Abstract
Epstein-Barr virus (EBV) nuclear antigen 3C (EBNA3C) is a known regulatory transcription factor that has been shown to interact with histone deacetylase 1 (HDAC1) when cotransfected in human cell lines and by in vitro binding experiments. Previous studies have shown that EBNA3C interacts with p300 and prothymosin alpha (ProTα) in EBV-infected cells and may be involved in recruiting acetyltransferases to the chromatin for acetylation of histones and transcriptional activation. EBNA3C has also been shown to function as a repressor of transcription when directed to promoters. In this report, we show that EBNA3C complexed with ProTα can also recruit deacetylase activity and associates in a complex that includes HDAC1 and HDAC2 in human B cells. A complex of EBNA3C and ProTα coimmunoprecipitated with HDAC1 and HDAC2 in cell lines stably expressing EBNA3C. Additionally, this complex associated with the mSin3A and NCoR corepressors in EBNA3C-expressing cell lines and may function in a complex with additional transcription factors known to be repressors of transcription. EBNA3C in complex with ProTα recruited deacetylase activity in cell lines stably expressing EBNA3C, and this activity was shown to be partially sensitive to trichostatin A (TSA). This suggests an association with other deacetylases that are insensitive to the general inhibitory effects of TSA, as the entire activity was not abolished in multiple assays. The association between EBNA3C and the corepressors as well as HDACs is likely to depend on the presence of ProTα in the complex. Immunoprecipitation with anti-ProTα antibody immunoprecipitated EBNA3C and the other repressors, whereas immunoprecipitation with anti-EBNA3C antibody resulted in little or no association with these molecules associated with transcription repression. Clearly, EBNA3C functions as a component of a number of dynamic complexes which function in repression and activation of transcription.
Regulators of cellular pathways are common targets usurped by specific proteins encoded by DNA tumor viruses (12, 18, 20, 25). Epstein-Barr virus (EBV) is a known human DNA tumor virus which targets B lymphocytes and epithelial cells and is tightly associated with a number of human cancers (19, 20, 25, 33). The initial discovery of EBV was linked to its association with Burkitt's lymphoma in the early 1960s, and the intense studies which followed led to identification of the viral genes expressed during latent infection and those that are essential for EBV-mediated transformation of primary B lymphocytes (6, 25). Of the EBV nuclear antigen 3 (EBNA3) family of proteins, EBNA3C was shown to be critical for the immortalization process and is expressed from the major latent Cp promoter, located approximately 110 kbp upstream of the open reading frame (31, 37).
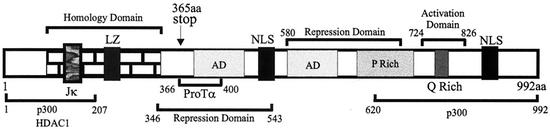
EBNA3C is an EBV-encoded transcription regulatory factor 992 amino acids in size, based on the sequence, and is localized in the nucleus, as shown with its punctate signals in immunofluorescence assays (13, 31). It is associated with a number of cellular factors involved in transcription regulation (Fig. 1) (9). Specific domains of the protein are involved in activation as well as repression of transcription and are capable of activating or repressing transcription when fused to the GAL4 DNA binding domain, which targets the fusion protein to GAL4-responsive elements (5, 35). The activation domain is rich in glutamines and prolines and is similar to the c-Jun/c-Fos family of transcription factors (5, 21). Other studies have shown that EBNA3C associates with the retinoblastoma protein in vitro, although this has not been demonstrated in EBV-infected cells or when cotransfected with EBNA3C (3). Additional studies have shown that EBNA3C also associates with the transcriptional repressor RBP-Jκ, also targeted by EBNA2, the known EBV activator of transcription (21, 27, 28). EBNA2 activates transcription of the major EBV latent promoters through its interaction with RBP-Jκ, one of the critical components derepressed at these major latent promoters (14, 15, 41).
FIG. 1.
Scheme showing the EBNA3C protein and the various domains. The homology domain is the region of highest homology to the other EBNA3 family members (26). The repression domains and activation domains are indicated. Regions that bind p300 at the amino terminus and carboxy terminus of EBNA3C are shown, and the interaction domains for ProTα and HDAC1 are shown within the amino-terminal 400 amino acids (aa) (9, 24). The RBP-Jκ binding site has also been mapped within the homology domain (27). LZ, leucine zipper; NLS, nuclear localization signal; AD, acidic domain.
Other factors known to associate with EBNA3C include the acetyltransferase p300, the DEAD box protein, the nuclear protein prothymosin alpha (ProTα), and the suppressor of metastasis Nm23-H1 (9, 35, 40). These interacting molecules are all involved in transcriptional regulation as activators or repressors of transcription and have been shown to have a direct effect on regulating the activity of EBNA3C on the major EBV latent promoters. Moreover, EBNA3C modulates the acetyltransferase activity of p300 when cotransfected in cells as well as in EBV-transformed B lymphocytes (9). These studies demonstrate that EBNA3C is also associated with coactivator complexes and that these activities are regulated through association with other factors that interact with the glutamine-rich domain known to be involved in transcriptional activation.
EBNA3C binds histone deacetylase 1 (HDAC1) in vitro in a region of the amino-terminal 200 amino acids known to associate with the transcription repressor RBP-Jκ (24). This interaction suggests that the association of EBNA3C with RBP-Jκ may also include complexing with HDACs and other molecules containing deacetylase activities in EBV-transformed cells.
In this report we show that EBNA3C complexed with ProTα associates with HDAC1 and HDAC2 and the corepressors mSin3A and NCoR. These complexes recruited histone deacetylase activity in EBNA3C-expressing cell lines. Additionally, the majority of the deacetylase activity was associated with EBNA3C and ProTα. However, the activity seen here may not be due only to the association with HDAC1, as EBNA3C and ProTα may be associated with other deacetylases yet to be characterized which are sensitive to trichostatin A (TSA) inhibition. The deacetylase activity was coimmunoprecipitated with complexes associated with ProTα. Moreover, in the presence of EBNA3C, this deacetylase activity immunoprecipitated by antibodies to ProTα was enhanced by at least 50% over the initial activity. Therefore, EBNA3C can influence the cellular deacetylase activity through interactions with ProTα in association with deacetylases and corepressors.
MATERIALS AND METHODS
Antibodies and cell lines.
EBNA3C rabbit polyclonal antibody and RBP-Jκ antibodies were provided by Elliott Kieff, Harvard Medical School, Boston, Mass. The A10 monoclonal antibody that recognizes EBNA3C was a gift from Martin Rowe (22). HDAC1, HDAC2, mSin3A, and NCoR antibodies were purchased from Santa Cruz Biotechnology, Inc.
BJAB cells are EBV-negative B cells isolated from a Burkitt's lymphoma patient and were provided by Elliott Kieff. Human embryonic kidney fibroblast 293 and 293T cells, transformed with E1A and T antigens, respectively, were obtained from Jon Aster, Brigham and Women's Hospital, Boston, Mass. Lymphoblastoid cell lines (LCLs) are EBV-transformed primary B lymphocytes infected with recombinant P3HR-1 rescued for EBNA2 and EBNALP (1, 8).
All B-cell lines were grown in RPMI 1640 containing 10% fetal bovine serum supplemented with 10 mM glutamine, 25 U of penicillin per ml, and 25 μg of streptomycin per ml (Invitrogen-Gibco, Gaithersburg, Md.). Adherent cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum supplemented with streptomycin and penicillin as above. All cell lines were grown at 37°C with 5% CO2 and passaged every 3 to 4 days.
Transfections.
BJAB and 293 cells were transfected by electroporation with a Bio-Rad Gene Pulser II electroporator; 10 million cells harvested in exponential phase were collected and washed in phosphate-buffered saline and then resuspended in 400 μl of RPMI with DNA for transfection. Resuspended cells were transferred to a 0.4-cm cuvette and electroporated at 975 μF and 220 V. The electroporated cells were then transferred to 10 ml of complete medium, followed by incubation at 37°C and 5% CO2. Transfections were harvested after 24 h and assayed for activity.
Deacetylase assays.
293 cells were transfected with CsCl-purified DNA with the appropriate constructs as described above. Cells were transfected with equivalent amounts of DNA for all transfections by normalizing with vector DNA. Transfection efficiencies were determined by cotransfecting green fluorescent protein (GFP) expression vector in each transfection and counting GFP-positive cells. Deacetylase assays were based on instructions obtained from the assay kit, purchased from Upstate Biotechnology Inc., and performed essentially as per the manufacturer's instructions. Briefly, transfected cells were incubated for 24 h posttransfection, harvested, and lysed in radioimmunoprecipitation assay (RIPA) buffer (0.5% NP-40, 10 mM Tris [pH 7.5], 2 mM EDTA, 150 mM NaCl, supplemented with protease inhibitors). Cell debris was removed by centrifugation, and the soluble lysate was transferred to a fresh tube. Rabbit polyclonal antibody against ProTα was used to immunoprecipitate complexes of ProTα and deacetylases for assays.
Briefly, bound complexes were incubated with tritium-labeled acetylated peptides corresponding to bovine histone H4 (amino acids 1 to 24). The reaction mix was incubated at 37°C for 2 h with gentle agitation. The reaction was stopped by addition of incubation buffer containing 50 mM Tris, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, and 2 mM phenylmethylsulfonyl fluoride supplemented with 1 M HCl and 0.16 M acetic acid. To extract free acetate, 500 μl of ethyl acetate was added; the sample was vortexed, and the organic phase was separated from the aqueous phase by centrifugation. The organic phase was removed and added to 2 ml of scintillation cocktail, the sample was vortexed, and the amount of tritium released was determined by liquid scintillation counting.
GST binding assays, Western blots, and immunofluorescence assays.
Glutathione S-transferase (GST) fusion proteins were purified from bulk Escherichia coli cultures following induction with isopropylthiogalactopyranoside (IPTG). Cells were lysed by sonication in NETN buffer (0.5% NP-40, 20 mM Tris [pH 8.0], 1 mM EDTA, and 100 mM NaCl) supplemented with Sarkosyl and dithiothreitol. Cell debris was removed by centrifugation, and Sarkosyl was neutralized with Triton X-100. GST fusion proteins were then purified from the supernatants by incubation with glutathione-Sepharose beads. Protein concentrations were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with subsequent Coomassie brilliant blue staining.
For GST pulldown assays, 5 × 107 cells were collected and washed in phosphate-buffered saline. Cells were lysed in RIPA buffer with 1% NP-40 and then cleared by centrifugation and transferred to a cold, sterile microcentrifuge tube. GST protein was added to the lysates for 30 min to preclear them, and then the GST fusion protein was added for 3 h with rotation at 4°C. Bound proteins were collected by centrifugation and then washed four times in RIPA buffer with protease inhibitors. Protein lysates were solubilized in SDS-lysis buffer and fractionated on SDS-polyacrylamide gels. Fractionated proteins were then transferred to 0.4-μm-pore nitrocellulose membranes, blocked in 5% nonfat milk, and incubated in primary antibody overnight at 4°C with moderate rocking. Primary antibodies were used typically at a dilution of 1:100 in phosphate-buffered saline. Protein-antibody complexes were detected with secondary antibody against mouse, rabbit, or goat immunoglobulin or protein A conjugated to horseradish peroxidase at the dilutions recommended by the manufacturer (Amersham Inc.).
Immunofluorescence analyses were performed essentially as described previously. Briefly, fixed cells were blocked in the appropriate serum and then incubated with the specific primary antibody for HDAC1, HDAC2, mSin3A, and NCoR (Santa Cruz Inc.) diluted at 1:200 or mouse anti-EBNA3C ascites diluted at 1:1,000 in phosphate-buffered saline for 1 h. Cells were washed and then further incubated with the appropriate secondary antibody conjugated to fluorescein isothiocyanate or Texas Red at 1:1,000 dilutions in phosphate-buffered saline for 1 h. Slides were washed and visualized with an Olympus XI70 inverted fluorescence microscope (Olympus Inc.) with an attached digital PixelFly camera and software (Cooke, Inc.).
RESULTS
EBNA3C enhances the deacetylase activity associated with ProTα when coexpressed in transient-transfection assays.
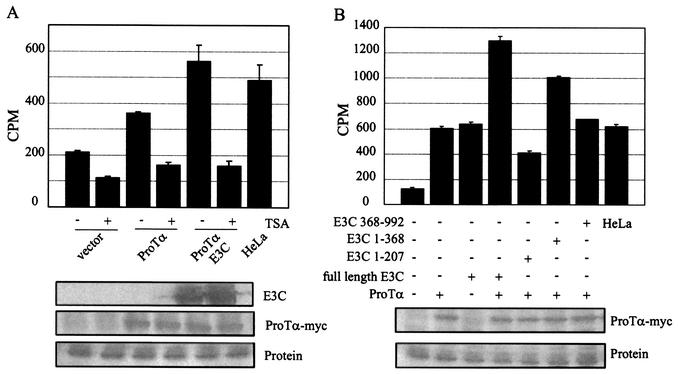
Previous studies indicated that EBNA3C is associated with deacetylase activity in transient assays (24). To investigate this further, we decided to determine if EBNA3C and ProTα were associated with deacetylase activity by evaluating the level of deacetylation when ProTα was immunoprecipitated alone or with EBNA3C. Therefore, we transfected a ProTα-expressing vector either alone or with EBNA3C-expressing constructs in 293 cells. Our results show that in the presence of EBNA3C, the level of deacetylation associated with ProTα increased to three times over that with the vector alone. This was similar to the levels seen with deacetylase activity from the HeLa control extract provided by Upstate Biotechnologies Inc., which was used as a positive control. Note that these control extracts were only used to determine activity and to validate our assay (Fig. 2A).
FIG. 2.
EBNA3C and ProTα recruit deacetylase activity in transient-transfection assays. (A) 293T cells were transfected with constructs containing ProTα or ProTα and EBNA3C with vector alone as the DNA control. Harvested lysates were immunoprecipitated with antibody against ProTα and assayed for deacetylase activity in the presence and absence of inhibitory concentrations of TSA. A positive-control HeLa lysate for deacetylase activity was provided by Upstate Biotechnology Inc. as part of the assay kit and was used to validate our results for deacetylase activity (panels A and B, right sample). ProTα immunoprecipitated deacetylase activity in this assay and was enhanced by the coexpression of EBNA3C (E3C). (B) Truncated regions of EBNA3C coexpressed with ProTα showed that the amino-terminal 368 amino acids enhanced deacetylase activity almost to the extent of wild-type EBNA3C. The carboxy terminus of EBNA3C showed increased deacetylase activity but not to the extent seen with the amino-terminal region when compared to the positive control used in our assays. Protein lysates were analyzed by Western blot for levels of expression of the transfected protein with the Myc monoclonal antibody to detect ProTα and A10 monoclonal antibody for detection of the EBNA3C protein. Protein lysate was used as a loading control and was stained with Ponceau S.
The level of deacetylase activity was about twofold over that with the vector alone when ProTα was expressed from the heterologous cytomegalovirus immediate-early promoter (Fig. 2A). The ProTα expression vector was monitored by Western blotting for the Myc epitope fused at the carboxy terminus. Western blots with the 9E10 Myc monoclonal antibody showed that similar levels of Myc-tagged ProTα protein were expressed in all transfected cells. EBNA3C levels were also confirmed with the A10 monoclonal antibody, which is reactive to EBNA3C (22).
In an effort to determine the effects of specific EBNA3C domains on the recruitment of deacetylase activity, we compared the domain shown to be involved in binding HDAC1, located within the amino-terminal 207-amino-acid region, with the amino-terminal region that includes the leucine zipper and the entire homology region for the EBNA3 family of proteins (26, 30, 31). In this deacetylase assay, we showed that ProTα and full-length EBNA3C recruited equivalent amounts of the deacetylase activity and that together the activity was doubled (Fig. 2B). When the amino-terminal 207 amino acids of EBNA3C were expressed along with ProTα, a limited amount of activity, approximately 60% of that seen with either ProTα or EBNA3C alone, was seen. However, when the region expressing the amino-terminal 368 amino acids was introduced in the system, the activity obtained was almost equivalent to that with the combination of ProTα and full-length EBNA3C (Fig. 2B). Assays evaluating the carboxy-terminal 630 amino acids also showed deacetylase activity but less than that seen with the 368-amino-acid amino-terminal region. Based on these results, we suggest that the amino-terminal and carboxy-terminal regions of EBNA3C may both contribute to the recruitment of deacetylase activity. This may include recruitment and binding to deacetylases HDAC1 and HDAC2 as well as other corepressors known to be associated with deacetylase activity.
HDAC1, HDAC2, mSin3A, and NCoR associate with EBNA3C and ProTα in EBNA3C-expressing Burkitt's lymphoma cell lines.
Previous studies showed that EBNA3C binds to HDAC1 in vitro and that the region of EBNA3C bound to HDAC1 is similar to that seen with RBP-Jκ, a known cellular transcription repressor associated with EBNA3C (24). However, there has been no direct evidence showing that HDAC1 associates with EBNA3C in EBNA3C-expressing cells or in EBV-transformed LCLs. Therefore, we wanted to determine if the association with the cellular deacetylases requires other factors that may function as stabilizers for active complexes which contain these deacetylases. Based on our previous data, we hypothesized that ProTα may be a critical component of this complex, as was previously demonstrated for acetylase complexes containing EBNA3C (9).
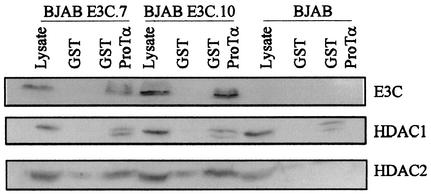
We utilized the GST-ProTα fusion protein to pull down complexes containing human deacetylases 1 and 2, existing as subunits of larger corepressor complexes containing EBNA3C. In cells stably expressing EBNA3C, we showed that EBNA3C as well as HDAC1 and HDAC2 were brought down in complex with the GST-ProTα fusion protein. Lysates from two isogenic EBNA3C-expressing BJAB cell lines were harvested from 50 million cells, lysed in RIPA buffer, and incubated with GST-ProTα bound to beads. Bound complexes were washed and solubilized in lysis buffer and then fractionated on SDS-PAGE gels. The results of Western blots for HDAC1 and HDAC2 indicated that EBNA3C and ProTα associated with both these deacetylases in EBNA3C-expressing cell lines. Interestingly, HDAC1 showed similar levels of association on visual inspection with GST-ProTα in EBNA3C and control cell lines. However, HDAC2 had little or no signal with ProTα in BJAB control cells but was clearly visible above background levels of association in BJAB cell lines stably expressing EBNA3C (Fig. 3, compare lanes 3, 6, and 9).
FIG. 3.
GST pulldown assays show that HDAC1 and HDAC2 associate with ProTα and EBNA3C in EBNA3C-expressing cell lines. BJAB E3C.7 and E3C.10 are cell lines that stably express EBNA3C under the control of a simian virus 40 promoter. GST protein was used as a control, and GST-ProTα fusion protein was used in the experimental lanes. All binding assays were done at 4°C for 3 h and then washed four times. Bound proteins were then solubilized and fractionated on SDS-8% PAGE. Proteins were transferred to nitrocellulose and blotted for EBNA3C with monoclonal antibody A10. The blot was then stripped and reprobed consecutively with specific antibodies against HDAC1 and HDAC2 (Santa Cruz Inc.).
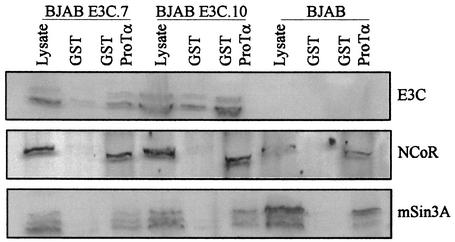
Complexes involved in repression of gene expression include known corepressor molecules such as NCoR and mSin3A. We wanted to determine if these complexes containing HDAC1 and HDAC2 may also contain these large corepressor proteins. In pulldown assays performed with the GST fusion of ProTα, we showed that mSin3A as well as NCoR associated with EBNA3C and ProTα in EBNA3C-expressing cell lines (Fig. 4). Additionally, we showed that these proteins also associated with ProTα in the absence of EBNA3C, suggesting that ProTα may be recruiting these large molecules to complexes and acting as a stabilizing component of these large transcriptional megacomplexes (Fig. 4). NCoR as well as mSin3A precipitated with the ProTα fusion protein, as seen for EBNA3C, suggesting that these molecules may exist in a complex that includes EBNA3C in EBNA3C-expressing cells. A faint background band in the GST control lanes was seen with this EBNA3C Western blot but was not typically seen in subsequent blots. In contrast to HDAC2, NCoR and mSin3A were pulled down with ProTα in the presence and in the absence of EBNA3C in the cell lines analyzed (Fig. 4).
FIG. 4.
GST pulldown assays show that corepressors NCoR and mSin3A associate with ProTα and EBNA3C in EBNA3C-expressing cell lines. BJAB E3C.7 and E3C.10 are cell lines that stably express EBNA3C under the control of a simian virus 40 promoter. As in Fig. 3, cells expressing EBNA3C were lysed and proteins were incubated with the GST-ProTα fusion. Bound proteins were solubilized and run on SDS-6% PAGE. The proteins were transferred to nitrocellulose and probed with antibodies against NCoR and mSin3A (Santa Cruz, Inc.).
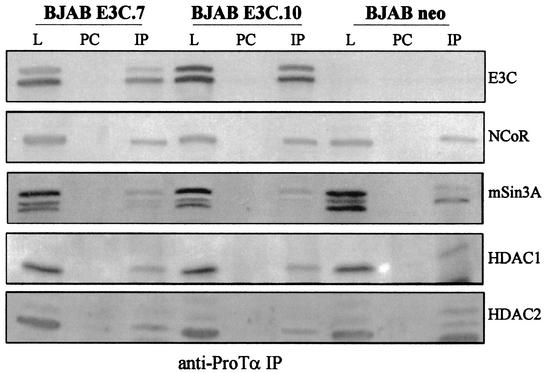
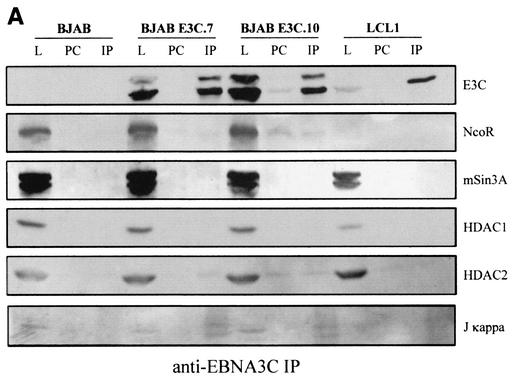
We next wanted to determine if these complexes seen with the GST fusion protein with ProTα were also seen with antibodies against ProTα. We used polyclonal rabbit antibody against ProTα and performed immunoprecipitation assays with Western blot analysis to determine if these complexes were stable to immunoprecipitation as well as GST pulldown. In these assays, we showed again that HDAC1 and HDAC2 coimmunoprecipitated with ProTα and EBNA3C (Fig. 5). The blots were stripped and reprobed each time with a different antibody to the HDAC and corepressor molecules. The data from these Western blots show that NCoR and mSin3A coimmunoprecipitated in EBNA3C-expressing cell lines as well as in the control cell line BJAB containing the vector alone (Fig. 5). Similar levels of proteins were seen in the immunoprecipitation lane for the EBNA3C-expressing cell lines and the negative control (Fig. 5).
FIG. 5.
Corepressors NCoR and mSin3A and HDAC1 and HDAC2 associate with EBNA3C complexed with ProTα in cell lines stably expressing EBNA3C. Immunoprecipitation analysis with ProTα rabbit antibody showed that the HDACs and corepressors were associated with ProTα and EBNA3C in EBNA3C-expressing cell lines. The blot was stripped and reprobed with specific antibodies to the antigens indicated. L, lysate; PC, preclear; IP, immunoprecipitate.
Complexes associated with EBNA3C and ProTα recruit histone deacetylase activity in B-cell lines stably expressing the EBNA3C protein.
Studies utilizing a transient system indicate that EBNA3C is associated with deacetylase activity and that it can associate with human HDAC1 when overexpressed by cotransfection (24). We wanted to determine if this activity may be associated with ProTα and EBNA3C, as EBNA3C has been shown to have both activating and repressive functions. ProTα was previously shown to associate with the acetyltransferase p300 and with EBNA3C (9). Therefore, we wanted to determine if ProTα may also be associated with deacetylases associated with transcription repression potentially regulated by EBNA3C.
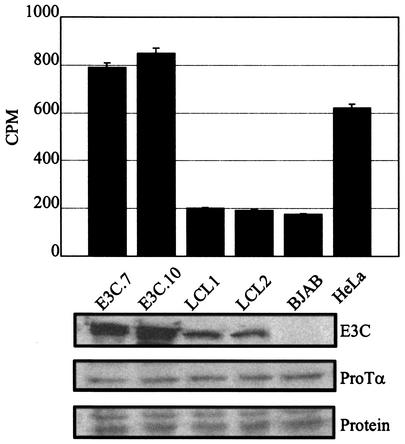
In this study, we coimmunoprecipitated complexes with ProTα antibody and tested for deacetylase activity. Western blotting with EBNA3C-specific monoclonal antibody A10 was used to determine the expression levels for EBNA3C in two cell lines stably expressing EBNA3C, E3C.7 and E3C.10, and in two recently transformed LCLs, LCL1 and LCL2. BJAB was used as a negative control. ProTα levels were monitored by Western blot analysis with the ProTα antibody. HeLa cell extract from the manufacturer was used as a positive control for deacetylase activity and was provided by Upstate Biotechnology Inc.
The results of these experiments showed that the cell lines expressing greater amounts of EBNA3C resulted in a greater association with deacetylase activity compared to LCLs expressing a lower amount of EBNA3C and other EBNA proteins, including EBNA2 and EBNALP (2, 25, 39) (Fig. 6). In fact, the amount of deacetylase activity was greater than that seen with the extract supplied from HeLa cells used as a positive control. The amount of ProTα immunoprecipitated was similar in each case, as demonstrated by Western blot and the protein loading control (Fig. 6, compare middle and lower panels).
FIG. 6.
EBNA3C and ProTα recruit deacetylase activity in cell lines stably expressing EBNA3C. HeLa cell extract was used as a control for deacetylase activity and was based on the deacetylase kit from Upstate Biotechnology Inc. LCLs showed deacetylase activity equivalent to that seen with the EBV-negative BJAB cell control. Immunoprecipitation of ProTα with rabbit polyclonal antibody to ProTα was followed by incubation with 14C-labeled acetylated peptides. The labeled molecules removed by deacetylation were counted in a scintillation counter and plotted. All experiments were done in triplicate, and the means of the experiments are shown. Western blots for detection of EBNA3C (E3C) and ProTα are shown below, as is a Ponceau S-stained gel showing the protein loading control.
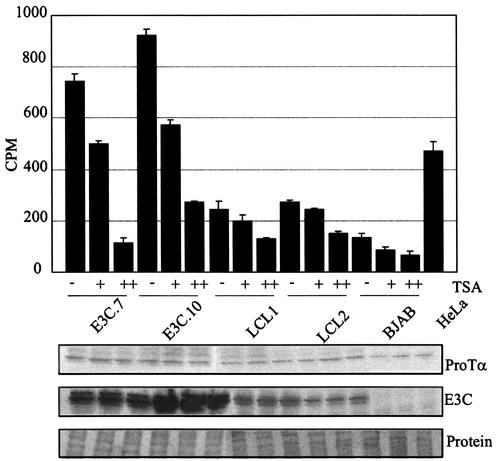
EBNA3C-associated deacetylase activity in stable BJAB cell lines is sensitive to TSA.
The above results suggest that EBNA3C recruits deacetylase activity when complexed with ProTα and that this activity is enhanced as EBNA3C levels increase in the cell. The level of deacetylase activity in LCLs are similar to that in the EBV-negative control. This may be a consequence of the expression of other EBNA proteins in EBV-transformed LCLs or simply indicate that a smaller amount of EBNA3C is present in LCLs. In order to determine if the deacetylase activity was inhibited by a general deacetylase inhibitor, we used TSA at concentrations of 10 mM and 50 mM. In this assay, we showed that TSA inhibited the deacetylase activity of EBNA3C-expressing cells; 10 mM TSA reduced the activity approximately 30%, and 50 mM reduced the levels to 75 to 90% of the initial amounts in EBNA3C-overexpressing lines (Fig. 7). The fact that the deacetylase activity associated with EBNA3C can be efficiently inhibited by TSA at low concentrations without affecting cell viability indicates that EBNA3C and ProTα may exist in repressor complexes with multiple partners to mediate repression of specific cellular and viral promoters. Based on these data, we decided to look for possible association of corepressors in the complex with EBNA3C in cell lines stably expressing EBNA3C as well as in EBV-transformed LCLs.
FIG. 7.
Deacetylation activity associated with EBNA3C and ProTα is inhibited by TSA. Deacetylation assays were performed as described for Fig. 6, and samples were incubated with increasing concentrations of TSA. Western blots show levels of ProTα and EBNA3C (E3C) as well as the protein loading control.
Association of complexes of HDACs and corepressors with EBNA3C is limited in EBV-transformed LCLs.
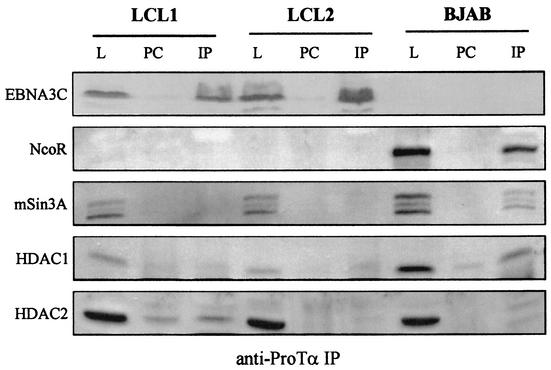
The association of EBNA3C with corepressors and HDACs in EBNA3C-expressing cell lines suggests that these molecules may function in regulating cellular and viral promoters in EBV-immortalized cells. It was previously shown that RBP-Jκ, a known cellular repressor, interacts in vitro with EBNA3C (28) and that HDAC1 binds to a similar region at the amino terminus of EBNA3C adjacent to the RBP-Jκ binding site, as determined by a series of in vitro binding studies (24). Here we found that, in contrast to cell lines stably expressing large amounts of EBNA3C, ProTα had little or no association with HDAC1, HDAC2, and the corepressors mSin3A and NCoR in EBV-transformed LCLs. Immunoprecipitation experiments from two independent transformed cell lines showed that there was no detectable signal above the background for the preimmune control lane with rabbit polyclonal ProTα antibody (Fig. 8). These results suggest that other competing molecules may sequester EBNA3C and ProTα away from these repressor complexes or that the limited number of molecules available for targeting deacetylases in LCLs is below the detection limit of our immunoprecipitation analysis. Other EBV nuclear antigens may also play a role in disrupting these complexes while functioning as regulators of gene expression from viral and cellular promoters required for maintenance of the transformed state. Additionally, it is possible that this specific antibody used to immunoprecipitate the ProTα complex may also destabilize some components of the complexes.
FIG. 8.
Immunoprecipitation analysis with anti-ProTα antibody in lysates from LCLs showed that there was little or no detectable association with HDAC1 and HDAC2 as well as mSin3A and NCoR. No NCoR signal was present in the LCLs even after prolonged exposure to film. Some HDAC1 and HDAC2 were detected, but the levels were barely above the background levels of control preimmune serum. The signal in the immune experimental lane was lower that that seen in the EBNA3C-expressing cell lines. L, lysate; PC, preclear; IP, immunoprecipitate.
One interesting observation made in these experiments was that in EBV-transformed LCLs, no NCoR signal was detected by Western blot repeated in multiple assays (Fig. 8). However, the signal was clearly seen in the lysate lane in EBV-negative BJAB cells. The signal was not seen in the lysate of LCL1 and LCL2 cells even after longer exposures (data not shown). These studies suggest that the expression of NCoR may be downregulated in primary B lymphocytes either directly or indirectly by an EBV-expressed latent antigen. However, it is also possible that NCoR is not expressed in primary B lymphocytes. This needs to be explored further, as the levels of NCoR in uninfected primary B cells have to be compared to those seen in LCLs and other Burkitt's lymphoma cell lines.
To determine if complexes might be differentially detected when immunoprecipitating with antibody reactive to EBNA3C rather than ProTα, we chose to use a rabbit polyclonal antibody specific for EBNA3C, which recognizes an epitope in the carboxy terminus. In these experiments, we again showed that little or no association detectable above background was seen (Fig. 9A). However, we did show that RBP-Jκ could be coimmunoprecipitated with the same antibody against EBNA3C in EBNA3C-expressing cell lines and LCLs (Fig. 9A). No RBP-Jκ signal was seen in the EBNA3C-negative BJAB cells but was seen in the EBNA3C-expressing cell lines and EBV-transformed LCL1 (Fig. 9A).
FIG. 9.
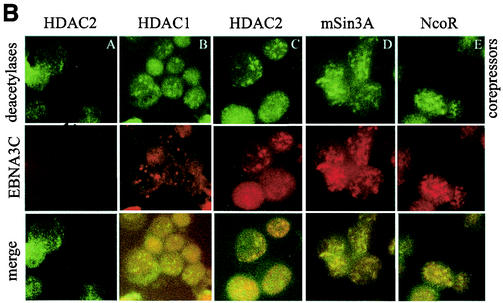
Immunoprecipitation analysis with rabbit anti-EBNA3C antibody showed that HDAC1, HDAC2, mSin3A, and NCoR were not directly immunoprecipitated with EBNA3C. However, RBP-Jκ was immunoprecipitated with anti-EBNA3C, as expected in EBNA3C-expressing lines and LCL1 (panel A). Immunofluorescence analysis showed that EBNA3C (E3C) was localized to the same nuclear compartment as HDAC1, HDAC2, mSin3A, and NCoR (panel B, A to E) in EBNA3C-expressing cells. L, lysate; PC, preclear; IP, immunoprecipitate.
EBNA3C may be localized in compartments similar to those of these cellular molecules. However, the association may be temporally regulated, based on the specific phase of the cell cycle. Therefore, we performed immunofluorescence analysis with HDAC1, HDAC2, mSin3A, and NCoR to determine if EBNA3C was in the same nuclear compartment. EBNA3C-expressing cells were fixed and probed with antibodies for the HDACs, mSin3A, and NCoR, followed by incubation with EBNA3C monoclonal antibody A10. Fluorescein isothiocyanate and Texas Red conjugated to the appropriate secondary antibody were used for detection. The results of these studies showed that EBNA3C was localized in the same nuclear compartments in B cells as HDAC1, HDAC2, mSin3A, and NCoR molecules (Fig. 9B).
DISCUSSION
The association of EBNA3C with deacetylases and corepressors has not been previously shown in cell lines or EBV-transformed LCLs. EBNA3C is known to have both activation and repression functions, and specific domains have been mapped for the specific functions of transcription repression and activation (21, 24, 26). We have shown that EBNA3C binds to the p300 coactivator and is associated with histone acetyltransferase activity in EBV-transformed LCLs (9). These studies indicate that ProTα may function as a displacement factor for histone H1, similar to the HMG1 factor that allows access to the promoter regions (7, 10, 11, 16, 23). Here we show that EBNA3C associated with HDAC1 and HDAC2 and also the corepressors mSin3A and NCoR in EBNA3C-expressing cells but had little association in EBV-transformed LCLs, as determined by our immunoprecipitation analyses.
Previous in vitro studies showed that HDAC1 interacts with EBNA3C at the amino-terminal region and that the deacetylase activity is associated with its amino-terminal region (24). However, to date no association has been demonstrated in EBV-transformed LCLs or in EBNA3C-expressing cells. In this report we show that HDAC1 and HDAC2 associate with EBNA3C in a complex with ProTα and that this is important for the in vivo association of EBNA3C and HDACs as well as other corepressor molecules, including mSin3A and NCoR. One interesting observation in our studies was that little association was seen when we immunoprecipitated directly with antibodies to the EBNA3C carboxy terminus. However, we were able to show association when we immunoprecipitated with anti-ProTα antibodies. These results suggest that ProTα is required to hold the complex together and that ProTα may be functioning to stabilize complexes with EBNA3C, the deacetylases HDAC1 and HDAC2, and the corepressors mSin3A and NCoR. Additionally, there is likely to be a level of regulation that is dependent on the posttranslational modification of ProTα that is yet to be determined in EBV-infected cells and is expected to be important for the association with numerous other cellular factors.
Deacetylase assays to determine the regions of EBNA3C involved in recruiting deacetylase activity showed that the domains at both the amino and carboxy terminus contain activity with approximately equivalent potency. This indicates that the regions of the molecules previously mapped to have repressive functions may independently recruit deacetylases. The fact that EBNA3C synergizes the activity seen with ProTα suggests that these molecules may be associated with a larger complex containing deacetylase activity, as no deacetylase activity has previously been assigned to either ProTα or EBNA3C. Large complexes containing deacetylases may require association with specific factors for triggering activity. Some molecules may have partial activity but becomes fully active in the presence of the required molecules recruited to the complex, which may include EBNA3C, ProTα, and other corepressors, including NCoR and mSin3A (Fig. 10).
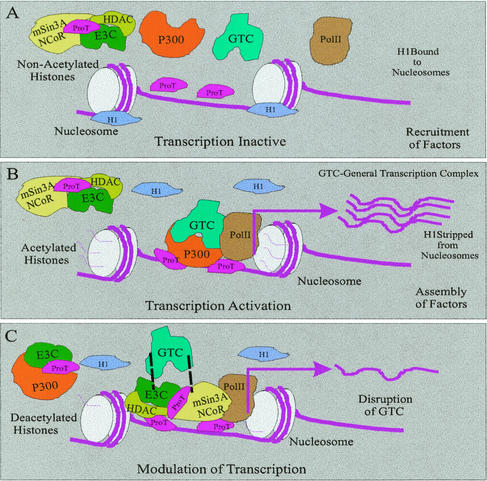
FIG. 10.
Schematic showing the putative role of EBNA3C in regulating chromatin acetylation and deacetylation, resulting in transcriptional control. EBNA3C associates with the acetylase p300, the deacetylases HDAC1 and HDAC2, and the corepressors mSin3A and NCoR. These complexes are stabilized by ProTα, which also acts in inhibiting histone H1 from binding to the nucleosome, allowing access of the large activation and repressor complexes. Therefore, EBNA3C is involved in regulating viral and cellular promoters, and this intrinsically requires the cellular molecule ProTα to modulate the associated activity.
The association of HDAC1 with ProTα as well as EBNA3C indicates that these molecules may be part of a larger repressor complex stabilized by ProTα (Fig. 10). HDAC2 seems to be more abundant in its association with ProTα and EBNA3C, as shown in the GST-ProTα pulldown experiments. Therefore, HDAC2 may be the more predominant deacetylase associated in complex with EBNA3C and ProTα, as HDAC2 had little or no association with ProTα in the absence of EBNA3C, suggesting a requirement for EBNA3C to recruit enhanced activity. In comparison, the corepressors mSin3A and NCoR associated to similar levels compared to that seen with EBNA3C.
It was clear from the immunoprecipitation data that EBNA3C was brought down in the same manner as NCoR, mSin3A, HDAC1, and HDAC2 in stable cell lines. Surprisingly, NCoR was not detected even in the lysates of EBV-transformed LCLs, suggesting a lack of expression of NCoR. This lack of expression may be dependent on the presence of EBV latent antigens, possibly the EBNAs. However, further studies will elucidate the possible regulation of NCoR expression by an EBV antigen. It is possible that EBNA2 and EBNALP or even EBNA3C itself may have a direct regulatory role on the NCoR promoter, and we will continue to address these studies with further experimentation. Clearly, in LCLs the deacetylase activity is reduced compared to that seen with the cell lines stably expressing EBNA3C. This may be a reflection of the levels of EBNA3C expressed in these LCLs compared to the stable cell lines or the result of expression of other latent genes, including EBNA2 and EBNALP, the coactivator of EBNA2, which may counter the overall effects at specific promoters recruiting deacetylases and corepressors.
The putative involvement of RBP-Jκ in these complexes is a reasonable question to address, as RBP-Jκ was previously shown to be a major player in EBV-mediated events and shown to be a repressor of both viral and cellular promoters (4, 14, 15). This is functionally reversed by EBNA2 and further regulated by EBNA3C (27). To address this directly, we performed immunoprecipitation and Western blotting for RBP-Jκ and, as expected, were able to show association with EBNA3C. This may indicate that RBP-Jκ is only one of the major regulators associated with the essential EBV latent antigens and that it may associate with other complexes with the EBNA proteins. It should be noted that to date, no direct association between RBP-Jκ and HDACs has been demonstrated in human cells or EBV-transformed LCLs. The interaction has only been shown by in vitro binding studies. It is possible that RBP-Jκ is outcompeted by other repressor molecules with a higher affinity for HDAC1 as well as other HDACs and that those associations are temporally regulated. The interaction of the essential EBV antigens with regulators of gene expression is expected to be tightly regulated and dependent on the presence of specific cellular factors required for maintaining the transformed phenotype of the infected cells.
Interestingly, in LCLs the association of HDAC1, HDAC2, and corepressors mSin3A and NCoR with EBNA3C and ProTα was negligible compared to the control background lanes. This was surprising, as we expected that the levels would be comparable to that in the EBNA3C cell lines and may be a function of the additional EBNAs expressed in EBV-transformed LCLs. Immunofluorescence analysis with antibodies against HDAC1 and HDAC2 as well as mSin3A and NCoR indicated that they were localized in the same nuclear compartments as EBNA3C. Additionally, the association requires ProTα and most likely other factors yet to be identified to stabilize the complex. As these factors localize in the same compartment but have different affinities for direct association, it is possible that their association with EBNA3C is dependent on the cell cycle, as it was previously shown that the expression of ProTα is upregulated during the S/G2 phase of the cell cycle (17, 36). ProTα is induced as the cell proliferates and at the S/G2 transition, as it may be required for stabilizing repression and activation complexes. The absence of ProTα results in a block to cell division and proliferation, suggesting that cellular molecules required for driving these cellular processes are intrinsically linked with ProTα (29, 32, 34, 38). The fact that EBNA3C binds ProTα clearly supports the hypothesis that EBNA3C is involved in regulating gene expression.
The association with p300 and acetylase activity as well as HDACs, corepressors, and deacetylase activity in B cells strengthens the argument for a role of EBNA3C in both activation and repression of transcription. In our model, EBNA3C is associated with corepressor complexes that include deacetylases (HDACs). This complex is recruited to promoters, resulting in downregulation of specific viral and cellular genes. ProTα stabilizes these complexes containing the acetylases and deacetylases and is critical for these complexes to be fully active (Fig. 10). EBNA3C seems to be in separate complexes with acetylases and deacetylases. The associated functions may then be dependent on cell cycle events as well as contributions from other cellular and viral antigens involved in regulating cell growth and proliferation. More studies dissecting these cellular processes and their regulation should shed light on the roles of ProTα and EBNA3C and their abilities to regulate these associated cellular events.
Acknowledgments
We thank Fernando Dominguez for the ProTα construct and Elliott Kieff for the EBNA3C expression construct. We thank Rama Orre for initial help with the deacetylase assays.
This work was supported by grants from the Leukemia and Lymphoma Society of America and the National Cancer Institute (CA072150 to E.S.R.). E.S.R. is a Leukemia and Lymphoma Society of America Scholar. J.S.K. is supported by NIH graduate training grant T32-AI-07632-02 at the University of Pennsylvania Medical School.
REFERENCES
- 1.Adams, A., G. Bjursell, C. Kaschka-Dierich, and T. Lindahl. 1977. Circular Epstein-Barr virus genomes of reduced size in a human lymphoid cell line of infectious mononucleosis origin. J. Virol. 22:373-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfieri, C., M. Birkenbach, and E. Kieff. 1991. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology 181:595-608. (Erratum, Virology 185:946.) [DOI] [PubMed] [Google Scholar]
- 3.Allday, M. J., and P. J. Farrell. 1994. Epstein-Barr virus nuclear antigen EBNA3C/6 expression maintains the level of latent membrane protein 1 in G1-arrested cells. J. Virol. 68:3491-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aster, J. C., E. S. Robertson, R. P. Hasserjian, J. R. Turner, E. Kieff, and J. Sklar. 1997. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jκ or nuclear localization sequences retain the ability to associate with RBP-Jκ and activate transcription. J. Biol. Chem. 272:11336-11343. [DOI] [PubMed] [Google Scholar]
- 5.Bain, M., R. J. Watson, P. J. Farrell, and M. J. Allday. 1996. Epstein-Barr virus nuclear antigen 3C is a powerful repressor of transcription when tethered to DNA. J. Virol. 70:2481-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burkitt, D. W. D. 1966. Geographical and tribal distribution of the African lymphoma in Uganda. Br. Med. J. 5487:569-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bustin, M. 2001. Chromatin unfolding and activation by HMGN(*) chromosomal proteins. Trends Biochem. Sci. 26:431-437. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, J. I., F. Wang, J. Mannick, and E. Kieff. 1989. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc. Natl. Acad. Sci. USA 86:9558-9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotter, M. A., 2nd, and E. S. Robertson. 2000. Modulation of histone acetyltransferase activity through interaction of Epstein-Barr nuclear antigen 3C with prothymosin alpha. Mol. Cell. Biol. 20:5722-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diaz-Jullien, C., A. Perez-Estevez, G. Covelo, and M. Freire. 1996. Prothymosin alpha binds histones in vitro and shows activity in nucleosome assembly assay. Biochim. Biophys. Acta 1296:219-227. [DOI] [PubMed] [Google Scholar]
- 11.Ding, H. F., M. Bustin, and U. Hansen. 1997. Alleviation of histone H1-mediated transcriptional repression and chromatin compaction by the acidic activation region in chromosomal protein HMG-14. Mol. Cell. Biol. 17:5843-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farrell, P. J., I. Cludts, and A. Stuhler. 1997. Epstein-Barr virus genes and cancer cells. Biomed. Pharmacother. 51:258-267. [DOI] [PubMed] [Google Scholar]
- 13.Fennewald, S., V. Van Santen, and E. Kieff. 1984. The nucleotide sequence of a messenger RNA transcribed in latent growth transforming virus infection indicates that it may encode a membrane protein. J. Virol. 51:411-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grossman, S. R., E. Johannsen, X. Tong, R. Yalamanchili, and E. Kieff. 1994. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc. Natl. Acad. Sci. USA 91:7568-7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henkel, T., P. D. Ling, S. D. Hayward, and M. G. Peterson. 1994. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science 265:92-95. [DOI] [PubMed] [Google Scholar]
- 16.Juan, L. J., R. T. Utley, M. Vignali, L. Bohm, and J. L. Workman. 1997. H1-mediated repression of transcription factor binding to a stably positioned nucleosome. J. Biol. Chem. 272:3635-3640. [DOI] [PubMed] [Google Scholar]
- 17.Karetsou, Z., R. Sandaltzopoulos, M. Frangou-Lazaridis, C. Y. Lai, O. Tsolas, P. B. Becker, and T. Papamarcaki. 1998. Prothymosin alpha modulates the interaction of histone H1 with chromatin. Nucleic Acids Res. 26:3111-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kieff, E. 1998. Current perspectives on the molecular pathogenesis of virus-induced cancers in human immunodeficiency virus infection and acquired immunodeficiency syndrome. J. Natl. Cancer Inst. Monogr. 23:7-14. [DOI] [PubMed] [Google Scholar]
- 19.Kieff, E. 1996. Epstein-Barr Virus and its replication, 3rd ed., vol. 2. Lippincott-Raven, Philadelphia, Pa.
- 20.Klein, E. 1998. The complexity of the Epstein-Barr virus infection in humans. Pathol. Oncol. Res. 4:3-7. [DOI] [PubMed] [Google Scholar]
- 21.Marshall, D., and C. Sample. 1995. Epstein-Barr virus nuclear antigen 3C is a transcriptional regulator. J. Virol. 69:3624-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maunders, M. J., L. Petti, and M. Rowe. 1994. Precipitation of the Epstein-Barr virus protein EBNA 2 by an EBNA 3c-specific monoclonal antibody. J. Gen. Virol. 75:769-778. [DOI] [PubMed] [Google Scholar]
- 23.Nightingale, K., S. Dimitrov, R. Reeves, and A. P. Wolffe. 1996. Evidence for a shared structural role for HMG1 and linker histones B4 and H1 in organizing chromatin. EMBO J. 15:548-561. [PMC free article] [PubMed] [Google Scholar]
- 24.Radkov, S. A., R. Touitou, A. Brehm, M. Rowe, M. West, T. Kouzarides, and M. J. Allday. 1999. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J. Virol. 73:5688-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rickinson, A., and E. Kieff. 1996. Epstein-Barr virus, 3rd ed., vol. 2. Lippincott-Raven, Philadelphia, Pa.
- 26.Robertson, E. S. 1997. The Epstein-Barr virus EBNA3 protein family as regulators of transcription. Epstein-Barr Virus Rep. 4:143-150. [Google Scholar]
- 27.Robertson, E. S., S. Grossman, E. Johannsen, C. Miller, J. Lin, B. Tomkinson, and E. Kieff. 1995. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein Jκ. J. Virol. 69:3108-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robertson, E. S., J. Lin, and E. Kieff. 1996. The amino-terminal domains of Epstein-Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJκ. J. Virol. 70:3068-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roson, E., R. Gallego, T. Garcia-Caballero, E. P. Heimer, A. M. Felix, and F. Dominguez. 1990. Prothymosin alpha expression is associated to cell division in rat testis. Histochemistry 94:597-599. [DOI] [PubMed] [Google Scholar]
- 30.Sample, C., and B. Parker. 1994. Biochemical characterization of Epstein-Barr virus nuclear antigen 3A and 3C proteins. Virology 205:534-539. [DOI] [PubMed] [Google Scholar]
- 31.Sample, J., L. Young, B. Martin, T. Chatman, E. Kieff, and A. Rickinson. 1990. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J. Virol. 64:4084-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sburlati, A. R., R. E. Manrow, and S. L. Berger. 1991. Prothymosin alpha antisense oligomers inhibit myeloma cell division. Proc. Natl. Acad. Sci. USA 88:253-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sixbey, J. W., J. G. Nedrud, N. Raab-Traub, R. A. Hanes, and J. S. Pagano. 1984. Epstein-Barr virus replication in oropharyngeal epithelial cells. N. Engl. J. Med. 310:1225-1230. [DOI] [PubMed] [Google Scholar]
- 34.Smith, M. R., A. al-Katib, R. Mohammad, A. Silverman, P. Szabo, S. Khilnani, W. Kohler, R. Nath, and M. G. Mutchnick. 1993. Prothymosin alpha gene expression correlates with proliferation, not differentiation, of HL-60 cells. Blood 82:1127-1132. [PubMed] [Google Scholar]
- 35.Subramanian, C., M. A. Cotter 2nd, and E. S. Robertson. 2001. Epstein-Barr virus nuclear protein EBNA-3C interacts with the human metastatic suppressor Nm23-H1: a molecular link to cancer metastasis. Nat. Med. 7:350-355. [DOI] [PubMed] [Google Scholar]
- 36.Szabo, P., D. Ehleiter, E. Whittington, and M. E. Weksler. 1992. Prothymosin alpha expression occurs during G1 in proliferating B or T lymphocytes. Biochem. Biophys. Res. Commun. 185:953-959. [DOI] [PubMed] [Google Scholar]
- 37.Tomkinson, B., E. Robertson, and E. Kieff. 1993. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J. Virol. 67:2014-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, C. L., A. L. Shiau, and C. S. Lin. 1997. Prothymosin alpha promotes cell proliferation in NIH 3T3 cells. Life Sci. 61:2091-2101. [DOI] [PubMed] [Google Scholar]
- 39.Young, L., C. Alfieri, K. Hennessy, H. Evans, C. O'Hara, K. C. Anderson, J. Ritz, R. S. Shapiro, A. Rickinson, E. Kieff, et al. 1989. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N. Engl. J. Med. 321:1080-1085. [DOI] [PubMed] [Google Scholar]
- 40.Zhao, B., and C. E. Sample. 2000. Epstein-Barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein-Barr virus nuclear antigen 2 through sequences encompassing an Spi-1/Spi-B binding site. J. Virol. 74:5151-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimber-Strobl, U., L. J. Strobl, C. Meitinger, R. Hinrichs, T. Sakai, T. Furukawa, T. Honjo, and G. W. Bornkamm. 1994. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-J kappa, the homologue of Drosophila Suppressor of Hairless. EMBO J. 13:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]