Abstract
An important step in the herpesvirus life cycle is the switch from latency to lytic reactivation. In order to study the life cycle of Kaposi's sarcoma-associated herpesvirus (KSHV), we developed a gene expression system in KSHV-infected primary effusion lymphoma cells. This system uses Flp-mediated efficient recombination and tetracycline-inducible expression. The Rta transcriptional activator, which acts as a molecular switch for lytic reactivation of KSHV, was efficiently integrated downstream of the Flp recombination target site, and its expression was tightly controlled by tetracycline. Like stimulation with tetradecanoyl phorbol acetate (TPA), the ectopic expression of Rta efficiently induced a complete cycle of viral replication, including a well-ordered program of KSHV gene expression and production of infectious viral progeny. A striking feature of Rta-mediated lytic gene expression was that Rta induced KSHV gene expression in a more powerful and efficient manner than TPA stimulation, indicating that Rta plays a central, leading role in KSHV lytic gene expression. Thus, our streamlined gene expression system provides a novel means not only to study the effects of viral gene products on overall KSHV gene expression and replication, but also to understand the natural viral reactivation process.
Kaposi's sarcoma-associated herpesvirus (KSHV), or human herpesvirus 8, is the potential etiological agent of Kaposi's sarcoma (KS) tumors (4, 21, 60), primary effusion lymphoma (PEL) (18), and some forms of multicentric Castleman's disease (85). KSHV is a gamma-2 herpesvirus that is closely related to herpesvirus saimiri and rhesus monkey rhadinovirus (2, 3, 28, 73, 80). Sequence analysis of the 140.5-kb KSHV genome revealed genes closely homologous to the viral replication and structural genes conserved among herpesviruses and other conserved homologous cellular genes that are thought to advance viral pathogenesis and contribute to the development of KSHV-associated neoplasms (73). These include a virus-encoded interleukin-6 (IL-6) (59, 62, 65), viral macrophage inflammatory protein (vMIP) (47, 66), a bcl-2 homolog (76), virus-encoded interferon regulatory factors (vIRFs) (16, 35, 49, 53, 72, 95), vCyclin (50, 77), vIL-8 receptor (8), viral FLICE-inhibitory protein (vFLIP) (10, 89), and vOX2 (25).
KSHV, a lymphotropic herpesvirus, infects CD19-positive B lymphocytes in vivo. These lymphocytes constitute a potential viral reservoir (12, 29) and can presumably sometimes develop into PEL cells. KSHV is also found in endothelial cells, thought to be the cell of origin for KS tumor cells (61). Like other herpesviruses, KSHV can produce latent infections (69). In KS lesions and PEL cells, the virus is primarily present in a latent state, with transcription restricted to a small set of viral genes and no detectable production of viral progeny (63). At present, there is no cell culture system that will efficiently support KSHV infection and replication, but treatment of latently infected PEL cell lines with inducing agents, such as phorbol esters or sodium butyrate, can induce the latently infected cells to complete the lytic infectious cycle, during which complex patterns of gene regulation can be observed (9, 19, 58, 71). KSHV also displays latent infection in vivo in KS lesions (36, 42).
Based on expression kinetics, herpesvirus genes can be categorized into four groups: latent, immediate early, early, and late (34). The genes of the immediate-early group usually encode regulatory proteins that govern the expression of various viral and cellular genes and therefore play a crucial role in the control of the herpesvirus life cycle. An important step in the herpesvirus life cycle is the switch from latency to lytic replication. The reactivation of Epstein-Barr virus (EBV) latency is controlled by two immediate-early genes, BZLF1 and BRLF1, whose products, ZTA and replication and transcription activator (Rta), respectively, are transcriptional activators that stimulate the expression of downstream viral target genes (24, 88, 92). KSHV Rta, the product of the KSHV open reading frame 50 (ORF50), is a homolog of the EBV Rta. KSHV Rta has been shown to play a central role in the switch from latency to lytic replication. Ectopic expression of Rta is sufficient to disrupt viral latency and activate lytic replication (37, 56, 86). Rta activates the expression of numerous viral genes in the KSHV lytic cycle, including the polyadenylated nuclear (PAN) RNA (also known as T1.1 or nut-1); Mta (ORF57), vOX2 (K14), viral G protein-coupled receptor (vGPCR) (ORF74), and vIRF-1 (K9); and its own promoter (22, 27, 30, 45, 74, 84, 93). Since some of the genes targeted by Rta are early genes and transactivators, this suggests that Rta may, in some way, lie at or near the apex of the KSHV regulatory cascade. The detailed mechanism of Rta-mediated transcriptional activation remains unclear, but several pieces of evidence suggest that Rta activates its target promoter activity through both direct binding to the specific sequence (20, 55, 83, 84) and interaction with various cellular transcriptional factors (22, 39, 40, 52, 74, 90). KSHV Rta thus appears to act as a critical point in the KSHV replication cycle. A careful delineation of the complete set of genes transactivated by Rta and the kinetics with which this transactivation occurs may provide a more comprehensive view of the earliest events in the KSHV replication cascade and the effects on the patterns of KSHV gene expression occurring later in the viral replication cycle.
To enhance our ability to investigate the involvement of individual KSHV genes in the viral replication cycle, we developed a system that permits the ready introduction of inducibly expressed genes into a PEL cell line latently infected with KSHV. This system uses efficient Flp-mediated recombination into the Flp recombination target (FRT) to allow the rapid and efficient introduction of test genes into the latently infected BCBL-1 PEL cell line (71) under the control of a tetracycline-inducible promoter (6, 13-15, 26, 38, 43, 78, 81). This system produced a derivative of the BCBL-1 cell line that inducibly expresses Rta. Rta was efficiently integrated downstream of the FRT site, and its expression successfully induced a complete cycle of KSHV replication, including a carefully ordered program of KSHV gene expression, shown by RNase protection assay (RPA) and viral microarray, and release of infectious viral progeny. The data provide additional information concerning the genomewide effects of a key KSHV regulatory gene and demonstrate the utility of a system that enables the rapid construction of PEL cell lines that can inducibly express selected KSHV gene products.
MATERIALS AND METHODS
Reagents and antibodies.
12-O-tetradecanoylphorbol 13-acetate (TPA) was purchased from Sigma Chemical (St. Louis, Mo.). Polyclonal anti-K8.1, anti-K-bZIP, anti-vIRF-1, anti-LANA, and anti-MIR1 antibodies were prepared as previously described (51). Anti-vIL-6 was obtained from Advanced Biotechnologies (Columbia, Md.), and mouse monoclonal antibody against KSHV Rta was a kind gift from Koichi Yamanishi (Osaka University, Osaka, Japan). Anti-actin antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.).
Cell culture.
KSHV-positive cell lines (BCBL-1 and its derivatives) were grown in RPMI 1640 medium (Invitrogen Life Technologies, Carlsbad, Calif.) supplemented with 20% heat-inactivated fetal bovine serum (FBS; Invitrogen) and 2 mM l-glutamine. The parental BCBL-1 cell line was a kind gift from Don Ganem. The 293 cells were grown in Dulbecco's modified Eagle medium (Invitrogen) supplemented with 10% FBS. All cells were maintained with 100 U of penicillin/ml and 100 μg of streptomycin/ml (both from Invitrogen) at 37°C under 5% CO2.
cDNA cloning and plasmid constructs.
To amplify full-length cDNA corresponding to KSHV Rta, total RNA was extracted from TPA-treated BCBL-1 cells for 48 h using TRIZOL (Invitrogen) according to the manufacturer's instructions. To synthesize single-stranded cDNA, 5 μg of total RNA was reverse transcribed with SuperScript II RNaseH− reverse transcriptase (RT; Invitrogen) in a 20-μl reaction mixture with oligo(dT) primer for 50 min at 42°C. The cDNA pool was employed as the template for the amplification of the Rta cDNA using the following primers, which contain an EcoRI site or an XbaI site (underlined): 5′-CGCGAATTCGCCGCCACCATGGCGCAAGATGACAAGG-3′ and 5′-CGCTCTAGAGTCTCGGAAGTAATTACGCCATTGG-3′. To construct pEF-Rta-myc-His, the PCR product was digested with EcoRI and XbaI and cloned into pEF1/Myc-His A (Invitrogen). To construct pcDNA/FRT/TO-Rta, the EcoRI/PmeI fragment of pEF-Rta-myc-His was blunted by T4 DNA polymerase and cloned into the EcoRV site of pcDNA5/FRT/TO. Plasmids pFRT/lacZeo, pcDNA6/TR, pcDNA5/FRT/TO, and pOG44 were obtained from Invitrogen.
Transfection.
Plasmid DNA for transfection was prepared by cesium chloride-ethidium bromide equilibrium centrifugation. For electroporation into BCBL-1 cells, ∼107 cells were pelleted by centrifugation, washed twice with RPMI 1640 without FBS, and resuspended in 400 μl of RPMI 1640 medium without FBS. The cells and plasmid DNA were placed in 0.4-cm-electrode gap electroporation cuvettes (Bio-Rad Laboratories, Hercules, Calif.), and electroporation was carried out at 250 V and 960 μF with a Gene Pulser II (Bio-Rad).
Development of a gene-inducible system in BCBL-1 cells.
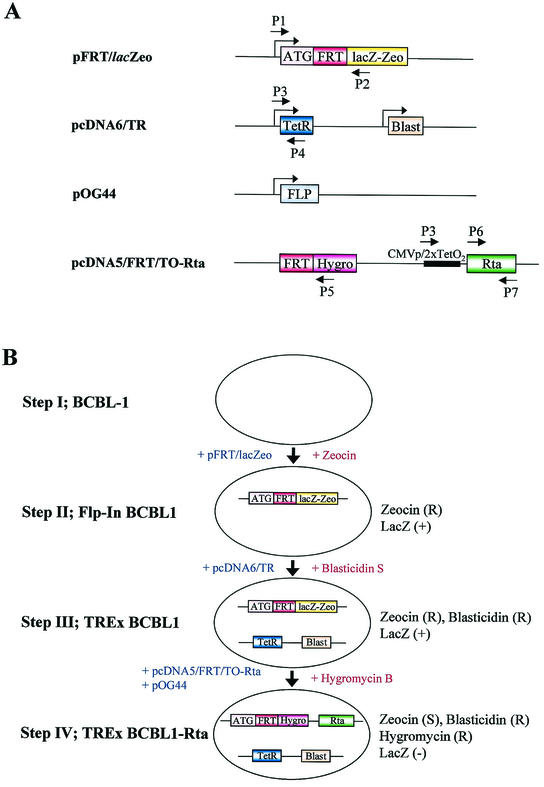
To develop a tetracycline-inducible system in BCBL-1 cells using Flp-mediated site-specific recombination, pFRT/lacZeo, which contains a Flp recombination target (FRT) site and a lacZ-Zeocin fusion gene, was electroporated into BCBL-1 cells. Forty-eight hours after transfection, the cells were incubated with medium containing 250 μg of Zeocin (Invitrogen)/ml in 96-well plates at 103 cells/well and selected for ∼6 weeks. Genomic DNAs of individual Zeocin-resistant clones were then extracted to assess the presence of the pFRT/lacZeo sequence by pFRT/lacZeo-specific PCR using primers P1 and P2 (Table 1). To measure the expression of lacZ-Zeocin fusion protein, a β-galactosidase assay (Promega, Madison, Wis.) was performed according to the manufacturer's protocol. To establish cell lines that constitutively expressed Tet repressor (TetR), pcDNA6/TR was electroporated into Flp-In BCBL1 cells. Forty-eight hours after transfection with pcDNA6/TR, Flp-In BCBL1 cells were selected with 250 μg of blasticidin S HCl (Invitrogen)/ml in 96-well plates. The integration of pcDNA6/TR in blasticidin-resistant clones was assessed by pcDNA6/TR-specific PCR using primers P3 and P4 (Table 1). To establish the cell lines which inducibly expressed Rta (TREx BCBL1-Rta), pcDNA/FRT/TO-Rta and pOG44, which constitutively expresses Flp recombinase, were cotransfected at a ratio of 1:9 (wt/wt) into 3 × 107 totalTREx BCBL1 cells by electroporation (Fig. 1). Forty-eight hours after electroporation, the cells were selected with 200 μg of hygromycin B (Invitrogen)/ml for 3 to 4 weeks.
TABLE 1.
Oligonucleotide primers used to establish TREx BCBL1-Rta cells
| Primer | Sequence (5′ → 3′) | Location |
|---|---|---|
| P1 | CAGCTGTGGAATGTGTGTCAGTTAG | SV40 promoter |
| P2 | GTGCTGCAAGGCGATTAAGTTGGG | lacZ-Zeocin |
| P3 | CGCAAATGGGCGGTAGGCGTG | CMV promoter |
| P4 | GGACCCACTTTCACATTTAAGTTG | Tet Repressor |
| P5 | GTAGTGTATTGACCGATTCCTTGC | Hygromycin |
| P6 | ATGGCGCAAGATGACAAGG | Rta |
| P7 | GTCTCGGAAGTAATTACGCCATTGG | Rta |
| P8 | CAGTGATCTCCTTCTGCATCCTGTC | β-Actin |
| P9 | GCTACGTCGCCCTGGACTTCGAG | β-Actin |
FIG. 1.
Summary of Flp-In/TetR system. (A) Schematic representation of plasmid constructs and positions of primers used for establishment of TREx BCBL1-Rta cells. P1 to P7 are primers, as detailed in Table 1. pFRT/lacZeo contains the FRT site for site-specific recombination downstream of the initiation codon (ATG) and expresses lacZ-Zeocin fusion protein (lacZ-Zeo) driven by the SV40 early promoter. pcDNA6/TR constitutively expresses TetR driven by the CMV promoter and also expresses the blasticidin resistance gene (Blast) driven by the SV40 promoter. pOG44 contains Flp recombinase (FLP), which mediates the FRT site-specific recombination. pcDNA5/FRT/TO-Rta contains the FRT site followed by the hygromycin resistance gene (Hygro), which carries neither its promoter nor its start codon. This vector also contains myc/His-tagged Rta cDNA (Rta) downstream of the CMV promoter and tetracycline operator sequences (CMVp/2xTetO2). (B) Schematic diagram of the establishment of TREx BCBL1-Rta cells. Plasmid vectors and drugs used to establish the system are depicted with blue and red letters, respectively. R, resistant; S, sensitive.
Multiprobe RPA.
Plasmids were constructed to make single-stranded RNA probes for RPA. The specific target sequences (Table 2) used to generate antisense riboprobes were amplified by PCR using genomic DNA of BCBL-1 cells as a template. Each PCR product was cloned into the EcoRI and BamHI cloning sites of pEF1/Myc-His A vector in an antisense orientation. For quantitation, two housekeeping genes coding for L32 and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were also amplified using the following primers, which contain an EcoRI or BamHI site (underlined): 5′-CGCGAATTCACCCAGAGGCATTGACAACAG-3′ and 5′-CGCGGATCCACTTCCAGCTCCTTGACGTTG-3′ for L32 and 5′-CGCGAATTCGTCTTCACCACCATGGAGAAG-3′ and 5′-CGCGGATCCATACTTCTCATGGTTCACAC-3′ for GAPDH. To prepare multiprobe RPA sets, 2 μg of each recombinant plasmid were mixed, linearized with XbaI, purified by phenol extraction, and reconstituted with Tris-EDTA (pH 8.0). One microgram of the plasmid mixture was used to synthesize an [α-32P]UTP-labeled antisense RNA probe set with T7 RNA polymerase using the Riboquant in vitro transcription kit (BD PharMingen, San Diego, Calif.).
TABLE 2.
Multiprobe RPA templates
| Template set | ORF | Genome coordinatesa | Probe size (bases) | Protected size (bases) |
|---|---|---|---|---|
| I | LANA | 127117-126748 | 452 | 370 |
| LANA2 | 90016-89676 | 423 | 341 | |
| ORF K1 | 325-634 | 392 | 310 | |
| ORF T1.1 or K7 | 28682-28961 | 362 | 280 | |
| vOX2 | 128284-128533 | 332 | 250 | |
| vGPCR | 129572-129791 | 302 | 220 | |
| Kaposin | 118101-117922 | 262 | 180 | |
| II | K-bZIP | 75000-75359 | 442 | 360 |
| Rta | 74184-74513 | 412 | 330 | |
| ORF45 | 68316-68017 | 382 | 300 | |
| Mta | 83017-83286 | 352 | 270 | |
| MIR1 | 19479-19240 | 322 | 240 | |
| vIRF-1 | 85109-84930 | 262 | 180 | |
| III | ORF29A | 54576-54227 | 432 | 350 |
| ORF36 | 56276-56595 | 402 | 320 | |
| ORF47 | 69815-69526 | 372 | 290 | |
| ORF K8.1 | 75995-76224 | 312 | 230 | |
| ORF58 | 95125-94926 | 282 | 200 |
According to GenBank accession no. U75698.
Cells were treated with 20 ng of TPA/ml (BCBL-1 cells) or 1 μg of doxycycline (Dox; BD Clontech, Palo Alto, Calif.)/ml (TREx BCBL1-Rta cells) and then harvested at various time points. Total RNA was extracted using TRIzol reagent. At each time point, 30 μg of total RNA was hybridized to the 32P-labeled RNA template at 90°C for 3 min and subsequently hybridized at 56°C for 16 h. RNase protection reactions were carried out using an RPA kit (BD PharMingen) according to the manufacturer's instructions. Following precipitation of the digested RNA, the samples were separated on a 5% denaturing acrylamide gel. The gels were analyzed using a phosphorimager (BAS 2000; Fuji Photo Film Co., Tokyo, Japan).
Immunoblot analysis.
Cells were lysed in lysis buffer (50 mM HEPES [pH 8.0], 150 mM NaCl, 1% NP-40) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 mM Na2VO3, 1 mM NaF, 1 μg of aprotinin/ml, 5 μg of leupeptin/ml, and 1 μg of pepstatin/ml). The cell lysates were resolved on sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis gels and transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore, Bedford, Mass.). After the membranes were blocked in 10% nonfat milk, primary antibodies were used at 1:500 to 1:2,000 dilutions, and secondary antibodies were used at 1:5,000 to 1:10,000 dilutions in 3% nonfat dry milk. After a final washing, the membranes were used for enhanced chemiluminescence assays (SuperSignal; Pierce, Rockford, Ill.) with a Fuji phosphorimager.
DNA microarray analysis.
A KSHV gene array was described by Paulose-Murphy et al. (68). We designed additional primer pairs to amplify KSHV fragments to produce detectors for the following ORFs: vMIP-II, vMIP-III, K4.2 (National Center for Biotechnology Information accession no. U93872), ORF58, K-bZIP (73), K8.1 (79), and K8.2 (94). To improve PCR efficiency, the primers were redesigned for KSHV ORF vMIP-I, ORF23, ORF62, ORF LANA, and ORF75 (73). The additional and replacement primers were synthesized by Invitrogen. The primer pairs (Table 3) were designed to amplify fragments of 100 to 300 bases from the appropriate viral ORF. BCBL-1 high-molecular-weight DNA was used as a template. All PCR products showed a single band of appropriate size by gel electrophoresis and were subsequently ethanol precipitated, washed twice with 70% ethanol, and resuspended in Tris-EDTA buffer. Microarrays were printed at the National Cancer Institute, Center for Cancer Research Advanced Technologies Center, using the OmniGrid arrayer (Gene Machines, San Carlos, Calif.). A total of 91 viral genes and a set of 96 cellular housekeeping genes (46) were provided by the National Human Genome Research Institute, National Institutes of Health (NIH). Each DNA fragment was diluted in 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) to a final concentration of 0.5 mg/ml. The diluted fragments were transferred to a 384-well plate prior to printing. Three control cellular gene subarrays were printed along with one viral subarray. Each DNA element was spotted in duplicate onto poly-l-lysine-coated slides using SMP-3 spotting pins (Telechem International, Sunnyvale, Calif.), UV cross-linked, and blocked as previously described (31, 82).
TABLE 3.
Supplemental primers for KSHV microarray
| ORF | Forward primer (5′) | Reverse primer (3′) |
|---|---|---|
| vMIP-II | GCTGACTGCCTTGCTTTGTTTG | GCACCTGTGGTAATGGTCTTTTCTG |
| vMIP-III | TTCCCTGTCCAGTGCCAGATTG | CCCGAGTGCGTTACAAAAATGAC |
| ORF K4.2 | GCTGGATTTTTGGACCGTGG | AACAACTTGACACAGGGGAAACAC |
| vMIP-I | TTTACCTGACGCCCACAGAAAG | GCCTAACCCAGTTTTTGGAAGG |
| K-bZIP | TACGGACAATACTGACGGTGGG | GCTGTTGCGAAATGTGTGGTC |
| ORF K8.1 | GCAATAGATGAATCGGGGTCG | GCGAGGGATTACAGTTGAAACG |
| ORF K8.2 | CGTGTGCCATTTTCTGCCAC | CACTATGTAGGGTTTCTTACGCCG |
| ORF23 | CGCATAAAGGTGTTTGACCCAGAG | AACTCCCGCTCGTGTGTTTTCG |
| ORF58 | GCTGCCCTTCATTGTCTTCTTG | AAGTGCCCTGTGGAGTTTTCG |
| ORF62 | GTTTATGCTCTCTGGAATGGAAGC | TATCACAAGGGAGGGTCAAGCC |
| LANA | CCCCAAGGTCCCTCTACACTAAAC | TCCTGCTGCTGTTGTGAACTTTG |
| ORF75 | GGCAGCAAGGAGTTCTTTACGC | GGGGTCTCTGAAACCATAAGTTCG |
Poly(A)+ RNA was isolated from 0.5 × 108 to 1.0 × 108 total BCBL-1 cells containing either the Rta expression vector (TREx BCBL1-Rta) or empty control vector (TREx BCBL1-vector) at 0, 2, 4, 8, 12, 16, 24, and 48 h poststimulation by Dox using the Fast Track 2.0 kit (Invitrogen) following the manufacturer's instructions.
Poly(A)+ RNAs isolated from TREx BCBL1-Rta cells and TREx BCBL1-vector cells were reverse transcribed using the Superscript II kit (Invitrogen) with an oligonucleotide (dT29V)-anchored primer and were labeled or reverse labeled, respectively, with Cy3- or Cy5-dUTP (Amersham Biosciences, Piscataway, N.J.). The paired reaction mixtures were combined, and unincorporated fluor-dUTPs were removed with Qiaquick PCR purification spin columns (Qiagen, Valencia, Calif.). Purified fluorescently labeled cDNA probe was mixed with human Cot-1 DNA (Boehringer Mannheim, Indianapolis, Ind.), poly(dA) (Amersham), yeast tRNA, and salmon sperm DNA (Sigma); applied to microarrays; and hybridized at 65°C for 16 to 18 h. Following hybridization, the slides were washed in 0.1% SDS-0.5× SSC followed by 0.01% SDS-0.5× SSC for 2 min each at 60°C, with a final wash in 0.06× SSC for 2 min at room temperature. The slides were dried by centrifugation and immediately scanned. Both normal and reverse-labeled hybridizations were repeated at least once. The data obtained using the different arrays and experiments were consistent.
The slides were scanned with the GenePix 4000 scanner (Axon Instruments, Inc., Foster City, Calif.) as previously described (68). The Cy5-Cy3 spot signal images of viral and cellular housekeeping gene spots were analyzed using GenePix Pro version 3.0 (Axon Instruments) and a custom designed gene array list file generated by the BioInformatics and Molecular Analysis Section at the Center for Information Technology, NIH. The localized raw ratios with background subtracted were subsequently uploaded into the Center for Cancer Research Microarray Data Base (mAdb) (http://nciarray.nci.nih.gov/) for normalization of the cellular housekeeping genes to establish a calibrated expression ratio (CalRatio) for the KSHV genes (11, 23, 68). The KSHV genes' CalRatios were sorted by sample time points and array replicates, log 2 transformed, and then clustered by Pearson's correlation using the hierarchical clustering algorithm (32) or by the self-organizing mapping (SOM) algorithm (48) using the mAdb analysis tools.
Electron microscopy.
Transmission electron microscopy was performed as previously described (51). TREx BCBL1-Rta and BCBL-1 cells were induced by Dox or TPA, respectively, for 72 h. The cells were rinsed twice in cacodylate buffer and placed in Karnovsky's fixative. The cell pellets were postfixed in 1% OsO4, dehydrated in a graded series of solutions of ethanol and propylene oxide, and embedded in Spurr's resin for thin sectioning. The thin sections were stained with uranyl acetate and lead citrate and examined on a Philips EM 300 electron microscope.
Infectivity assay.
TREx BCBL1-Rta cells (3 × 105/ml) were incubated for 96 h with or without 1 μg of Dox/ml. The culture medium was then centrifuged at 770 × g for 5 min, and cell-free supernatants were obtained by filtration through a 0.45-μm-pore-size membrane. The supernatants were incubated with 293 cells in six-well plates for 24 h, followed by total RNA extraction. cDNA was synthesized by SuperScript II RNaseH− RT in a 20-μl reaction mixture with random hexamers for 50 min at 42°C. ORF29 cDNA was then assayed by PCR, as described previously (70).
RESULTS
Development of a gene-inducible system in KSHV-infected PEL cells.
To study the life cycle of KSHV and the contributions of selected viral genes to the viral life cycle, BCBL-1 cells persistently infected by KSHV were engineered to establish a Flp-In/TetR-inducible system. This system streamlines the generation of stable, tetracycline-inducible mammalian expression cell lines by taking advantage of a yeast DNA recombination system which uses Flp recombinase and site-specific recombination to facilitate integration of the gene of interest into the genome. To generate an Flp-In- and TetR-inducible BCBL-1 cell line, we sequentially transfected two plasmids (pFRT/LacZeo and pcDNA6/TR) into BCBL-1 cells (Fig. 1A). After the first round of transfection, multiple Zeocin-resistant clones were assayed for pFRT/lacZeo DNA by PCR, Southern blotting, and β-galactosidase activity assay (Fig. 1B, step I). The β-galactosidase assay showed similar levels of activity in individual Zeocin-resistant clones, indicating that the pFRT/lacZeo plasmid was integrated at transcriptionally active loci (data not shown). We selected one clone, called Flp-In BCBL1, to introduce the pcDNA6/TR plasmid vector, which expresses the tetracycline repressor protein (TetR) (Fig. 1B, step II). After selection with blasticidin B, individual blasticidin-resistant clones were used to assay for the presence of the pcDNA6/TR plasmid by PCR (data not shown). We chose one clone, called TREx BCBL1, for further study (Fig. 1B, step III).
Generation of tetracycline-inducible Rta expression in BCBL-1 cells using Flp-In recombination system.
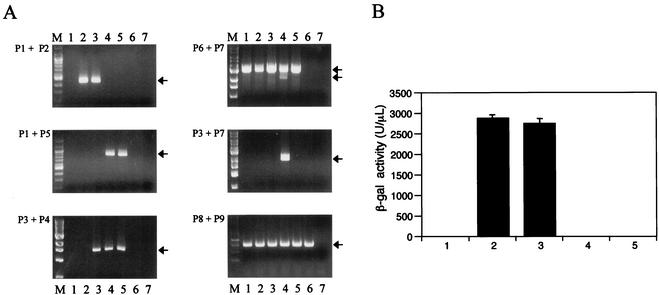
To develop a system in which the KSHV Rta could be expressed under the control of a tetracycline-inducible promoter, we introduced cDNA encoding Rta into TREx BCBL1 cells. The expression plasmids pcDNA5/FRT/TO-Rta, containing the myc/His-tagged full-length Rta, and pOG44, containing the Flp gene, were electroporated into TREx BCBL1 cells, followed by selection with 200 μg of hygromycin B/ml for 3 weeks (Fig. 1B, step IV). In addition, the control vector pcDNA5/FRT/TO was included to generate TREx BCBL1-vector cells as a control. PCR and β-galactosidase assays confirmed the presence of the vectors within the cells (Fig. 1 and Table 1). The lacZ-Zeocin fusion gene was detected in the Flp-In BCBL1 and TREx BCBL1 cells, but not in TREx BCBL1-Rta and TREx BCBL1-vector cells (Fig. 2A, top left). The 3.0-kb full-length Rta genomic sequence was readily amplified from parental BCBL-1 and BCBL-1-derived cells, whereas the 2.1-kb spliced Rta cDNA sequence was detected only in TREx BCBL1-Rta cells (Fig. 2A, top right). Furthermore, the Rta cDNA sequence downstream of the cytomegalovirus (CMV)/tetO2 promoter was detected only in TREx BCBL1-Rta cells and not in other cells (Fig. 2A, middle right). Finally, Flp-In BCBL1 and TREx BCBL1 cells displayed strong β-galactosidase activities, whereas TREx BCBL1-Rta and TREx BCBL1-vector cells showed an absence of β-galactosidase activity (Fig. 2B). These results indicated that the lacZ-Zeocin fusion gene was replaced by the hygromycin resistance gene through the Flp-In recombination system and that the myc/His-tagged Rta cDNA was accurately integrated downstream of the CMV/tetO2 promoter sequence.
FIG. 2.
Survey of FRT site-specific recombination. (A) PCR analysis. Genomic DNAs from BCBL-1 (lanes 1), Flp-In BCBL1 (lanes 2), TREx BCBL1 (lanes 3), TREx BCBL1-Rta (lanes 4), TREx BCBL1-vector (lanes 5), BJAB (lanes 6), and H2O (lanes 7) were used for PCR amplification. PCR primers P1 to P9 are shown in Fig. 1 and Table 1. The arrows indicate appropriate PCR products. M, molecular size marker. (B) β-Galactosidase assay of BCBL-1 (bar 1), Flp-In BCBL1 (bar 2), TREx BCBL1 (bar 3), TREx BCBL1-Rta (bar 4), and TREx BCBL1-vector (bar 5). The values represent averages of three individual experiments, with the error bars showing standard deviations.
Expression of KSHV viral proteins induced by Rta.
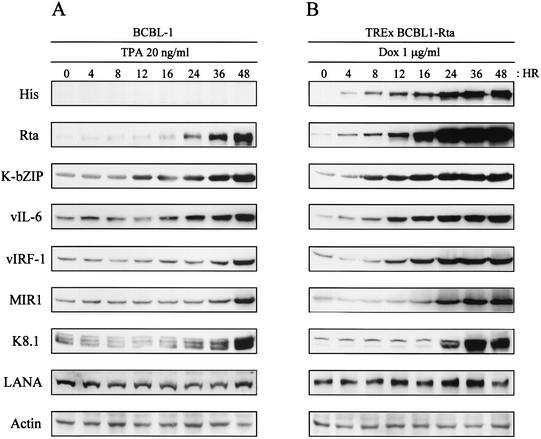
KSHV Rta expression is sufficient to end viral latency and activate lytic replication to completion (37, 56, 86). To assess the ability of the Dox-induced Rta to activate the expression of other viral genes, TREx BCBL1-Rta cells were treated with 1 μg of Dox/ml for various periods and harvested for immunoblot analysis with antibodies against His, Rta, K-bZIP, vIL-6, vIRF-1, MIR1, K8.1, and LANA. In addition, TPA-stimulated parental BCBL-1 cells were included as controls. Anti-His and anti-Rta antibodies readily detected the His-tagged Rta protein derived from the CMV/tetO2 promoter and/or authentic Rta derived from its own viral promoter after 4 h of Dox treatment (Fig. 3). Upon Dox treatment, an increase in immediate-early lytic K-bZIP and vIL-6 protein expression was also detected somewhat earlier than that of early lytic vIRF-1 and MIR1 proteins in TREx BCBL1-Rta cells. Finally, late K8.1 viral envelope protein was detected after 24 h of Dox treatment. This indicates that upon Dox treatment, Rta is able to induce the expression of several viral genes in a well-ordered, kinetically appropriate manner. A striking feature of Rta-mediated lytic replication was that the expression of Rta increased much more rapidly and to much higher levels following induction with Dox than with TPA. Increased Rta expression was detected only after 24 h of TPA stimulation, whereas it was detected within 4 h of Dox treatment (Fig. 3). Increased expression of other viral proteins was also detected at much earlier time points after induction in Dox-treated TREx BCBL1-Rta cells than in TPA-stimulated BCBL-1 cells (Fig. 3). These results indicate that the ectopic expression of Rta was capable of inducing KSHV gene expression in a more powerful and efficient manner than TPA stimulation.
FIG. 3.
KSHV protein expression in TPA-induced BCBL-1 cells (A) or Dox-induced TREx BCBL1-Rta cells (B). At the indicated times after stimulation with TPA (20 ng/ml) or Dox (1 μg/ml), equal amounts of total proteins were analyzed by immunoblotting with KSHV-specific antibodies. Anti-actin antibody was used to monitor protein amounts.
Detection of KSHV gene expression by multiprobe RPA.
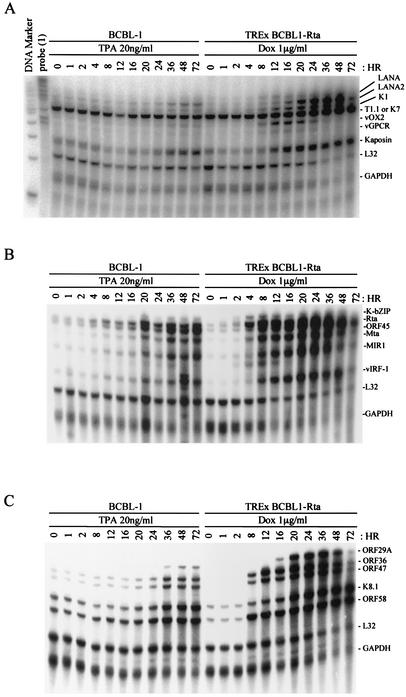
Having confirmed that Dox induction leads to high levels of Rta expression and the expression of several other gene products, we developed a multiprobe RPA to characterize the kinetics of a series of KSHV RNAs following Dox induction. The multiprobe RPA allows the relative expression levels of multiple viral genes to be compared in an individual sample. We chose to analyze a panel of genes that were expressed (i) during the immediate-early lytic cycle (K-bZIP, ORF45, Rta, and Mta genes), (ii) during the early lytic cycle (T1.1, K1, MIR1, vOX2, vIRF-1, vGPCR, and Kaposin genes), (iii) during the late lytic cycle (ORF29A, ORF36, ORF47, ORF58, and K8.1 genes), and (iv) during latency (LANA and LANA2 genes). These 18 genes were divided into three multiprobe sets (Table 2). Also included in each set were probes for the genes encoding human ribosomal protein L32 and GAPDH, which served as housekeeping genes to correct for sample variability.
Total RNA was isolated from Dox-treated TREx BCBL1-Rta cells and TPA-treated BCBL-1 cells at time points from 0 to 72 h. Upon Dox treatment, TREx BCBL1-Rta cells showed strong, rapid induction of K1, LANA2, and Kaposin and a lesser induction of LANA and T1.1 (Fig. 4A). Both vOX2 and vGPCR in TREx BCBL1-Rta cells became highly expressed 8 h after Dox stimulation, reached a maximum at 16 to 20 h, and declined thereafter (Fig. 4A). In contrast, vOX2 and vGPCR expression weakly and continuously increased following TPA stimulation (Fig. 4A). This indicates that the kinetics of vOX2 and vGPCR in TREx BCBL1-Rta cells is distinct from that in TPA-induced BCBL-1 cells. Using the second set of probes, we found that a drastic increase in Rta mRNA was detected 4 h after Dox treatment in TREx BCBL1-Rta cells, followed by a rapid increase in K-bZIP, ORF45, Mta, and MIR1 mRNAs (Fig. 4B). While both TPA and Rta expression efficiently induced the expression of late genes, Rta-induced expression of late genes was greater than that induced by TPA (Fig. 4C). It is also interesting that, as seen with the replication of many herpesviruses, L32 and GAPDH cellular expression was greatly reduced during Rta-induced lytic replication, whereas it was not affected during TPA-induced lytic replication (Fig. 4). Thus, these results reinforce the fact that the Dox-induced expression of Rta is capable of inducing high-level viral gene expression in TREx BCBL1-Rta cells in a kinetically appropriate manner.
FIG. 4.
Multiprobe RPA of KSHV genes. Total RNAs (30 μg) extracted from TPA-induced BCBL-1 cells and Dox-induced TREx BCBL1-Rta cells at the indicated time points were subjected to RPA. The protected RNA fragments were visualized with a phosphorimager.
Effect of Rta expression on viral gene expression.
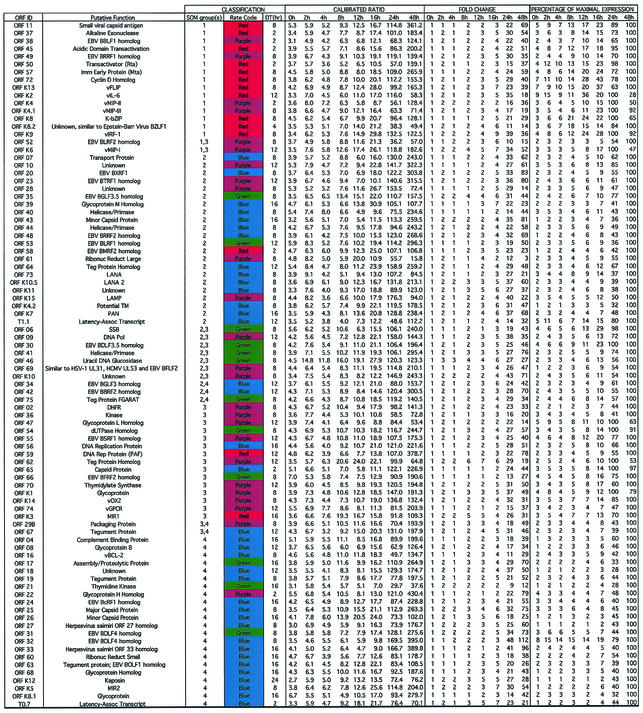
Array technology can be used to provide a comprehensive view of viral gene expression patterns during the viral replication cycle. To further delineate the effects of overexpressing Rta on KSHV gene expression, we utilized a recently described KSHV DNA microarray (68). This viral array, which contains nearly every known KSHV ORF, allowed us to monitor the global changes in KSHV gene expression following induction of Rta expression. RNAs from Dox-treated TREx BCBL1-vector (control) and Dox-treated TREx BCBL1-Rta (Rta-expressing) cells were reverse transcribed, and the labeled cDNAs (Cy3-TREx BCBL1-vector and Cy5-TREx BCBL1-Rta) were hybridized to the viral array. The array was scanned, and the expression ratio of the viral genes in the Dox-treated TREx BCBL1-Rta cells to that in the Dox-treated TREx BCBL1-vector cells was obtained. This expression ratio was normalized to a set of housekeeping genes to obtain a CalRatio. This CalRatio is tabulated for each of the viral genes in Table 4, together with the putative function of each ORF, the expression class assigned for each gene using three different classification methods (see below), the change in expression (n-fold) for each gene above the baseline value, and the percentage of the maximum expression observed for each gene at each time point.
TABLE 4.
Summary of analyzed data for Rta-induced KSHV genesa
Viral genes are listed according to SOM group order. Duplicate spotted genes’ calibrated ratios that were split between two groups were placed near the closest SOM group. The color codes represent the gene’s respective rate profiles described in the text and shown in Fig. 7. DT, time of first expressional doubling with respect to baseline expression at time zero. The quantitative data (from left to right) represent the respective average calibrated Rta/empty-vector ratios for each gene at each hour collected post-Dox addition; the change measured by each gene’s expression at each time point (CalRatio) divided by its time zero baseline expression (CalRatio); and the percentage of maximum expression measured by each gene’s change at each time point divided by its respective maximum change (set at 100).
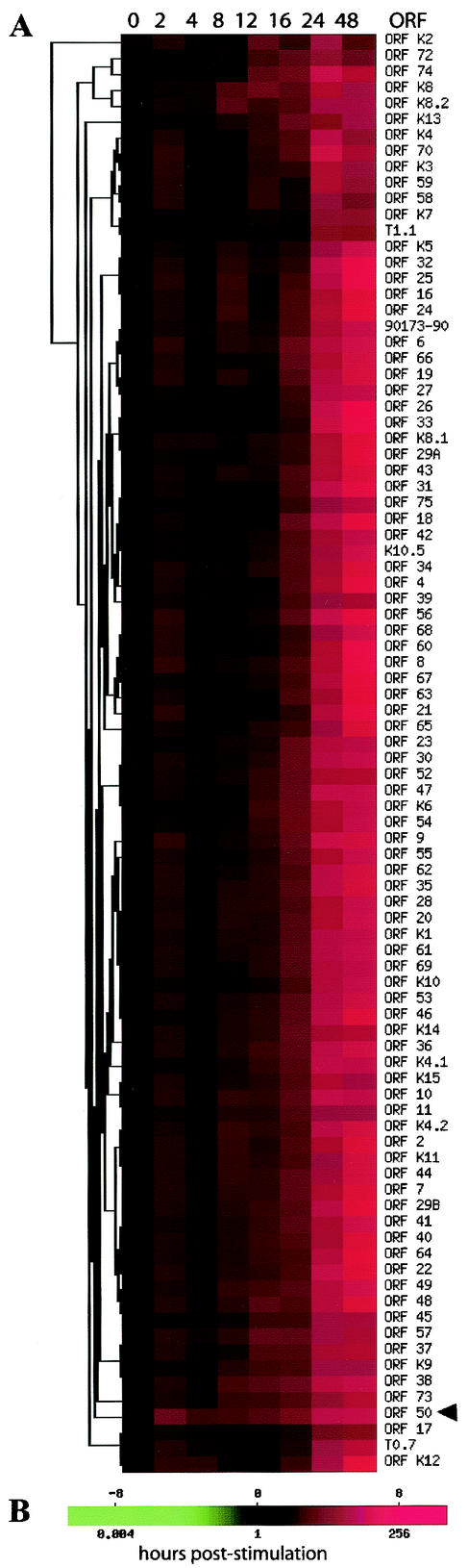
Overall, it is apparent that expression of Rta led to a very powerful induction of all the viral ORFs, with the changes in expression observed following induction ranging up to and beyond 100-fold by 24 to 48 h. The results generally agree with those seen using the RPA, and this result is far greater than that previously observed with induction of KSHV replication following induction with TPA (63). The data were subjected to a hierarchical clustering analysis (Fig. 5). Here, it can be seen that the expression of the induced Rta gene (ORF50) increases at the earliest time points, followed by increases in expression of the other viral genes. Even though all the viral genes were very strongly induced, the increases in expression of some viral genes can be seen to occur at earlier time points than others.
FIG. 5.
Hierarchical clustering of KSHV gene expression induced by Rta. (A) Color-coded expression scale. Expression of KSHV genes was strongly induced by Rta up to 48-fold by 24 h. (B) Color scale representing the degree of expression based on a calibrated ratio of Dox-stimulated Cy3-dUTP-labeled viral cDNA signal intensity over empty vector control Cy5-dUTP-labeled cDNA signal intensity. A calibrated ratio of 1, represented by black, indicates no change in expression. A calibrated ratio much greater than 1, represented by red, indicates a very strong increase in expression. Note the early increase in expression after 2 h of Rta induction (ORF 50), indicated by the arrowhead. While all genes were strongly overexpressed following Rta induction, it is apparent that certain genes show increases in expression earlier than others. The top scale is log 2 (CalRatio).
The replication cycle of herpesviruses involves a carefully ordered pattern of gene expression, classically including immediate-early, early, middle, and late genes (5, 41, 57, 67). To better appreciate the different expression patterns of the various viral genes, the CalRatio values obtained at each time point were normalized by dividing by the maximal expression and multiplying by 100 to yield a percentage of maximal expression. This normalization aids in comparing the expression patterns of abundantly expressed and scarcely expressed genes. The maximum-normalized expression data are presented in Table 4. To better understand the regulation of viral gene expression, it may be helpful to determine when during the viral replication cycle increases in expression of a given gene first begin. In one approach to this question, we determined when the expression of each gene first increased twofold over its baseline value. To eliminate the baseline expression due to vector “leakiness,” each gene's CalRatio was first normalized by dividing by its expression at zero hour. The time at which a doubling in expression was first observed and the time the expression levels were normalized to the value at the time of maximum expression are included in Table 4. Some genes, such as the known early genes and activators ORF45, ORF K8 (K-bZIP), and ORF57 (Mta), show rapid increases after the induction of Rta expression. Other genes, such as those involved in viral DNA replication, show increases in expression later, while still others, such as those for the virion structural proteins, ORF25 (major capsid protein), ORF26 (minor capsid protein), and ORF17 (involved in virion assembly and proteolytic processing), show notable increases in expression only much later. Following Rta induction, many of the cellular homologs (“pirated cellular genes”) appear to be expressed relatively early. Although there are certainly exceptions, the viral gene expression generally appears to follow patterns that would be predicted given their known functions and homologies.
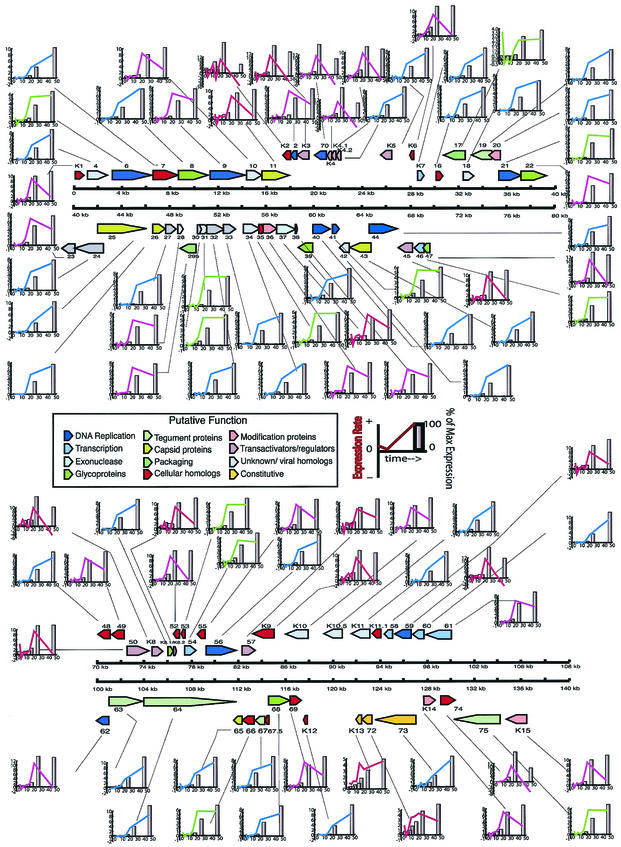
While the time at which a gene first shows a twofold increase in expression offers one way of assessing the onset of expression, that analysis may be subject to some distortions. We therefore used additional analytic approaches to assess changes in KSHV gene expression and to group the viral genes into different expression classes. In one analysis, we examined each gene on a more qualitative level based on its expression rate characteristics. We defined the expression rate as the change in normalized expression over change in time, or Δ[(Ex/Ex-0)/Ex-max]/Δt, where Ex is the calibrated ratio for gene X, Ex-0 is the calibrated ratio for gene X at time zero, Ex-max is gene X's maximal calibrated ratio, and Δt is the difference between corresponding consecutive time points. The rates for each gene were graphed and manually sorted into four different groups. The assignment of each gene into expression rate groups is listed in Table 4 based on pattern similarity. The color-coded rate plots are presented in the context of the viral ORF map in Fig. 6. The red group's rate profile was defined by a change in expression rate of ≥2-fold prior to 12 h, typically with a strong peak by 24 h, followed by rapid decline until 48 h. The purple group's rate profile was defined by a rate that was near zero until after 16 h, when it increased to >2-fold and then rapidly declined from 24 to 48 h. The green group's rate was near zero until after 16 h, when the rate exceeded twofold and then remained steady up to 48 h. The blue group's rate hovered near zero until after 16 h, when the rate exceeded twofold and continued to rise until 48 h. This analysis indicated that each color group principally represented immediate-early, early, early-late, and late gene expression of KSHV.
FIG. 6.
Expression rate profiling of KSHV gene expression induced by Rta. The colored lines represent the kinetic classes defined by the rate or change in expression over change in time (see the text for more detail). The bars represent the percentages of maximal expression, set at 100% (tallest bar), normalized to time zero.
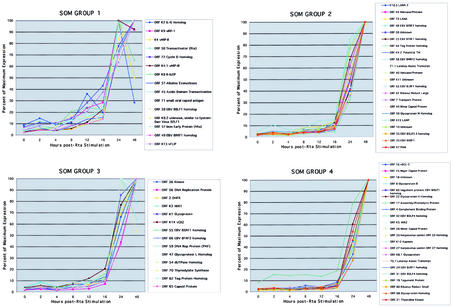
To further characterize the kinetics of KSHV gene expression following Rta induction in groups that may be biologically pertinent, we grouped the KSHV genes into four distinct kinetic classes using SOM, a self-learning algorithm, to assign the genes to distinct sets. SOM analysis classified the genes into different expression groups (Fig. 7). The assignment of genes to each group generally accords with their putative functions. The genes clustered in SOM group 1 started to be expressed 2 to 4 h after Dox treatment, typically with a strong peak by 24 h, followed by rapid decline. This group consisted of immediate-early lytic genes, such as the Rta gene, ORF57 (Mta), ORF K8 (K-bZIP), and K8.2 (Fig. 7). The cluster of genes in SOM groups 2 and 3 showed a peak in expression 20 h after Dox treatment, but the cluster in SOM group 3 showed a more gradual fall in comparison to the cluster in SOM group 2 (Fig. 7). These two groups largely consisted of DNA replication proteins, along with additional transcription factors, including ORF44, ORF40, ORF54, ORF56, ORF59, and ORF61. Finally, the genes assigned to SOM group 4 consisted mainly of late lytic structural and assembly genes, gB, K8.1, ORF19, ORF25, and ORF68 (Fig. 7). Although some genes were not placed in the same groups by all the different analyses, most of the assignments to expression class groups were reasonably consistent. Genes that were placed in SOM group 1 generally were placed in the red expression rate group and had early expression-doubling times. Many of these genes were known immediate-early genes and transactivators. Conversely, many of the genes in SOM group 4 were also placed in the blue expression rate group and had later expression-doubling times. Thus, the KSHV expression pattern following induction by Rta generally conforms to the pattern expected of a herpesvirus. Also as expected, the gene expression patterns provide evidence for coordinated regulation of expression across the KSHV genome. That is, genes with like functions have similar expression patterns, even though they lie far apart on the genome and are under the control of different promoters. It is interesting that the kinetics of induction of individual genes does not necessarily depend on precise mechanisms that Rta uses to mediate the changes in expression. For example, ORF K7 (PAN) is directly targeted by Rta, but its increases in expression come only relatively late following Rta induction (78). Other genes not known to be directly targeted by Rta show large increases in expression soon after induction by Rta.
FIG. 7.
SOM analysis of KSHV gene expression. SOM was used to analyze KSHV gene expression upon Rta expression. A 2-by-2 SOM grid with 250,000 iterations was used to group the KSHV expression patterns into four groups by similarity (Pearson's correlation). The graphs represent each SOM grouping of gene patterns normalized to the percentage of maximum expression.
Thus, our viral microarray results demonstrate that Rta strongly induces activation of viral lytic gene expression and that, unlike TPA stimulation, which leads to mostly linear lytic gene expression, Rta-induced lytic gene expression is strictly ordered.
Release of infectious viral progeny by Rta expression.
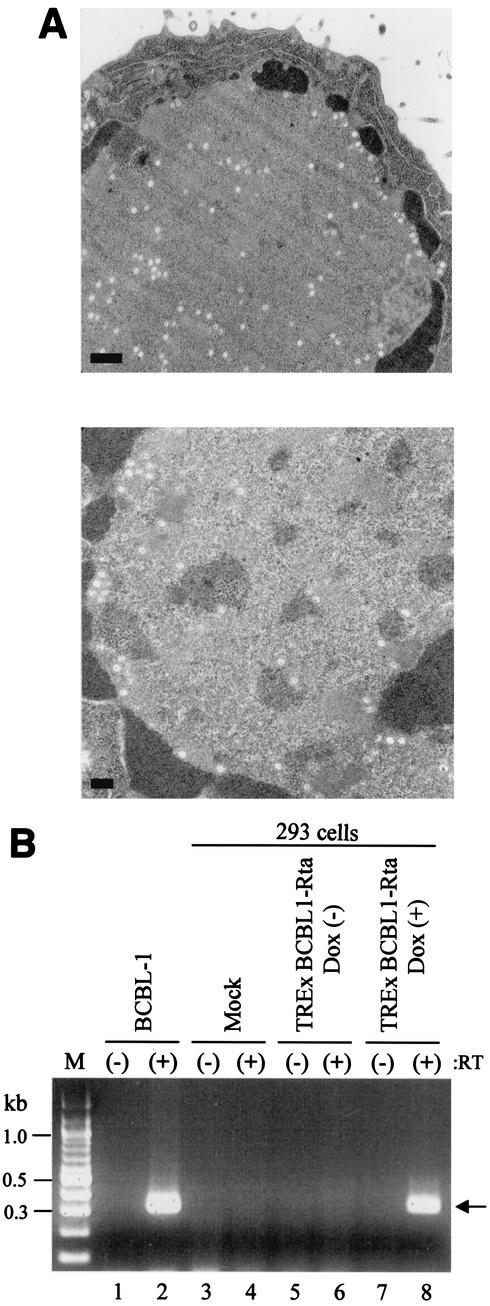
While it has been generally accepted that Rta expression is sufficient to trigger the completion of a full KSHV replication cycle, this has not been formally demonstrated. To determine whether Rta expression was sufficient to trigger the completion of the KSHV replication cycle, we further examined KSHV virion production following induction of Rta expression. TREx BCBL1-Rta cells and parental BCBL-1 cells were induced by Dox and TPA, respectively, for 72 h and subjected to transmission electron microscopy. This experiment showed that TREx BCBL1-Rta cells produced virion particles upon Dox treatment at a level similar to that of TPA-stimulated BCBL-1 cells (Fig. 8A).
FIG. 8.
Assays for release of infectious viral progeny. (A) Electron microscopy to detect virion particles. TPA-treated BCBL-1 cells (top) and Dox-treated TREx BCBL1-Rta cells (bottom) were subjected to transmission electron microscopy. Bars, 500 (top) and 200 (bottom) nm. (B) ORF29-specific RT-PCR analysis to examine KSHV infectivity. cDNA synthesis reactions were performed with (+) or without (−) RT and amplified with ORF29 primers. Lanes: M, molecular size markers; 1 and 2, BCBL-1 cells; 3 and 4, 293 cells with mock infection; 5 and 6, 293 cells with the supernatant from uninduced TREx BCBL1-Rta cells; 7 and 8, 293 cells with the supernatant from Dox-induced TREx BCBL1-Rta cells. The arrow indicates the ORF29 cDNA fragment.
To test the infectivity of KSHV virion progeny produced following induction of Rta, we used an assay developed by Renne et al. (70). This assay used RT-PCR analysis of ORF29 to discriminate viral expression from input virus and to provide information about the nature of the infection in the recipient cell. Cultured 293 cells were infected with cell-free supernatants of TREx BCBL1-Rta cells with or without Dox treatment. After adsorption, the cells were washed twice with fresh culture medium. Total cellular RNA was extracted and used for RT-PCR analysis as previously described (70). No PCR product was obtained from 293 cells that were mock infected or infected with the supernatant of TREx BCBL1-Rta cells without Dox treatment (Fig. 8B, lanes 3, 4, 5, and 6). By contrast, 293 cells exposed to the supernatant of TREx BCBL1-Rta cells with Dox treatment showed a band that comigrated with the PCR product obtained by amplifying cDNA from TPA-stimulated BCBL-1 cells (Fig. 8B, lanes 1, 2, 7, and 8). These data indicate that Rta expression initiates the completion of KSHV lytic replication to produce infectious viral progeny, which are capable of entering 293 cells, delivering the viral genome to the nucleus, and allowing transcription of genes such as ORF29.
DISCUSSION
Herpesviruses replicate through a carefully ordered program of gene expression. In order to fully understand the KSHV replication cycle, the details of the KSHV gene expression program must be defined. Unfortunately, no fully satisfactory in vitro replication model exists for KSHV. To provide a useful means to study KSHV replication and reactivation, we developed the Flp-In-mediated recombination and tetracycline-inducible expression system in KSHV-infected PEL cells. This Flp-In/TetR system has several advantages. (i) The target gene is efficiently integrated into the host genome downstream of the simian virus 40 (SV40) promoter after the replacement of the lacZ-Zeocin fusion gene. Our PCR analysis and β-galactosidase assay indicate that Flp recombinase precisely and effectively induces DNA recombination at the FRT site. As a consequence, the entire lacZ-Zeocin fusion gene was replaced with the target gene downstream of the SV40 early promoter region in an efficient manner, and this replacement was verified by β-galactosidase assay. (ii) The single locus for gene insertion minimizes the genetic variation that is often associated with the random integration of target genes. Specifically, this would be an important issue for the comparative study of the functions of different viral genes or mutants. (iii) KSHV-infected PEL cells, including BCBL-1 cells, have been shown to have a transfection efficiency of <5%. Because of this inability to support efficient transfection, it is technically challenging to conduct studies that investigate the effects of selected genes upon the virus within the latently infected cells. It may also sometimes be difficult to interpret these studies because the investigator must distinguish the actions of the transfected gene upon the small fraction of transfected cells from the background observations due to the large majority of untransfected cells. These distinctions may be particularly difficult because ∼3 to 5% of untreated or uninduced BCBL-1 cells are engaged in a KSHV spontaneous lytic replication cycle at any given time (56). Thus, the Flp-In/TetR system allows us to overcome poor transfection efficiency and the difficulties that result from spontaneous lytic replication and also provides a homogenous population of KSHV-infected BCBL-1 cells for the study of individual gene functions in viral replication.
To date, studies of KSHV replication have generally employed latently infected PEL cell lines induced to undergo lytic replication by treatment with TPA or sodium butyrate (17, 58, 64, 71, 75, 87, 91, 94). Three published studies have described a genomewide transcriptional pattern of KSHV during a lytic replication cycle (33, 44, 68). While treatment of latently infected PEL cells with TPA or butyrate can trigger a complete KSHV replication cycle, the agents are nonphysiological, relatively promiscuous activators that broadly target the protein kinase C pathway and the AP-1 family of transcription factors (7, 96) in the case of TPA or histone acetylation for butyrate (54). A viral replication cycle triggered by these inducing agents may differ from physiological replication to a certain extent because the widespread targeting of signal transduction by TPA and butyrate may bypass the more complicated and subtle regulatory pathways that are engaged during a physiological viral replication cycle. Here, we demonstrate that Rta expression induces a carefully ordered, genomewide program of viral gene expression, leading to the production of infectious virions. Shortly after the induction of Rta, the expression of a set of genes that includes many well-known transactivators increases. Somewhat later, the expression of genes involved in viral DNA replication increases. At the latest time points following Rta induction, the expression of viral structural genes increases. These results indicate that Rta efficiently induces a complete cycle of viral replication, including a well-ordered program of KSHV gene expression and production of infectious viral progeny.
While the general features of the genomewide expression patterns that characterize the KSHV lytic replication cycle appeared to be similar following induction by exogenously added TPA and induction by the KSHV Rta transactivator, certain features of the expression patterns distinguished the viral gene expression patterns following the two modes of induction. For example, Rta induces lytic gene expression to higher levels than TPA stimulation, indicating that Rta may be a much more potent global inducer of KSHV gene expression than TPA. This might, perhaps, be expected, since Rta is the physiological transactivator responsible for the induction of KSHV gene expression during the course of the viral life cycle and is believed to be one of the key genes controlling the exit from latency and entry into a lytic infection cycle. Furthermore, the patterns of expression of certain individual genes differed in the TPA-treated and the Rta-induced cells. For example, a carefully ordered program of Rta-induced viral gene expression was clear for the vOX2 and vGPCR genes, which are derived from a single bicistronic transcript. The expression of both transcripts strongly increased upon Rta expression, reached a maximum at 16 to 20 h, and declined afterward, whereas their expression increased weakly and continuously upon TPA stimulation (Fig. 4A). Other genes that showed notable differences in expression patterns from microarray analysis following Rta and TPA induction (68) were ORF K13 (vFLIP), ORF72 (vCyclin), and ORF K9 (vIRF-1). Increases in their expression occurred at earlier time points in Rta induction than in TPA induction (68). Another interesting difference is that ORF73 (LANA) showed a substantial increase in expression following Rta induction, while its expression was relatively constant following TPA induction (68). In fact, ORF73 expression of mouse herpesvirus 68 has also been shown to increase during lytic replication (1). Thus, an overall appreciation of the expression program following Rta induction reinforces the notion that Rta is the physiological transactivator and that it plays a central, controlling role in the KSHV replication cycle.
To provide general views of the gene expression kinetics following induction of Rta, we used several complementary approaches to organizing and analyzing the expression data: hierarchical clustering, clustering using SOM analysis, classification according to maximal change in the rate of expression, and classification according to when the genes first show a twofold increase over baseline. The hierarchical clustering and SOM group the genes according to their expression profiles over the course of the experiment. The SOM grouping into four clusters may be helpful, given that herpesvirus gene expression patterns have often been considered to fall into four general classes (immediate early, delayed early, middle, and late). The grouping according to the maximal rate of change of expression and the doubling time analysis may be helpful because, when the systems that control gene expression are considered, the time at which a gene is first induced may be of particular interest. Each of these approaches has drawbacks, however. The doubling time analysis may be confusing for genes that have unusually low or high baseline levels of expression, while the rate-of-change analysis may be perturbed by noise in the measurements. Nevertheless, the different approaches show general agreement, although some details may differ. For example, many genes in SOM group 1 were also assigned to the red group according to their rates and showed doubling of expression at the earliest time points during the experiment. These groups included several transactivators and regulatory genes. Conversely, genes in SOM group 4 were generally placed in the blue group by the rate analysis and generally had later doubling times. While no analytical approach may provide a perfect view of the expression program, a combination of approaches may offer additional insights and help guide future studies of individual gene functions.
The generation of a cell line latently infected with KSHV and containing an inducible viral transactivator that, upon induction, can trigger a well-ordered viral lytic gene expression program culminating in the production of infectious virions should be of significant help in understanding KSHV replication and pathogenesis. Since the herpesviruses use similar infection and replication strategies, this approach should be useful not only for KSHV but also for other γ-herpesviruses, like EBV, for which a cell line truly permissive for viral replication do not exist.
Acknowledgments
We especially thank K. Yamanishi and D. Ganem for Rta antibody and BCBL-1 cells, M. Li for critical discussion, J. Zarycki for excellent technical help, J. MacKey for electron microscopy analysis, and J. Macke for manuscript editing. We also thank E. Chuang, M. Tsai, H. Yao, and S. Zhao at the ATC, NCI, for assisting us in preparing the viral arrays; J. Powell and BIMAS, CIT, NIH, for providing us with the viral gene array lists and web-based tools on mAdb for our analyses; and Mike Bittner, Paul Meltzer, and Jeff Trent of the Cancer Genetics Branch, NHGRI, for providing clones containing cellular housekeeping genes.
This work was partly supported by Public Health Service grants CA82057, CA86841, CA91819, AI38131, and RR00168; American Cancer Society grant RPG001102; the NIH Intramural AIDS Targeted Antiviral Program; and an Intramural Research Award from the Center for Cancer Research, NCI. J. U. Jung is a Leukemia and Lymphoma Society Scholar.
REFERENCES
- 1.Ahn, J. W., K. L. Powell, P. Kellam, and D. G. Alber. 2002. Gammaherpesvirus lytic gene expression as characterized by DNA array. J. Virol. 76:6244-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht, J. C., J. Nicholas, D. Biller, K. R. Cameron, B. Biesinger, C. Newman, S. Wittmann, M. A. Craxton, H. Coleman, B. Fleckenstein, et al. 1992. Primary structure of the herpesvirus saimiri genome. J. Virol. 66:5047-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander, L., L. Denekamp, A. Knapp, M. R. Auerbach, B. Damania, and R. C. Desrosiers. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J. Virol. 74:3388-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambroziak, J. A., D. J. Blackbourn, B. G. Herndier, R. G. Glogau, J. H. Gullett, A. R. McDonald, E. T. Lennette, and J. A. Levy. 1995. Herpes-like sequences in HIV-infected and uninfected Kaposi's sarcoma patients. Science 268:582-583. [DOI] [PubMed] [Google Scholar]
- 5.Anderson, K. P., R. H. Costa, L. E. Holland, and E. K. Wagner. 1980. Characterization of herpes simplex virus type 1 RNA present in the absence of de novo protein synthesis. J. Virol. 34:9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrews, B. J., G. A. Proteau, L. G. Beatty, and P. D. Sadowski. 1985. The FLP recombinase of the 2 micron circle DNA of yeast: interaction with its target sequences. Cell 40:795-803. [DOI] [PubMed] [Google Scholar]
- 7.Angel, P., M. Imagawa, R. Chiu, B. Stein, R. J. Imbra, H. J. Rahmsdorf, C. Jonat, P. Herrlich, and M. Karin. 1987. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell 49:729-739. [DOI] [PubMed] [Google Scholar]
- 8.Arvanitakis, L., E. Geras-Raaka, A. Varma, M. C. Gershengorn, and E. Cesarman. 1997. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature 385:347-350. [DOI] [PubMed] [Google Scholar]
- 9.Arvanitakis, L., E. A. Mesri, R. G. Nador, J. W. Said, A. S. Asch, D. M. Knowles, and E. Cesarman. 1996. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood 88:2648-2654. [PubMed] [Google Scholar]
- 10.Belanger, C., A. Gravel, A. Tomoiu, M. E. Janelle, J. Gosselin, M. J. Tremblay, and L. Flamand. 2001. Human herpesvirus 8 viral FLICE-inhibitory protein inhibits Fas-mediated apoptosis through binding and prevention of procaspase-8 maturation. J. Hum. Virol. 4:62-73. [PubMed] [Google Scholar]
- 11.Bittner, M., P. Meltzer, Y. Chen, Y. Jiang, E. Seftor, M. Hendrix, M. Radmacher, R. Simon, Z. Yakhini, A. Ben-Dor, N. Sampas, E. Dougherty, E. Wang, F. Marincola, C. Gooden, J. Lueders, A. Glatfelter, P. Pollock, J. Carpten, E. Gillanders, D. Leja, K. Dietrich, C. Beaudry, M. Berens, D. Alberts, and V. Sondak. 2000. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature 406:536-540. [DOI] [PubMed] [Google Scholar]
- 12.Bosing, T., F. Bellos, F. W. Cremer, C. Gemmel, G. Moldenhauer, A. D. Ho, H. Goldschmidt, and M. Moos. 2000. CD19+ and CD20+ B cells from the peripheral blood of patients with multiple myeloma are not infected with human herpesvirus 8. Leukemia 14:1330-1331. [DOI] [PubMed] [Google Scholar]
- 13.Broach, J. R., V. R. Guarascio, and M. Jayaram. 1982. Recombination within the yeast plasmid 2μ circle is site-specific. Cell 29:227-234. [DOI] [PubMed] [Google Scholar]
- 14.Broach, J. R., and J. B. Hicks. 1980. Replication and recombination functions associated with the yeast plasmid, 2μ circle. Cell 21:501-508. [DOI] [PubMed] [Google Scholar]
- 15.Buchholz, F., L. Ringrose, P. O. Angrand, F. Rossi, and A. F. Stewart. 1996. Different thermostabilities of FLP and Cre recombinases: implications for applied site-specific recombination. Nucleic Acids Res. 24:4256-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burysek, L., and P. M. Pitha. 2001. Latently expressed human herpesvirus 8-encoded interferon regulatory factor 2 inhibits double-stranded RNA-activated protein kinase. J. Virol. 75:2345-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cannon, J. S., D. Ciufo, A. L. Hawkins, C. A. Griffin, M. J. Borowitz, G. S. Hayward, and R. F. Ambinder. 2000. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi's sarcoma herpesvirus-containing supernatant. J. Virol. 74:10187-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 19.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 20.Chang, P. J., D. Shedd, L. Gradoville, M. S. Cho, L. W. Chen, J. Chang, and G. Miller. 2002. Open reading frame 50 protein of Kaposi's sarcoma-associated herpesvirus directly activates the viral PAN and K12 genes by binding to related response elements. J. Virol. 76:3168-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 22.Chen, J., K. Ueda, S. Sakakibara, T. Okuno, and K. Yamanishi. 2000. Transcriptional regulation of the Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor gene. J. Virol. 74:8623-8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen, Y., E. R. Dougherty, and M. L. Bittner. 1997. Ratio-based decisions and the quantitative analysis of cDNA microarray images. J. Biomed. Optics 2:364-374. [DOI] [PubMed] [Google Scholar]
- 24.Chevallier-Greco, A., E. Manet, P. Chavrier, C. Mosnier, J. Daillie, and A. Sergeant. 1986. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 5:3243-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung, Y. H., R. E. Means, J. K. Choi, B. S. Lee, and J. U. Jung. 2002. Kaposi's sarcoma-associated herpesvirus OX2 glycoprotein activates myeloid-lineage cells to induce inflammatory cytokine production. J. Virol. 76:4688-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Craig, N. L. 1988. The mechanism of conservative site-specific recombination. Annu. Rev. Genet. 22:77-105. [DOI] [PubMed] [Google Scholar]
- 27.Deng, H., A. Young, and R. Sun. 2000. Auto-activation of the rta gene of human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus. J. Gen. Virol. 81:3043-3048. [DOI] [PubMed] [Google Scholar]
- 28.Desrosiers, R. C., V. G. Sasseville, S. C. Czajak, X. Zhang, K. G. Mansfield, A. Kaur, R. P. Johnson, A. A. Lackner, and J. U. Jung. 1997. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J. Virol. 71:9764-9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dittmer, D., C. Stoddart, R. Renne, V. Linquist-Stepps, M. E. Moreno, C. Bare, J. M. McCune, and D. Ganem. 1999. Experimental transmission of Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) to SCID-hu Thy/Liv mice. J. Exp. Med. 190:1857-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duan, W., S. Wang, S. Liu, and C. Wood. 2001. Characterization of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF57 promoter. Arch. Virol. 146:403-413. [DOI] [PubMed] [Google Scholar]
- 31.Duggan, D. J., M. Bittner, Y. Chen, P. Meltzer, and J. M. Trent. 1999. Expression profiling using cDNA microarrays. Nat. Genet. 21:10-14. [DOI] [PubMed] [Google Scholar]
- 32.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fakhari, F. D., and D. P. Dittmer. 2002. Charting latency transcripts in Kaposi's sarcoma-associated herpesvirus by whole-genome real-time quantitative PCR. J. Virol. 76:6213-6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fields, B. N., D. M. Knipe, and P. M. Howley (ed.). 2002. Fields virology, 4th ed. Raven Press, New York, N.Y.
- 35.Gao, S. J., C. Boshoff, S. Jayachandra, R. A. Weiss, Y. Chang, and P. S. Moore. 1997. KSHV ORF K9 (vIRF) is an oncogene which inhibits the interferon signaling pathway. Oncogene 15:1979-1985. [DOI] [PubMed] [Google Scholar]
- 36.Gao, S. J., Y. J. Zhang, J. H. Deng, C. S. Rabkin, O. Flore, and H. B. Jenson. 1999. Molecular polymorphism of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent nuclear antigen: evidence for a large repertoire of viral genotypes and dual infection with different viral genotypes. J. Infect. Dis. 180:1466-1476. [DOI] [PubMed] [Google Scholar]
- 37.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gronostajski, R. M., and P. D. Sadowski. 1985. Determination of DNA sequences essential for FLP-mediated recombination by a novel method. J. Biol. Chem. 260:12320-12327. [PubMed] [Google Scholar]
- 39.Gwack, Y., H. Byun, S. Hwang, C. Lim, and J. Choe. 2001. CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi's sarcoma-associated herpesvirus open reading frame 50. J. Virol. 75:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gwack, Y., S. Hwang, C. Lim, Y. S. Won, C. H. Lee, and J. Choe. 2002. Kaposi's Sarcoma-associated herpesvirus open reading frame 50 stimulates the transcriptional activity of STAT3. J. Biol. Chem. 277:6438-6442. [DOI] [PubMed] [Google Scholar]
- 41.Honess, R. W., and B. Roizman. 1974. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 14:8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horenstein, M. G., R. G. Nador, A. Chadburn, E. M. Hyjek, G. Inghirami, D. M. Knowles, and E. Cesarman. 1997. Epstein-Barr virus latent gene expression in primary effusion lymphomas containing Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8. Blood 90:1186-1191. [PubMed] [Google Scholar]
- 43.Jayaram, M. 1985. Two-micrometer circle site-specific recombination: the minimal substrate and the possible role of flanking sequences. Proc. Natl. Acad. Sci. USA 82:5875-5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeong, J., J. Papin, and D. Dittmer. 2001. Differential regulation of the overlapping Kaposi's sarcoma-associated herpesvirus vGCR (orf74) and LANA (orf73) promoters. J. Virol. 75:1798-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khan, J., L. H. Saal, M. L. Bittner, Y. Chen, J. M. Trent, and P. S. Meltzer. 1999. Expression profiling in cancer using cDNA microarrays. Electrophoresis 20:223-229. [DOI] [PubMed] [Google Scholar]
- 47.Kledal, T. N., M. M. Rosenkilde, F. Coulin, G. Simmons, A. H. Johnsen, S. Alouani, C. A. Power, H. R. Luttichau, J. Gerstoft, P. R. Clapham, I. Clark-Lewis, T. N. Wells, and T. W. Schwartz. 1997. A broad-spectrum chemokine antagonist encoded by Kaposi's sarcoma-associated herpesvirus. Science 277:1656-1659. [DOI] [PubMed] [Google Scholar]
- 48.Kohonen, T. (ed.). 2001. Self-organizing maps, 3rd ed. Springer, New York, N.Y.
- 49.Li, M., H. Lee, J. Guo, F. Neipel, B. Fleckenstein, K. Ozato, and J. U. Jung. 1998. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor. J. Virol. 72:5433-5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li, M., H. Lee, D. W. Yoon, J. C. Albrecht, B. Fleckenstein, F. Neipel, and J. U. Jung. 1997. Kaposi's sarcoma-associated herpesvirus encodes a functional cyclin. J. Virol. 71:1984-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li, M., J. MacKey, S. C. Czajak, R. C. Desrosiers, A. A. Lackner, and J. U. Jung. 1999. Identification and characterization of Kaposi's sarcoma-associated herpesvirus K8.1 virion glycoprotein. J. Virol. 73:1341-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liang, Y., J. Chang, S. J. Lynch, D. M. Lukac, and D. Ganem. 2002. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jκ (CSL), the target of the Notch signaling pathway. Genes Dev. 16:1977-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lubyova, B., and P. M. Pitha. 2000. Characterization of a novel human herpesvirus 8-encoded protein, vIRF-3, that shows homology to viral and cellular interferon regulatory factors. J. Virol. 74:8194-8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luka, J., B. Kallin, and G. Klein. 1979. Induction of the Epstein-Barr virus (EBV) cycle in latently infected cells by n-butyrate. Virology 94:228-231. [DOI] [PubMed] [Google Scholar]
- 55.Lukac, D. M., L. Garibyan, J. R. Kirshner, D. Palmeri, and D. Ganem. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 75:6786-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 57.Mackem, S., and B. Roizman. 1980. Regulation of herpesvirus macromolecular synthesis: transcription-initiation sites and domains of alpha genes. Proc. Natl. Acad. Sci. USA 77:7122-7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller, G., L. Heston, E. Grogan, L. Gradoville, M. Rigsby, R. Sun, D. Shedd, V. M. Kushnaryov, S. Grossberg, and Y. Chang. 1997. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J. Virol. 71:314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moore, P. S., C. Boshoff, R. A. Weiss, and Y. Chang. 1996. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science 274:1739-1744. [DOI] [PubMed] [Google Scholar]
- 60.Moore, P. S., and Y. Chang. 1995. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection. N. Engl. J. Med. 332:1181-1185. [DOI] [PubMed] [Google Scholar]
- 61.Moses, A. V., K. N. Fish, R. Ruhl, P. P. Smith, J. G. Strussenberg, L. Zhu, B. Chandran, and J. A. Nelson. 1999. Long-term infection and transformation of dermal microvascular endothelial cells by human herpesvirus 8. J. Virol. 73:6892-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neipel, F., J. C. Albrecht, A. Ensser, Y. Q. Huang, J. J. Li, A. E. Friedman-Kien, and B. Fleckenstein. 1997. Human herpesvirus 8 encodes a homolog of interleukin-6. J. Virol. 71:839-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neipel, F., J. C. Albrecht, and B. Fleckenstein. 1999. Human herpesvirus 8: is it a tumor virus? Proc. Assoc. Am. Physicians 111:594-601. [DOI] [PubMed] [Google Scholar]
- 64.Nicholas, J., V. Ruvolo, J. Zong, D. Ciufo, H. G. Guo, M. S. Reitz, and G. S. Hayward. 1997. A single 13-kilobase divergent locus in the Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genome contains nine open reading frames that are homologous to or related to cellular proteins. J. Virol. 71:1963-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nicholas, J., V. R. Ruvolo, W. H. Burns, G. Sandford, X. Wan, D. Ciufo, S. B. Hendrickson, H. G. Guo, G. S. Hayward, and M. S. Reitz. 1997. Kaposi's sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat. Med. 3:287-292. [DOI] [PubMed] [Google Scholar]
- 66.Nicholas, J., J. C. Zong, D. J. Alcendor, D. M. Ciufo, L. J. Poole, R. T. Sarisky, C. J. Chiou, X. Zhang, X. Wan, H. G. Guo, M. S. Reitz, and G. S. Hayward. 1998. Novel organizational features, captured cellular genes, and strain variability within the genome of KSHV/HHV8. J. Natl. Cancer Inst. Monogr. 23:79-88. [DOI] [PubMed] [Google Scholar]
- 67.O'Hare, P., and G. S. Hayward. 1984. Expression of recombinant genes containing herpes simplex virus delayed-early and immediate-early regulatory regions and trans activation by herpesvirus infection. J. Virol. 52:522-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paulose-Murphy, M., N. K. Ha, C. Xiang, Y. Chen, L. Gillim, R. Yarchoan, P. Meltzer, M. Bittner, J. Trent, and S. Zeichner. 2001. Transcription program of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 75:4843-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ploegh, H. L. 1998. Viral strategies of immune evasion. Science 280:248-253. [DOI] [PubMed] [Google Scholar]
- 70.Renne, R., D. Blackbourn, D. Whitby, J. Levy, and D. Ganem. 1998. Limited transmission of Kaposi's sarcoma-associated herpesvirus in cultured cells. J. Virol. 72:5182-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 72.Rivas, C., A. E. Thlick, C. Parravicini, P. S. Moore, and Y. Chang. 2001. Kaposi's sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. J. Virol. 75:429-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sakakibara, S., K. Ueda, J. Chen, T. Okuno, and K. Yamanishi. 2001. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 75:6894-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sarid, R., O. Flore, R. A. Bohenzky, Y. Chang, and P. S. Moore. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 72:1005-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sarid, R., T. Sato, R. A. Bohenzky, J. J. Russo, and Y. Chang. 1997. Kaposi's sarcoma-associated herpesvirus encodes a functional bcl-2 homologue. Nat. Med. 3:293-298. [DOI] [PubMed] [Google Scholar]
- 77.Sarid, R., J. S. Wiezorek, P. S. Moore, and Y. Chang. 1999. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J. Virol. 73:1438-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sauer, B. 1994. Site-specific recombination: developments and applications. Curr. Opin. Biotechnol. 5:521-527. [DOI] [PubMed] [Google Scholar]
- 79.Seaman, W. T., D. Ye, R. X. Wang, E. E. Hale, M. Weisse, and E. B. Quinlivan. 1999. Gene expression from the ORF50/K8 region of Kaposi's sarcoma-associated herpesvirus. Virology 263:436-449. [DOI] [PubMed] [Google Scholar]
- 80.Searles, R. P., E. P. Bergquam, M. K. Axthelm, and S. W. Wong. 1999. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 73:3040-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Senecoff, J. F., R. C. Bruckner, and M. M. Cox. 1985. The FLP recombinase of the yeast 2-micron plasmid: characterization of its recombination site. Proc. Natl. Acad. Sci. USA 82:7270-7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shalon, D., S. J. Smith, and P. O. Brown. 1996. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 6:639-645. [DOI] [PubMed] [Google Scholar]
- 83.Song, M. J., H. J. Brown, T. T. Wu, and R. Sun. 2001. Transcription activation of polyadenylated nuclear RNA by Rta in human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:3129-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Song, M. J., X. Li, H. J. Brown, and R. Sun. 2002. Characterization of interactions between RTA and the promoter of polyadenylated nuclear RNA in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 76:5000-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 86.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sun, R., S. F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Takada, K., N. Shimizu, S. Sakuma, and Y. Ono. 1986. trans activation of the latent Epstein-Barr virus (EBV) genome after transfection of the EBV DNA fragment. J. Virol. 57:1016-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thome, M., P. Schneider, K. Hofmann, H. Fickenscher, E. Meinl, F. Neipel, C. Mattmann, K. Burns, J. L. Bodmer, M. Schroter, C. Scaffidi, P. H. Krammer, M. E. Peter, and J. Tschopp. 1997. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature 386:517-521. [DOI] [PubMed] [Google Scholar]
- 90.Wang, S., S. Liu, M. H. Wu, Y. Geng, and C. Wood. 2001. Identification of a cellular protein that interacts and synergizes with the RTA (ORF50) protein of Kaposi's sarcoma-associated herpesvirus in transcriptional activation. J. Virol. 75:11961-11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu, Y., J. B. Black, C. S. Goldsmith, P. J. Browning, K. Bhalla, and M. K. Offermann. 1999. Induction of human herpesvirus-8 DNA replication and transcription by butyrate and TPA in BCBL-1 cells. J. Gen. Virol. 80:83-90. [DOI] [PubMed] [Google Scholar]
- 92.Zalani, S., E. Holley-Guthrie, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. USA 93:9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhong, W., and D. Ganem. 1997. Characterization of ribonucleoprotein complexes containing an abundant polyadenylated nuclear RNA encoded by Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8). J. Virol. 71:1207-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu, F. X., T. Cusano, and Y. Yuan. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5556-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zimring, J. C., S. Goodbourn, and M. K. Offermann. 1998. Human herpesvirus 8 encodes an interferon regulatory factor (IRF) homolog that represses IRF-1-mediated transcription. J. Virol. 72:701-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.zur Hausen, H., F. J. O'Neill, U. K. Freese, and E. Hecker. 1978. Persisting oncogenic herpesvirus induced by the tumour promotor TPA. Nature 272:373-375. [DOI] [PubMed] [Google Scholar]