d and e (Fig. 1; Table 1). DNA was amplified with PfuTurbo Hotstart DNA polymerase (Stratagene) using the following conditions: 1 cycle of 94°C for 4 min; 30 cycles of 94°C for 15 s, 54°C for 30 s, and 72°C for 1 min; and 1 cycle of 72°C for 5 min. PCR products were visualized by ethidium staining, tailed with Taq polymerase (Invitrogen), and cloned into a PCR cloning vector (pCR-TOPO-II; Invitrogen). Inserts were sequenced by automated cycle sequencing using either SP6 or T7 primers.
Results from the clinical specimens, along with selected tumor isolates, are shown in Table 2. Met129 was found in clinical specimens harboring the B958a-B958 strain (the strain from the B95-8 cell line) and in specimens with the closely related substrain B958b-F2. Interestingly, Jijoye BL cells harbored the closely related substrain B958a-F2, but were Met129 negative (Table 2). Strain 1-G from specimen 28 was Met129 positive, whereas Raji cells (also strain 1-G) were Met129 negative (Table 2). B958a-B958 and 1-G strains share the repeat insertion in LMP-1's carboxy terminus, amino acids 276 to 280, and are evolutionarily closely related (50). These results suggest that Met129 does not always correlate with the B958 or 1-G strain identities.
Strains 2, 3, 4, and 5 are evolutionarily related and distinct from B958 and strain 1 (50). Strains 2a-F1 and 3c-D were variably associated with Met129. Two clinical specimens (numbers 6 and 18) in this group were Met129 positive and one was Met129 negative (number 2). The single sample harboring strain 3c-C (number 20) was Met129 positive. Akata, CAO, NPC3, NPC6, Xeno-2117, and NPC10 were strain 2a-F1 (all Met129 negative). AG876 was strain 2a-C (Met129 negative). C15 was strain 2c-C (Met129 negative), and NPC1510 was strain 2b-F1 (Met129 negative). Thus, it is not possible to predict the presence of Met129 within this evolutionarily related group of EBV strains.
Strains 7, 8, and 9 are the third major evolutionary group and the most evolutionarily distant from the B958 and 1-G group (50). The single clinical sample from this group (number 14), strain 8-F3, was Met129 positive. NPC30 harbored strain 7-H and was Met129 negative. These results again demonstrate that Met129 cannot be predicted by strain identity within this closely related group of EBV strains.
The two peripheral blood mononuclear cell (PBMC) specimens from healthy individuals, four of six healthy saliva specimens, and the single saliva specimen from an IM patient were Met129 positive. The HIV+ PBMC specimen was Met129 positive, as were the four HIV+ normal tongue specimens. Interestingly, four of six strains from HIV+ HLP specimens harboring lytic EBV infection were Met129 negative. Of the three immunodeficiency-associated lymphoma samples, two were Met129 positive and one was Met129 negative. None of these three lymphomas harbored lytic EBV infection (data not shown).
The lyLMP-1 promoter, pED-L1A, is conserved in samples lacking Met129.
Sequences from a number of samples represented in Fig. 3 and Table 2 were examined for the presence of the lyLMP-1 promoter pED-L1A. The lyLMP-1 transcript and the cap site and TATA box (TATTACA) comprising the ED-L1A promoter have been mapped in B95-8 cells (25). Interestingly, in all cases shown in Fig. 3 and Table 2 for which the nucleotide sequence, rather than amino acid sequence, is available (all our clinical samples and many of the published sequences [i.e., Akata, Raji, NPC1510, and CAO]), both Met129 positive and Met129 negative, the TATTACA sequence (B95-8 coordinates 169,201 to 169,195, within the first intron of the LMP-1 gene) in the ED-L1A promoter is conserved (data not shown). Thus, mutation of the lyLMP-1 initiator in Met129-negative cases occurred independently of the lyLMP-1 promoter, suggesting that the lyLMP-1 transcript is, or can be, expressed. This has been shown to be true for Akata BL cells, which lack Met129 but nonetheless express the lyLMP-1 transcript (48).
Variations in LMP-1's carboxy terminus (including 30- and 69-bp deletions) and the loss of the XhoI site at the 5′ end of the LMP-1 ORF have been used to characterize EBV variants (4, 7, 21, 31, 33, 34, 44). EBV variants lacking the XhoI site and having the 30-bp carboxy-terminal deletion are found in NPC tumor biopsies and EBV-dependent lymphoproliferative disorders (6-9, 23, 30-32). Thus, specific polymorphisms within the LMP-1 gene have been suggested to be predictive of the tumorigenic potential of EBV variants (21-24, 29-31, 44, 49). Substitution of codon 129, resulting in a change from methionine to isoleucine/threonine, differs from previously reported sequence variations in the LMP-1 gene because, in addition to having the potential to alter the activity of the LMP-1 protein, this substitution knocks out the lyLMP-1 ORF and thus has dramatic effects on the expression of the lyLMP-1 protein (Fig. 2).
Our finding that Met129 is not peculiar to any specific strain of EBV demonstrates that the Met129-negative status of tumor isolates does not simply reflect the prevalence of parental virus present in the geographical area from which the tumor originated. Furthermore, we have identified Met129-negative samples from healthy donors in Switzerland, from individuals in the United States (HIV+, healthy donors, and an IM patient), and from tumors originating in Africa and Asia. Thus, Met129 does not appear to be geographically restricted. Met129 is strain independent and clearly is not restricted to the B95-8 strain.
Absence of the lyLMP-1 ORF in tumor virus isolates raises the intriguing possibility that lyLMP-1 expression may be incompatible with EBV-dependent tumor progression. Development and/or maintenance of many types of EBV-associated tumors in vivo requires LMP-1 expression. Erickson and Martin have shown that NF-κB activation by LMP-1 is inhibited by coexpressed lyLMP-1 (14). Consistent with this finding are recent results showing that NF-κB activation by LMP-1 in EBV-positive cell lines plays a role in preventing lytic cycle reactivation (1).
lyLMP-1 is a lytic cycle protein (3, 25). We have evidence that lyLMP-1 is incorporated into virus particles as they bud through nuclear and cytoplasmic membranes (G. Vazirabadi, T. Geiger, W. F. Coffin III, and J. M. Martin, submitted for publication). These findings, together with our results that the lyLMP-1 ORF is not encoded by tumor-associated virus isolates, suggest a role for lyLMP-1 in the productive phase of EBV's life cycle and are consistent with the existence of selective pressures affecting Met129 in EBV strains in tumors.
The HH514 cell line is a descendant of Jijoye, via the subcloning of a Jijoye subclone (P3HR1) (41). Jijoye BL cells exhibit a latent phenotype and are Met129 negative, whereas HH514 cells are permissive for virus replication and are Met129 positive. This observation suggests that HH514 represents a Met129-positive EBV isolate selected by in vitro pressures and is consistent with the hypothesis that Met129 serves a function during productive infection but is incompatible with the latency state required for tumorigenesis. In the same context, there may be a correlation between lyLMP-1 expression and the cell's ability to support and complete the lytic cycle. For example, the Met129-positive B95-8 and HH514 cell lines are well known for their spontaneous entry into the lytic cycle in vitro. Akata cells (Met129 negative) will sometimes also spontaneously enter the lytic cycle in vitro, albeit at low levels, as do P3HR1 cells. However, several EBV strains replicating in HLP in vivo were found to be Met129 positive. It is interesting that all the Met129-negative NPCs reported in Fig. 3 contain 2a/b/c-C/F1 strains, yet we have found 2a-F1 specimens (not from NPC) that are Met129 positive. This observation suggests, but does not prove, the existence of in vivo selection against Met129 in the pathogenesis of NPC specifically.
In summary, Met129 is common in B95-8-like strains but is neither universal in, nor unique to, B95-8-like strains. Met129 has likely independently evolved, and reverted, many times in many different EBV isolates. It is plausible that positive and negative selective pressures exist for Met129, but we don't yet know what these may be, nor do we know if they might exist in vivo, in vitro, or both.
Acknowledgments
This work was supported in part by NIH grants CA-64610 and AI-01537 to J. M. Martin and by NIH grant RO1-DE12323 to D. M. Walling.
We thank Brad Olwin and Tin Tin Su for critical reading of the manuscript and Shawn Keil for help with the in vitro translation.
REFERENCES
- 1.Adler, B., E. Schaadt, B. Kempkes, U. Zimber-Strobl, B. Baier, and G. W. Bornkamm. 2002. Control of Epstein-Barr virus reactivation by activated CD40 and viral latent membrane protein 1. Proc. Natl. Acad. Sci. USA 99:437-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. [DOI] [PubMed] [Google Scholar]
- 3.Baichwal, V. R., and B. Sugden. 1987. Posttranslational processing of an Epstein-Barr virus-encoded membrane protein expressed in cells transformed by Epstein-Barr virus. J. Virol. 61:866-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger, C., D. van Baarle, M. J. Kersten, M. R. Klein, A. S. Al-Homsi, B. Dunn, C. McQuain, R. van Oers, and H. Knecht. 1999. Carboxy-terminal variants of Epstein-Barr virus encoded latent membrane protein 1 during long term HIV infection: reliable markers for individual strain identification. J. Infect. Dis. 179:240-244. [DOI] [PubMed] [Google Scholar]
- 5.Biggin, M., M. Bodescot, M. Perricaudet, and P. Farrell. 1987. Epstein-Barr virus gene expression in P3HR1-superinfected Raji cells. J. Virol. 61:3120-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, Y. S., P. J. Chung, C. J. Shu, C. K. Ng, S. J. Wu, and S. T. Liu. 1995. Detection of an Epstein-Barr-virus variant in T-cell-lymphoma tissue identical to the distinct strain observed in nasopharyngeal carcinoma in the Taiwanese population. Int. J. Cancer 62:673-677. [DOI] [PubMed] [Google Scholar]
- 7.Chen, M. L., C. N. Tsai, C. L. Liang, C. H. Shu, C. R. Hang, D. Sulitzeanu, S. T. Liu, and Y. S. Chang. 1992. Cloning and characterization of the latent membrane protein (LMP) of a specific Epstein-Barr virus variant derived from the nasopharyngeal carcinoma in the Taiwanese population. Oncogene 7:2131-2140. [PubMed] [Google Scholar]
- 8.Cheung, S.-T., S.-F. Leung, K.-W. Lo, K. W. Chiu, J. S. L. Tam, T. F. Fok, P. J. Johnson, J. C. K. Lee, and D. P. Huang. 1998. Specific latent membrane protein 1 gene sequences in type 1 and type 2 Epstein-Barr virus from nasopharyngeal carcinoma in Hong Kong. Int. J. Cancer 76:399-406. [DOI] [PubMed] [Google Scholar]
- 9.Cheung, S.-T., K.-W. Lo, S. F. Leung, W.-Y. Chan, P. H. K. Chio, P. J. Johnson, J. C. K. Lee, and D. P. Huang. 1996. Prevalence of LMP1 deletion variant of Epstein-Barr virus in nasopharyngeal carcinoma and gastric tumors in Hong Kong. Int. J. Cancer 66:711-712. [DOI] [PubMed] [Google Scholar]
- 10.Desgranges, C., G. de The, H. Wolf, and H. zur Hausen. 1975. Further studies on the detection of the Epstein-Barr virus DNA in nasopharyngeal carcinoma biopsies from different parts of the world. IARC Sci. Publ. 11:191-193. [PubMed] [Google Scholar]
- 11.Edwards, R. H., F. Seillier-Moiseiwitsch, and N. Raab-Traub. 1999. Signature amino acid changes in latent membrane protein 1 distinguish Epstein-Barr virus strains. Virology 261:79-95. [DOI] [PubMed] [Google Scholar]
- 12.Eliopoulos, A. G., and A. B. Rickinson. 1998. Epstein-Barr virus: LMP1 masquerades as an active receptor. Curr. Biol. 8:R196-R198. [DOI] [PubMed] [Google Scholar]
- 13.Erickson, K. D., and J. M. Martin. 1997. Early detection of the lytic LMP-1 protein in EBV-infected B-cells suggests its presence in the virion. Virology 234:1-13. [DOI] [PubMed] [Google Scholar]
- 14.Erickson, K. D., and J. M. Martin. 2000. The late lytic LMP-1 protein of Epstein-Barr virus can negatively regulate LMP-1 signaling. J. Virol. 74:1057-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farrell, P. J. 1998. Signal transduction from the Epstein-Barr virus LMP-1 transforming protein. Trends Microbiol. 6:175-178. [DOI] [PubMed] [Google Scholar]
- 16.Fennewald, S., S. van Santen, and E. Kieff. 1984. Nucleotide sequence of an mRNA transcribed in latent growth-transforming virus infection indicates that it may encode a membrane protein. J. Virol. 51:411-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fielding, C. A., K. Sandvej, A. Mehl, P. Brennan, M. Jones, and M. Rowe. 2001. Epstein-Barr virus LMP-1 natural sequence variants differ in their potential to activate cellular signaling pathways. J. Virol. 75:9129-9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammerschmidt, W., B. Sugden, and V. R. Baichwal. 1989. The transforming domain alone of the latent membrane protein of Epstein-Barr virus is toxic to cells when expressed at high levels. J. Virol. 63:2469-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatfull, G., A. T. Bankier, B. G. Barrell, and P. J. Farrell. 1988. Sequence analysis of Raji Epstein-Barr virus DNA. Virology 164:334-340. [DOI] [PubMed] [Google Scholar]
- 20.Henle, G., W. Henle, G. Klein, P. Gunven, P. Clifford, R. H. Morrow, and J. L. Ziegler. 1971. Antibodies to early Epstein-Barr virus-induced antigens in Burkitt's lymphoma. J. Natl. Cancer Inst. 46:861-871. [PubMed] [Google Scholar]
- 21.Hu, L., F. Chen, F. Zheng, I. Ernberg, S. Cao, B. Christensson, G. Klein, and G. Winberg. 1993. Clonability and tumorigenicity of human epithelial cells expressing the EBV-encoded membrane protein LMP1. Oncogene 8:1575-1583. [PubMed] [Google Scholar]
- 22.Hu, L., B. Troyanovsky, X. Zhang, I. Ernberg, and G. Klein. 2000. Differences in the immunogenicity of latent membrane protein 1 (LMP1) encoded by Epstein-Barr virus genomes derived from LMP1-positive and -negative nasopharyngeal carcinoma. Cancer Res. 60:5589-5593. [PubMed] [Google Scholar]
- 23.Hu, L.-F., E. R. Zabarovsky, F. Chen, S.-L. Cao, I. Ernberg, G. Klein, and G. Winberg. 1991. Isolation and sequencing of the Epstein-Barr virus BNLF-1 gene (LMP-1) from a Chinese nasopharyngeal carcinoma. J. Gen. Virol. 72:2399-2409. [DOI] [PubMed] [Google Scholar]
- 24.Hu, L. F., F. Chen, Q.-F. Zhen, Y.-W. Zhang, Y. Luo, X. Zheng, G. Winberg, I. Ernberg, and G. Klein. 1995. Differences in the growth pattern and clinical course of EBV-LMP1 expressing and nonexpressing nasopharyngeal carcinomas. Eur. J. Cancer 31A:658-660. [DOI] [PubMed] [Google Scholar]
- 25.Hudson, G. S., P. J. Farrell, and B. G. Barrell. 1985. Two related but differentially expressed potential membrane proteins encoded by the EcoRI Dhet region of Epstein-Barr virus B95-8. J. Virol. 53:528-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huen, D. S., S. A. Henderson, D. Croom-Carter, and M. Rowe. 1995. The Epstein-Barr virus latent membrane protein (LMP1) mediates activation of NF-κB and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene 10:549-560. [PubMed] [Google Scholar]
- 27.Khanim, F., Q. Y. Yao, G. Niedobitek, S. Sihota, A. B. Rickinson, and L. S. Young. 1996. Analysis of Epstein-Barr virus gene polymorphisms in normal donors and in virus-associated tumors from different geographic locations. Blood 88:3491-3501. [PubMed] [Google Scholar]
- 28.Kieff, E. 1996. Epstein-Barr virus and its replication, 3rd ed. Raven Press, Ltd., Philadelphia, Pa.
- 29.Kingma, D. W., W. B. Weiss, E. S. Jaffe, S. Kumar, K. Frekko, and M. Raffeld. 1996. Epstein-Barr virus latent membrane protein-1 oncogene deletions: correlation with malignancy in Epstein-Barr virus-associated lymphoproliferative disorders and malignant lymphomas. Blood 88:242-251. [PubMed] [Google Scholar]
- 30.Knecht, H., E. Bachmann, P. Brousset, S. Rothenberger, J. Einsele, V. S. Lestou, G. Delsol, F. Bachmann, P. F. Ambros, and B. F. Odematt. 1995. Mutational hot spots within the carboxy terminal region of the LMP1 oncogene of Epstein-Barr virus are frequent in lymphoproliferative disorders. Oncogene 10:523-528. [PubMed] [Google Scholar]
- 31.Knecht, H., E. Bachmann, P. Brousset, K. Sandvej, D. Nadal, F. Bachmann, B. F. Odermatt, G. Delsol, and G. Pallesen. 1993. Deletions within the LMP1 oncogene of Epstein-Barr virus are clustered in Hodgkin's disease and identical to those observed in nasopharyngeal carcinoma. Blood 82:2937-2942. [PubMed] [Google Scholar]
- 32.Knecht, H., F. Matius, E. Bachman, T. Hoffman, D. R. Zimmerman, S. Rothenberger, K. Sandvej, W. Wegman, N. Hurwitz, B. F. Odermatt, H. Kummer, and G. Pallesen. 1995. A deletion mutant of the LMP-1 oncogene of Epstein-Barr virus is associated with evolution of angioimmunoblastic lymphadenopathy into B immunoblastic lymphoma. Leukemia 9:458-465. [PubMed] [Google Scholar]
- 33.Larcher, C., C. McQuain, C. Berger, M. Mitterer, P. J. Quesenberry, H. P. Huemer, and H. Knecht. 1997. Epstein-Barr virus-associated persistent polyclonal B-cell lymphocytosis with a distinct 69 base pair deletion in the LMP1 oncogene. Ann. Hematol. 74:23-38. [DOI] [PubMed] [Google Scholar]
- 34.Leung, S. Y., S. T. Yuen, L. P. Chung, A. S. Y. Chan, and M. P. Wong. 1997. Prevalence of mutations and 30 bp deletion in the C-terminal region of Epstein-Barr virus latent membrane protein-1 oncogene in reactive lymphoid tissue and non-nasopharyngeal EBV-associated carcinomas in Hong Kong Chinese. Int. J. Cancer 72:225-230. [DOI] [PubMed] [Google Scholar]
- 35.Liebowitz, D., J. Mannick, K. Takada, and E. Kieff. 1992. Phenotypes of Epstein-Barr virus LMP1 deletion mutants indicate transmembrane and amino-terminal cytoplasmic domains necessary for effects in B-lymphoma cells. J. Virol. 66:4612-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, G., and M. Lipman. 1973. Release of infectious Epstein-Barr virus by transformed marmoset leukocytes. Proc. Natl. Acad. Sci. USA 70:190-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, W. E., R. H. Edwards, D. M. Walling, and N. Raab-Traub. 1994. Sequence variation in the Epstein-Barr virus latent membrane protein 1. J. Gen. Virol. 75:2729-2740. [DOI] [PubMed] [Google Scholar]
- 38.Mitchell, T., and B. Sugden. 1995. Stimulation of NF-κB-mediated transcription by mutant derivatives of the latent membrane protein of Epstein-Barr virus. J. Virol. 69:2968-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moorthy, R., and D. Thorley-Lawson. 1993. Biochemical, genetic, and functional analyses of the phosphorylation sites on the Epstein-Barr virus-encoded oncogenic latent membrane protein LMP-1. J. Virol. 67:2637-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pallesen, G., S. J. Hamilton-Dutolt, M. Rowe, and I. S. Young. 1991. Expression of Epstein-Barr virus latent gene products in tumour cells of Hodgkin's disease. Lancet 337:320-322. [DOI] [PubMed] [Google Scholar]
- 41.Rabson, M., L. Heston, and G. Miller. 1983. Identification of a rare Epstein-Barr virus variant that enhances early antigen expression in Raji cells. Proc. Natl. Acad. Sci. USA 80:2762-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rowe, D. T. 1999. Epstein-Barr virus immortalization and latency. Front. Biosci. 4:D346-D371. [DOI] [PubMed] [Google Scholar]
- 43.Sample, J., E. F. Kieff, and E. D. Kieff. 1994. Epstein-Barr virus types 1 and 2 have nearly identical LMP-1 transforming genes. J. Gen. Virol. 75:2741-2746. [DOI] [PubMed] [Google Scholar]
- 44.Sandvej, K., S. C. Peh, B. S. Anderson, and G. Pallesen. 1994. Identification of potential hot spots in the carboxy-terminal part of the Epstein-Barr virus (EBV) BNLF-1 gene in both malignant and benign EBV-associated diseases: high frequency of a 30 bp deletion in Malaysian and Danish peripheral T-cell lymphomas. Blood 84:4053-4060. [PubMed] [Google Scholar]
- 45.Sugden, B. 1984. Immune deficiency and cancer, p. 165-177. In D. T. Purtilo (ed.), Expression of virus-associated functions in cells transformed in vitro by Epstein-Barr virus: Epstein-Barr virus cell surface antigen and virus-release from transformed cells. Plenum Medical Book Co., New York, N.Y.
- 46.Sugden, B. 1994. Latent infection of B lymphocytes by Epstein-Barr virus. Semin. Virol. 5:197-205. [Google Scholar]
- 47.Takada, K. 1984. Cross-linking of cell surface immunoglobulin induces Epstein-Barr virus in Burkitt lymphoma lines. Int. J. Cancer 33:27-32. [DOI] [PubMed] [Google Scholar]
- 48.Torii, T., K. Konishi, J. Sample, and K. Takada. 1998. The truncated form of the Epstein-Barr virus LMP-1 is dispensable or complementable by the full-length form in virus infection and replication. Virology 251:273-278. [DOI] [PubMed] [Google Scholar]
- 49.Trivedi, P., L. F. Hu, F. Chen, B. Christensson, M. G. Masucci, G. Klein, and G. Winberg. 1994. The Epstein-Barr virus (EBV)-encoded membrane protein LMP1 from a nasopharyngeal carcinoma is nonimmunogenic in a murine model system, in contrast to a B-cell-derived homologue. Eur. J. Cancer 30A:84-88. [DOI] [PubMed] [Google Scholar]
- 50.Walling, D. M., N. Shebib, S. C. Weaver, C. M. Nichols, C. M. Flaitz, and J. Webster-Cyriaque. 1999. The molecular epidemiology and evolution of Epstein-Barr virus: sequence variation and genetic recombination in the latent membrane protein-1 gene. J. Infect. Dis. 179:763-764. [DOI] [PubMed] [Google Scholar]
- 51.Wang, D., D. Liebowitz, and E. Kieff. 1988. The truncated form of the Epstein-Barr virus latent-infection membrane protein expressed in virus replication does not transform rodent fibroblasts. J. Virol. 62:2337-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, D., D. Liebowitz, F. Wang, C. Gregory, A. Rickinson, R. Larson, T. Springer, and E. Kieff. 1988. Epstein-Barr virus latent infection membrane protein alters the human B-lymphocyte phenotype: deletion of the amino terminus abolishes activity. J. Virol. 62:4173-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weigel, R., and G. Miller. 1983. Major EB virus-specific cytoplasmic transcripts in a cellular clone of the HR-1 Burkitt's lymphoma line during latency and after induction of virus replicative cycle by phorbol esters. J. Virol. 125:287-298. [DOI] [PubMed] [Google Scholar]
- 54.Wilson, G., and G. Miller. 1979. Recovery of Epstein-Barr virus from nonproducer neonatal human lymphoid cell transformants. Virology 95:351-358. [DOI] [PubMed] [Google Scholar]