Abstract
The fusion (F) glycoproteins of measles virus strains Edmonston (MV-Edm) and wtF (MV-wtF) confer distinct cytopathic effects and strengths of hemagglutinin (H) interaction on a recombinant MV-Edm virus. They differ in just two amino acids, V94 and V101 in F-Edm versus M94 and F101 in F-wtF, both of which lie in the relatively uncharacterized F2 domain. By comparing the sequence of MV F with those of the parainfluenza virus SV5 and Newcastle disease virus (NDV) F proteins, the structures of which are known, we show that MV F2 also possesses a potential heptad repeat (HR) C domain. In NDV, the N-terminal half of HR-C interacts with HR-A in F1 while the C-terminal half is induced to kink outward by a central proline residue. We found that this proline is part of an LXP motif conserved in all three viruses. Folding and transport of MV F require this motif to be intact and also require covalent interaction of cysteine residues that probably support the potential HR-A-HR-C interaction. Amino acids 94 and 101, both located in “d” positions of the HR-C helical wheel, lie in the potentially outwardly kinked region. We demonstrate that their effect on MV fusogenicity and glycoprotein interaction is mediated solely by amino acid 94. Substitutions at position 94 with polar or charged amino acids are tolerated poorly or not at all, while changes to smaller and more hydrophilic amino acids are tolerated in both transiently expressed F protein and recombinant virus. MV F V94A and MV F V94G viruses induce extensive syncytium formation and are relatively, or almost completely, resistant to a known inhibitor of MV glycoprotein-induced fusion. We propose that the conformational changes in MV F protein required to expose the fusion peptide involve the C-terminal half of the HR-C helix, specifically amino acid 94.
The Paramyxovirinae subfamily of enveloped negative-stranded RNA viruses encompasses a number of important human and animal pathogens including human parainfluenza viruses, mumps virus, canine distemper virus, Newcastle disease virus (NDV), parainfluenza virus SV5, and measles virus (MV) (22), which remains responsible for more than 1 million deaths per year (40).
The attachment of Paramyxovirinae to host cell receptors is mediated by the envelope protein hemagglutinin (H), hemagglutinin-neuraminidase (HN), or glycoprotein (G), depending on the genus of virus (26). Subsequent to receptor binding, these proteins provide fusion support for the fusion (F) glycoprotein, which is responsible for mediating virus-cell fusion (21). F, a type I transmembrane protein, is synthesized as an inactive precursor, F0, that trimerizes in the endoplasmic reticulum (14, 28) and is cleaved by furin in the late Golgi apparatus and trans-Golgi network to yield active complexes containing covalently linked F1 and F2 subunits (33). The newly generated N terminus possesses a hydrophobic “fusion peptide” that is inserted into the target membrane during fusion.
Structural studies on a number of viral fusion proteins, particularly that of influenza virus, have suggested that prior to interaction with the host cell, fusion proteins exist in a metastable state that is triggered to undergo conformational changes resulting in exposure of the fusion peptide and its insertion into the host cell membrane (8, 12, 39). For the Paramyxovirinae, this trigger is believed to be receptor binding (21), analogous to the proposed mechanism of human immunodeficiency virus (HIV) fusion (11). The postfusion states of many viral proteins and proteins involved in vesicular fusion share a six-helix coiled-coil bundle in which the fusion peptide and transmembrane anchor are in close proximity (1, 5, 6, 10, 16, 19, 24, 35, 37, 38). This six-helix core comprises a trimer of heterodimers of helical heptad repeat (HR) A and B domains (35) and is conserved in SV5 F (1), respiratory syncytial virus F (24), and NDV F (41).
Crystallization of the prefusion NDV F protein has shown it to possess a structure distinct from that of influenza virus hemagglutinin (13). The central triple-stranded HR-A coiled-coil that forms part of the six-helix core is oriented in the opposite direction with respect to the viral membrane in the two molecules. For the respiratory syncytial virus (23, 24) and NDV (13) F proteins, a third helical region, called HR-3 or HR-C, respectively, has been identified in addition to the HR-A and HR-B domains; its function is unclear. The NDV F crystal structure reveals an HR-C helix that lies within F2 immediately N-terminal to the furin processing site and is covalently linked through a disulfide bridge to the central HR-A coiled-coil of F1. The N-terminal half of this HR-C helix was shown to pack in a parallel fashion into the groove of the HR-A coiled-coil (13). Downstream of a central proline residue, HR-C is described as bending outward and extending upward along the widening neck of the F protein (13).
In previous work, it was shown that the strength of MV glycoprotein interaction may be a determinant of viral cytopathicity, with decreased H and F complex stability being associated with more-extensive fusogenicity (29). This finding extends to primary isolates of MV; MV Edmonston (MV-Edm) carrying the F glycoprotein of the primary viral isolate MV-wtF is significantly less fusogenic (18), which coincides with a strong interaction between H and F (29). F proteins derived from MV-Edm and MV-wtF differ in only two residues located in F2. In this study we investigated the molecular basis for the phenotypic differences between recombinant MV-Edm and MV-Edm F-wtF. By comparison with known structural features of NDV F (13), we localized the residues differing between F-Edm and F-wtF to a potential HR-C domain in the MV F protein structure, and we explored the role of these residues in viral fusogenicity. Furthermore, we investigated the importance of the disulfide bridge between HR-A and HR-C and a proline-induced kink in HR-C for folding, intracellular transport, and fusion activity of MV F. We determined that amino acid 94 accounts solely for the observed phenotypic differences between F-Edm and F-wtF, and we investigated the effect on fusion of changes of F-94 to polar and weakly basic, acidic, or smaller and more hydrophilic amino acids. Lastly, we assessed the impact of more-hydrophilic substitutions at position F-94 on fusion induced by recombinant viruses.
MATERIALS AND METHODS
Cell culture, transfection, and production of MV stocks.
Vero (African green monkey kidney) cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, penicillin, and streptomycin at 37°C and 5% CO2. Stably transfected 293-3-46 helper cells (30) were grown in the presence of 1.2 μg of Geneticin/ml. For transient transfection, Lipofectamine 2000 (Gibco BRL) was used, and cells were analyzed 18 to 24 h posttransfection. Where indicated, fusion inhibitory peptide (FIP) (Bachem), originally derived from a region within the MV-F fusion peptide (31, 32), was added 4 h posttransfection at a final concentration of 50 μM.
To prepare virus stocks, Vero cells were infected with the relevant virus at a multiplicity of infection (MOI) of 0.01 PFU/cell and incubated at 37°C. Cells were scraped in Opti-MEM (Gibco BRL), and particles were released by three freeze-thaw cycles. Titers were determined by 50% tissue culture infective dose (TCID50) titration on Vero cells according to the Spearmann-Karber method.
Plasmid construction.
Parental plasmids for mutagenesis and all experiments were pCG-H, pCG-F, and pCG-wtFBF, encoding MV-Edm H and F (9) and MV-wtF F (18), respectively, under the control of the cytomegalovirus promoter. Site-directed mutagenesis was performed by using the QuikChange system (Stratagene) and confirmed in all cases by DNA sequencing and Western blot analysis. The following primers were used to introduce the indicated mutations in the F gene (nucleotides coding for altered amino acids are boldfaced): F-C68S, 5′-GCCCAATATAACTCTCCTCAATAACTCCACGAGGGTAGAGATTGCAGAATAC; F-C195S, 5′-GATACCGTCTATGAACCAACTATCTTCTGATTTAATCGGCCAGAAGCTCGGG; F-L84P, 5′-GAATACAGGAGACTACTGAGAACTGTTCCAGAACCAATTAGAGATGCACTTAATGC; F-L84A, 5′-GAATACAGGAGACTACTGAGAACTGTTGCAGAACCAATTAGAGATGCACTTAATGC; F-P86L, 5′-GGAGACTACTGAGAACTGTTTTGGAATTGATTAGAGATGCACTTAATGCAGTGACC; F-P86A, 5′-GGAGACTACTGAGAACTGTTTTGGAAGCAATTAGAGATGCACTTAATGCAGTGACC; F-L84A P86A, 5′-GAATACAGGAGACTACTGAGAACTGTTGCAGAAGCAATTAGAGATGCACTTAATGCAGTGACC; F-L84P P86L, 5′-GAATACAGGAGACTACTGAGAACTGTTCCAGAATTGATTAGAGATGCACTTAATGCAGTGACC; F-V94 M, 5′-GAACCAATTAGAGATGCACTTAATGCAATGACCCAGAATATAAGACCGTTTC; F-V94E, 5′-GAACCAATTAGAGATGCACTTAATGCAGAGACCCAGAATATAAGACCGGTTCAG;F-V94N, 5′-GAACCAATTAGAGATGCACTTAATGCAAACACCCAGAATATAAGACCGTTTC; F-V94A, 5′-GAACCAATTAGAGATGCACTTAATGCAGCAACCCAGAATATAAGACCGGTTC; F-V94G, 5′-GAACCAATTAGAGATGCACTTAATGCAGGTACCCAGAATATAAGACCGGTTCAG; and F-F101V, 5′-GCAATGACCCAGAATATAAGACCGGTTCAGAGTGTAGCTTCAAGTAGGAG.
To transfer mutated F variants into a DNA copy of the MV genome, NarI PacI fragments of pCG-F containing the F open reading frame were cloned into NarI PacI-digested p(+)MV-NSe (34) by adhering to the reported “rule of six” (7).
Western blot analysis.
Cells (5 × 105) were infected at an MOI of 0.1 PFU/cell and 36 h later were washed in phosphate-buffered saline (PBS), lysed for 10 min at 4°C in lysis buffer (50 mM Tris [pH 8.0], 62.5 mM EDTA, 0.4% deoxycholate, 1% Igepal [Sigma]) containing protease inhibitors (Complete mix [Roche]) and 1 mM phenylmethylsulfonyl fluoride (PMSF), and centrifuged at 5,000 × g for 10 min at 4°C. Total protein concentrations of postnuclear supernatants were determined by using the DC Protein-Assay kit (Bio-Rad). Unless otherwise stated, 2.5 μg of total protein was mixed with urea buffer (200 mM Tris [pH 6.8], 8 M urea, 5% sodium dodecyl sulfate, 0.1 mM EDTA, 0.03% bromphenol blue, 1.5% dithiothreitol) for 25 min at 50°C. Samples were fractionated on sodium dodecyl sulfate-polyacrylamide gels, blotted to polyvinylidene difluoride membranes (Millipore), and subjected to enhanced chemiluminescence detection (Amersham Pharmacia Biotech) using antibodies specific for the cytosolic tail of F (27). For statistical analysis, Western blots were scanned to yield arbitrary densitometric units. Results were normalized to values for the control cells in each experiment.
Surface biotinylation.
Cells were transfected with 3 μg of plasmid DNA encoding MV F variants as indicated. After a wash in cold PBS, cells were incubated in PBS with 0.5 mg of NHS-SS-Biotin (Pierce)/ml for 20 min at 4°C, followed by washing and quenching for 5 min at 4°C in Dulbecco's modified Eagle's medium. Cells were scraped in immunoprecipitation buffer (10 mM HEPES [pH 7.4], 50 mM sodium pyrophosphate, 50 mM sodium fluoride, 50 mM sodium chloride, 5 mM EDTA, 5 mM EGTA, 100 μM sodium vanadate, 1% Triton X-100) containing protease inhibitors (Complete mix) and 1 mM PMSF, and lysates were cleared by centrifugation for 20 min at 20,000 × g and 4°C. As an internal standard, 30 μl of total protein was mixed with urea buffer. Biotinylated proteins were absorbed to Sepharose-coupled streptavidin (Amersham Pharmacia Biotech) for 90 min at 4°C, washed first in buffer 1 (100 mM Tris [pH 7.6], 500 mM lithium chloride, 0.1% Triton X-100) and then in buffer 2 (20 mM HEPES [pH 7.2], 2 mM EGTA, 10 mM magnesium chloride, 0.1% Triton X-100), incubated in urea buffer for 25 min at 50°C, and subjected to Western blot analysis using antibodies specific for the MV-F tail.
Hemifusion assay.
Vero cells were cotransfected with 3 μg each of plasmid DNA encoding MV H and F variants as indicated and were incubated in the presence of 200 μM FIP. Sixteen hours posttransfection, cells were washed repeatedly in PBS and overlaid with washed and R18 (Molecular Probes)-labeled African green monkey-derived erythrocytes (Bio Whittaker). Subsequent to binding of labeled erythrocytes for 1 h at 4°C, cells were washed extensively and incubated at 37°C for 30 to 45 min, and the red fluorescence of Vero cells, indicating dye transfer, was assessed.
Coimmunoprecipitation.
Cells were cotransfected with 2.5 μg each of plasmid DNA encoding MV H and F variants as indicated. After a wash in PBS, cells were scraped in immunoprecipitation buffer containing protease inhibitors (Complete mix) and 1 mM PMSF. Lysates were cleared by centrifugation for 25 min at 20,000 × g and 4°C, and 300 μg of total protein was incubated with antibodies directed against MV H (Chemicon) for 90 min at 4°C. As an internal standard, 5 μg of total protein was mixed with urea buffer. Immune complexes were adsorbed to protein G-agarose (Gibco BRL) for 90 min at 4°C, washed first in buffer 1 and then in buffer 2, and incubated in urea buffer for 25 min at 50°C. The amount of coprecipitated F protein was determined by Western blot analysis using antibodies specific for the MV-F tail.
Quantitative fusion assay.
Cells were cotransfected with 1.5 μg each of plasmid DNAs encoding MV-H and F and 3 μg of plasmid DNA encoding the β-galactosidase reporter gene under the control of the T7 promoter. Six hours posttransfection, 1.5 × 105 cells were mixed with cells previously infected with vaccinia virus encoding T7 polymerase. After 12 h of incubation, cells were lysed and β-galactosidase activity was assessed by use of a β-Gal assay kit (Invitrogen) according to the manufacturer's instructions. Average activities were calculated on the basis of four independent experiments, each performed in duplicate.
Recovery of recombinant viruses.
Recombinant MVs were generated essentially as described previously (30) Briefly, the helper cell line 293-3-46, stably expressing MV N, MV P, and T7 polymerase, was transfected by calcium phosphate precipitation by using the ProFection kit (Promega) with a DNA copy of the relevant MV genome and MV polymerase L. Helper cells were overlaid on Vero cells 76 h posttransfection, and resulting infectious centers were passaged on Vero cells. In all cases, the integrity of recombinant viruses was confirmed by reverse transcription-PCR and DNA sequencing of the modified genes.
Virus growth kinetics.
Vero cells (5 × 105 per time point) were infected at an MOI of 0.03 PFU/cell. At the indicated time points, supernatants were cleared by centrifugation, cells were scraped in Opti-MEM (Gibco BRL) and subjected to three freeze-thaw cycles, and cell-associated titers were determined by TCID50 titration on Vero cells.
RESULTS
MV-F contains a putative HR-C domain.
To better understand the structural basis for the observed differences in phenotype between MV-Edm containing F-Edm and MV-Edm containing F-wtF, we first used the Clustal W algorithm (36) to align MV F with F glycoproteins from SV5 and NDV, for which structural information is available. The comparison is shown in Fig. 1 for the F2 subunits of strains NDV-B1-Hitchner/47, SV5-W3, and MV-Edmonston-tag (30); the consensus sequence is given where possible.
FIG. 1.
Alignment of F2 sequences derived from strains NDV B1-Hitchner/47, SV5 W3, and MV-Edmonston-tag. The predicted consensus sequence is given. Backgrounds are red for residues conserved in all three strains, blue for residues identical in two strains, green for residues with similar biophysical properties, and yellow for the approximate locations of heptad repeat HR-C. The hydrophobic “a” and “d” positions of the postulated HR-C helical wheel are indicated.
Consistent with previous reports (3, 20, 25), we found the areas containing the furin cleavage site and the adjacent fusion peptides to be highly conserved among the three proteins (data not shown). Also well conserved are heptad repeats HR-A and HR-B, located downstream of the fusion peptide and upstream of the transmembrane domain, respectively. With some minor variations at the immediate N terminus and within the transmembrane domains and cytosolic tails, cysteine residues are highly conserved among the three proteins (25) (data not shown), further suggesting similar structural frameworks based on the formation of disulfide bridges at comparable positions in the F ectodomains.
The HR-C region identified in NDV F lies in the C-terminal part of the F2 domain. Our alignment (Fig. 1) proved the properties of amino acids in this region to be well conserved among NDV, MV, and SV5 F proteins; the respective “a” and “d” positions in the helical wheel structure are indicated. We found only a tyrosine residue located toward the N terminus and a central LXP motif to be identical in this area in all three proteins. Remarkably, both amino acid differences between F-Edm and F-wtF, V94M and V101F, are located in the same “d” position of the HR-C helical wheels, downstream of the central LXP motif.
The disulfide bridge between F1 and F2 is essential for correct folding.
The fact that we have found a potential HR-C region in MV F2 raises the possibility that an HR-A-HR-C interaction occurs between the MV F1 and F2 subunits, similar to that described for NDV. There, the N-terminal half of the F2 HR-C domain is thought to form an α-helical coiled-coil with HR-A. Given that the positions of cysteine residues in the paramyxovirus F ectodomain are highly conserved, and given that for the related Sendai virus the disulfide bond between paramyxovirus F subunits has been shown to form between the cysteine residue in F2 and the most N-terminal cysteine of F1 (17), an interaction between the MV HR-A and HR-C domains might be stabilized through a disulfide bridge between C68, located immediately upstream of HR-C (Fig. 1), and, most likely, C195, located within the HR-A region. This raises the question whether the hydrophobic HR-A-HR-C interaction alone can be sufficient for proper folding of nascent F polypeptides, or whether the increased stability provided by the covalent interaction of both domains is indispensable. Not surprisingly, when C68 of MV F was exchanged to serine, the residue with biophysical properties closest to cysteine, no proteolytic cleavage of the F0 precursor, indicating export from the endoplasmic reticulum through Golgi compartments, could be observed (data not shown).
Since an unpaired cysteine residue might interact strongly with endoplasmic reticulum-resident oxidoreductases, resulting in intracellular retention (15), we changed both residues C68 and C195 to serines. When analyzed by surface biotinylation and Western blot analysis, the double mutant also proved to be retained intracellularly, strongly suggesting a necessity of the covalent interaction between the HR-A and HR-C domains for acquisition of a native, transport-competent conformation (Fig. 2). Consistent with their intracellular retention, neither F mutant induced cell-to-cell fusion when cotransfected with H-Edm, whereas F-Edm induced extensive syncytia (data not shown) and was processed efficiently, as shown by the presence of the F1 subunit (Fig. 2).
FIG. 2.
F2-C68 and F1-C195 residues are required for F processing. Biotinylated F protein displayed at the cell surface (SF) and total cell lysates (TL) were detected by Western blot analysis using antibodies directed against the F cytosolic tail. As a control (Co), biotinylation of cells transfected with equal amounts of F-Edm was omitted.
The central kink in HR-C is required for folding of MV F.
The NDV F structure reveals an outward kink of the C-terminal half of the HR-C helix, induced by a central proline residue. Our alignment reveals corresponding proline residues in MV and SV5 that lie within a highly conserved LXP motif in the center of HR-C. Furthermore, the leucine residue within this motif, L84 for MV, is located in a central “a” position of the HR-C leucine zipper.
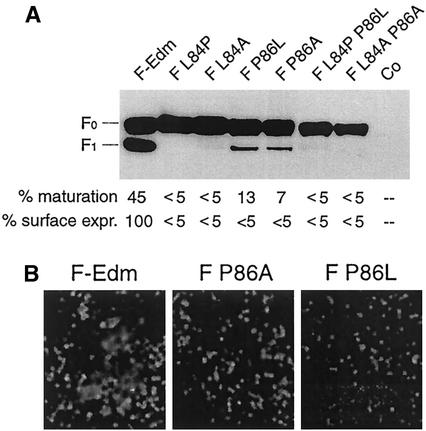
To study the effects of both the leucine and proline residues on MV F function, we mutated the conserved amino acids L84 and P86 singly and in combination to alanine and also interchanged their positions, resulting in mutants carrying PTP, LTL, and PTL residues, disturbing the zipper structure. As shown in Fig. 3A, all mutant proteins were expressed with F-Edm-like steady-state levels, but only mutants F P86L and F P86A displayed some processing from F0 to F1. Approximately 10% of the total F protein was found in the F1 fraction for these mutants, compared to 45% for F-Edm (Fig. 3A), while mutants F L84P and F L84A remained fully unprocessed. Unlike F-Edm, all of these mutants failed to induce cell-to-cell fusion when coexpressed with H-Edm (data not shown). Consistent with these findings, the double mutants F L84P P86L and F L84A P86A also were not processed efficiently to F1 (Fig. 3A) and failed to induce syncytium formation.
FIG. 3.
The central LXP motif in MV HR-C is important for proper F folding. (A) Western blot analysis of F mutants using antibodies directed against the F cytosolic tail. As a control (Co), cells were mock transfected with equal amounts of empty plasmid DNA. The relative percentage of the F1 fraction in relation to the total F protein (% maturation) and the relative percentage of F1 detected in surface biotinylation (described in the legend to Fig. 2) in relation to F-Edm F1 (% surface expr.) are given below the gel. (B) Hemifusion assay subsequent to transient transfection of cells with equal amounts of plasmid DNA encoding MV-H and F constructs as indicated, and overlay with R18-labeled red blood cells. Representative fields of view are shown.
To assess whether the partially processed P86L and P86A mutants reached the cell surface, we performed a surface biotinylation experiment. While F-Edm was efficiently expressed at the cell surface, both F P86L and F P86A were expressed at less than 5% of the F-Edm surface level (Fig. 3A), indicating intracellular retention. The limited maturation of these mutants suggests that they acquire a conformation allowing a low degree of furin cleavage in the early secretory system, as reported for some intracellularly retained proteins (2). Since we could not exclude the possibility that a very small fraction of the processed F P86L or F P86A material reached the surface, we analyzed the hemifusion capacities of these mutants based on the transfer of R18 lipid dye. If hemifusion occurs, the dye becomes distributed between unlabeled MV-H- and F-expressing cells and labeled receptor-positive African green monkey erythrocytes, while in the absence of hemifusion the glycoprotein-expressing cells remain unstained. Subsequent to coexpression of F P86L and F P86A with MV-H, no dye transfer could be detected, while cells coexpressing F-Edm and MV-H showed extensive dye uptake following incubation with labeled receptor-positive cells (Fig. 3B). Together, these observations indicate that the P86 residue and thus most likely the outward kink of the HR-C helix are highly important for proper MV F folding.
F residue 94 solely accounts for phenotypic differences between F-Edm and F-wtF.
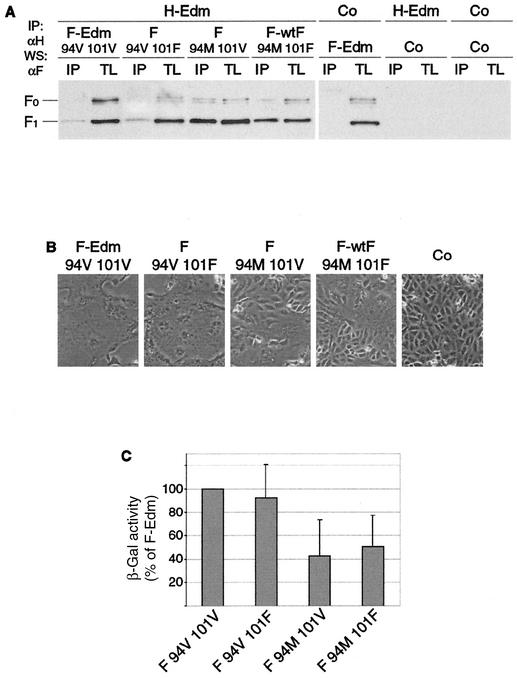
Since both residues differing between F-Edm and F-wtF, V94M and V101F, are located in the potential MV HR-C domain, we addressed whether both contribute to the observed alteration in phenotype. Through site-directed mutagenesis, we constructed two F chimeras, F V94 F101 and F M94 V101. Together with the parental plasmids containing F-Edm (F V94 V101) and F-wtF (F M94 F101), this resulted in four F variants comprising all combinations of the two mutations. We subjected cells transiently expressing these constructs and MV-H to coimmunoprecipitation experiments, in which MV H was precipitated with specific antibodies, and the amount of coprecipitated F protein was determined by immunoblotting using an anti-F antiserum (Fig. 4A). We found that only the residue at position 94 determined the efficiency of F coprecipitation with MV-H, i.e., V94 conferred a weak F-Edm-like interaction with MV-H, and M94 conferred a strong F-wtF-like interaction, while whether position 101 was valine or phenylalanine had no measurable effect on the strength of the interaction. Furthermore, F-Edm and F-wtF homooligomerization capacities were found to be virtually identical within the limitations of a sucrose gradient fractionation assay (data not shown), suggesting that the effect of amino acid differences between F-Edm and F-wtF on fusion phenotype is not mediated by an indirect effect on F trimerization efficiency.
FIG. 4.
The residue at F position 94 accounts solely for the differences in phenotype between F-Edm and F-wtF. (A) Coimmunoprecipitation of F-Edm, F-wtF, and chimeric constructs with H following transient expression in Vero cells. F protein from coimmunoprecipitated (IP) samples and total cell lysates (TL) was detected by Western blot analysis (WS) using specific antibodies directed against the cytosolic F tail. Control (Co) transfection was done with equal amounts of noncoding plasmid DNA. (B) Representative fields of view of Vero cells transfected with equal amounts of plasmid DNA encoding MV-H or MV-F variants as indicated. Syncytium formation was assessed 20 h posttransfection. (C) Quantification of fusion activity of MV-F variants coexpressed with MV-H. The relative percentage of β-galactosidase activity in relation to the activity induced by F-Edm is given.
The fact that residue 101 does not affect the efficiency of H-F interaction suggests that it also does not account for any differences in fusion phenotype. To test this hypothesis, cells were cotransfected with MV-H and either F-Edm, F-wtF, or the chimeric constructs, and their cytopathic effects were assessed 16 h posttransfection (Fig. 4B). These experiments demonstrated that the amount of cell-to-cell fusion was significantly increased to F-Edm-like levels when cells expressed F V94 F101. In contrast, expression of F M94 V101 resulted in an F-wtF-like fusion phenotype, i.e., small syncytia with limited lateral spread. Consistent with these data, a quantitative fusion assay based on expression of a β-galactosidase reporter gene revealed that F-wtF and F M94 V101, when coexpressed with MV H, induced β-galactosidase activity to about 50% of the level induced by F-Edm or F V94 F101 (Fig. 4C).
Confirming our coprecipitation data, these findings underline the fact that the difference in cytopathicity between F-Edm and F-wtF is due solely to the point mutation at position 94.
Smaller, more hydrophilic amino acids are tolerated at F residue 94.
Despite their difference in size, valine and methionine side chains have comparable biophysical properties: both are noncharged and mildly hydrophobic. To assess the effects of more-drastic changes at this position on F folding and activity, we introduced charged, polar, and more-hydrophilic amino acids at this position. In the F-Edm background, V94 was mutated to acidic (glutamate) (F V94E), polar (asparagine) (F V94N), and smaller (alanine and glycine) (F V94A and F V94G) residues. For all constructs, steady-state levels and surface expression were determined by Western blot analysis and surface biotinylation (Fig. 5A), and fusion capacity was assessed microscopically subsequent to cotransfection with equal amounts of MV-H plasmid (Fig. 5B) and quantitatively by using the quantitative fusion assay based on β-galactosidase reporter gene expression (Fig. 5C).
FIG. 5.
Charged or polar amino acids at F position 94 abolish or significantly reduce F fusion activity, while substitutions to more-hydrophilic residues are tolerated. (A) Surface expression of F variants determined as described in the legend to Fig. 2. Relative percentages of processed (% maturation) and transported (% surface expr.) F protein, as described in the legend to Fig. 3A, are given. (B) Representative fields of view after overlay of transiently transfected cells with R18-labeled red blood cells and syncytium formation assessed 20 h posttransfection. (C) Quantification of fusion activity as described in the legend to Fig. 4C.
While all mutants displayed F-Edm-like steady-state levels, indicating unchanged protein stability, F V94E was not proteolytically processed and was completely intracellularly retained, suggesting compromised folding of this variant (Fig. 5A); consequently, this mutant did not induce any cell-to-cell fusion (Fig. 5B and C). In contrast, both changes to smaller and more-hydrophilic amino acids (F V94A and F V94G) were better tolerated. While surface expression of F V94A was identical to that of F-Edm, transport of F V94G was considerably reduced, indicating impaired folding. The latter might be due to interference of the glycine residue with the HR-C α-helical structure. F V94N also was not completely retained but revealed a surface steady-state level comparable to that of F V94G (Fig. 5A), indicating less effect on proper protein folding than the acidic V94E side chain.
Consistent with its reduced surface expression, F V94G revealed some reduction in the onset of syncytium formation when cotransfected with H-Edm, while the cell-to-cell fusion capacity of F V94A was virtually indistinguishable from that of F-Edm (Fig. 5B, bottom panel, and 5C). Although the amount of proteolytic maturation and cell surface steady-state levels of F V94N were very similar to those of F V94G, the polar asparagine side chain nearly completely abolished syncytium formation (Fig. 5B, bottom panel, and 5C), further supporting our notion that this position is of high importance for the fusion process. When the F V94N construct was further analyzed in a lipid mixing assay, we observed only limited R18 dye transfer, indicating partial hemifusion (Fig. 5B, top panel).
Thus, smaller and more hydrophilic amino acid substitutions at position 94 do not abolish F activity, whereas polar or acidic side chains at this position strongly interfere with F folding and/or activity.
Alanine and glycine residues at F position 94 confer resistance to a FIP.
Previous studies confirmed that MV glycoprotein-induced syncytium formation can be effectively suppressed through incubation of transfected or infected cells with the FIP, a tripeptide derived from a region within the MV fusion peptide (31, 32). Surprisingly, we observed that cells cotransfected with MV-H and F V94A or F V94G showed strong resistance to FIP inhibition. Formation of large syncytia was clearly detectable in the presence of 50 μM FIP, while cells expressing MV-H and F-Edm did not fuse under these conditions (Fig. 6A). Quantification of cell-to-cell fusion using the β-galactosidase-based fusion assay confirmed these microscopic observations (Fig. 6B). Fusion of cells cotransfected with MV-H and F-Edm was reduced by approximately 80% in the presence of FIP, while F V94A displayed only a 40% reduction under these conditions and F V94G was virtually unaffected. Although the molecular basis for the mechanism of FIP inhibition is unknown to date, this finding further suggests that the residue at position 94 directly participates in F-induced cell-to-cell fusion.
FIG. 6.
F V94A and F V94G mutants show resistance to suppression of fusion by a peptide inhibitor. (A) Syncytium formation after incubation of transfected cells in the presence or absence of 50 μM FIP for 20 h. (B) Quantification of remaining fusion activity in the presence of 50 μM FIP based on the expression of a β-galactosidase reporter construct. 100% corresponds to the β-galactosidase activity in the absence of FIP.
Mutations V94A and V94G influence F activity in the context of viral infection.
Given that our studies so far were mostly based on transient coexpression of the different F variants with MV-H, it was important to assess whether the presence of other viral proteins during infection would further influence the fusion phenotype, i.e., whether the F V94A and F V94G substitutions would also result in FIP resistance in cells infected by recombinant MV virions. We therefore generated MV-Edm-based viruses carrying F V94A and F V94G instead of F-Edm. Subsequent to recovery of both recombinant viruses, their molecular nature was confirmed through reverse transcription of viral RNA from infected cells and DNA sequencing (data not shown).
When comparing the growth of MV-Edm, MV F V94A, and MV F V94G in Vero cells, we observed that only MV F V94A reached a maximum titer of cell-associated infectious particles similar to that of MV-Edm (Fig. 7A). Its more rapid drop in titer starting 32 h postinfection most likely reflects a slightly higher extent of syncytium induction, resulting in earlier complete lysis and detachment of target cells (Fig. 7B). In contrast, MV F V94G replicated to maximal titers approximately 2.5 log units lower, and its maximal growth rate was also reduced, as indicated by the shallower slope of the curve in the logarithmic-growth phase (Fig. 7A). This delay in viral growth is consistent with our observation that in cells transiently expressing F V94G protein, intracellular transport of the protein and the onset of syncytium induction are slightly delayed. Despite its lower final titer, MV F V94G virus did not appear substantially compromised in its ability to induce syncytium formation at later time points (Fig. 7B).
FIG. 7.
Phenotype of recombinant MV particles with more hydrophilic substitutions at F position 94. (A) Growth kinetics of MV-Edm, MV F V94A, and MV F V94G in Vero cells. Cells were infected at an MOI of 0.03 PFU/cell. At the indicated time points, titers of cell-associated viral particles were determined in duplicate. (B) Representative fields of view of Vero cells infected with MV-Edm, MV F V94A, or MV F V94G, at an MOI of 0.03 PFU/cell, in the presence or absence of 75 μM FIP at the indicated times postinfection.
Importantly, when cells were infected with the three viruses in the presence of 75 μM FIP, MV F V94G was almost unaffected in its ability to induce syncytia, and MV F V94A was slightly affected but still induced extensive syncytium formation by 40 h postinfection, while syncytium formation in MV-Edm-infected cells was completely suppressed (Fig. 7B). Thus, our data regarding the extent of syncytium formation and FIP resistance in infected or transfected cells are fully consistent.
DISCUSSION
We have observed by sequence alignment that the amino acid properties of the NDV HR-C region are well conserved in MV and SV5 F2 glycoproteins, strongly suggesting that an HR-C region is a common feature of paramyxovirus fusion proteins. A central LXP motif with the leucine residue in the “a” position of the HR-C helical wheel is also highly conserved among NDV, SV5, and MV, and based on the structure of NDV F, its proline residue is predicted to induce an outward kink of the downstream half of the HR-C helix. Indeed, both leucine and proline residues in this central motif were found to be required for proper folding of MV F. This suggests that the outward kink induced by P86 in the HR-C helix may be of major importance for F functionality. Amino acid 94 lies within this outwardly kinked part of the MV F2 HR-C region and is a determinant of the F fusion phenotype, further supporting the importance of this region for F activity.
Our identification of an HR-C helix in the MV F protein predicts the existence of an interaction between the HR-A helix of F1 and the HR-C helix of F2 similar to that described for the NDV F protein (13). Our findings indicate that the covalent interaction between the F1 and F2 subunits that most likely stabilizes the contact between HR-A and HR-C is required for MV F to be exported from the endoplasmic reticulum. This suggests that the postulated hydrophobic interaction between the two helices alone is not sufficient to maintain a proper intersubunit association that enables the nascent chain to acquire a conformation suitable for further transport to the plasma membrane. In the absence of the covalent interaction, hydrophobic patches of the two helices that are usually buried in the cores of the interacting subunits most likely become surface exposed and are targeted by molecular chaperones in the endoplasmic reticulum that mediate F retention. Not surprisingly, interactions between MV F protein and calnexin, calreticulin, and grp78 have previously been documented (4).
When assessing the molecular basis for the phenotypic differences previously observed between recombinant MV F-Edm and MV F-wtF, we found amino acid 94, which lies in a “d” position of the HR-C helical wheel in F2, to be solely responsible for the altered fusion phenotype. F-Edm carrying the methionine found at position 94 in F-wtF induced syncytia poorly, like F-wtF, and the reciprocal change of methionine to valine in F-wtF induced extensive, F-Edm-like syncytium formation. Amino acid 101, the other point mutation between the F variants, had no effect on fusion or syncytium phenotype. Coimmunoprecipitation of these F constructs with MV-H corroborated these findings; the molecular nature of amino acid 94 alone determined whether MV-F trimers interacted strongly (F-wtF-like) or weakly (F-Edm-like) with the H protein.
One explanation for our coimmunoprecipitation experiments might be that the HR-C domain surrounding position 94 is in direct contact with MV-H. At present, however, we have no further evidence for a physical interaction between this F region and H oligomers. Alternatively, it might well be that mutations of residue 94 alter the conformation of more-distant F regions, which then triggers an impaired interaction with H. Our results confirm that amino acid 94 cannot mediate its effects on glycoprotein interaction and fusion by indirect effects resulting from reduced homooligomerization of F, since we found that F-Edm and F-wtF formed trimers with equal efficiency.
While a V94E substitution resulted in intracellular retention and impaired maturation of the F protein, a polar, weakly basic amino acid was better tolerated at this position. Surface expression and maturation of F carrying a V94N substitution was impaired compared to those of F-Edm, but not abolished. When coexpressed with MV-H, F V94N did not cause extensive syncytium formation and was able to mediate only very moderate lipid dye transfer, indicating significantly compromised hemifusion activity. Replacing the valine residue at amino acid 94 with smaller but more-hydrophilic residues generated F proteins able to induce cell-to-cell fusion, although the V94G substitution caused reductions in F maturation and surface expression similar to those with the V94N mutation.
Surprisingly, both mutations V94A and V94G conferred significant resistance to fusion inhibition by the inhibitory tripeptide FIP, while replacing V94 with the methionine of MV-wtF did not induce any FIP resistance. The basis for the inhibitory effect of this tripeptide, which was originally derived from part of the F fusion peptide itself (31, 32), has yet to be solved. Potentially, resistance to FIP may reflect a facilitated ability of the F V94A and F V94G variants to undergo conformational changes that result in exposure of the fusion peptide. In this scenario, smaller residues at position 94 might lower the activation energy barrier that must be overcome in order for F conformational changes to occur; in this way these F variants could nonspecifically overcome FIP inhibition. If FIP, however, specifically binds and stabilizes F in a prefusion conformation, its binding site could be in close proximity to the HR-C domain. Under these circumstances, mutations in position 94 may directly alter the pocket recognized by FIP. It was previously found that the strength of the MV H-F interaction was not influenced by FIP (28), ruling out the possibility that FIP interferes with heterooligomerization of F and H and with fusion support provided by MV-H upon receptor binding.
Importantly, our findings concerning FIP resistance were not restricted to transient expression of MV glycoproteins but were confirmed in the context of infection by generating recombinant viruses MV F V94A and MV F V94G. Cells infected with either virus in the presence of inhibitory concentrations of FIP are recruited into large syncytia. Consequently, the FIP resistance phenotype is not subject to modulation by other viral components in the context of MV infection.
The fact that amino acid 94 can modulate syncytium formation in this manner, coupled with the demonstration that the proline residue at position 86 and hence the outward kink in the HR-C domain is important for F folding, implicates this region of MV F2 in membrane fusion. Our data suggest a model in which the outward kink in the C-terminal half of HR-C, induced by proline 86, is essential for facilitating the correct conformational presentation of the critical residue 94, most likely in the context of other, as yet unidentified amino acids. The sensitivity of position 94 not only to charge or polarity but also to the size of the amino acid side chain may suggest that this residue stands in close context with other amino acids of F2 or F1 which together mediate the observed effects on fusion. Amino acid 94 may well be one of the residues directly involved in initiating and mediating the conformational changes that occur in the MV glycoproteins for exposure of the fusion peptide. Future identification of neighboring amino acids contacting F residue 94 and structural consequences of mutations in this area might provide further insights into the functional role of the F2 HR-C domain for membrane fusion.
Acknowledgments
We thank M. A. Billeter for providing plasmids encoding the full-length MV-Edm genome; R. Cattaneo for MV-H- and MV-F-encoding plasmids and antibodies directed against the MV-F tail; S. Schneider-Schaulies for the MV F-wtF-encoding plasmid; and A. L. Hammond for critical reading of the manuscript.
This work was supported by NIH grant CA18611 (to R.W.C.) and a Feodor Lynen fellowship from the Alexander von Humboldt foundation (to R.K.P.).
REFERENCES
- 1.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 2.Bass, J., C. Turck, M. Rouard, and D. F. Steiner. 2000. Furin-mediated processing in the early secretory pathway: sequential cleavage and degradation of misfolded insulin receptors. Proc. Natl. Acad. Sci. USA 97:11905-11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg, M., A. C. Bergvall, M. Svenda, A. Sundqvist, J. Moreno-Lopez, and T. Linne. 1997. Analysis of the fusion protein gene of the porcine rubulavirus LPMV: comparative analysis of paramyxovirus F proteins. Virus Genes 14:55-61. [DOI] [PubMed] [Google Scholar]
- 4.Bolt, G. 2001. The measles virus (MV) glycoproteins interact with cellular chaperones in the endoplasmic reticulum and MV infection upregulates chaperone expression. Arch. Virol. 146:2055-2068. [DOI] [PubMed] [Google Scholar]
- 5.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37-43. [DOI] [PubMed] [Google Scholar]
- 6.Caffrey, M., M. Cai, J. Kaufman, S. J. Stahl, P. T. Wingfield, D. G. Covell, A. M. Gronenborn, and G. M. Clore. 1998. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 17:4572-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calain, P., and L. Roux. 1993. The rule of six, a basic feature for efficient replication of Sendai virus defective-interfering RNA. J. Virol. 67:4822-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carr, C. M., and P. S. Kim. 1993. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell 73:823-832. [DOI] [PubMed] [Google Scholar]
- 9.Cathomen, T., C. J. Buchholz, P. Spielhofer, and R. Cattaneo. 1995. Preferential initiation at the second AUG of the measles virus F mRNA: a role for the long untranslated region. Virology 214:628-632. [DOI] [PubMed] [Google Scholar]
- 10.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 11.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 12.Chen, J., K. H. Lee, D. A. Steinhauer, D. J. Stevens, J. J. Skehel, and D. C. Wiley. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95:409-417. [DOI] [PubMed] [Google Scholar]
- 13.Chen, L., J. J. Gorman, J. McKimm-Breschkin, L. J. Lawrence, P. A. Tulloch, B. J. Smith, P. M. Colman, and M. C. Lawrence. 2001. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure (Cambridge) 9:255-266. [DOI] [PubMed] [Google Scholar]
- 14.Dutch, R. E., T. S. Jardetzky, and R. A. Lamb. 2000. Virus membrane fusion proteins: biological machines that undergo a metamorphosis. Biosci. Rep. 20:597-612. [DOI] [PubMed] [Google Scholar]
- 15.Ellgaard, L., and A. Helenius. 2001. ER quality control: towards an understanding at the molecular level. Curr. Opin. Cell Biol. 13:431-437. [DOI] [PubMed] [Google Scholar]
- 16.Fass, D., S. C. Harrison, and P. S. Kim. 1996. Retrovirus envelope domain at 1.7 angstrom resolution. Nat. Struct. Biol. 3:465-469. [DOI] [PubMed] [Google Scholar]
- 17.Iwata, S., A. C. Schmidt, K. Titani, M. Suzuki, H. Kido, B. Gotoh, M. Hamaguchi, and Y. Nagai. 1994. Assignment of disulfide bridges in the fusion glycoprotein of Sendai virus. J. Virol. 68:3200-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnston, I. C., V. ter Meulen, J. Schneider-Schaulies, and S. Schneider-Schaulies. 1999. A recombinant measles vaccine virus expressing wild-type glycoproteins: consequences for viral spread and cell tropism. J. Virol. 73:6903-6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobe, B., R. J. Center, B. E. Kemp, and P. Poumbourios. 1999. Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose-binding protein chimera reveals structural evolution of retroviral transmembrane proteins. Proc. Natl. Acad. Sci. USA 96:4319-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komada, H., H. Bando, M. Ito, H. Ohta, M. Kawano, M. Nishio, M. Tsurudome, N. Watanabe, N. Ikemura, S. Kusagawa, et al. 1995. Sequence analyses of human parainfluenza virus type 4A and type 4B fusion proteins. J. Gen. Virol. 76:3205-3210. [DOI] [PubMed] [Google Scholar]
- 21.Lamb, R. A. 1993. Paramyxovirus fusion: a hypothesis for changes. Virology 197:1-11. [DOI] [PubMed] [Google Scholar]
- 22.Lamb, R. A., and D. Kolakofsky. 1996. Paramyxoviridae: the viruses and their replication, p. 577-604. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fundamental virology. Lippincott-Raven, Philadelphia, Pa.
- 23.Lambert, D. M., S. Barney, A. L. Lambert, K. Guthrie, R. Medinas, D. E. Davis, T. Bucy, J. Erickson, G. Merutka, and S. R. Petteway, Jr. 1996. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc. Natl. Acad. Sci. USA 93:2186-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matthews, J. M., T. F. Young, S. P. Tucker, and J. P. Mackay. 2000. The core of the respiratory syncytial virus fusion protein is a trimeric coiled coil. J. Virol. 74:5911-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGinnes, L. W., and T. G. Morrison. 1986. Nucleotide sequence of the gene encoding the Newcastle disease virus fusion protein and comparisons of paramyxovirus fusion protein sequences. Virus Res. 5:343-356. [DOI] [PubMed] [Google Scholar]
- 26.Morrison, T. G., and A. Portner. 1991. Structure, function, and intracellular processing of the glycoproteins of Paramyxoviridae, p. 347-382. In D. W. Kingsbury (ed.), The Paramyxoviruses. Plenum Press, New York, N.Y.
- 27.Plemper, R. K., A. L. Hammond, and R. Cattaneo. 2000. Characterization of a region of the measles virus hemagglutinin sufficient for its dimerization. J. Virol. 74:6485-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plemper, R. K., A. L. Hammond, and R. Cattaneo. 2001. Measles virus envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J. Biol. Chem. 276:44239-44246. [DOI] [PubMed] [Google Scholar]
- 29.Plemper, R. K., A. L. Hammond, D. Gerlier, A. K. Fielding, and R. Cattaneo. 2002. Strength of envelope protein interaction modulates cytopathicity of measles virus. J. Virol. 76:5051-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radecke, F., P. Spielhofer, H. Schneider, K. Kaelin, M. Huber, C. Dotsch, G. Christiansen, and M. A. Billeter. 1995. Rescue of measles viruses from cloned DNA. EMBO J. 14:5773-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richardson, C. D., and P. W. Choppin. 1983. Oligopeptides that specifically inhibit membrane fusion by paramyxoviruses: studies on the site of action. Virology 131:518-532. [DOI] [PubMed] [Google Scholar]
- 32.Richardson, C. D., A. Scheid, and P. W. Choppin. 1980. Specific inhibition of paramyxovirus and myxovirus replication by oligopeptides with amino acid sequences similar to those at the N-termini of the F1 or HA2 viral polypeptides. Virology 105:205-222. [DOI] [PubMed] [Google Scholar]
- 33.Scheid, A., and P. W. Choppin. 1974. Identification of biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis, and infectivity of proteolytic cleavage of an inactive precursor protein of Sendai virus. Virology 57:475-490. [DOI] [PubMed] [Google Scholar]
- 34.Singh, M., R. Cattaneo, and M. A. Billeter. 1999. A recombinant measles virus expressing hepatitis B virus surface antigen induces humoral immune responses in genetically modified mice. J. Virol. 73:4823-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skehel, J. J., and D. C. Wiley. 1998. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell 95:871-874. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weissenhorn, W., A. Carfi, K. H. Lee, J. J. Skehel, and D. C. Wiley. 1998. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell 2:605-616. [DOI] [PubMed] [Google Scholar]
- 38.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 39.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289:366-373. [DOI] [PubMed] [Google Scholar]
- 40.World Health Organization. 2000. World Health Report 2000. World Health Organization, Geneva, Switzerland.
- 41.Yu, M., E. Wang, Y. Liu, D. Cao, N. Jin, C. W. Zhang, M. Bartlam, Z. Rao, P. Tien, and G. F. Gao. 2002. Six-helix bundle assembly and characterization of heptad repeat regions from the F protein of Newcastle disease virus. J. Gen. Virol. 83:623-629. [DOI] [PubMed] [Google Scholar]