Abstract
17q23 is a frequent site of gene amplification in breast cancer. Several lines of evidence suggest the presence of multiple amplicons on 17q23. To characterize distinct amplicons on 17q23 and localize putative oncogenes, we screened genes and expressed sequence tags (ESTs) in existing physical and radiation hybrid maps for amplification and overexpression in breast cancer cell lines by semiquantitative duplex PCR, semiquantitative duplex RT-PCR, Southern blot, and Northern blot analyses. We identified two distinct amplicons on 17q23, one including TBX2 and another proximal region including RPS6KB1 (PS6K) and MUL. In addition to these previously reported overexpressed genes, we also identified amplification and overexpression of additional uncharacterized genes and ESTs, some of which suggest potential oncogenic activity. In conclusion, we have further defined two distinct regions of gene amplification and overexpression on 17q23 with identification of new potential oncogene candidates. Based on the amplification and overexpression patterns of known and as of yet unrecognized genes on 17q23, it is likely that some of these genes mapping to the discrete amplicons function as oncogenes and contribute to tumor progression in breast cancer cells.
Keywords: 17q23 amplification, TBX2, RPS6KB1, MUL, breast cancer
Introduction
Gene amplification is a common mechanism through which protooncogenes are overexpressed and contribute to tumor progression. Comparative genomic hybridization (CGH) and fluorescent in situ hybridization studies (FISH) have revealed several regions of amplification on chromosome 17q in breast cancer [1,2]. One well-known amplicon on 17q21 is associated with overexpression of the ERBB2 (HER-2/NEU) protooncogene. Other regions of 17q amplifications distal to and excluding the ERBB2 (HER-2/NEU) region have also been reported [3,4]. 17q23 is of particular interest because it is a gene-rich region and amplification of 17q23 has been frequently described in breast cancer. Amplification and overexpression of 17q23 genes (RAD51C, RPS6KB1 (PS6K), NACA, PAT1, SIGMA1B, and TBX2) have been reported in breast cancer [4–6]. Independently, Rouillard et al. [7] recently identified a discrete 228-kb amplified region that contains 200 copies of the TBX2 gene in a primary breast tumor sample by using a novel virtual genome scanning method to detect regions of amplification, deletion, or aberrant methylation. Recent functional studies of TBX2 demonstrate that it acts as a potent immortalizing gene by downregulating p19ARF [8]. Another interesting gene that is amplified and overexpressed on 17q23 is RPS6KB1, which is involved in G1-S progression [9,10]. Thus, independent lines of evidence suggest that 17q23 has several strong functional candidates as amplification targets.
To further characterize and to better define the boundaries of 17q23 amplicon(s) and find other potential candidate oncogenes, we extended the screen of the expressed sequence tags (ESTs) and known genes in physical and radiation hybrid maps on 17q23 for amplification and overexpression in 24 breast cancer cell lines. Thus, in this study we delineated two distinct amplicons on 17q23 including possible targets: MUL, RPS6KB1, and TBX2. Moreover, we identified additional potential targets of gene amplification and overexpression in these amplicons that may serve as oncogenes in the malignant progression of some breast tumors.
Materials and Methods
Cancer Cell Lines and Tumor DNA
All breast cancer cell lines, except SUM and human papilloma virus immortalized nontumorigenic mammary cell lines (HPV), were obtained from American Type Culture Collection (ATCC, Manassas, VA) and grown under recommended conditions (http://www.atcc.org/). SUM and HPV lines were developed and provided by S. P. Ethier at The University of Michigan Comprehensive Cancer Center [11–14]. Information about these cell lines is available at www.cancer.med.umich.edu/breast_cell/clines/clines.html.
Primary breast tumor DNA (T-200) has a well-defined 228-kb amplicon including TBX2 on 17q23 [7] and was used as a positive control for amplification in our experiments (use of anonymous human samples was approved by the University of Michigan Institutional Review Board).
DNA and RNA Isolation
Genomic DNA was isolated from breast cell lines by treatment with 200 µg/ml of proteinase K, 1% SDS, 500 mM Tris, 20 mM EDTA, 10 mM NaCl pH 9.0, and extracted with 1:1 phenol/chloroform using standard methods. Ethanol-precipitated DNA was resuspended in 1xTE (0.5 M EDTA, 1M Tris, pH 8.0). Total RNA was isolated from cell lines grown to 70% confluency after cell lysis using Trizol reagent according to manufacturer's instructions (Gibco BRL, Rockville, MD) and resuspended in nuclease-free water.
Semiquantitative Duplex PCR
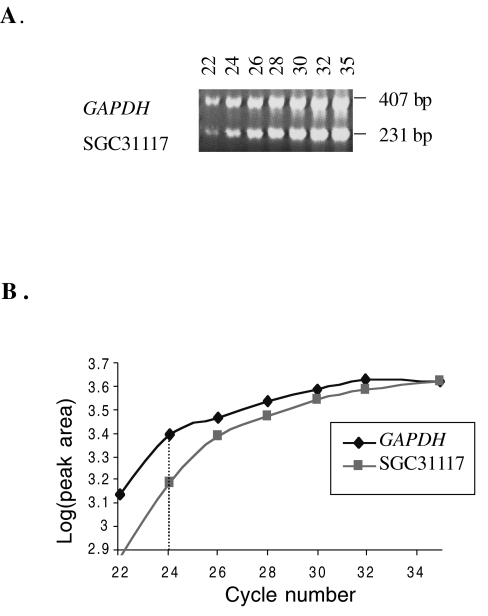
Primers for the ESTs and genes on 17q23 in physical (Human Genome Sequencing, http://www.ncbi.nlm.nih.gov/genome/seq) and radiation hybrid maps (Gene-Map'99, http://www.ncbi.nlm.nih.gov/genemap99) were designed by using the Primer program (Whitehead Institute for Biomedical Research, Version 0.5) (Table 1). Genes and ESTs were coamplified in duplex PCR reactions using titrated primer concentrations of an internal control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and/or thymidine kinase 1 (TK1) to generate equal amounts of PCR products. The exponential phase of amplification was determined for duplexed primer sets by analyzing PCR products after completion of cycles 22, 24, 26, 28, 30, 32, and 35. The products were resolved and analyzed on ethidium bromide-stained 3% agarose gel (Figure 1A). Band intensities were quantified by densitometry (Alpha Innotech IS-1000 Digital Imaging System, version 2.00). The exponential increase range of amplification was determined by plotting the log (peak area) versus PCR cycle number graph (Figure 1B). DNA samples from cell lines and the primary breast tumor sample were coamplified with the duplexed primers using the number of cycles that is in the exponential range of amplification curve. PCR products were quantified by densitometry. The ratios of amplification between gene products and controls (GAPDH and/or TK1) were compared to the ratio calculated for normal human genomic DNA samples.
Table 1.
Designed Primers for ESTs and Genes Mapped to 17q23.
| STS/Gene Name | RH | Size (bp) | Sense Primer | Antisense Primer |
| SGC34835 | GB4, G3 | 148 | AGAGTAGCACAAAAGTCCCAGA | TGGGAGAGATGATGTGCCTG |
| stsG22003 | GB4 | 282 | TTGTTTATTTTCCTCCCAAGC | TGGGGATTTCTTGGTTTGTG |
| RPS6KB1 | G3 | 129 | GTCTTTGCTTCCATTTTGCT | TCTGATTGTGTTGAAGAAGGG |
| SHGC-24306 | GB4, G3 | 221 | ACTAAATGCCCTCAAAGCCC | TCTGTGGGATTTCATCTTAGGG |
| stSG52524 | GB4 | 196 | CTACTCATCCAAGGGAGGTCA | GGTTGTGCTTCCTTACTGACTG |
| TBX2 | GB4 | 188 | GGTGCAGACAGACAGTGCGT | AGGCCAGTAGGTGACCCATG |
| FB10A2 | GB4 | 153 | ATGACAACTCGCACCCCAC | GCCAGAGGGATAGGTGGAGT |
| Zs29c11.r1 | - | 337 | TTGCTTCCATTTTGAGACATT | AGCCAGGTGTAGAGCCCAAG |
| stsG46879 | GB4 | 91 | TGCTGTTGGTACTGGTATTGTC | TCCACTTTAACAAAGCAGCC |
Other primer sequences that are not in this list are from databases (http://www.ncbi.nlm.nih.gov/genemap99). PCR conditions are available on request.
Figure 1.
Semiquantitative duplex PCR optimization of SGC31117 and GAPDH primers. (A) Exponential range of amplification was detected by analyzing PCR products for duplexed primer sets after completion of cycles 22, 24, 26, 28, 30, 32, and 35. The products were resolved on an ethidium bromide-stained 3% agarose gel. Band intensities were quantified by densitometry. (B) Log (peak area) versus cycle number graph reveals the linear range of amplification. For these primers, 24 cycles were chosen for semiquantitative duplex PCR screening of breast cancer cell lines for amplification.
Semiquantitative Duplex RT-PCR
RNA was treated with RNase-free DNase1 (Boehringer Mannheim, Indianapolis, IN) to eliminate DNA contamination. Five micrograms RNA was used to generate cDNA with Oligo(dT) primers using the Superscript Preamplification system (Gibco BRL). Duplexing and quantification was done as described above for the semiquantitative duplex PCR. The ratios of amplification were compared to human papilloma virus immortalized nontumorigenic mammary cell lines (HPV11-21 and/or HPV11-6).
Southern Blot Analysis
Five micrograms of genomic DNA from each cell line was digested to completion with excess restriction endonuclease MspI under standard reaction conditions (Gibco BRL). Digested DNA was separated on 0.9% agarose gels in 1xTBE and transferred to Hybond N+ membranes (Amersham, Piscataway, NJ) in 20xSSC. Membranes were fixed in 0.4 N NaOH and rinsed in 5xSSC. Probes were generated by random priming [15] with [α-32P]dCTP and preblocked with 50 µg human Cot1-DNA at 60°C to mask any repetitive elements. Hybridizations were performed overnight in 0.5 M NaH2PO4, 1 mM EDTA, 7% SDS buffer [16]. After hybridization, blots were washed at 60°C in 2xSSC, 0.1% SDS and exposed to Kodak XAR-5 film to obtain various exposures. Control probes GAPDH (407 bp, F: GGGAGCCAAAAGGGTCATCA, R:TTTCTAGACGGCAGGTCAGGT) or β-actin (340bp, F: GGCACCACACCTTCTACAATG R: CTCCTTAATGTCACGCACGA) were amplified from normal human genomic DNA.
Northern Blot Analysis
Ten micrograms of total RNA sample was fractionated on 1.5% denaturing agarose gels and transferred to HybondxL membranes (Amersham) in 20xSSC. Hybridization was performed as described previously. The same control probes were used as described in Southern blot analysis. Ethidium bromide staining of ribosomal RNA on agarose gels was consistent with GAPDH hybridization patterns as a loading control (data not shown).
Results
To further delineate the 17q23 amplicon(s), we used semiquantitative duplex PCR as an initial screening tool to identify amplifications in 24 breast cancer cell lines and in one previously described 17q23 amplification positive primary breast tumor sample (T-200, see Materials and Methods section). Primers for genes and ESTs that mapped to 17q23 on physical [17] and radiation hybrid maps (http://www.ncbi.nlm.nih.gov/genemap99/) were duplexed with the control primers. The linear range of amplification was detected for each reaction (Figure 1). The 17q23 amplification positive primary breast tumor sample (T-200) and MCF7 cell line have been previously shown to have amplification and overexpression of TBX2 by semiquantitative duplex PCR [7]. In this study, we subsequently detected the same region to be amplified in SUM-52 and MDA-MB361 cell lines, as well. We detected amplification of TBX2 in SUM-52 (5-fold) by semiquantitative duplex PCR and in MDA-MB361 (2-fold) by Southern blot analysis (Figure 2).
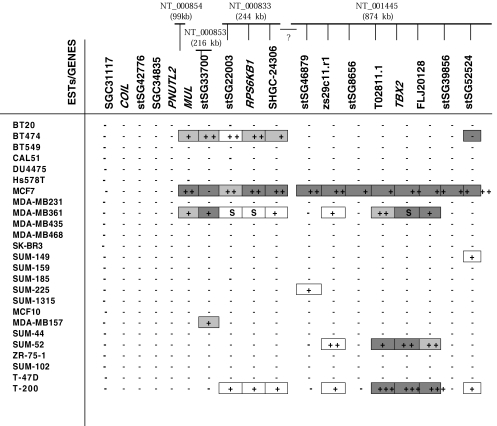
Figure 2.
Amplification and overexpression patterns of ESTs and genes in 24 breast cancer cell lines and one primary breast tumor sample (T-200) (detected by semiquantitative duplex PCR and semiquantitative duplex RT-PCR except the boxes marked as “S” that are detected only by Southern blot analysis). Markers in the databases are listed proximal to distal on 17q23 based on the physical map of Monni et al. [17]. The screened contigs in the databases are shown at the top of corresponding EST/gene names. Plus sign (+) indicates the level of amplification; +:2 to 2.5-fold, + +: 2.5 to 5-fold, + + +: more than 5-fold, S: amplification detected only by Southern analysis. Light gray box ( ) indicates overexpression 1.0 to 2.5 times. Darker gray box (■) indicates overexpression more than 2.5 times. Minus sign (-) indicates no amplification and no overexpression. Gap between the contigs is marked with a question mark.
) indicates overexpression 1.0 to 2.5 times. Darker gray box (■) indicates overexpression more than 2.5 times. Minus sign (-) indicates no amplification and no overexpression. Gap between the contigs is marked with a question mark.
TBX2 maps to contig NT_001445 (http://www.ncbi.nlm.nih.gov/genome/seq/) that contains six overlapping BAC and PAC clones. We screened seven ESTs for amplification and overexpression in this contig. In addition to TBX2, FLJ20128, a protein of unknown function, was also significantly amplified and overexpressed in the primary breast tumor sample (>20-fold and 4.5-fold), in SUM-52 (4-fold, 1.6-fold), in MDA-MB361 (2.5-fold, 4.5-fold), and in MCF7 (3.4-fold, 10-fold) compared to normal human genomic DNA and HPV immortalized nontumorigenic mammary cell line (Figure 3). An EST, T02811.1, was also amplified and overexpressed in T-200 (>20-fold, 5.5-fold), in SUM-52 (2.5-fold, 5.5-fold), MCF7 (2.5-fold, 2.9-fold), and in MDA-MB361 (3.5-fold, 1.2-fold). No conserved domains or homology to known genes for FLJ20128 and T02811.1 was found in comprehensive database searches, ExPASy Proteomics tools (Swiss Institute of Bioinformatics), [18] and RPSBLAST [19].
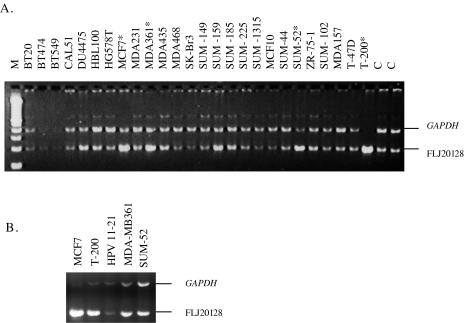
Figure 3.
Amplification and overexpression of a gene (FLJ20128) that encodes a protein of unknown function distal to TBX2. (A) Semiquantitative duplex PCR for FLJ20128. Amplified samples are shown with asterisks. The ratios of amplification compared to the normal human control (C) is MCF7: 3.4, MDA-MB361: 2.5, SUM-52: 4, and T-200: more than 20-folds of amplification (exact ratio was not measured due to high amplification and saturation). (B) Semiquantitative duplex RT-PCR for FLJ20128. The ratios of overexpression compared to HPV11-21 is MCF7: 10, MDA-MB361: 4.5, SUM-52: 1.6, and T-200: 4.5-folds. HPV11-21 is a human papilloma virus immortalized nontumorigenic mammary cell line and is used as a control.
The more proximal (stSG8656) and distal EST markers (stSG39856) that flank the small TBX2 amplicon did not show any amplification in SUM-52 and MDA-MB361. Lack of amplification was also in agreement with semiquantitative RT-PCR results indicating no overexpression for T-200, MDA-MB361, and SUM-52 for those flanking EST markers. Interestingly, however, MCF7 had a region of amplification and overexpression larger than the amplicon detected in MDA-MB361, SUM-52, and the primary breast tumor sample (T-200). The presence of the same amplicon boundaries for SUM-52, MDA-MB361, and the amplification positive primary breast tumor sample (T-200) led us to further evaluate the presence of other discrete amplicons in the region.
Because there are no overlapping contigs with NT_001445 in the databases (http://www.ncbi.nlm.nih.gov/genome/seq/), we screened the genes and ESTs mapping to contigs that are proximal to NT_001445, which includes contigs: NT_000833, NT_000853, and NT_000854. NT_000833 contains RPS6KB1, which is amplified in MCF7 independent of ERBB2 amplicon, in BT474, and in MDA-MB361 cell lines [6]. Our results confirmed this pattern as well as amplification and overexpression of flanking ESTs and MUL (Mulibrey nanism). MUL was 2.5-fold overexpressed in MCF7, 2-fold in BT474, and 1.7-fold in MDA-MB361 as detected by semiquantitative duplex RT-PCR (Figure 2). Northern blot analysis also confirmed the overexpression patterns in these cell lines (Figure 4). The same region was also amplified in the primary breast tumor (T-200) but no overexpression was detected in T-200. Interestingly, BT474 demonstrated 2.9-fold amplification for the EST stSG22003 but no overexpression was detected by semiquantitative duplex RT-PCR, whereas 2.5-fold overexpression was detected for the EST stsG52524, which had no genomic amplification. Despite the lack of genomic amplification in BT474, overexpression of the EST stsG52524 was consistent with the low-level amplification and overexpression in primary breast tumor T-200 (2-fold, 4-fold). Additional proximal ESTs and genes (SGC31117, COIL, stSG42776, SGC34835, and PNUTL2) did not show any amplification by PCR in any of the 24 cell lines tested.
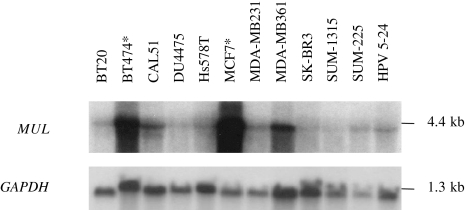
Figure 4.
Overexpression of MUL in MCF7 and BT474 (marked with an asterisk). Low-level overexpression in MDA-MB361 cell line was also in agreement with low-level overexpression detected by semiquantitative duplex RT-PCR. Hybridization of GAPDH probe was used to assess equal loading of samples. Ethidium bromide staining of ribosomal RNA on agarose gels was consistent with both running and loading patterns of the samples assessed by GAPDH hybridization (data not shown).
Discussion
In this study, we screened EST and gene-derived PCR markers and probes mapping to 17q23 contigs, NT_000833 (244179bp), NT_000854 (99411bp), NT_000853 (216981bp), and NT_001445 (874273bp) to determine the oncogenic targets and to refine the boundaries of 17q23 amplicon(s) in breast cancer cells. We identified two distinct amplification regions within 17q23. MCF7 showed a more continuous amplification and overexpression pattern for ESTs and genes in NT_001445 and NT_000833, covering both proximal and distal regions, whereas BT474 had significant amplification and overexpression localized to NT_000833 and SUM-52 localized to NT_001445. Monni et al. [17] reported a continuous region of amplification in BT474 and two distinct regions in MCF7 on 17q23. Although our results also confirm lack of amplification for stSG33700 in MCF7 cells, our data suggest that this EST is, in fact, overexpressed threefold. This supports the growing body of evidence that the degree of amplification and overexpression might not always be linearly related. Usually, a high level of amplification is associated with overexpression but our data suggest that some genes might be grossly overexpressed even though they are not significantly amplified suggesting that other factors rather than increased gene copy number are associated with overexpression. Similarly, 2.5-fold overexpression was detected for the EST stsG52524 in BT474 whereas no significant genomic amplification was detected. According to our data, the critical and significant region of amplification for BT474 is only localized to ESTs and genes mapping to NT_000833 as we did not detect significant amplification or overexpression in ESTs and genes mapping to NT_001445 in BT474 by PCR, Southern, and Northern analyses. It is possible that BT474 has very low amplification localized to NT_001445 as 1.2-fold amplification was detected for TBX2 in BT474 previously [1], but the lack of significant overexpression suggests that the genes in this distal region may not be potent oncogenes in BT474 cells. Thus, our results show that the critical amplicon for BT474 resides in the proximal region whereas MCF7 has amplification and overexpression in both proximal and distal regions. MDAMB361 had a more discontinuous pattern of amplification throughout both of the proximal and distal amplicons and SUM-52 had low-level amplification and overexpression only at the distal region including TBX2. We did not detect amplification in the proximal region in MDA-MB361 cells by PCR alone but Southern blot analysis revealed around two-fold amplification of ESTs tested for this cell line. In spite of the ease of semiquantitative duplex PCR as an initial screening tool, our studies support that Southern blot analysis should be used to confirm PCR results in cases where amplification is no more than 2-to 2.5-fold. In other cases, Southern and Northern blot results were in agreement with PCR results.
The significant region of amplification for the primary breast tumor sample resides only in the distal, several hundred kilobases region in NT_001445 covering a hypothetical protein (FLJ20128), TBX2, and an EST (T02811.1). We confirmed the presence of this distinct 17q23 amplicon in two other breast cancer cell lines. We also detected a more proximal region of amplification in the primary breast tumor sample but the ratio of amplification in the proximal amplicon was around two-fold with no significant overexpression, whereas overexpression was more than five-fold in the distal TBX2 amplification region. This suggests that the genes in the proximal 17q23 region are unlikely candidates for potential oncogenes in this specific primary breast tumor sample.
Identification of the distinct amplicons confirms the importance of the present candidate genes including TBX2, RPS6KB1, and MUL and suggests potential coactivity of different amplification targets. MUL (Mulibrey nanism) is an interesting gene that encodes a protein with a RING-B-box-Coiled coil domain of zinc finger proteins. This group of proteins is involved in the diverse cellular functions such as developmental patterning and oncogenesis as 4% of patients with Mulibrey nanism develop Wilm's tumor [20]. In addition, we report here amplification and overexpression of ESTs that are also potential oncogene candidates. As 17q23 is a gene-rich region and there are some ESTs not yet well-characterized, it will be interesting to further study these ESTs to understand their potential roles in cancer progression. For example, FLJ20128 is a protein of unknown function with no known domains or homology to any genes in the databases and it is significantly amplified and overexpressed in three breast cancer cell lines and the primary breast tumor sample.
It is also possible that there are other small amplicons in the region. We have only screened a small subset of a very gene-rich region. Thus, as more sequence becomes available, the boundaries and the sizes of the amplicons reported here and other possible new small amplicons will be identified more precisely. In addition, there are orientation discrepancies of markers within the contigs between the literature [17] and the databases. This discrepancy is due to lack of sequenced overlapping contigs in the databases. Thus, as the gaps between the 17q23 contigs are sequenced, the discrepancy will be resolved. We present the markers in the order defined in the recent physical map of the region [17]. This discrepancy does not affect the results reported here because only orientation of markers in contigs might be changed (NT_001445), but not the genomic regions of amplification and overexpression.
In summary we have delineated two distinct amplicons on 17q23 in breast cancer cell lines. The presence of two critical amplicons suggests that the known strong candidate genes or as of yet uncharacterized genes in the region might be functioning together in the complex process of tumor progression. Further analysis on primary tumors is also required to reveal the significance of these amplifications and functional analyses will determine the roles and interactions of overexpressed candidate oncogenes in these amplicons on 17q23.
Acknowledgements
We thank L. M. Kalikin and K. A. Myrie for the critical reading of the manuscript and valuable advice and S. P. Ethier for the cell lines.
Footnotes
This work was supported in part by National Cancer Institute grants R01 CA72877 (E.M.P., S.K.D.) and K08 CA66613 (E.M.P.). A. E. E. was supported by the Higher Education Council of Turkey. B. L. N. was supported in part by National Institute of General Medical Sciences grant T32 GM07863. J. M. R. was supported by a grant from the Association pour la Recherche sur le Cancer.
References
- 1.Barlund M, Monni O, Kononen J, Cornelison R, Torhorst J, Sauter G, Kallioniemi O, Kallioniemi A. Multiple genes at 17q23 undergo amplification and overexpression in breast cancer. Cancer Res. 2000;60:5340–5344. [PubMed] [Google Scholar]
- 2.Barlund M, Tirkkonen M, Forozan F, Tanner MM, Kallioniemi O, Kallioniemi A. Increased copy number at 17q22–q24 by CGH in breast cancer is due to high-level amplification of two separate regions. Genes Chromosomes Cancer. 1997;20:372–376. doi: 10.1002/(sici)1098-2264(199712)20:4<372::aid-gcc8>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 3.Doyle GA, Bourdeau-Heller JM, Coulthard S, Meisner LF, Ross J. Amplification in human breast cancer of a gene encoding a c-myc mRNA-binding protein. Cancer Res. 2000;60:2756–2759. [PubMed] [Google Scholar]
- 4.Wu GJ, Sinclair CS, Paape J, Ingle JN, Roche PC, James CD, et al. 17q23 amplifications in breast cancer involve the PAT1, RAD51C, PS6K, and SIGma1B genes. Cancer Res. 2000;60:5371–5375. [PubMed] [Google Scholar]
- 5.Barlund M, Forozan F, Kononen J, Bubendorf L, Chen Y, Bittner ML, Torhorst J, Haas P, Bucher C, Sauter G, Kallioniemi OP, Kallioniemi A. Detecting activation of ribosomal protein S6 kinase by complementary DNA and tissue microarray analysis. J Natl Cancer Inst. 2000;92:1252–1259. doi: 10.1093/jnci/92.15.1252. [DOI] [PubMed] [Google Scholar]
- 6.Couch FJ, Wang XY, Wu GJ, Qian J, Jenkins RB, James CD. Localization of PS6K to chromosomal region 17q23 and determination of its amplification in breast cancer. Cancer Res. 1999;59:1408–1411. [PubMed] [Google Scholar]
- 7.Rouillard JM, Erson AE, Kuick R, Asakawa J, Wimer K, Muleris M, Petty EM, Hanash SM. Virtual genome scan: A tool for restriction landmark based scanning of the human genome. Genome Res. 2001;11:1453–1459. doi: 10.1101/gr.181601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobs JJ, Keblusek P, Robanus-Maandag E, Kristel P, Lingbeek M, Nederlof PM, van Welsem T, van de Vijver MJ, Koh EY, Daley GQ, van Lohuizen M. Senescence bypass screen identifies TBX2, which represses Cdkn2a (p19ARF) and is amplified in a subset of human breast cancers. Nat Genet. 2000;26:291–299. doi: 10.1038/81583. [DOI] [PubMed] [Google Scholar]
- 9.Lane HA, Fernandez A, Lamb NJ, Thomas G. p70s6k function is essential for G1 progression. Nature. 1993;363:170–172. doi: 10.1038/363170a0. [DOI] [PubMed] [Google Scholar]
- 10.Reinhard C, Fernandez A, Lamb NJ, Thomas G. Nuclear localization of p85s6k: Functional requirement for entry into S phase. EMBO J. 1994;13:1557–1565. doi: 10.1002/j.1460-2075.1994.tb06418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ethier SP, Kokeny KE, Ridings JW, Dilts CA. erbB family receptor expression and growth regulation in a newly isolated human breast cancer cell line. Cancer Res. 1996;56:899–907. [PubMed] [Google Scholar]
- 12.Ethier SP, Mahacek ML, Gullick WJ, Frank TS, Weber BL. Differential isolation of normal luminal mammary epithelial cells and breast cancer cells from primary and metastatic sites using selective media. Cancer Res. 1993;53:627–635. [PubMed] [Google Scholar]
- 13.Ignatoski KM, Ethier SP. Constitutive activation of pp125fak in newly isolated human breast cancer cell lines. Breast Cancer Res Treat. 1999;54:173–182. doi: 10.1023/a:1006135331912. [DOI] [PubMed] [Google Scholar]
- 14.Sartor CI, Dziubinski ML, Yu CL, Jove R, Ethier SP. Role of epidermal growth factor receptor and STAT-3 activation in autonomous proliferation of SUM-102PT human breast cancer cells. Cancer Res. 1997;57:978–987. [PubMed] [Google Scholar]
- 15.Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 16.Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monni O, Barlund M, Mousses S, Kononen J, Sauter G, Heiskanen M, Paavola P, Avela K, Chen Y, Bittner ML, Kallioniemi A. Comprehensive copy number and gene expression profiling of the 17q23 amplicon in human breast cancer. Proc Natl Acad Sci USA. 2001;98:5711–5716. doi: 10.1073/pnas.091582298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Appel RD, Bairoch A, Hochstrasser DF. A new generation of information retrieval tools for biologists: The example of the ExPASy WWW server. Trends Biochem Sci. 1994;19:258–260. doi: 10.1016/0968-0004(94)90153-8. [DOI] [PubMed] [Google Scholar]
- 19.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zheng Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avela K, Lipsanen-Nyman M, Idanheimo N, Seemanova E, Rosengren S, Makela TP, Perheentupa J, Chapelle Ad, Lehesjoki AE. Gene encoding a new RING-B-box-Coiled-coil protein is mutated in Mulibrey nanism. Nat Genet. 2000;25:298–301. doi: 10.1038/77053. [DOI] [PubMed] [Google Scholar]