Abstract
The genome of the coronavirus transmissible gastroenteritis virus (TGEV) has been engineered as an expression vector with an infectious cDNA. The vector led to the efficient (>40 μg/106 cells) and stable (>20 passages) expression of a heterologous gene (green fluorescent protein [GFP]), driven by the transcription-regulating sequences (TRS) of open reading frame (ORF) 3a inserted in the site previously occupied by the nonessential ORFs 3a and 3b. Expression levels driven by this TRS were higher than those of an expression cassette under the control of regulating sequences engineered with the N gene TRS. The recombinant TGEV including the GFP gene was still enteropathogenic, albeit with a 10- to 102-fold reduction in enteric tissue growth. Interestingly, a specific lactogenic immune response against the heterologous protein has been elicited in sows and their progeny. The engineering of an additional insertion site for the heterologous gene between viral genes N and 7 led to instability and to a new genetic organization of the 3′ end of the recombinant viruses. As a consequence, a major species of subgenomic mRNA was generated from a TRS with the noncanonical core sequence 5′-CUAAAA-3′. Extension of the complementarity between the TRS and sequences at the 3′ end of the viral leader was associated with transcriptional activation of noncanonical core sequences. The engineered vector led to expression levels as high as those of well-established vectors and seems very promising for the development of vaccines and, possibly, for gene therapy.
Transmissible gastroenteritis virus (TGEV) is a member of the Coronaviridae family, composed of enveloped viruses of medical and veterinary importance causing disease in humans and animals. The Coronaviridae, Arteriviridae, and Ronaviridae families are included in the Nidovirales order and, despite significant differences in their genome size, have the same polycistronic genome organization and regulation of gene expression, leading to a nested set of subgenomic mRNAs (sgmRNAs) (7, 8, 15). The TGEV genome is a single-stranded, positive-sense 28.5-kb RNA. About two-thirds of the entire RNA comprises open reading frames (ORFs) 1a and 1b, encoding the replicase. The 3′ one-third of the genome comprises the genes encoding the structural and nonstructural proteins (13). Sequences preceding each gene represent signals for the discontinuous transcription of sgmRNAs (26, 44). These are the transcription-regulating sequences (TRSs), which include a highly conserved core sequence, 5′-CUAAAC-3′, that is identical in all TGEV genes, and the core sequence 5′ upstream (5′ TRS) and 3′ downstream (3′ TRS) flanking sequences (3). The high conservation of the core sequence suggests that this sequence may be particularly relevant in the virus life cycle.
The recent construction of a full-length cDNA clone of TGEV (2, 54) has created the possibility of specifically engineering the TGEV genome to study fundamental viral processes and to develop expression vectors. Coronaviruses have several advantages over other viral expression systems as vectors (for a review, see reference 14). For instance, these viruses have the largest RNA virus genome and, in principle, have room for the insertion of large foreign genes (14, 32). Since coronaviruses, in general, infect the respiratory and enteric mucosal surfaces, they may be used to target the antigen to these areas to induce a pleiotropic secretory immune response that includes lactogenic immunity (16).
Two types of coronavirus-derived expression systems have been developed. One type, the helper-dependent expression systems, permits the production of significant levels of heterologous genes (2 to 8 μg/106 cells), although with limited stability (4). Another corresponds to single-genome coronavirus vectors, which were obtained first for murine hepatitis virus (MHV) by targeted recombination (17, 21, 32) and, recently, for TGEV and human coronavirus 229E (HCoV-229E) by the construction of an infectious cDNA clone (9, 45). However, the wide potential of coronaviruses as vectors has not yet been systematically investigated.
In this report, the infectious cDNA clone obtained for TGEV as a bacterial artificial chromosome (2) has been engineered as a vector to express high levels of the heterologous green fluorescent protein (GFP) gene with transcription-regulating sequences of the nonessential gene 3a (9, 16, 27, 34, 38, 52) or regulating sequences engineered from the N gene TRS. The TGEV-derived virus vector elicited an efficient lactogenic immune response in swine against both the vector and the heterologous gene, showing its potential to provide protection to piglets against mucosal infections.
MATERIALS AND METHODS
Cells and viruses.
The TGEV PUR46-MAD strain (43) was grown and titrated as previously described (24). Baby hamster kidney cells (BHK-21) stably transformed with the gene coding for porcine aminopeptidase N (BHK-APN) (11) were grown in Dulbecco's modified Eagle's medium supplemented with 5% fetal calf serum and geneticin (G418; 1.5 mg/ml) as a selection agent. Viruses were grown in swine testis cells (33).
Plasmid constructs.
To delete the nonessential genes 3a and 3b from the TGEV genome (GenBank accession number AJ271965), the 872-bp PpuMI-BlpI fragment comprising nucleotides 24822 to 25693 was removed from the intermediate plasmid pSL-SC11-3EMN7C8-BGH (19), which was subsequently blunt-ended with T4 DNA polymerase and religated, generating plasmid pSL-SC11-Δ3EMN7C8-BGH. The 2,029-bp AvrII-AvrII fragment, including the ORF 3a and 3b deletion, was inserted into the corresponding sites (nucleotides 22967 and 25867 of the TGEV genome) of plasmid pBAC-TGEVΔClaI (2), leading to plasmid pBAC-TGEV-Δ3ΔClaI. Insertion of the ClaI-ClaI restriction fragment (nucleotides 4417 to 9615) into ClaI-linearized pBAC-TGEV-Δ3ΔClaI led to plasmid pBAC-TGEV-Δ3.
The mammalian codon-optimized version of the GFP gene was amplified from plasmid pGL-1 (Gibco-BRL) with a forward oligonucleotide (5′-GCCAGGTCCTGTATGAGCAAGGGCGAGG-3′) and a reverse oligonucleotide (5′-GGCGCTAAGCTCACTTGTACAGCTCG-3′), which included PpuMI and BlpI restriction endonuclease sites, respectively (bold nucleotides). The GFP initiation and stop codons are underlined. The GFP gene was cloned at the PpuMI and BlpI sites of plasmid pSL-SC11-3EMN7C8-BGH, replacing the dispensable TGEV ORFs 3a and 3b. Transfer of the 2,752-bp AvrII-AvrII fragment including the GFP gene into the corresponding sites of plasmid pBAC-TGEVΔClaI and subsequent insertion of the ClaI-ClaI fragment led to plasmid pBAC-TGEV-Δ3-TRS3a-GFP, encoding the GFP gene downstream of the transcription-regulatory sequences of ORF 3a.
The transcription efficacy of an engineered TRS including the 5′ TRS from the N gene (TRSn) at the position of the deleted ORFs 3a and 3b was also analyzed. The TRSn preceding the GFP gene was synthesized by overlap extension PCR. A forward oligonucleotide (5′-GCACTTGGTGGAGGCGCCGTGGCTATACCTTTTGC-3′) including the restriction site NarI (bold nucleotides) and a reverse oligonucleotide (5′-CCGAGGACCTGTTTAGTTATACCATATGTAATAATAATTTTAAATTTAATGGACGTGCACTTTTTCAATTGG-3′) containing the restriction site PpuMI (bold nucleotides), the core sequence (underlined nucleotides), and 22 nucleotides from the 5′-flanking sequences of the N gene (italic nucleotides) were used to amplify a PCR product comprised of nucleotides 23443 to 24712 of the TGEV genome. The GFP gene was independently PCR amplified from plasmid pGL-1 with a forward oligonucleotide (5′-GGCAGGTCCTGCCATGAGCAAGGGCGAGGAAC-3′) and a reverse oligonucleotide (5′-GGCGCTAAGCTCACTTGTACAGCTCG-3′) including restriction sites for PpuMI and BlpI, respectively (bold nucleotides). The GFP initiation and stop codons are underlined.
Both PCR products were gel purified and used as templates for a third PCR, performed with the two outer oligonucleotides as primers. The final PCR product was digested with restriction endonucleases NarI and BlpI and cloned into the corresponding sites of plasmid pSL-SC11-3EMN7C8-BGH. Transfer of the AvrII-AvrII fragment to the corresponding sites of plasmid pBAC-TGEVΔClaI and subsequent insertion of the ClaI-ClaI fragment, as described above, led to plasmid pBAC-TGEV-Δ3-TRSn-GFP, encoding the GFP gene downstream of the N gene 5′ TRS at the position of deleted ORFs 3a and 3b.
The effect on transcription of the insertion site within the TGEV genome was analyzed by introducing an expression cassette containing the 5′ TRS from the N gene and the GFP gene either at the position of deleted ORFs 3a and 3b (see above) or between the N and 7 genes. The TRSn preceding the GFP gene was synthesized by PCR-directed mutagenesis with the plasmid pSL-SC11-Δ3EMN-AscI-7C8-BGH, including a unique AscI restriction site separating the N and 7 genes, as the template. The forward oligonucleotide (5′-CAGAGCAAGATGTGGTACCTGATGC-3′) including the KpnI restriction site (bold nucleotides) and the reverse oligonucleotide (5′-GCCTTGGCGCGCCGTTTAGTTATACCATATGTAATAATTTTTTAGTTCGTTACCTCATCAATTATCTCAACCTGTGT-3′) containing the AscI restriction site (bold nucleotides), the core sequence (underlined nucleotides), and 22 nucleotides from the 5′-flanking sequences of the N gene (italic nucleotides) were used to amplify a PCR product comprised of nucleotides 27971 to 28028 of the TGEV genome. The PCR product was digested with restriction endonucleases KpnI and AscI and cloned into the corresponding sites of plasmid pSL-SC11-Δ3EMN-AscI-7C8-BGH, leading to pSL-SC11-Δ3EMN-TRSn-AscI-7C8-BGH.
The GFP gene was amplified from plasmid pGL-1 with a forward oligonucleotide (5′-TTGGCGCGCCATGAGCAAGGGCGAG-3′) and a reverse oligonucleotide (5′-TTGGCGCGCCTCACTTGTACAGCTCG-3′) including the AscI restriction site (bold nucleotides). The GFP initiation and stop codons are underlined. The PCR product was digested with restriction endonuclease AscI and cloned into the corresponding site of plasmid pSL-SC11-Δ3EMN-TRSn-AscI-7C8-BGH, leading to pSL-SC11-Δ3EMN-TRSn-GFP-7C8-BGH. Transfer of the 5,018-bp NarI-BamHI fragment to the corresponding sites of plasmid pBAC-TGEVΔClaI and subsequent insertion of the ClaI-ClaI fragment, as described above, led to plasmid pBAC-TGEV-Δ3-N-TRSn-GFP-7, encoding the GFP gene downstream of the 5′ TRS from the N gene inserted between the N and 7 genes. To ensure that the expected plasmids were generated, the constructs were sequenced at the cloning junctions with an Applied Biosystems 373 DNA sequencer.
Transfection and recovery of infectious TGEV from cDNA clones.
BHK-APN cells were grown to 60% confluence in 35-mm-diameter plates and transfected with 10 μg of either pBAC-TGEV-Δ3, pBAC-TGEV-Δ3-TRS3a-GFP, pBAC-TGEV-Δ3-TRSn-GFP, or pBAC-TGEV-Δ3-N-TRSn-GFP-7 plasmid and 15 μg of Lipofectin (Life Technologies, Gibco) according to the manufacturer's specifications. The efficiency of this transfection system was 8%. Cells were incubated at 37°C for 6 h, and then the transfection medium was replaced with fresh Dulbecco's modified Eagle's medium containing 10% (vol/vol) fetal bovine serum. After an incubation period of 2 days, the cell supernatants (referred to as passage 0) were harvested and passaged four times on fresh swine testis cell monolayers. Viruses present in the cell supernatant were quantified by plaque titration. After four passages, viruses were cloned by three plaque purification steps. Unstable recombinant TGEV (rTGEV) viruses were cloned from passage 0 supernatant.
RNA analysis by Northern blotting.
Total intracellular RNA was extracted at 16 h postinfection from virus-infected swine testis cells with the Ultraspec RNA isolation system (Biotecx) according to the manufacturer's instructions. RNAs were separated in denaturing 1% agarose-2.2 M formaldehyde gels and blotted onto nylon membranes (Duralon-UV; Stratagene) as described before (3). Northern hybridizations were performed with hybridization buffer containing [α-32P]dATP-labeled probe synthesized with a random-priming procedure (Strip-EZpec DNA; Ambion) according to the manufacturer's instructions. The 3′ untranslated region-specific single-stranded DNA probe was complementary to nucleotides 28300 to 28544 of the TGEV strain PUR46-MAD genome (40). After hybridization, RNA was analyzed with a Personal FX molecular imager (Bio-Rad).
RT-PCR.
Detection of GFP and 7 sgmRNAs was performed by reverse transcription (RT)-PCR. Total intracellular RNA was extracted (see above) at 16 h postinfection from swine testis cells infected with rTGEV viruses. cDNAs were synthesized at 42°C for 1 h with Moloney murine leukemia virus reverse transcriptase (Ambion) and antisense primers GFP2 (5′-GGCGCTAAGCTCACTTGTACAGCTCG-3′), complementary to nucleotides 702 to 717 of the GFP gene; GFP-AscI (5′-TTGGCGCGCCTTACTTGTACAGCTCG-3′), complementary to nucleotides 705 to 720 of the GFP gene; or X3-136 (5′-TCTGGTTTCTGCTAAACTCC-3′), complementary to nucleotides 136 to 155 of gene 7. The cDNAs generated were used as templates for sgmRNA-specific PCRs (leader-body PCR). A virus sense primer, leader 15+ (5′-GTGAGTGTAGCGTGGCTATATCTCTTC-3′), complementary to nucleotides 15 to 41 of the TGEV leader sequence, and the reverse-sense primers described for the RT reaction were used for the PCR.
Optimization of RT-PCRs for semiquantitative analysis was carried out by normalization to the viral genomic RNA (gRNA) in each sample and the use of serial dilutions of the cDNAs. To determine the relative amount of gRNA, the forward oligonucleotide Orf1a 4310 (5′-CTTTTATCAGGGTGCTTTGG-3′) and the reverse oligonucleotide Orf1a 4829 (5′-AACAGACACACGTTCATGG-3′), complementary to TGEV nucleotides 4310 to 4329 and 4811 to 4829, respectively, were used. PCR was performed with a GeneAmp PCR system 9600 thermocycler (Perkin-Elmer) for 35 cycles. Each cycle comprised 30 s of denaturation at 94°C, 45 s of annealing at 53°C, and 1.5 min of extension at 72°C. The 35 cycles were followed by a 10-min incubation at 72°C. RT-PCR products were separated by electrophoresis in 0.8% agarose gels, purified, and used for direct sequencing with the oligonucleotide leader 15+ and the same reverse oligonucleotide used for PCR.
Western blot analysis.
Protein expression in cells infected with rTGEV viruses was analyzed at different passages by Western blot. Cell lysates were separated by gradient sodium dodecyl sulfate-polyacrylamide (5 to 20%) gel electrophoresis (SDS-PAGE). Proteins were transferred to a nitrocellulose membrane with a Bio-Rad Mini Protean II electroblotting apparatus at 150 mA for 2 h in 25 mM Tris-192 mM glycine buffer (pH 8.3) containing 20% methanol. Membranes were blocked for 1 h with 5% dried skimmed milk in TBS (20 mM Tris-HCl [pH 7.5], 150 mM NaCl). The membranes were then incubated with monoclonal antibodies specific for the GFP protein (Roche) or the TGEV N protein (3D.C10), diluted 1:1,000 in TTBS buffer (TBS with 0.1% Tween 20). Bound antibody was detected with horseradish peroxidase-conjugated rabbit anti-mouse immunoglobulin diluted 1:3,000 in TTBS buffer and the ECL detection system (Amersham Pharmacia Biotech). The amount of GFP protein expressed was determined by Western blot, with standard calibration curves generated by using purified GFP (Roche). TGEV N protein was used as an internal standard.
Flow cytometry analysis.
Swine testis cells grown in 35-mm-diameter dishes were infected with the recombinant virus rTGEV-Δ3-TRS3a-GFP, rTGEV-Δ3-TRSn-GFP, or rTGEV-Δ3-N-TRSn-GFP (multiplicity of infection, 1). At 16 h postinfection, cells were washed and resuspended in 500 μl of phosphate-buffered saline. GFP expression in mock-infected and infected cells was analyzed on a flow cytometry system (Epics XL-MCL; Coulter).
Virulence assay.
The in vivo growth and virulence of TGEV recombinant viruses were determined as described before (42). Briefly, groups of 14 2- to 4-day-old swine obtained by crossing Large White and Belgium Landrace pigs were oronasally and intragastrically inoculated with 108 PFU per route of rPUR-MAD-SC11, rTGEV-Δ3, or rTGEV-Δ3-TRS3a-GFP virus in a biosafety level 3 containment facility. The virus titers in lung, jejunum, and ileum were determined 2 days after infection.
Immunization of swine.
Two pregnant sows seronegative for TGEV, as tested by enzyme-linked immunosorbent assay (ELISA), were immunized with the recombinant virus rTGEV-Δ3-TRS3a-GFP by the nasal and intramuscular routes (108 PFU in 5 ml of Dulbecco's modified Eagle's medium per route) at days 35 and 75 of gestation. Serum from the sows and their progeny (nine and seven piglets) was collected at days 2, 7, 19, and 27 after delivery. Colostrum was collected from the sows on the day of delivery. The antibody response against TGEV and GFP was evaluated by ELISA in serum and colostrum.
ELISA.
Antibodies generated against GFP and TGEV were detected by ELISA as described before (41). ELISA was performed with purified TGEV (0.2 μg per well) or GFP protein (75 ng per well) as the antigen, bound to 96-well microplates, saturated with 5% bovine serum albumin in phosphate-buffered saline for 2 h at 37°C, and incubated with serial dilutions of the serum sample in phosphate-buffered saline-0.1% bovine serum albumin for 3 h at room temperature. Microplates were washed six times with 0.1% bovine serum albumin and 0.1% Tween 20 in phosphate-buffered saline, and bound antibodies were detected by incubation with peroxidase-conjugated protein A diluted 1:2,000 in phosphate-buffered saline with 0.1% bovine serum albumin. ELISAs were developed with phenylenediamine dihydrochloride (Sigma FAST) as the peroxidase substrate for 15 min at room temperature. Reactions were stopped with 1.5 M H2SO4, and the absorbance was read at 492 nm.
RESULTS
Construction of recombinant TGEV vector expressing GFP.
In order to increase the cloning capacity of the TGEV single genome, the nonessential ORFs 3a and 3b were eliminated from the full-length cDNA clone, creating a deletion of 872 bp (nucleotides 24822 to 25693) in the TGEV sequence (GenBank accession number AJ271965), leading to a cDNA encoding the recombinant virus rTGEV-Δ3. This deletion preserves the TRS of TGEV ORF 3a, consisting of the native 5′ TRS, the core sequence (5′-CUAAAC-3′), and 19 out of the 23 nucleotides of the 3′ TRS. The last 177 nucleotides of TGEV ORF 3b were also conserved to maintain the 5′ TRS of the next gene (ORF E).
Swine testis cells were transfected with the cDNA, and the rTGEV-Δ3 virus was recovered 48 h posttransfection. Intracellular RNAs isolated from cells infected with the wild-type and rTGEV-Δ3 viruses were analyzed by Northern blot (Fig. 1). Results from several experiments showed that differences in the relative amounts of each of the six mRNAs (genomic RNA and mRNAs of viral genes S, E, M, N, and 7) were not significant, indicating that the engineered deletion did not affect viral transcription. An additional subgenomic mRNA (sgmRNA Δ3) 872 nucleotides smaller than the viral mRNA 3a was generated from the remaining TRS of ORF 3a at similar levels as the mRNA 3a from the wild-type TGEV (Fig. 1), indicating that the TRS of TGEV ORF 3a was functional in the new viral context. The mRNA Δ3 was amplified by RT-PCR with primer pairs complementary to the leader RNA sequence and to sequences downstream of the deletion and subsequently sequenced, confirming the synthesis of the new transcript from the ORF 3a core sequence (data not shown).
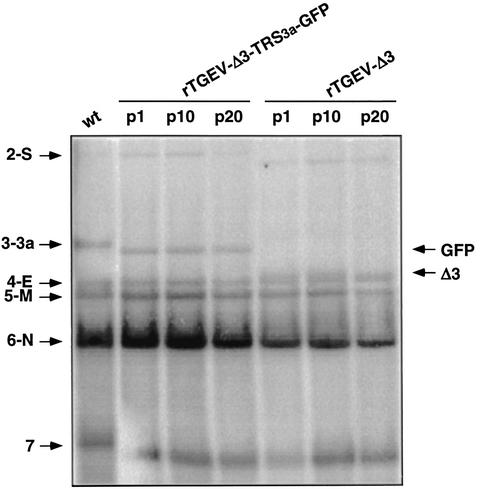
FIG. 1.
Stability of rTGEV-Δ3 and rTGEV-Δ3-TRS3a-GFP recombinant viruses. Northern blot analysis of intracellular RNAs extracted at passages 1 (p1), 10 (p10), and 20 (p20) from cells infected with the wild-type (wt), rTGEV-Δ3, or rTGEV-Δ3-TRS3a-GFP virus. Hybridization was performed with a probe complementary to the 3′ untranslated region of the genome. Numbers and letters on the left indicate the position of TGEV mRNAs. S, spike; 3a, gene 3a; E, envelope; M, membrane; N, nucleoprotein; 7, gene 7. The positions of the new sgmRNAs generated from recombinant viruses are indicated by arrows on the right side of the figure. GFP, sgmRNA encoding GFP. Δ3, sgmRNA synthesized from the TRS of ORF 3a in the deletion mutant rTGEV-Δ3. The mobility shift noted in mRNA 7 of wild-type virus is a product of running the gel and does not represent size differences in the sgmRNA. The transfer efficiency of mRNAs during the Northern blotting is highly dependent on mRNA size, and some variability may be observed, especially in the transfer efficiency of RNAs of the larger (gRNA) or smaller (mRNA 7) size. Genomic RNA is not shown because in this experiment, the transfer conditions of RNAs during Northern blotting were set up to optimize the transfer of non-genome-size mRNAs.
The heterologous GFP gene (717 nucleotides) was inserted in cDNA constructs, replacing the deleted TGEV ORFs 3a and 3b (nucleotides 24822 to 25693 of PUR46-MAD). The reporter gene GFP was selected to facilitate analysis of expression at the cellular level. The resulting virus vector encoding GFP was designated rTGEV-Δ3-TRS3a-GFP (Fig. 2A). The GFP gene was located just downstream of the native TRS of TGEV ORF 3a, comprising the 5′ TRS, the core sequence, and 23 nucleotides of the 3′ TRS preceding the translation initiation codon (AUG). This transcriptional unit, including the GFP gene, was designed to avoid the duplication of genetic information and to reduce the probability of losing the foreign gene by homologous recombination. Swine testis cells were transfected with the cDNA encoding the rTGEV-Δ3-TRS3a-GFP genome, and infectious virus was recovered 48 h posttransfection. Intracellular RNAs from cell cultures infected with rTGEV-Δ3-TRS3a-GFP virus were analyzed by Northern blot, showing the synthesis of a GFP mRNA of the expected size (3,700 nucleotides) and at levels similar to those of mRNA 3a in the wild-type virus (Fig. 1). The six viral mRNAs (genomic RNA and mRNAs of viral genes S, E, M, N, and 7) were synthesized with a ratio similar to that of the wild-type virus mRNAs.
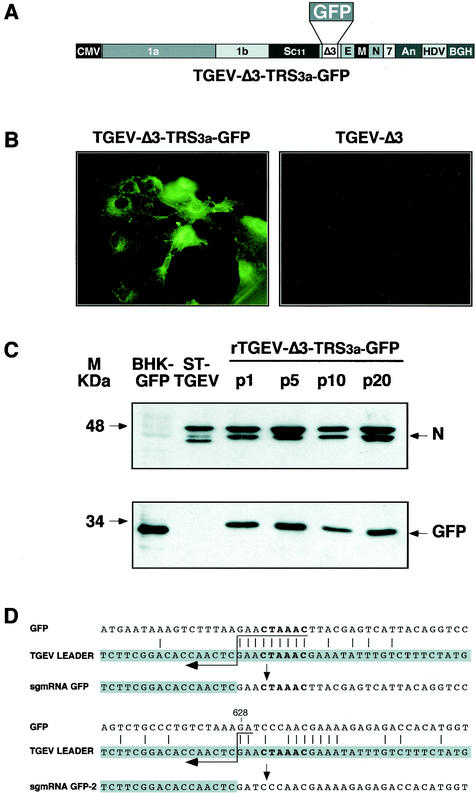
FIG. 2.
Analysis of GFP protein expressed by recombinant virus rTGEV-Δ3-TRS3a-GFP. (A) Schematic structure of the cDNA encoding the rTGEV-Δ3-TRS3a-GFP RNA with the GFP gene under the TRS of ORF 3a. The numbers and letters inside the rectangles indicate the viral genes. CMV, cytomegalovirus immediate-early promoter; An, poly(A); HDV, hepatitis delta virus ribozyme; BGH, bovine growth hormone termination and polyadenylation signals. (B) GFP expression observed by fluorescence microscopy in rTGEV-Δ3-TRS3a-GFP- but not in rTGEV-Δ3-infected swine testis cells. (C) Western blot analysis of GFP protein expressed by rTGEV-Δ3-TRS3a-GFP in infected swine testis cells. Cell lysates from passages 1 (p1), 5 (p5), 10 (p10), and 20 (p20) were separated by SDS-PAGE under reducing conditions and probed with monoclonal antibodiesspecific for the viral N protein (upper panel) or GFP (lower panel). BHK-GFP, extracts from BHK cells expressing the GFP protein from the noncytopathic Sindbis virus replicon were used as a positive control. ST-TGEV, extracts from TGEV-infected swine testis cells. The molecular mass is indicated on the left. (D) Nucleotide identity between the leader RNA of TGEV and the genomic sequences of rTGEV-Δ3-TRS3a-GFP surrounding the core sequence of TRS3a (upper scheme) or the noncanonical junction site in the 3′ half of the GFP gene (lower scheme). The leader region (shaded nucleotides) includes the canonical core sequence (in bold type) and the 20 immediately upstream and downstream nucleotides. Leader-body common nucleotides are indicated by vertical bars. Postulated polymerase strand switching during minus-strand synthesis is indicated by the arrow. The jump during discontinuous transcription could have occurred anywhere within the consecutive nucleotides underlined by the tail of the arrow. The number above the genomic sequence indicates the position relative to the start of the GFP gene.
Expression of GFP at 16 h postinfection was observed in 70% of the cells infected with rTGEV-Δ3-TRS3a-GFP by fluorescence microscopy (Fig. 2B) and flow cytometry (results not shown). Expression of 40 μg of GFP protein per 106 cells was shown by Western blot with a GFP-specific monoclonal antibody (Fig. 2C), with standard calibration curves generated with purified GFP (Roche).
Cytoplasmic RNAs from cells infected with rTGEV-Δ3-TRS3a-GFP were analyzed by RT-PCR with primer pairs complementary to the leader sequence and to the 3′ end of the GFP gene. The expected RT-PCR amplification product corresponding to the complete GFP gene (0.8 kb), and another minor amplification product (0.2 kb) were detected (data not shown). The larger amplified cDNA corresponded to the GFP mRNA generated by a canonical leader-to-body fusion at the hexanucleotide 5′-CUAAAC-3′ into the TRS of ORF 3a, as determined by sequencing (Fig. 2D). Sequencing of the smaller amplification product, sgmRNA GFP-2, showed that this new transcript was generated by a leader-to-body joining at an internal noncanonical GFP site (Fig. 2D) preceded by the sequence 5′-CUAAAGA-3′, showing the presence of cryptic transcription signals within the 3′ end of the GFP gene and the striking observation that a canonical core sequence is not essential for transcription in TGEV.
Stability and growth of recombinant viruses rTGEV-Δ3 and rTGEV-Δ3-TRS3a-GFP in cell culture and in vivo.
The stability of recombinant viruses rTGEV-Δ3 and rTGEV-Δ3-TRS3a-GFP was studied for 20 passages in cell culture. Northern blot analysis of intracellular RNAs extracted from swine testis cells at passages 1, 10, and 20 showed no differences either in the expected RNA pattern or in the abundance of GFP mRNA (Fig. 1). Analysis of the cytoplasmic extracts by Western blot showed that the rTGEV-Δ3-TRS3a-GFP virus expressed similar levels of the GFP protein after 20 passages in cell culture (Fig. 2C). These results indicate that expression of heterologous genes in this context of the viral genome is very stable.
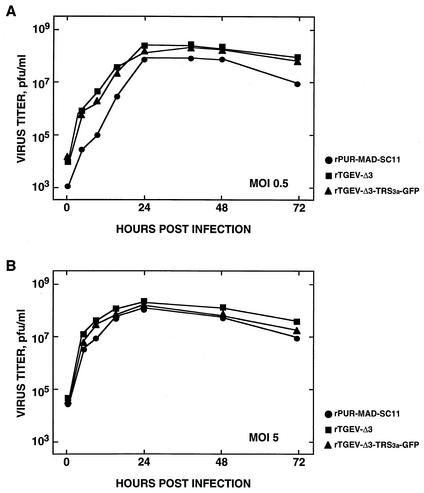
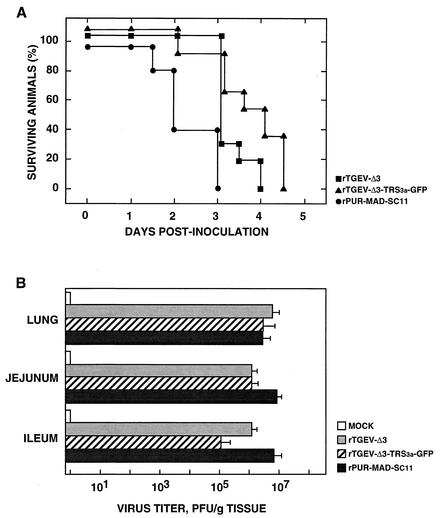
The recombinant viruses rTGEV-Δ3 and rTGEV-Δ3-TRS3a-GFP showed growth kinetics in cell culture similar to those of the parental virus rPUR-MAD-SC11 at both low and high (0.5 and 5, respectively) multiplicities of infection (Fig. 3). The cytopathic effect included induction of cell fusion and formation of large plaques (3-mm diameter) as described for the parental virus (rPUR-MAD-SC11) carrying the S gene from strain PUR-C11 (2). The in vivo properties of the rTGEV-Δ3 and rTGEV-Δ3-TRS3a-GFP viruses were also similar to those of the parental virus rPUR-MAD-SC11, showing a virulence (100% mortality, 14 of 14 piglets) comparable to that of the PUR-C11 strain of TGEV for breast-fed newborn animals. However, a delay of 1 day (3.25 ± 0.42) or 1.5 days (3.75 ± 0.42) in the mean day of death was observed for recombinant viruses rTGEV-Δ3 and rTGEV-Δ3-TRS3a-GFP, respectively, compared to rPUR-MAD SC11 (mean day of death, 2.25 ± 0.63) (Fig. 4A). The P values associated with comparison of the mean day of death between the wild-type and recombinant viruses were, in both cases, near the significance level.
FIG. 3.
Growth kinetics of recombinant viruses rTGEV-Δ3 and rTGEV-Δ3-TRS3a-GFP in cell culture. Growth of rPUR-MAD-SC11, rTGEV-Δ3, and rTGEV-Δ3-TRS3a-GFP viruses on swine testis cells after infection at (A) low and (B) high multiplicities of infection (0.5 and 5, respectively). Mean values of three experiments are indicated. The standard deviation was less than 30% in all cases (not shown).
FIG. 4.
Growth of recombinant viruses rTGEV-Δ3 and rTGEV-Δ3-TRS3a-GFP in swine. Groups of 14 2- to 4-day-old piglets were oronasally and intragastrically inoculated with 2 × 108 PFU/animal of rPUR-MAD-SC11, rTGEV-Δ3, or rTGEV-Δ3-TRS3a-GFP. (A) Number of surviving piglets at different times postinoculation. Results of a representative experiment of two that gave similar results are shown. The significance of the results was analyzed by the Student t test (see Results section). (B) Growth of rPUR-MAD-SC11, rTGEV-Δ3, and rTGEV-Δ3-TRS3a-GFP viruses in the indicated tissues. Values are the means for representative tissue samples from three animals sacrificed 2 days after inoculation. Error bars represent the standard deviation.
The recombinant viruses rTGEV-Δ3 and rTGEV-Δ3-TRS3a-GFP grew with high titers both in the enteric tract (>105 PFU/g of tissue) and in the lungs (>106 PFU/g of tissue) of infected animals. Virus titers in the lungs were similar to those obtained for rPUR-MAD-SC11. However, the growth of recombinant viruses in enteric tissues decreased 10- to 102-fold compared with rPUR-MAD-SC11 (Fig. 4B). These results demonstrate that deletion of ORFs 3a and 3b of TGEV and insertion of the heterologous gene GFP had no apparent effect in cell culture but did have a limited effect on the virus's behavior in swine.
Lactogenic immunity induced in pigs by TGEV-derived virus vector rTGEV-Δ3-TRS3a-GFP.
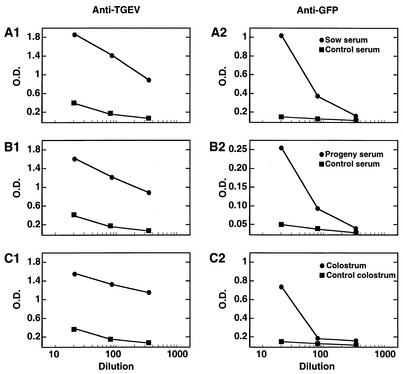
In order to evaluate the potential of the TGEV-derived virus vector to provide protection against enteric and respiratory infections by the expression of antigens, pregnant sows were immunized twice (at days 35 and 75 of gestation) with the virus rTGEV-Δ3-TRS3a-GFP. The antibody response (immunoglobulin A [IgA] plus IgG) against the vector and the heterologous GFP protein was determined by ELISA with the sera of the sows and their progeny and in the colostrum. TGEV-specific antibodies reached titers ranging from 1 × 103 to 5 × 103 in the serum of both the sows and their progeny (Fig. 5A1 and 5B1). GFP-specific antibodies were detected in the serum of sows and their progeny with titers of around 3 × 102 and 1 × 102, respectively (Fig. 5A2 and 5B2). Interestingly, the colostrum from day 1 of lactation contained TGEV- and GFP-specific antibodies, with titers of around 2 × 103 and 1 × 102, respectively (Fig. 5C1 and 5C2). The presence of significant levels of antibodies specific for TGEV and GFP in the serum of the piglets demonstrated that vector rTGEV-Δ3-TRS3a-GFP had elicited lactogenic immunity.
FIG. 5.
Lactogenic immune response induced in swine by recombinant rTGEV-Δ3-TRS3a-GFP virus. Two pregnant sows seronegative for TGEV were immunized with the recombinant virus rTGEV-Δ3-TRS3a-GFP. The virus (2 × 108 PFU per animal) was administered by the nasal and intramuscular routes at days 35 and 75 of gestation. Serum from the sows and their progeny was collected at days 2, 7, 19, and 27 after delivery. Colostrum was collected from sows on the day of delivery. The antibody response against TGEV (A1, B1, and C1) and GFP (A2, B2, and C2) in the serum of the sows (A) and the progeny (B) and in the colostrum (C) was evaluated by ELISA. Values represented are the maximum level detected in the serum of sows (days 19 and 27 after delivery) and in the serum of the progeny (day 2 after delivery).
Expression of GFP gene with 5′ TRS derived from N gene.
Previous results obtained with the minigenome expression system (3) delimited the extension of the essential TRS required for efficient expression of heterologous genes in TGEV. To compare the sgmRNA expression efficacy of an engineered TRS and the native ORF 3a TRS in the context of the single-genome vector, the heterologous gene GFP was inserted at the position of the deleted TGEV ORFs 3a and 3b under the control of a TRS (TRSn) consisting of 22 nucleotides of the 5′ TRS derived from the viral N gene, the core sequence (5′-CUAAAC-3′), and a 10-nucleotides sequence of nonviral origin flanking the 3′ end of the core sequence (Fig. 6A). This artificial 3′ TRS, 5′-AGGTCCTGCC-3′, included a unique restriction endonuclease site and an optimized Kozak sequence and was similar to another one assayed in the minigenome system that showed high expression levels for the heterologous gene (3).
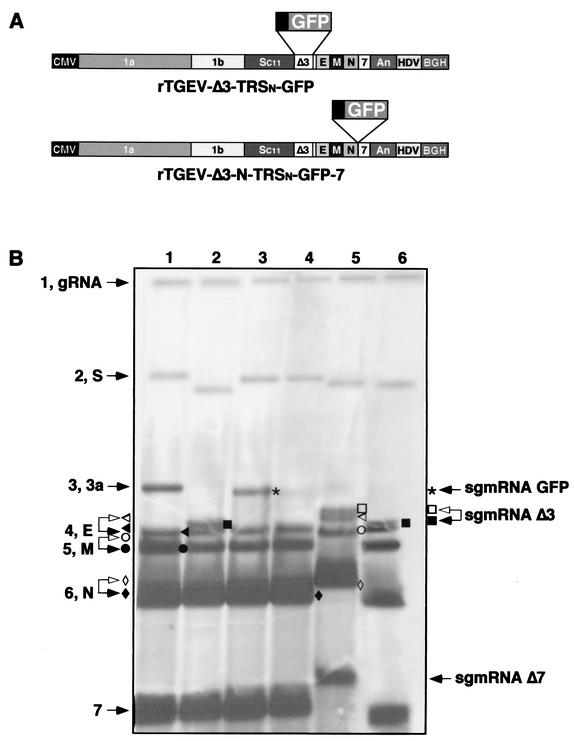
FIG. 6.
Northern blot analysis of recombinant TGEV viruses carrying the GFP gene under different TRSs and at different positions of the viral genome. (A) Schematic structure of the cDNAs encoding the rTGEV-Δ3-TRSn-GFP-7 and rTGEV-Δ3-N-TRSn-GFP-7 RNAs with the GFP gene under the 5′ TRS of the N gene inserted at place where ORFs 3a and 3b were deleted or between genes N and 7, respectively. The numbers and letters inside the rectangles indicate the viral genes. CMV, cytomegalovirus immediate-early promoter; An, poly(A); HDV, hepatitis delta virus ribozyme; BGH, bovine growth hormone termination and polyadenylation signals. (B) Northern blot analysis of intracellular RNAs extracted at passage 2 from cells infected with the wild-type TGEV virus (lane 1), rTGEV-Δ3 (lane 2), rTGEV-Δ3-TRS3a-GFP (lane 3), rTGEV-Δ3-TRSn-GFP-7 (lane 4), rTGEV-Δ3-N-TRSn-GFP-7 clone 3 (lane 5), or rTGEV-Δ3-N-TRSn-GFP-7 clone 12 (lane 6). Hybridization was performed with a probe complementary to the 3′ untranslated region of the genome. Numbers and letters on the left indicate the position of the genomic (gRNA) and viral mRNAs. S, spike; 3a, gene 3a; E, envelope; M, membrane; N, nucleoprotein; 7, gene 7. The positions of the new sgmRNAs generated from recombinant viruses are indicated by arrows on the right side of the figure. sgmRNA-GFP, sgmRNA encoding GFP gene; sgmRNA-Δ3, sgmRNA synthesized from the TRS of ORF 3a in the deletion mutant rTGEV-Δ3; sgmRNA-Δ7, sgmRNA synthesized by rTGEV-Δ3-N-TRSn-GFP-7 clone 3 from a noncanonical junction site. Open arrowheads and symbols indicate sgmRNAs of rTGEV-Δ3-N-TRSn-GFP-7 clone 3 with detectable mobility differences relative to the corresponding sgmRNAs of wild-type TGEV or rTGEV-Δ3 (solid arrowheads and symbols).
TGEV ORF 3a TRS was deleted and replaced by the TRSn. The expression cassette was inserted just downstream of the S gene stop codon (nucleotides 24722 to 25693 of PUR46-MAD virus), leading to the recombinant virus rTGEV-Δ3-TRSn-GFP, which showed growth kinetics similar to those of the parental virus rPUR-MAD-SC11 in cell culture at both low and high multiplicities of infection (data not shown). RNAs from cells infected with rTGEV-Δ3-TRSn-GFP showed (Fig. 6B) the new mRNA TRSn-GFP (3,623 nucleotides) and the expected viral mRNAs by Northern blot, and there were no differences in their relative amounts and sizes. Although the amounts of each sgmRNA are very reproducible in the same experiment, some differences may be observed between experiments in the ratio of low- to high-molecular-weight sgmRNAs as a consequence of differences in the efficiency of RNA transfer.
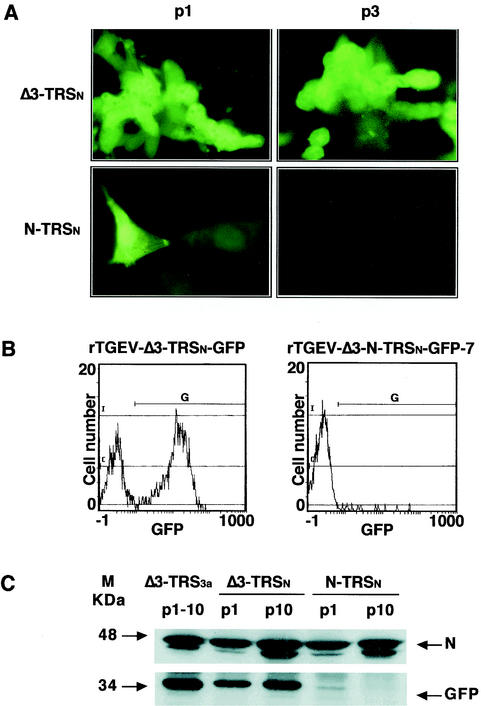
The GFP mRNA was amplified by RT-PCR with primer pairs complementary to the leader RNA sequence and to the 3′ end of the GFP gene and sequenced, confirming the synthesis of the new transcript from the core sequence of the TRSn. Heterologous GFP mRNA transcription levels under the control of TRS3a and TRSn were compared by Northern blot and by semiquantitative RT-PCR with twofold dilutions of the expressed mRNA (data not shown). mRNA synthesis from the TRS3a was 20-fold more efficient than from the TRSn, suggesting that, in this viral context, transcription is more efficiently driven by the native TRS3a than by the artificial TRSn. Formation of large syncytia and the expression of GFP at 16 h postinfection were observed in 70% of the cells infected with rTGEV-Δ3-TRSn-GFP by fluorescence microscopy (Fig. 7A) and by flow cytometry (Fig. 7B). Quantification of the GFP protein by Western blot with purified GFP as a standard showed that the expression levels obtained from TRS3a were fourfold higher than from the TRSn (Fig. 7C). Together, these data indicate that protein expression in these vectors is dependent on both transcriptional and translational regulation. The heterologous GFP protein was stably expressed by rTGEV-Δ3-TRSn-GFP for at least 10 passages in cell culture (Fig. 7C).
FIG. 7.
Analysis of GFP protein expressed by recombinant viruses rTGEV-Δ3-TRSn-GFP and rTGEV-Δ3-N-TRSn-GFP-7. (A) GFP expression observed by fluorescence microscopy at passages 1 (p1) and 3 (p3) in cell cultures infected with rTGEV-Δ3-TRSn-GFP (Δ3-TRSn) or rTGEV-Δ3-N-TRSn-GFP-7 (N-TRSn). (B) Flow cytometry analysis of GFP expression in swine testis cells infected (passage 1) with rTGEV-Δ3-TRSn-GFP or rTGEV-Δ3-N-TRSn-GFP-7. G represents the GFP-positive cell population. Green fluorescence intensity (GFP) is revealed on the x axis. (C) Western blot analysis of the GFP protein expressed by rTGEV-Δ3-TRSn-GFP (Δ3-TRSn) or rTGEV-Δ3-N-TRSn-GFP-7. (N-TRSn) in infected swine testis cells. Cell lysates from passages 1 (p1) and 10 (p10) were separated by SDS-PAGE under reducing conditions and probed with monoclonal antibodies specific for the viral N protein (upper panel) or GFP (lower panel). Δ3-TRS3a, GFP expressed in extracts from rTGEV-Δ3-TRS3a-GFP-infected swine testis cells from passages 1 to 10 (p1 to p10). The molecular mass is indicated on the left.
GFP expression efficiency from the 3′ end of the TGEV genome.
In order to evaluate whether the expression efficiency of the GFP gene was affected by its position in the viral genome, a new recombinant virus carrying the expression cassette at the 3′ end of the genome, between genes N and 7, was constructed. The expression cassette included the GFP gene under the control of an engineered TRS similar to the TRSn described above, with the unique restriction endonuclease site AscI at the 3′ TRS. This expression cassette was inserted downstream of the N gene stop codon (nucleotide 28066 of PUR46-MAD), preceding the TRS of gene 7, leading to the recombinant virus rTGEV-Δ3-N-TRSn-GFP-7 (Fig. 6A). In the TGEV viral genome, the N gene stop codon (UAA) is included within the core sequence (5′-CUAAAC-3′) of gene 7. To maintain the 5′ TRS of gene 7 after insertion of the expression cassette, a duplication of 15 nucleotides consisting of the last nucleotides of the N gene was introduced immediately upstream of the core sequence preceding gene 7.
After transfection of swine testis cells with the corresponding cDNA, only two (clones 3 and 12) of the 12 plaque-purified viral clones selected at passage zero contained the expression cassette, as determined by RT-PCR. These clones expressed the GFP protein, as determined by direct fluorescence (data not shown). After infection with the recombinant virus rTGEV-Δ3-N-TRSn-GFP-7, only a small percentage (around 5%) of the infected cells expressed GFP, although at high levels, as observed by fluorescence microscopy (Fig. 7A) and by flow cytometry (Fig. 7B). Overall expression levels obtained with TRSn at this insertion site were 20-fold lower than those from TRSn at the 3a site, as determined by Western blot analysis at passage one, probably as a consequence of the instability (Fig. 7C). This expression was lost after three passages in cell culture, indicating that the insertion of GFP at the 3′ end of the genome was unstable.
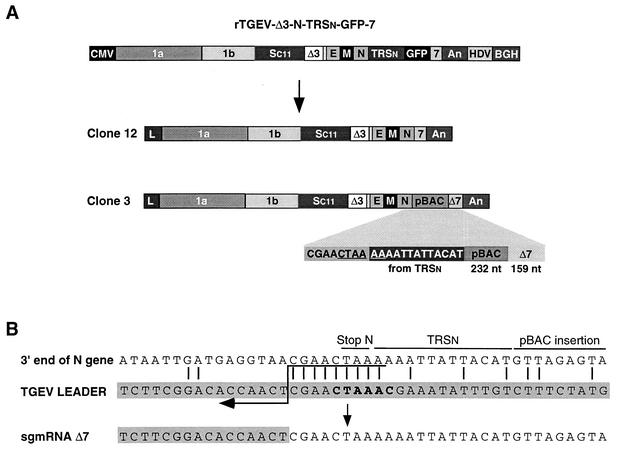
The RNAs extracted at passage one from cells infected with the two plaque-purified clones (3 and 12) of rTGEV-Δ3-N-TRSn-GFP-7 were analyzed by Northern blot (Fig. 6B and Fig. 8A), and the expected GFP mRNA (1,300 nucleotides) was below the detection limit of the technique. To further characterize the RNA composition of the resulting viruses, the genomic region and N and 7 mRNAs were amplified by RT-PCR and sequenced. The mRNA pattern of one of the plaque-purified viruses (clone 12) corresponded to a TGEV-Δ3 genome that had completely lost the GFP gene (Fig. 6B), confirming that the insertion of heterologous sequences at this position was unstable. Sequencing of the 3′-end genomic RNA from clone 12 of rTGEV-Δ3-N-TRSn-GFP-7 (Fig. 8A) amplified by RT-PCR confirmed that the expression cassette had been completely deleted, leaving the wild-type sequence and suggesting that the mechanism involved in the deletion is homologous recombination.
FIG. 8.
Genetic structure of recombinant viruses derived from rTGEV-Δ3-N-TRSn-GFP-7. (A) Schematic structure of the cDNA encoding rTGEV-Δ3-N-TRSn-GFP-7 RNA and the resulting viral clones 12 and 3, showing the new organization at the 3′ end of the genome. Below the shaded area of clone 3, the last nucleotides of the N gene, the 5′-most 13 nucleotides of TRSn, a 232-nucleotide insertion derived from the pBAC plasmid, and the 3′-end 159 nucleotides of gene 7 are represented. The noncanonical core sequence generated at the 3′ end of the N gene is underlined. The numbers and letters inside the rectangles indicate the viral genes. CMV, cytomegalovirus immediate-early promoter; An, poly(A); HDV, hepatitis delta virus ribozyme; BGH, bovine growth hormone termination and polyadenylation signals. pBAC, insertion derived from the pBAC plasmid. Δ7, gene 7 with a 78-nucleotide deletion at the 5′ end. (B) Nucleotide identity between the leader RNA of TGEV and the genomic sequences of rTGEV-Δ3-N-TRSn-GFP-7 clone 3 flanking the noncanonical junction site at the 3′ end of the N gene that led to sgmRNA Δ7. Letters above genomic sequences indicate the origin of nucleotides. The leader region (shaded nucleotides) includes the canonical core sequence (in bold type) and the 20 immediately upstream and downstream nucleotides. Leader-body common nucleotides are indicated by vertical bars. Postulated polymerase strand switching during minus-strand synthesis is indicated by the arrow. The jump during discontinuous transcription could have occurred anywhere within the consecutive nucleotides underlined by the tail of the arrow.
The other plaque-purified virus (clone 3) showed a new pattern of mRNAs apparently compatible with partial loss of the heterologous GFP gene (500 out of the 700 nucleotides of the gene) except for the anomalous size (800 nucleotides instead of the expected 600 nucleotides) of the gene 7 mRNA (mRNA Δ7) (Fig. 6B). Sequencing of the RNAs derived from rTGEV-Δ3-N-TRSn-GFP-7 viral clone 3 showed a new genetic structure for the 3′-end viral region, consisting of 13 out of the 22 nucleotides of TRSn included in the expression cassette, a 232-nucleotides insertion derived from the pBAC plasmid vector, and the last 159 nucleotides of gene 7 (Fig. 8A). These data indicate that, although the GFP gene had been completely deleted, the presence of other heterologous sequences was not deleterious to the new TGEV virus. The two recombinant virus rTGEV-Δ3-N-TRSn-GFP-7 clones (3 and 12) showed growth kinetics in cell culture similar to those of the parental virus at low and high multiplicities of infection, probably as a consequence of rapid rearrangements in virus genomes reverting virus growth to wild-type levels.
Interestingly, the sequencing of viral mRNA Δ7 transcribed from clone 3 of rTGEV-Δ3-N-TRSn-GFP-7 revealed that the leader-body junction in this new transcript took place in a noncanonical core sequence (5′-CUAAAA-3′) (Fig. 8B) located around the N gene stop codon. This finding demonstrates for the first time efficient transcription from a core sequence different from the canonical one (5′-CUAAAC-3′) in TGEV. Since the core sequence and the first 78 nucleotides of gene 7 had also been deleted, neither the corresponding mRNA nor the protein was synthesized during infection, demonstrating that the product of ORF 7 was not essential for virus growth in cell culture, as recently described by our laboratory (39).
DISCUSSION
With a TGEV infectious cDNA clone, we have shown the engineering of a TGEV-derived single genome vector that stably expresses high levels of the heterologous gene GFP from the ORF 3a TRS. GFP expression with a recombinant TGEV has been reported (9). In this paper we have made relevant progress over this initial observation, with a coronavirus-derived vector engineered as a bacterial artificial chromosome, by showing that (i) the TGEV single genome led to high expression levels of the heterologous protein (40 μg/106 cells), (ii) the virus vector stability is very high, (iii) vector tropism and virulence are present in swine, as is the induction of a lactogenic immune response, (iv) different TRSs and insertion sites have different efficacies, and (v) transcription may take place from TRSs with a noncanonical core sequence, demonstrating that complementarity between core sequence-flanking sequences and the leader TRS can compensate for the absence of a canonical core sequence.
TGEV ORFs 3a and 3b are nonessential for virus growth (9, 16, 27, 34, 53). Therefore, it was tempting to assume that insertion of heterologous genes into the TGEV genome replacing these genes would not affect the behavior of the virus either in cell culture or in vivo. In fact, we have shown in this paper that rTGEV-Δ3 virus kept the infectivity, replication efficiency, and tropism of the wild-type virus with a very small reduction in its virulence, demonstrating that these properties were influenced very little by genes 3a and 3b. Expression levels obtained with this TGEV-derived vector are of the same order as those described for vectors derived from other positive-strand RNA viruses such as Sindbis virus (50 μg/106 cells) (1, 18) or DNA expression systems such as adenovirus type 5 (10 to 100 μg/106 cells) (31, 36).
With this virus vector, a good antibody response specific for viral antigens and for the heterologous GFP protein has been demonstrated in the serum of immunized animals. Interestingly, the TGEV-derived vector elicited lactogenic immunity against the virus and the GFP by the production of specific antibodies in the colostrum of sows and the transfer of maternal antibodies to the piglets. The limited antibody response against the heterologous GFP protein compared with the high titers obtained against the TGEV vector is probably due to the known low immunogenicity of GFP (48, 49). The induction of lactogenic immunity indicates that TGEV-derived vectors are very promising for the development of vaccines to protect against mucosal infections. In fact, other antigens relevant in protection against viral infections, such as the porcine respiratory and reproductive syndrome virus ORF 5, have been efficiently expressed with a virus vector and elicited partial protection against porcine respiratory and reproductive syndrome virus-induced disease (S. Alonso, I. Sola, S. Zuñiga, J. Plana-Durán, and L. Enjuanes, submitted for publication).
A correlation has been observed between the proximity to the 3′ end of the genome and the relative efficiency of sgRNA synthesis from a given TRS in several viral systems, including coronaviruses, such as MHV and TGEV (3, 20, 47), and the Mononegavirales (22, 50). In order to increase heterologous gene expression levels, an expression cassette was inserted at the 3′ end of the genome. Unfortunately, insertion of the expression cassette between TGEV genes N and 7 in the recombinant virus rTGEV-Δ3-N-TRSn-GFP-7 resulted in an unstable virus, leading to complete deletion of the additional transcriptional unit TRSn-GFP. Since we have shown above that TGEV ORFs 3a and 3b can be stably replaced by the GFP gene, the location of the insertion, not the nature of the gene, was most likely responsible for the instability. The instability of expression cassettes inserted at the 3′ end of the genome, between genes N and 7, seems a more general phenomenon, since, in addition to the results presented in this paper with the GFP gene, we have shown (S. Alonso, I. Sola, S. Zuñiga, J. Plana-Durán, and L. Enjuanes, submitted for publication) that several expression cassettes with the reporter gene β-glucuronidase were also unstable at this position of the genome but not at the ORF 3a site. Furthermore, in another coronavirus (MHV), insertion of other sequence fragments (i.e., 3′-end 141 nucleotides of the N gene or 717 nucleotides of GFP) between the N gene and the 3′ untranslated region also produced genomic instability (21).
In some TGEV recombinant viruses, such as clone 12 of TGEV-Δ3-N-TRSn-GFP-7, the origin of the instability was the homologous recombination promoted by the presence of duplicated viral sequences, while in other viruses (clone 3 of TGEV-Δ3-N-TRSn-GFP-7) the instability was due to nonhomologous recombination yielding a virus that had lost the GFP gene and also the 5′ end of gene 7. Therefore, in addition to similarity-essential recombination, similarity-nonessential recombination (37) may also lead to instability in these viruses. The insertion of pBAC sequences in the viral genome most likely occurred during the passages in bacteria. Possibly, this process was irrelevant for the mammalian system that we are interested in and not related to the virus life cycle except to confirm the flexibility of the virus in accepting unrelated sequences in this region.
A novel transcriptional unit with a noncanonical core sequence (5′-CUAAAA-3′) that led to a major unexpected sgmRNA species was found downstream of the N gene. Along the TGEV wild-type genome, eight noncanonical core sequences (5′-CUAAAA-3′) identical to the novel transcriptional motif that did not drive transcription of detectable sgmRNAs were found. Interestingly, the noncanonical core sequence active in transcription was the only one with the 5′-flanking sequence 5′-CGAA-3′ that increased the complementarity with the leader sequence, suggesting that the extended leader-body homology upstream of the core sequence compensates for the incomplete complementarity between the leader and the core sequence and determines which core sequence-like sequence leads to RNA transcription.
Junction of the leader to another noncanonical core sequence (5′-CUAAAGA-3′) located at the 3′ end of the GFP gene, upstream of a sequence providing extended complementarity to the leader RNA, was observed during infection with rTGEV-Δ3-TRS3a-GFP. Comparison between anomalous sgmRNAs generated from the GFP gene in the TGEV vector and those documented in the coronavirus MHV (17) and the arterivirus equine arthritis virus (10) showed no coincidence at the junction sites except for their location at the 3′ half of the gene. In MHV, anomalous junction sites within the GFP gene are explained by long-range RNA or ribonucleoprotein interactions independent of the core sequence-like motif, whereas in equine arthritis virus they are attributed to the presence of core sequence-like sequences.
In TGEV, in addition to the presence of a core sequence-like sequence (5′-CUAAAG-3′) just upstream of the junction site, we observed the presence of potential extra leader-body base pairing immediately downstream of the leader core sequence. Therefore, we believe that this extension of the complementarity between the leader TRS and the sequence complementary to the TRS that precedes each gene is an important determinant for sgRNA synthesis. The effect of sequences flanking the core sequence in a TRS is in agreement with previous results with MHV (5, 21, 23, 25, 28, 29, 46, 47) showing that flanking sequences may contribute through base pairing in similarity-assisted homologous recombination that leads to the production of coronavirus mRNAs (6).
The occurrence of sgmRNA heterogeneity in the region of the leader-body junction site, leading to the detection of minor sgmRNA species synthesized from noncanonical core sequences during infection, has been described for the coronavirus MHV (17, 30, 55) and in arteriviruses (12, 35). Transcription in TGEV apparently behaved with extraordinary accuracy, since no heterogeneity has been detected in the production of sgmRNAs (40). Furthermore, no mRNA was detected even from ORF 3b in viral strains with a mutated core sequence (5′-CUAAAU-3′) (3, 51). In contrast, we report in this paper the synthesis of abundant novel sgmRNA species from a noncanonical core sequence as a response to modification of the virus genomes. The existence of alternatives to the canonical core sequence might be an evolutionary resource to maintain virus viability.
Acknowledgments
We thank F. Almazán and J. Ortego for critically reading the manuscript.
This work was supported by grants from the Comisión Interministerial de Ciencia y Tecnología (CICYT), La Consejería de Educación y Cultura de la Comunidad de Madrid, Fort Dodge Veterinaria, and the European Communities (Frame V, Key Action 2, Control of Infectious Disease Projects). I.S., S.A., and S.Z. received postdoctoral fellowships from the Community of Madrid and the European Union (Frame V, Key Action 2, Control of Infectious Disease Projects QLRT-1999-00002, QLRT-1999-30739, and QLRT-2000-00874).
REFERENCES
- 1.Agapov, E. V., I. Frolov, B. D. Lindenbach, B. M. Pragai, S. Schlesinger, and C. M. Rice. 1998. Noncytopathic Sindbis virus RNA vectors for heterologous gene expression. Proc. Natl. Acad. Sci. USA 95:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almazán, F., J. M. González, Z. Pénzes, A. Izeta, E. Calvo, J. Plana-Durán, and L. Enjuanes. 2000. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 97:5516-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alonso, S., A. Izeta, I. Sola, and L. Enjuanes. 2002. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus. J. Virol. 76:1293-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alonso, S., I. Sola, J. Teifke, I. Reimann, A. Izeta, M. Balach, J. Plana-Durán, R. J. M. Moormann, and L. Enjuanes. 2002. In vitro and in vivo expression of foreign genes by transmissible gastroenteritis coronavirus-derived minigenomes. J. Gen. Virol. 83:567-579. [DOI] [PubMed] [Google Scholar]
- 5.An, S., and S. Makino. 1998. Characterizations of coronavirus cis-acting RNA elements and the transcription step affecting its transcription efficiency. Virology 243:198-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brian, D. A., and W. J. M. Spaan. 1997. Recombination and coronavirus defective interfering RNAs. Semin. Virol. 8:101-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowley, J. A., C. M. Dimmock, K. M. Spann, and P. J. Walker. 2000b. Gill-associated virus of Penaeus monodon prawns: molecular evidence for the first invertebrate nidovirus. In E. Lavi, S. Weiss, and S. T. Hingley (ed.), Nidoviruses, in press. Plenum Press, New York, N.Y. [PubMed]
- 8.Cowley, J. A., and P. J. Walker. 2002. The complete genome sequence of gill-associated virus of Penaeus monodon prawns indicates a gene organisation unique among nidoviruses. Arch. Virol. 147:1977-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curtis, K. M., B. Yount, and R. S. Baric. 2002. Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J. Virol. 76:1422-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Vries, A. A. F., A. L. Glaser, M. J. B. Raamsman, and P. J. M. Rottier. 2001. Recombinant equine arteritis virus as an expression vector. Virology 284:259-276. [DOI] [PubMed] [Google Scholar]
- 11.Delmas, B., J. Gelfi, H. Sjöström, O. Noren, and H. Laude. 1994. Further characterization of aminopeptidase-N as a receptor for coronaviruses. Adv. Exp. Med. Biol. 342:293-298. [DOI] [PubMed] [Google Scholar]
- 12.Den Boon, J. A., M. F. Kleijnen, W. J. M. Spaan, and E. J. Snijder. 1996. Equine arteritis virus subgenomic mRNA synthesis: analysis of leader-body junctions and replicative-form RNAs. J. Virol. 70:4291-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enjuanes, L., D. Brian, D. Cavanagh, K. Holmes, M. M. C. Lai, H. Laude, P. Masters, P. Rottier, S. G. Siddell, W. J. M. Spaan, F. Taguchi, and P. Talbot. 2000. Coronaviridae, p. 835-849. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carsten, M. K. Estes, S. M. Lemon, D. J. McGeoch, J. Maniloff, M. A. Mayo, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Academic Press, New York, N.Y.
- 14.Enjuanes, L., I. Sola, F. Almazán, J. Ortego, A. Izeta, J. M. González, S. Alonso, J. M. Sánchez-Morgado, D. Escors, E. Calvo, C. Riquelme, and C. M. Sánchez. 2001. Coronavirus derived expression systems. J. Biotechnol. 88:183-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enjuanes, L., W. Spaan, E. Snijder, and D. Cavanagh. 2000. Nidovirales, p. 827-834. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carsten, M. K. Estes, S. M. Lemon, D. J. McGeoch, J. Maniloff, M. A. Mayo, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Academic Press, New York, N.Y.
- 16.Enjuanes, L., and B. A. M. Van der Zeijst. 1995. Molecular basis of transmissible gastroenteritis coronavirus epidemiology, p. 337-376. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, N.Y.
- 17.Fischer, F., C. F. Stegen, C. A. Koetzner, and P. S. Masters. 1997. Analysis of a recombinant mouse hepatitis virus expressing a foreign gene reveals a novel aspect of coronavirus transcription. J. Virol. 71:5148-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frolov, I., T. A. Hoffman, B. M. Prágai, S. A. Dryga, H. V. Huang, S. Schlesinger, and C. M. Rice. 1996. Alphavirus-based expression vectors: Strategies and applications. Proc. Natl. Acad. Sci. USA 93:11371-11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.González, J. M., Z. Penzes, F. Almazán, E. Calvo, and L. Enjuanes. 2002. Stabilization of a full-length infectious cDNA clone of transmissible gastroenteritis coronavirus by the insertion of an intron. J. Virol. 76:4655-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiscox, J. A., K. L. Mawditt, D. Cavanagh, and P. Britton. 1995. Investigation of the control of coronavirus subgenomic mRNA transcription by with T7-generated negative-sense RNA transcripts. J. Virol. 69:6219-6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsue, B., and P. S. Masters. 1999. Insertion of a new transcriptional unit into the genome of mouse hepatitis virus. J. Virol. 73:6128-6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iverson, L. E., and J. K. Rose. 1981. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell 23:477-484. [DOI] [PubMed] [Google Scholar]
- 23.Jeong, Y. S., J. F. Repass, Y.-N. Kim, S.-M. Hwang, and S. Makino. 1996. Coronavirus transcription mediated by sequences flanking the transcription consensus sequence. Virology 217:311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiménez, G., I. Correa, M. P. Melgosa, M. J. Bullido, and L. Enjuanes. 1986. Critical epitopes in transmissible gastroenteritis virus neutralization. J. Virol. 60:131-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joo, M., and S. Makino. 1995. The effect of two closely inserted transcription consensus sequences on coronavirus transcription. J. Virol. 69:272-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai, M. M. C., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laude, H., D. Rasschaert, B. Delmas, M. Godet, J. Gelfi, and C. Bernard. 1990. Molecular biology of transmissible gastroenteritis virus. Vet. Microbiol. 23:147-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makino, S., and M. Joo. 1993. Effect of intergenic consensus sequence flanking sequences on coronavirus transcription. J. Virol. 67:3304-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makino, S., M. Joo, and J. K. Makino. 1991. A system for study of coronavirus messenger RNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J. Virol. 65:6031-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makino, S., L. H. Soe, C. K. Shieh, and M. C. C. Lai. 1988. Discontinuous transcription generates heterogeneity at the leader fusion sites of coronavirus mRNAs. J. Virol. 62:3870-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mason, B. B., A. R. Davis, B. M. Bhat, M. Ghengalvala, M. D. Lubeck, G. Zandle, B. Kostek, S. Cholodofsky, S. Dheer, K. Molnar-Kimber, S. Mizutani, and P. P. Hung. 1990. Adenovirus vaccine vectors expressing hepatitis B surface antigen: importance of regulatory elements in the adenovirus major late intron. Virology 177:452-461. [DOI] [PubMed] [Google Scholar]
- 32.Masters, P. S. 1999. Reverse genetics of the largest RNA viruses. Adv. Virus Res. 53:245-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McClurkin, A. W., and J. O. Norman. 1966. Studies on transmissible gastroenteritis of swine. II. Selected characteristics of a cytopathogenic virus common to five isolates from transmissible gastroenteritis. Can. J. Comp. Med. Vet. Sci. 30:190-198. [PMC free article] [PubMed] [Google Scholar]
- 34.McGoldrick, A., J. P. Lowings, and D. J. Paton. 1999. Characterisation of a recent virulent transmissible gastroenteritis virus from Britain with a deleted ORF 3a. Arch. Virol. 144:763-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meulenberg, J. J. M., E. J. de Meijer, and R. J. M. Moormann. 1993. Subgenomic RNAs of Lelystad virus contain a conserved leader body junction sequence. J. Gen. Virol. 74:1697-1701. [DOI] [PubMed] [Google Scholar]
- 36.Mittal, S. K., J. B. Andrew, L. Prevec, and F. L. Graham. 1995. Foreign gene expression by human adenovirus type 5-based vectors studied with firefly luciferase and bacterial β-galactosidase genes as reporters. Virology 210:226-230. [DOI] [PubMed] [Google Scholar]
- 37.Nagy, P. D., and A. E. Simon. 1997. New insights into the mechanisms of RNA recombination. Virology 235:1-9. [DOI] [PubMed] [Google Scholar]
- 38.Ortego, J., D. Escors, H. Laude, and L. Enjuanes. 2002. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 76:11518-11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ortego, J., I. Sola, F. Almazan, J. E. Ceriani, C. Riquelme, M. Balach, J. Plana-Durán, and L. Enjuanes. 2003. Transmissible gastroenteritis coronavirus gene 7 is not essential but influences in vivo virus replication and virulence. Virology, in press. [DOI] [PMC free article] [PubMed]
- 40.Penzes, Z., J. M. González, E. Calvo, A. Izeta, C. Smerdou, A. Mendez, C. M. Sánchez, I. Sola, F. Almazán, and L. Enjuanes. 2001. Complete genome sequence of transmissible gastroenteritis coronavirus PUR46-MAD clone and evolution of the Purdue virus cluster. Virus Genes 23:105-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 42.Sánchez, C. M., A. Izeta, J. M. Sánchez-Morgado, S. Alonso, I. Sola, M. Balasch, J. Plana-Durán, and L. Enjuanes. 1999. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 73:7607-7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sánchez, C. M., G. Jiménez, M. D. Laviada, I. Correa, C. Suñé, M. J. Bullido, F. Gebauer, C. Smerdou, P. Callebaut, J. M. Escribano, and L. Enjuanes. 1990. Antigenic homology among coronaviruses related to transmissible gastroenteritis virus. Virology 174:410-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sawicki, S. G., and D. L. Sawicki. 1998. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 440:215-220. [DOI] [PubMed] [Google Scholar]
- 45.Thiel, V., J. Herold, B. Schelle, and S. Siddell. 2001. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 82:1273-1281. [DOI] [PubMed] [Google Scholar]
- 46.van der Most, R. G., R. J. De Groot, and W. J. M. Spaan. 1994. Subgenomic RNA synthesis directed by a synthetic defective interfering RNA of mouse hepatitis virus: a study of coronavirus transcription initiation. J. Virol. 68:3656-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Marle, G., W. Luytjes, R. G. Van der Most, T. van der Straaten, and W. J. M. Spaan. 1995. Regulation of coronavirus mRNA transcription. J. Virol. 69:7851-7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walsh, E. P., M. D. Baron, J. Anderson, and T. Barrett. 2000. Development of a genetically marked recombinant rinderpest vaccine expressing green fluorescent protein. J. Gen. Virol. 81:709-718. [DOI] [PubMed] [Google Scholar]
- 49.Walsh, E. P., M. D. Baron, L. F. Rennie, P. Monaghan, J. Anderson, and T. Barrett. 2000. Recombinant rinderpest vaccines expressing membrane-anchored proteins as genetic markers: evidence of exclusion of marker protein from the virus envelope. J. Virol. 74:10165-10175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wertz, G. W., V. P. Perepelitsa, and L. A. Ball. 1998. Gene rearrangement attenuates expression and lethality of a nonsegmented negative strand RNA virus. Proc. Natl. Acad. Sci. USA 95:3501-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wesley, R. D., A. K. Cheung, D. M. Michael, and R. D. Woods. 1989. Nucleotide sequence of coronavirus TGEV genomic RNA: evidence of 3 mRNA species between the peplomer and matrix protein genes. Virus Res. 13:87-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wesley, R. D., I. V. Wesley, and R. D. Woods. 1991. Differentiation between transmissible gastroenteritis virus and porcine respiratory coronavirus with a cDNA probe. J. Vet. Diagn. Investig. 3:29-32. [DOI] [PubMed] [Google Scholar]
- 53.Wesley, R. D., R. D. Woods, and A. K. Cheung. 1991. Genetic analysis of porcine respiratory coronavirus, an attenuated variant of transmissible gastroenteritis virus. J. Virol. 65:3369-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yount, B., K. M. Curtis, and R. S. Baric. 2000. Strategy for systematic assembly of large RNA and DNA genomes: the transmissible gastroenteritis virus model. J. Virol. 74:10600-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, X., and M. M. C. Lai. 1994. Unusual heterogeneity of leader-mRNA fusion in a murine coronavirus: implications for the mechanism of RNA transcription and recombination. J. Virol. 68:6626-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]