Abstract
The abundant nuclear enzyme poly(ADP-ribose) polymerase (PARP) functions in DNA damage surveillance and repair and at the decision between apoptosis and necrosis. Here we show that PARP binds to simian virus 40 (SV40) capsid proteins VP1 and VP3. Furthermore, its enzymatic activity is stimulated by VP3 but not by VP1. Experiments with purified mutant proteins demonstrated that the PARP binding domain in VP3 is localized to the 35 carboxy-terminal amino acids, while a larger peptide of 49 amino acids was required for full stimulation of its activity. The addition of 3-aminobenzamide (3-AB), a known competitive inhibitor of PARP, demonstrated that PARP participates in the SV40 life cycle. The titer of SV40 propagated on CV-1 cells was reduced by 3-AB in a dose-dependent manner. Additional experiments showed that 3-AB did not affect viral DNA replication or capsid protein production. PARP did not modify the viral capsid proteins in in vitro poly(ADP-ribosylation) assays, implying that it does not affect SV40 infectivity. On the other hand, it greatly reduced the magnitude of the host cytopathic effects, a hallmark of SV40 infection. Additional experiments suggested that the stimulation of PARP activity by VP3 leads the infected cell to a necrotic pathway, characterized by the loss of membrane integrity, thus facilitating the release of mature SV40 virions from the cells. Our studies identified a novel function of the minor capsid protein VP3 in the recruitment of PARP for the SV40 lytic process.
Simian virus 40 (SV40) is a papovavirus, with a small, double-stranded, circular DNA genome of 5.2 kb (37a). The viral capsid, surrounding the viral minichromosome, is composed of three virus-encoded proteins, VP1, VP2, and VP3. Seventy-two pentamers of VP1 form the outer shell, while VP2 and VP3 bridge between the VP1 shell and the chromatin core. VP3 translation initiates from an internal AUG within the VP2 coding sequence, utilizing the same translational frame. Thus, except for the amino part that is unique to VP2, the two proteins are identical and are commonly referred to as VP2/3.
The VP1 monomer is composed of an antiparallel β-strand core with a jelly roll topology, with a short N-terminal arm that contains the DNA-binding domain and nuclear localization signal (21) and a long C-terminal arm (22). VP1 monomers are tightly bound in pentamers, forming structures of fivefold symmetry with an inward-facing cavity (22, 35). The VP1 pentamers are tied together in the capsid through their carboxy-terminal arms. The five arms extending from each pentamer are inserted into the neighboring pentamers in three distinct kinds of interactions. This unique type of bonding underlies the variability in contacts between the identical building blocks, allowing the flexibility required for the packing geometry (22).
A single molecule of VP2 or VP3 is firmly anchored in the inner cavity of each VP1 pentamer (VP15) by hydrophobic interactions (3, 15), through a region near, but not at, the carboxy terminus of VP2/3 (see Fig. 3). The amino part and the carboxy terminus of VP2/3 protrude from the cavity of the VP1 pentamer, remaining exposed in the complex. The carboxy terminus contains the DNA-binding domain (5) and the nuclear localization signal (6). The VP15-VP2/3 complex is formed in the cytoplasm soon after translation (11, 19). In the nucleus VP15-VP2/3 is recruited by transcription factor Sp1 to the packaging signal of the SV40 minichromosome, ses. The Sp1 interactive domain of VP2/3 is also present at the carboxy terminus, overlapping the DNA-binding domain (14). It was proposed that the recruitment complex forms a nucleation center at ses (15) and that the first step in capsid assembly is the formation of a fivefold symmetric complex with a “pentavalent” pentamer at its center (35). Assembly is thought to occur by gradual addition and organization of capsid building blocks around the SV40 chromatin (2, 13).
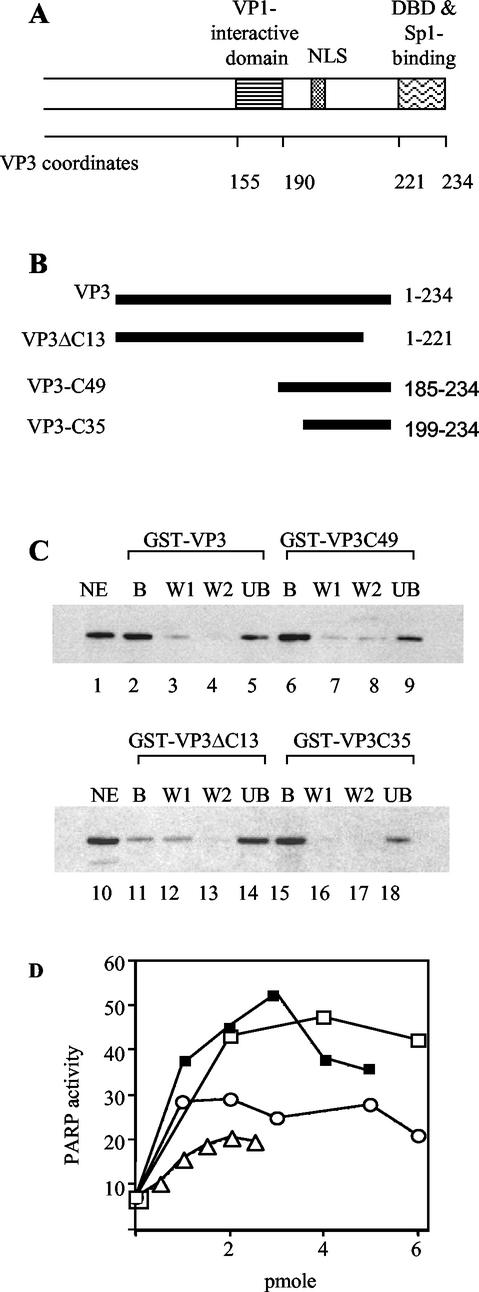
FIG. 3.
Binding and stimulation of PARP activity by VP3 and VP3 mutants. (A) Schematic representation of the carboxy part of SV40 VP3 (which is shared with VP2). Amino acid coordinates (Swiss-Protaccession number P03093) are shown below the map. The VP1 interactive domain of VP3 (3), nuclear localization signal (NLS) (19), DNA-binding domain (DBD) (5), and Sp1-binding domain (14, 15) are designated above. (B) Schematic representation of VP3 truncation mutants. The heavy lines represent the amino acids included in the mutant proteins. The VP3 amino acids are designated at the right. (C) Binding of the VP3 mutants to PARP. GST pulldown assays were performed with GST-VP3, GST-VP3C49, GST-VP3ΔC13, and GST-VP3C35, immobilized on glutathione agarose, and incubated with nuclear extracts of K562 cells that contain PARP. The bound proteins (B), two washes (W1 and W2), and supernatant containing unbound material (UB) were analyzed by SDS-PAGE and Western blotting with anti-PARP antibody. The aliquots shown in each of the lanes correspond to equivalent amounts of the input nuclear extract (NE) shown in the first lane in each panel. (D) Stimulation of PARP enzymatic activity by VP3 mutants. Incorporation of [32P]ADP-ribose was assayed by the poly(ADP-ribosylation) assay followed by PAGE and PhosphorImager analysis. □, GST-VP3; ▪, GST-VP2/3C49; ○, GST-VP2/3C35; and ▵, GST-VP3ΔC13.
SV40 uses host factors for many stages in its life cycle, such as viral entry and DNA replication and transcription. Late in the infection cycle, SV40 usurps cellular transcription factor Sp1 for the recruitment of the capsid proteins to the viral packaging signal, ses, conferring specific DNA recognition upon the proteins, which otherwise bind to DNA nonspecifically. Thus, the capsid building blocks overcome the problem of recognizing the SV40 minichromosome in the presence of excess cellular chromatin within the nucleus (14, 15). In this study we discovered a second host factor, the nuclear enzyme poly(ADP-ribose) polymerase (PARP), that participates at late stages of the SV40 life cycle.
PARP is an abundant nuclear enzyme that is highly conserved and constitutively expressed in most eukaryotic cells investigated. It modifies proteins by catalyzing the formation of extensive branched polymers of poly(ADP-ribose) by using NAD+ as a substrate (reviewed in reference 20). PARP has several targets that are primarily DNA-binding proteins, such as histones and transcription factors, including Sp1 and YY1 (29), but is most active in automodification. Because of the high negative charge on ADP-ribose polymers, poly(ADP-ribosylated) proteins lose their affinity for DNA (reviewed in reference 30).
Several functional domains of PARP have been defined and well studied (Fig. 1A). The DNA-binding domain located at the amino terminus is composed of two zinc fingers and binds to DNA strand breaks, DNA loops, and cruciform structures (reviewed in reference 37). Adjacent to the DNA-binding domain is the glutamic acid-rich automodification domain. When PARP is modified, it no longer binds to DNA. The poly(ADP-ribosyl) catalytic domain is at the C-terminal end (reviewed in reference 8). The catalytic activity of PARP is stimulated >500-fold upon binding to DNA strand breaks.
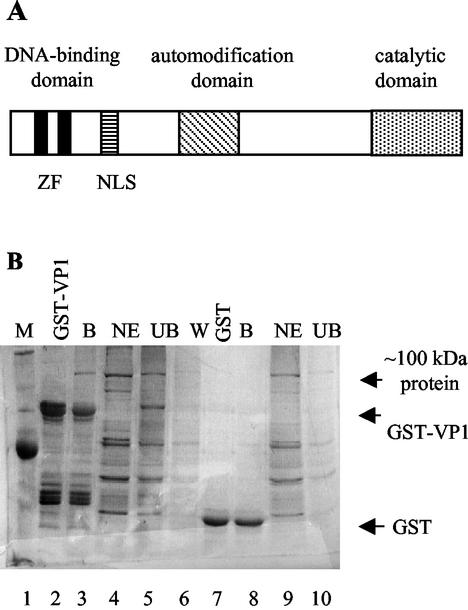
FIG. 1.
VP1 binds PARP. (A) Schematic representation of PARP. The DNA-binding domain, automodification domain, and catalytic domain are designated above the map. The two zinc fingers in the DNA-binding domain (ZF) and the nuclear localization signal (NLS) are designated below. (B) GST pulldown assay. GST-VP1 (lane 2) and GST (lane 7) immobilized on glutathione agarose were incubated with nuclear extracts of K562 cells. The bound proteins (B), nuclear extract (NE), supernatant containing unbound material (UB), and wash (W) were analyzed by SDS-PAGE and silver staining. A distinct band of ∼100 kDa is present in the GST-VP1-bound fraction (B, lane 3, indicated by an arrow) but not in the GST-VP1 alone bound to GSH-agarose (lane 2). This protein is not found in the GST-bound fraction (B, lane 8). The band was excised and analyzed by mass spectrometry. M, molecular mass marker.
The function of PARP has been implicated in several key cellular processes: repair of DNA damage, recombination, telomer length and chromosome stability, apoptosis, transcription regulation, and histone shuttling (28, 30). If limited damage to DNA occurs, PARP acts as a survival factor by rapidly activating DNA repair pathways, through recruitment of the base excision repair complex. In addition, early in apoptosis PARP poly(ADP-ribosylates) p53. Subsequent degradation of the poly(ADP-ribose) chains coincides with the activation of caspase 3 and the cleavage of PARP (33). PARP is cleaved by caspase 3 at a single site, separating its DNA-binding domain from the catalytic domain and thus inactivating its enzymatic activity. PARP cleavage has been shown in almost all forms of apoptosis and is generally used as a hallmark of apoptosis. Whenever apoptosis is inhibited, PARP cleavage is also blocked (30).
Under genotoxic stress massive DNA damage overactivates PARP, leading to the depletion of cellular stores of NAD+, ATP depletion, and necrotic cell death. The failure of the cell to cleave PARP may contribute to the depletion of cell energy during necrosis (30). In addition PARP activates NF-κB, a key player in inflammatory response. Furthermore, excessive poly(ADP-ribosylation) of p53 reduces its DNA-binding activity. Thus, PARP functions at the crossroads of cell survival, apoptosis, and necrosis (4).
PARP was reported to poly(ADP-ribosylate) SV40 large T antigen (1). It plays a role in retroviral infection, facilitating integration of the provirus into the host genome (12). Likewise, it is also required for efficient human immunodeficiency virus type 1 integration (17). It modifies core proteins V and VII of adenovirus, both of which are intimately associated with the viral genome and act in its organization inside the virion particle (9). During Sindbis virus infection PARP is rapidly activated, leading to apoptotic cell death (27).
In the present study we found that PARP enzymatic activity is stimulated by the SV40 minor structural protein VP3 and that PARP assists SV40 release to the medium, presumably by channeling the infected cell to necrosis.
MATERIALS AND METHODS
Cells and virus.
CV-1 cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. K562 cells (K562 is a human erythroleukemia cell line) were cultures in RPMI media with 10% fetal bovine serum. SV40 was propagated on CV-1 cells. The virus was harvested by repeated freeze-thaw followed by sonication. Titration was performed on CV-1 cells either by the plaque assay or by scoring for infective centers by using in situ hybridization as previously described (7).
Protein purification.
PARP, purified by DNA-cellulose chromatography, was purchased from ICN.
Glutathione transferase (GST)-VP3, GST-VP3C35, and GST-VP3C49, expressed from pGEX-VP3 (14), pGEX-VP3C35, and pGEX-VP3C49 (15), were purified on glutathione (GSH)-agarose resin as recommended by the manufacturer with minor variations as previously described (15). GST-VP3ΔC13 was expressed from pGEX-VP3ΔC13 (5), kindly provided by Harumi Kasamatsu.
His-VP1 and His-VP1ΔC60, expressed from pQE-VP1 (38) and pQE-VP1ΔC60 (15), were purified on Ni-nitrilotriacetic acid chromatography as recommended by the manufacturer (Qiagen) with minor variations as previously described (15). His-VP1ΔC60 (34 kDa) is a VP1 mutant with a deletion of 60 amino acids that comprise the C-terminal arm. It forms pentamers and cannot assemble into capsids.
GST pulldown assay.
GST fusion proteins were bound to GSH-agarose beads as described above. Fifty microliters of beads loaded with fusion protein was incubated with approximately 20 μg of K562 nuclear extract in a solution of 0.2 ml of phosphate-buffered saline (PBS) for 20 min in a rotary shaker at 4°C. After centrifugation, the supernatant containing the unbound protein was collected and the beads were washed several times with PBS. The bound proteins were eluted by boiling in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. The fractions were resolved by SDS-PAGE (NuPage; Invitrogen), and the bands were visualized by Western blotting with anti-PARP antibody (Santa Cruz). Equivalent amounts of bound and unbound proteins were loaded on the gel. Pretreatment with ethidium bromide was carried out by incubating both the GST-VP1 immobilized on GSH-agarose and the K562 nuclear extract with 3 μM ethidium bromide for 15 min on ice.
Analysis of SV40 DNA replication.
Infected CV-1 cells were harvested using the Hirt procedure (17a). The Hirt supernatants were treated with RNase and underwent phenol extraction and ethanol precipitation. The level of SV40 DNA was determined by gel electrophoresis and Southern blot hybridization (34a) with SV40 DNA as a probe. Quantification was performed by using the PhosphorImager.
In vitro poly(ADP-ribosylation) assay.
The enzymatic reaction was initiated by the addition of 1 to 200 ng of purified PARP (ICN) to a reaction mixture containing 10 mM Tris-HCl, pH 8.0, 7 mM MgCl2, 50 μM ZnCl2, 1% bovine serum albumin, 3 μM [32P]NAD+, and 0.1 μg of sonicated salmon sperm DNA (or poly[dIdC])/μl in a volume of 15 μl. The capsid proteins or their derivatives were added to the reaction mixture prior to the addition of PARP, as specified. The reaction was incubated for 20 min on ice and was stopped by the addition of SDS-PAGE sample buffer. The proteins were separated on SDS-PAGE, analyzed by autoradiography, and quantified by phosphorimaging. For the production of long poly(ADP-ribose) chains, excess unlabeled NAD+ (1 mM) was added after the initial 20 min of incubation with [32P]NAD and the reaction was allowed to continue for an additional 20 min.
Staining with AO and PI.
Infected and uninfected CV-1 cells were cultured on cover slides in 24-well plates (105 CV-1 cells/well). Infection with SV40 was at a multiplicity of infection (MOI) of 10. Slides were harvested daily. The cells were washed once with PBS and stained with acridine orange (AO) (40 μg/ml) or with AO (40 μg/ml) and propidium iodide (PI) (5 μg/ml). As a positive control for apoptosis, uninfected CV-1 cells were cultured with and without etoposide (15 μM) and stained with AO or with AO and PI after 3 days.
RESULTS
Identification of a host protein that binds to the SV40 capsid proteins.
In a search for host proteins that potentially participate in the SV40 life cycle, we looked for proteins that interact with the major capsid protein VP1. GST-VP1 immobilized on GSH-agarose resin was used as “bait” to specifically “fish” such proteins. The results (Fig. 1B) showed a clear, major ∼100-kDa protein that bound to VP1 (Fig. 1B, lane 3, shown beside the arrow as “∼100 kDa protein”). The majority of the proteins in the nuclear extract did not bind to VP1 (compare the unbound proteins in lane 5 [UB] to the total nuclear extracts [NE] in lane 4). The ∼100-kDa protein did not bind to the GST control (Fig. 1B, lane 8). The band was excised from the gel and analyzed by mass spectrometry, and the protein was identified as PARP, with a molecular mass of 116 kDa. Identification of the protein was confirmed by Western blotting with anti-PARP specific antibody (not shown).
Both VP1 and PARP bind DNA, raising the possibility that the interactions between VP1 and PARP were mediated by a DNA contaminant in the nuclear extract or in the GST-VP1 preparation. We therefore conducted an experiment similar to the one shown in Fig. 1B, except that both the GST-VP1 immobilized on the GSH-agarose and the K562 nuclear extract were preincubated with 3 μM ethidium bromide for 15 min on ice, to disrupt possible protein-DNA interactions. The results (not shown) indicated that ethidium bromide did not disrupt PARP-VP1 binding, suggesting that the interaction between the proteins was not mediated by DNA.
GST-VP3 immobilized on GSH-agarose also pulled down a ∼100-kDa protein from nuclear extracts of K562 cells. Western blotting with an anti-PARP antibody identified this protein as PARP (see experiment shown in Fig. 3C, lanes 2 to 5). Since the coding sequence of VP3 is completely contained within VP2, it may be extrapolated that PARP may also bind to VP2. As both PARP and VP3 have nonspecific DNA-binding activities, we tested whether the interactions observed in this assay were mediated by a DNA contaminant. The zinc finger DNA binding of PARP was disrupted by treatment with 50 mM EDTA. At this concentration, PARP enzymatic activity, which depends on DNA binding, was significantly inhibited (Fig. 2D). GST-VP3 pulldown assays showed that pretreatment with EDTA did not affect the binding between PARP and VP3 (not shown), indicating that PARP-VP3 binding was not mediated by DNA.
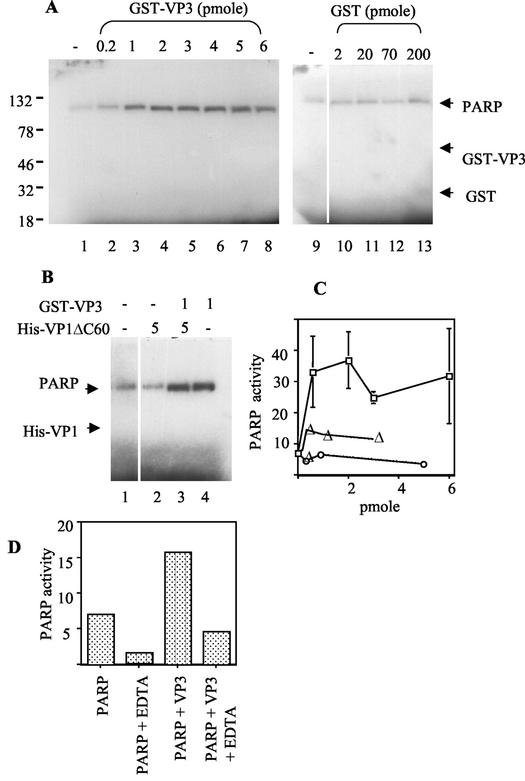
FIG. 2.
PARP enzymatic activity is stimulated by VP3. (A) Poly(ADP-ribosylation) assay. PARP (1 ng ∼ 0.01 pmol) was incubated with a limiting concentration (3 μM) of [32P]NAD+ and sheared salmon sperm DNA (1.5 μg/reaction). The reaction was stopped after 20 min by the addition of SDS-PAGE sample buffer, and the proteins were separated on SDS-PAGE and analyzed by autoradiography. PARP was incubated alone (lane 1) or in the presence of increasing amounts of GST-VP3 or GST (as indicated above). Positions of molecular weight markers are shown on the left. The position of [32P]ADP-ribosylated PARP and the expected positions of GST-VP3 and GST are designated by the arrows. (B) The poly(ADP-ribosylation) assay was performed with PARP (1 ng) alone (lane 1) or in the presence of His-VP1ΔC60 (5 pmol), GST-VP3 (1 pmol), or both proteins preincubated at a 5:1 molar ratio, as indicated (in pmoles) above the autoradiogram. (C) Quantitative analysis of PARP enzymatic activity. The autoradiogram in panel A and several additional experiments were quantified by PhosphorImager analysis for the incorporation of [32P]NAD+. The results for each experiment were normalized to the level of PARP activity in the absence of the capsid proteins. □, GST-VP3; ▵, His-VP1; and ○, His-VP1ΔC60. (D) The effect of EDTA on PARP enzymatic activity and on the stimulation by VP3. In vitro poly(ADP-ribosylation) assays were carried out as described above, with and without preincubation of PARP with 50 mM EDTA. The incorporation of [32P]NAD+ was quantified by PhosphorImager analysis.
SV40 VP3 stimulates PARP enzymatic activity.
PARP enzymatic activity is assayed by in vitro poly(ADP-ribosylation), SDS-PAGE analysis, and phosphorimaging. When PARP is incubated with a limiting concentration of [32P]NAD+ (3 μM), it is [32P]ADP-ribosylated (Fig. 2A, lane 1). As PARP activity depends on the presence of DNA, the reaction is routinely performed in the presence of sonicated salmon sperm DNA or poly(dIdC). Interestingly, the addition of GST-VP3 to the assay consistently resulted in a substantial stimulation of PARP enzymatic activity, as measured by auto-poly(ADP-ribosylation) (Fig. 2A, lanes 2 to 8, and B, lane 4; also see Fig. 6, lanes 2, 5, and 8). GST alone had no effect, even when added at much higher concentrations than GST-VP3 (Fig. 2A, lanes 10 to 13). In contrast to VP3, purified His-VP1 or the mutant His-VP1ΔC60, which forms pentamers but cannot assemble into capsids, had almost no effect on PARP activity (Fig. 2B, lane 2; see Fig. 6, lanes 3, 6, and 9). These results, which are quantified in Fig. 2C, demonstrate that VP3 stimulates PARP enzymatic activity, while VP1 does not. The stimulation in various experiments was three- to fivefold and reached a plateau at 0.5 to 2 pmol of GST-VP3.
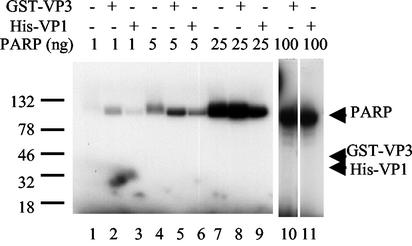
FIG. 6.
VP1 and VP3 are not ADP-ribosylated by PARP. Poly(ADP-ribosylation) assay was performed with PARP, at four different concentrations as shown above the gel. PARP was incubated alone (lanes 1, 4, and 7) or with either GST-VP3 (lanes 2, 5, 8, and 10) or His-VP1 (lanes 3, 6, 9, and 11). Lanes 2, 5, and 8 have 600 ng of GST-VP3, and lane 10 has 3 μg. Lanes 3, 6, and 9 have 200 ng of His-VP1, and lane 11 has 1 μg. The arrows designate the expected positions of GST-VP3 (54 kDa) and His-VP1 (41 kDa). Note that only PARP is poly(ADP-ribosylated) in these assays.
Soon after synthesis, a pentamer of VP1 associates with a monomer of VP2 or VP3 to form a VP15-VP2/3 complex, the basic building block of the SV40 capsid (3, 15, 19). Is PARP enzymatic activity stimulated by the VP15-VP3 building block as well? To address this question, we tested for PARP stimulation following preincubation of His-VP1ΔC60 with GST-VP3 at a 5:1 ratio, at conditions that allow the formation of VP15-VP3 complexes (15). The results showed that VP15-VP3 stimulated PARP enzymatic activity (Fig. 2B, lane 3) to the same extent as GST-VP3 alone (lane 4) and confirmed that VP1 alone did not stimulate PARP (lane 2). These results suggest that the VP3 moiety in the building block mediates PARP stimulation and that the stimulation is not hindered by the presence of VP15.
PARP activity is stimulated >500-fold upon binding to DNA strand breaks (reviewed in reference 28). To determine whether the stimulation by VP3 also requires DNA binding, we pretreated PARP with EDTA to disrupt the zinc finger DNA-binding domain. Figure 2D shows that as expected, PARP activity is significantly reduced by treatment with EDTA. The stimulation of PARP by GST-VP3 is also reduced by the EDTA treatment. This indicates that although VP3 and PARP bind one another in the absence of DNA, PARP must bind to DNA for full enzymatic stimulation by VP3.
Mapping the region of VP3 that binds to PARP and stimulates its activity.
A VP2/3 monomer is bound to the inner cavity of a VP1 pentamer (VP15) through a region near, but not at, its C terminus (Fig. 3A, amino acids 155 to 190 of VP3) (3, 15). Both the carboxy and amino termini of VP2/3 emerge from the VP15 cavity and extend inward to the center of the virion. One can predict that these domains are free to interact with PARP. To map the PARP interactive domain, we performed GST pulldown experiments of K562 nuclear extracts containing PARP by using the following GST-VP3 mutants (Fig. 3B): GST-VP3ΔC13, with deletion of the C-terminal 13 amino acids that comprise the DNA-binding domain (5), and GST-VP3C49 and GST-VP3C35 (15), containing the 49 and 35 C-terminal amino acids, respectively (amino acids 185 to 234 and 199 to 234). The results showed that both GST-VP3C49 and GST-VP3C35 bound PARP to the same extent as the full-length protein (Fig. 3C, compare lanes 6 and 15 with lane 2), while the binding of GST-VP3ΔC13 to PARP was poor (Fig. 3C, lane 11). Thus, the C-terminal 35 amino acids of VP2/3 are sufficient for PARP binding.
To map the region of VP3 that is required to stimulate PARP enzymatic activity, we carried out in vitro poly(ADP-ribosylation) assays by using the same GST-VP3 mutants. The results, quantified in Fig. 3D, showed that GST-VP3C49 stimulated PARP to the same extent as the full-length GST-VP3. Stimulation by GST-VP3C35 was lower, and GST-VP3ΔC13 stimulated PARP activity even less. These results show that the C-terminal 49 amino acids of VP3 are sufficient for both binding and stimulation of PARP enzymatic activity.
PARP participates in the SV40 life cycle in vivo.
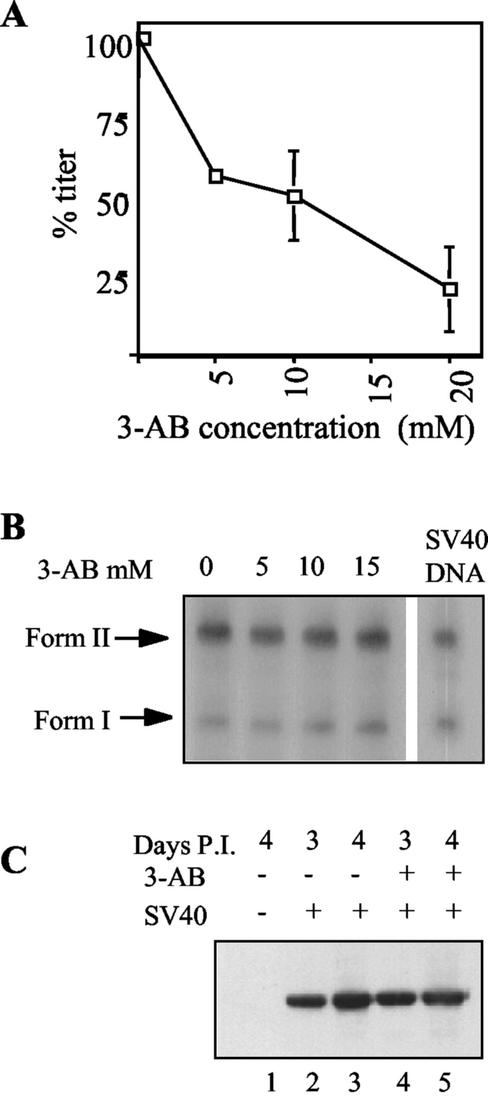
The interaction between PARP and the capsid proteins and its stimulation by VP15-VP3 suggested that PARP has a role in the SV40 life cycle. The competitive inhibitor of PARP, 3-aminobenzamide (3-AB), is an analogue of NAD+ that has a high affinity to PARP but cannot be used as a substrate; thus, in its presence PARP binds to DNA but is catalytically inactive (reviewed in reference 28). We found that 3-AB consistently and reproducibly reduced the SV40 titer in a dose-dependent manner (Fig. 4A). At the highest concentration of 3-AB used, 20 mM, the titer was reduced to 20%. We concluded that PARP participates in the SV40 life cycle.
FIG. 4.
The effect of the PARP inhibitor 3-AB on different steps in the SV40 life cycle. (A) 3-AB inhibits SV40 production. The graph shows SV40 titer as a function of 3-AB concentration. Subconfluent CV-1 cells were infected with SV40 (an MOI of 3 to 10 in different experiments), grown in the presence of increasing concentrations of 3-AB, and harvested after 5 days by repeated freeze-thaw and sonication. Viral titer was assayed by in situ hybridization (7, 32). At each concentration of 3-AB, the titer is shown as a percentage of the titer obtained in the absence of 3-AB in the same experiment. Each point represents data from at least two experiments. The error bar at 5 mM 3-AB is too small to be seen on this scale. (B) 3-AB does not inhibit SV40 DNA replication. Viral DNA replication assay was performed by Southern blot analysis (34a) of Hirt supernatant (17a) of cells infected in the presence and absence of increasing concentrations of 3-AB 2 days postinfection. SV40 DNA control is shown on the right. Duplicate cultures were used to count the number of cells, and the aliquots analyzed (∼0.1 of the Hirt supernatants) were normalized accordingly. The blot was hybridized to 32P-labeled SV40 DNA. Form I, supercoiled SV40 DNA; and form II, relaxed SV40 DNA. (C) 3-AB does not inhibit VP1 production. Nuclear extract of CV-1 cells infected in the absence (lanes 2 and 3) and presence (lanes 4 and 5) of 20 mM 3-AB was analyzed by SDS-PAGE and Western blotting with anti-VP1 antibody. Nuclear proteins were harvested 3 (lanes 2 and 4) and 4 (lanes 1, 3, and 5) days postinfection (P.I.). The aliquots were normalized to the amounts of total proteins (assayed by the Bradford reaction) in each of the extract.
At what point in the SV40 life cycle does PARP participate?
We tested SV40 grown in the presence of 3-AB for different steps in the viral life cycle. SV40 DNA replication was analyzed by measuring the amount of viral DNA 2 days postinfection at 5, 10, and 15 mM 3-AB (Fig. 4B) and in another experiment at 20 mM (not shown). An equal amount of DNA was found with and without the PARP inhibitor. In another experiment the effect on DNA replication was tested 4 days postinfection. Again the level of DNA per cell was not reduced (20 mM 3-AB). Therefore, PARP is not likely to participate in SV40 DNA replication or at any stage prior to replication, such as initial infection or T antigen expression.
As PARP has a role in transcription control (29, 30), we asked whether PARP affects capsid protein expression. We assayed for VP1 in nuclear extracts of cells infected with SV40 in both the presence and absence of 20 mM 3-AB by Western blotting with anti-VP1 antibody. A typical experiment is shown in Fig. 4C (compare lanes 4 and 5 and 2 and 3). The results consistently showed that the expression of VP1 and of VP2 and VP3 (not shown) was not affected by the PARP inhibitor. We concluded that PARP participates at a late stage of the virus life cycle, after late gene expression.
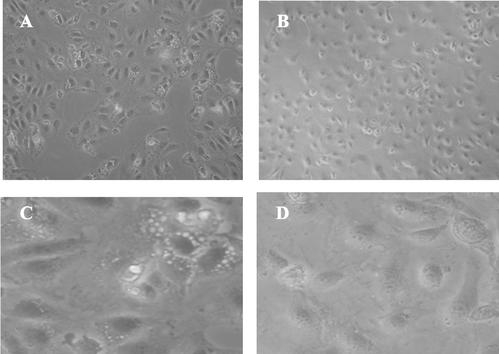
Cytopathic effects (CPE) are a hallmark of SV40 infection. Figure 5 depicts a typical experiment of cells infected with SV40 in the absence (Fig. 5A and C) and presence (Fig. 5B and D) of 20 mM 3-AB. In the absence of 3-AB, CPE start to appear at 3 days (not shown) and are readily visible at 4 days postinfection (Fig. 5A and C). In contrast, in the presence of 3-AB, there is very little, if any, CPE (Fig. 5B and D) at 4 days postinfection. We found that the PARP inhibitor reduced the magnitude and delayed the onset of CPE in SV40-infected cells. The inhibition of CPE was also observed at 5 and 10 mM 3-AB (not shown).
FIG. 5.
The PARP inhibitor 3-AB reduces the magnitude of SV40 CPE. Subconfluent CV-1 cells were infected with SV40 and grown for 4 days in the absence (A and C) or presence (B and D) of 20 mM 3-AB.
SV40 capsid proteins are not modified by PARP.
The protein-protein interactions between PARP and the capsid proteins, taken together with the stimulation of PARP activity by VP15-VP3, implicates a functional interaction between PARP and the capsid proteins. The simplest hypothesis is that PARP poly(ADP)-ribosylates the capsid proteins, modifying the surface of the virion in a way that is both critical for infectivity and might determine viral host range. As seen in Fig. 2A and B, no poly(ADP-ribosylation) of either His-VP1 (molecular mass, 41 kDa) or GST-VP3 (molecular mass, 54 kDa) was observed, indicating that they were not modified by PARP. We performed additional experiments that used higher concentrations of PARP. We have never observed the modification of GST-VP3 or His-VP1, using PARP at levels varying from 1 to 100 ng/reaction, although PARP was enzymatically active as demonstrated by its consistent automodification (Fig. 6). These data indicate that PARP does not modify VP1 or VP3 at a detectable level. The results do not exclude the possibility that the N terminus that is unique to VP2 is modified by PARP.
SV40-infected CV-1 cells do not undergo apoptosis.
PARP plays a role in early steps of apoptosis (33) and later undergoes caspase-induced cleavage and becomes inactive (23). As PARP's enzymatic activity is stimulated by the SV40 capsid proteins, it was of interest to test whether infected cells undergo apoptosis. We first examined whether PARP undergoes cleavage in SV40- infected CV-1 cells. Caspase-specific cleavage product (89 kDa) was not observed (not shown), suggesting that SV40-infected CV-1 cells do not undergo apoptosis.
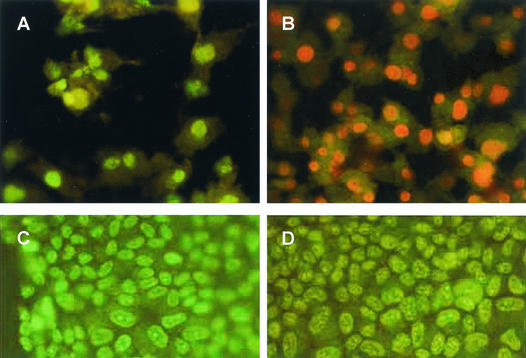
We have also tested for apoptosis by staining with AO and PI. AO penetrates the plasma membrane and stains DNA in living cells. The nuclei of apoptotic cells have highly condensed chromatin and are brightly stained by AO. The membranes of apoptotic cells retain their integrity and thus are not permeable to PI, while dead cells are readily stained by PI. CV-1 cells were infected at an MOI of 10 and were stained with AO and by AO and PI at several time points following infection. Classic apoptotic morphology was not observed at any time point postinfection. Figure 7 shows that 5 days postinfection most of the infected cells are dead, as seen by PI staining, with many of the cells floating in the medium. In contrast, classical apoptotic cells were observed in a control experiment, in which CV-1 cells were treated with etoposide, an agent known to cause apoptosis in those cells (18). Clusters of bright, spherical bodies that are smaller than the cell nucleus, with no discernible cytoplasm, were observed. As expected, the etoposide-treated CV-1 cells were not permeable to PI (not shown).
FIG. 7.
SV40-infected CV-1 cells do not undergo apoptosis. CV-1 cells that were either infected by SV40 (A and B) or mock infected (C-D) at 5 days postinfection. (A and C) AO staining. (B and D) AO and PI costaining.
PARP facilitates the release of SV40 to the medium.
The preservation of intact plasma membranes in apoptotic cells is probably incompatible with the virus lytic infection and release of mature virions to the environment. We hypothesize that, by stimulating PARP activity, the virus ensures that the cells will undergo necrosis rather than apoptosis. It is possible that in the 3-AB-treated cells some virus is trapped in the cell. To test this hypothesis, we infected CV-1 cells in the presence and absence of 3-AB and measured separately, by titration, the virus present in the medium and virus trapped in the cells (harvested by freeze-thaw) after the medium had been removed. As seen in the representative experiment shown in Table 1, virus release from the cells was reduced from 45% in the absence of 3-AB to <20% in its presence, indicating that PARP facilitates SV40 release. It may be noted that the total titer was also reduced (see also Fig. 4A), suggesting the existence of additional mechanisms, direct or indirect, by which PARP affects the SV40 life cycle.
TABLE 1.
The PARP inhibitor 3-AB interferes with SV40 release from infected CV-1 cellsa
| Inhibitor used | Total virus | Virus in cells | Virus in medium | % Released |
|---|---|---|---|---|
| Control | 6.2 × 108 | 3.5 × 108 | 2.7 × 108 | 44 |
| 20 mM 3-AB | 2.1 × 108 | 1.7 × 108 | 0.4 × 108 | 19 |
The values represent SV40 titer measured as infectious units/ml (7).
DISCUSSION
The mechanism by which mature virions are released from infected cells is poorly understood. The present study shows that in SV40 infection VP3 triggers the process by stimulating the activity of PARP, leading the cell to die by necrosis. Our results demonstrate that PARP binds to and is activated by the VP15-VP3 capsid building blocks. The titer of SV40, CPE, and release to the medium are significantly reduced by a competitive inhibitor of PARP. DNA replication and late protein expression are not affected, indicating that PARP participates at a late stage in the viral life cycle. The capsid proteins are not poly(ADP)-ribosylated by PARP, suggesting that PARP does not affect SV40 infectivity.
The minor capsid protein of SV40, VP3, stimulates PARP enzymatic activity, although it is not itself modified by PARP. PARP interacts with many cellular proteins. Several of its partners, such as transcription factor YY1 (29), are both modified by PARP and stimulate PARP activity. In contrast, other target proteins (i.e., histones) are ADP-ribosylated but have no significant effect on the activity of PARP (16). Other partners, such as XRCC1 (24), are not modified by PARP, although they inhibit PARP activity. This spectrum of interactive patterns may reflect the participation of PARP in widely different cellular processes. To the best of our knowledge, VP3 is the first virus protein shown to stimulate PARP activity and the first protein shown to stimulate its activity without being itself modified.
VP2 and VP3 are structural proteins that were also implicated in delivery of the SV40 minichromosome to the nucleus during viral infection and entry (26) and in specific recognition of the viral minichromosome through interaction with Sp1 during assembly (14, 15). The present study demonstrates that these structural proteins also function in the stimulation of PARP activity as part of the mechanism that leads SV40-infected cells to necrotic death and lysis.
Our findings demonstrate that the C-terminal 35 amino acids of VP2/3 are sufficient for binding PARP but that maximum stimulation of PARP activity requires additional amino acids present in GST-VP3C49. Thus, the PARP-stimulating domain and the PARP binding domain of VP3 do not fully overlap. This implies that VP3 binding per se is not sufficient for stimulation of PARP activity. Our data show that the terminal 13 amino acids of VP2/3 participate in PARP binding and stimulation only to a very low extent. Hence, most of the PARP interactive domain resides between amino acids 185 and 221 of VP3, within the C-terminal tail of VP2/3, which emerges from the inner cavity of the VP1 pentamer. VP1 also binds to PARP, implying the presence of additional contacts between PARP and the VP15-VP3 building block. Although these interactions do not lead to higher stimulation of PARP enzymatic activity, they most likely act in stabilizing the PARP-VP15-VP3 complex.
SV40 titer was reduced by 80% by the PARP inhibitor 3-AB, and SV40 release to the medium fell by ∼50%. While these reductions are significant, they are only partial. A possible explanation is the presence of functional redundancy in the regulation of the cellular decision among DNA damage repair, apoptosis, and necrosis. Indeed a number of PARP homologues have been identified in recent years: PARP-2, PARP-3, Vault-PARP, and tankyrase, as well as an alternatively spliced species of PARP that lacks the DNA-binding domain (reviewed in reference 34). The similarity between these proteins is limited to a 40-kDa fragment at the C terminus of PARP-1 that is sufficient for its catalytic activity and contains the NAD+ binding site. Thus, there is a wide divergence between PARP homologues, suggesting that certain inhibitors will be specific to each.
Cell death may occur by apoptosis or by necrosis or any intermediate pathway. During apoptosis nuclear chromatin becomes condensed and fragmented, the cytoplasm undergoes shrinkage and condensation, and the integrity of the plasma membrane is preserved. Finally the cell breaks down into membrane-enclosed apoptotic bodies. On the other hand, in necrotic cells the plasma membrane loses its physiological integrity, which leads to swelling, loss of isotonic balance, and massive breakup of cell components, culminating in a complete dissolution of the cell into debris and release of the internal contents.
Our study shows that cell death following SV40 infection is by necrosis rather than apoptosis: (i) PARP is not cleaved during the course of SV40 infection. (ii) Staining by AO and PI demonstrates that SV40-infected cells do not undergo apoptosis. This is consistent with previous electron microscopy studies that showed that the ultrastructural alterations associated with SV40 lytic infection include swelling of the endoplasmic reticulum and condensation of the mitochondria, followed by swelling and disintegration of the membranes. In that study, cell death was found to resemble that caused by anoxia and ischemia (10).
Viruses modulate the expression of the host factors required for their life cycles. T antigen modulates transcriptional regulation of cellular genes (reviewed in reference 25) and modifies cell cycle control and growth regulation by interacting with Rb and with p53 (reviewed in reference 36). Sp1, which is needed for the activity of the early and late promoters as well as for recruitment of the capsid proteins during assembly, is upregulated in SV40-infected cells (31). The preservation of intact plasma membranes in apoptic cells is probably incompatible with the virus lytic infection and release of mature virions to the environment. We suggest that, by stimulating PARP activity, the virus ensures that the cells will undergo necrosis rather than apoptosis. The stimulation of PARP enzymatic activity by the SV40 building block is an additional example of usurpation of a cellular factor that the virus needs for successful completion of its life cycle.
Acknowledgments
We thank Amos Oppenheim for a critical review of the manuscript, Orly Ben-Nun-Shaul for helpful discussions, Vered Roitman for assistance in some of the experiments, Boaz Nahmias for advice on the AO and PI staining, and Harumi Kasamatsu for kindly providing us with pGEX-VP3ΔC13.
This research was supported by The Israel Science Foundation founded by The Academy of Sciences and Humanities.
REFERENCES
- 1.Baksi, K., H. Alkhatib, and M. Smulson. 1987. In vivo charaterization of the poly(ADP-ribosylation) of SV40 chromatin and large T antigen by immunofractionation. Exp. Cell Res. 172:110-123. [DOI] [PubMed] [Google Scholar]
- 2.Bina, M. 1986. Simian virus 40 assembly. Comments Mol. Cell. Biophys. 4:55-62. [Google Scholar]
- 3.Chen, X. S., T. Stehle, and S. C. Harrison. 1998. Interaction of polyomavirus internal protein VP2 with the major capsid protein VP1 and implications for participation of VP2 in viral entry. EMBO J. 17:3233-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiarugi, A. 2002. Poly(ADP-ribose) polymerase: killer or conspirator? The ′suicide hypothesis' revisited. Trends Pharmacol. Sci. 23:122-129. [DOI] [PubMed] [Google Scholar]
- 5.Clever, J., D. Dean, and H. Kasamatsu. 1993. Identification of a DNA binding domain in simian virus 40 capsid proteins VP2 and VP3. J. Biol. Chem. 268:20877-20883. [PubMed] [Google Scholar]
- 6.Clever, J., and H. Kasamatsu. 1991. Simian virus 40 VP2/3 small structural proteins harbor their own nuclear transport signal. Virology 181:78-90. [DOI] [PubMed] [Google Scholar]
- 7.Dalyot-Herman, N., O. Ben-nun-Shaul, A. Gordon-Shaag, and A. Oppenheim. 1996. The simian virus 40 packaging signal ses is composed of redundant DNA elements which are partly interchangeable. J. Mol. Biol. 259:69-80. [DOI] [PubMed] [Google Scholar]
- 8.de Murcia, G., V. Schreiber, M. Molinete, B. Saulier, O. Poch, M. Masson, C. Niedergang, and J. Menissier de Murcia. 1994. Structure and function of poly(ADP-ribose) polymerase. Mol. Cell. Biochem. 138:15-24. [DOI] [PubMed] [Google Scholar]
- 9.Dery, C. V., G. de Murcia, D. Lamarre, N. Morin, G. G. Poirier, and J. Weber. 1986. Possible role of ADP-ribosylation of adenovirus core proteins in virus infection. Virus Res. 4:313-329. [DOI] [PubMed] [Google Scholar]
- 10.Eggleton, K. H., and L. C. Norkin. 1981. Cell killing by simian virus 40: the sequence of ultrastructural alterations leading to cellular degeneration and death. Virology 110:73-86. [DOI] [PubMed] [Google Scholar]
- 11.Forstova, J., N. Krauzewicz, S. Wallace, A. J. Street, S. M. Dilworth, S. Beard, and B. E. Griffin. 1993. Cooperation of structural proteins during late events in the life cycle of polyomavirus. J. Virol. 67:1405-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaken, J. A., M. Tavassoli, S. U. Gan, S. Vallian, I. Giddings, D. C. Darling, J. Galea-Lauri, M. G. Thomas, H. Abedi, V. Schreiber, J. Menissier-de Murcia, M. K. Collins, S. Shall, and F. Farzaneh. 1996. Efficient retroviral infection of mammalian cells is blocked by inhibition of poly(ADP-ribose) polymerase activity. J. Virol. 70:3992-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garber, E. A., M. M. Seidman, and A. J. Levine. 1980. Intracellular SV40 nucleoprotein complexes: synthesis to encapsidation. Virology 107:389-401. [DOI] [PubMed] [Google Scholar]
- 14.Gordon-Shaag, A., O. Ben-Nun-Shaul, H. Kasamatsu, A. Oppenheim, and A. Oppenheim. 1998. The SV40 capsid protein VP3 cooperates with the cellular transcription factor Sp1 in DNA-binding and in regulating viral promoter activity. J. Mol. Biol. 275:187-195. [DOI] [PubMed] [Google Scholar]
- 15.Gordon-Shaag, A., O. Ben-Nun-Shaul, V. Roitman, Y. Yosef, and A. Oppenheim. 2002. Cellular transcription factor Sp1 recruits simian virus 40 capsid proteins to the viral packaging signal, ses. J. Virol. 76:5915-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griesenbeck, J., M. Ziegler, N. Tomilin, M. Schweiger, and S. L. Oei. 1999. Stimulation of the catalytic activity of poly(ADP-ribosyl) transferase by transcription factor Yin Yang 1. FEBS Lett. 443:20-24. [DOI] [PubMed] [Google Scholar]
- 17.Ha, H. C., K. Juluri, Y. Zhou, S. Leung, M. Hermankova, and S. H. Snyder. 2001. Poly(ADP-ribose) polymerase-1 is required for efficient HIV-1 integration. Proc. Natl. Acad. Sci. USA 98:3364-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 18.Inman, M., G. C. Perng, G. Henderson, H. Ghiasi, A. B. Nesburn, S. L. Wechsler, and C. Jones. 2001. Region of herpes simplex virus type 1 latency-associated transcript sufficient for wild-type spontaneous reactivation promotes cell survival in tissue culture. J. Virol. 75:3636-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishii, N., A. Nakanishi, M. Yamada, M. H. Macalalad, and H. Kasamatsu. 1994. Functional complementation of nuclear targeting-defective mutants of simian virus 40 structural proteins. J. Virol. 68:8209-8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeggo, P. A. 1998. DNA repair: PARP—another guardian angel? Curr. Biol. 8:R49-R51. [DOI] [PubMed] [Google Scholar]
- 21.Li, P. P., A. Nakanishi, D. Shum, P. C.-K. Sun, A. M. Salazar, C. F. Fernandez, S.-W. Chan, and H. Kasamatsu. 2001. Simian virus 40 Vp1 DNA-binding domain is functionally separable from the overlapping nuclear localization signal and is required for effective virion formation and full viability. J. Virol. 75:7321-7329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liddington, R., Y. Yan, J. Moulai, R. Sahli, T. Benjamin, and S. Harrison. 1991. Structure of simian virus 40 at 3.8-A resolution. Nature 354:278-284. [DOI] [PubMed] [Google Scholar]
- 23.Los, M., M. Mozoluk, D. Ferrari, A. Stepczynska, C. Stroh, A. Renz, Z. Herceg, Z. Q. Wang, and K. Schulze-Osthoff. 2002. Activation and caspase-mediated inhibition of PARP: a molecular switch between fibroblast necrosis and apoptosis in death receptor signaling. Mol. Biol. Cell 13:978-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masson, M., C. Niedergang, V. Schreiber, S. Muller, J. Menissier-de Murcia, and G. de Murcia. 1998. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 18:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moens, U., O. M. Seternes, B. Johansen, and O. P. Rekvig. 1997. Mechanisms of transcriptional regulation of cellular genes by SV40 large T- and small T-antigens. Virus Genes 15:135-154. [DOI] [PubMed] [Google Scholar]
- 26.Nakanishi, A., J. Clever, M. Yamada, P. P. Li, and H. Kasamatsu. 1996. Association with capsid proteins promotes nuclear targeting of simian virus 40 DNA. Proc. Natl. Acad. Sci. USA 93:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nargi-Aizenman, J. L., C. M. Simbulan-Rosenthal, T. A. Kelly, M. E. Smulson, and D. E. Griffin. 2002. Rapid activation of poly(ADP-ribose) polymerase contributes to Sindbis virus and staurosporine-induced apoptotic cell death. Virology 293:164-171. [DOI] [PubMed] [Google Scholar]
- 28.Oei, S. L., J. Griesenbeck, and M. Schweiger. 1997. The role of poly(ADP-ribosyl)ation. Rev. Physiol. Biochem. Pharmacol. 131:127-173. [DOI] [PubMed] [Google Scholar]
- 29.Oei, S. L., J. Griesenbeck, M. Schweiger, and M. Ziegler. 1998. Regulation of RNA polymerase II-dependent transcription by poly(ADP-ribosyl)ation of transcription factors. J. Biol. Chem. 273:31644-31647. [DOI] [PubMed] [Google Scholar]
- 30.Oliver, F. J., J. Menissier-de Murcia, and G. de Murcia. 1999. Poly(ADP-ribose) polymerase in the cellular response to DNA damage, apoptosis, and disease. Am. J. Hum. Genet. 64:1282-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saffer, J. D., S. P. Jackson, and S. J. Thurston. 1990. SV40 stimulates expression of the transacting factor Sp1 at the mRNA level. Genes Dev. 4:659-666. [DOI] [PubMed] [Google Scholar]
- 32.Sandalon, Z., N. Dalyot-Herman, A. B. Oppenheim, and A. Oppenheim. 1997. In vitro assembly of SV40 virions and pseudovirions: vector development for gene therapy. Hum. Gene Ther. 8:843-849. [DOI] [PubMed] [Google Scholar]
- 33.Simbulan-Rosenthal, C. M., D. S. Rosenthal, S. Iyer, A. H. Boulares, and M. E. Smulson. 1998. Transient poly(ADP-ribosyl)ation of nuclear proteins and role of poly(ADP-ribose) polymerase in the early stages of apoptosis. J. Biol. Chem. 273:13703-13712. [DOI] [PubMed] [Google Scholar]
- 34.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31-40. [DOI] [PubMed] [Google Scholar]
- 34a.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 35.Stehle, T., S. J. Gamblin, Y. Yan, and S. C. Harrison. 1996. The structure of simian virus 40 refined at 3.1 A resolution. Structure 4:165-182. [DOI] [PubMed] [Google Scholar]
- 36.Sullivan, C. S., and J. M. Pipas. 2002. T antigens of simian virus 40: molecular chaperones for viral replication and tumorigenesis. Microbiol. Mol. Biol. Rev. 66:179-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tong, W. M., U. Cortes, and Z. Q. Wang. 2001. Poly(ADP-ribose) polymerase: a guardian angel protecting the genome and suppressing tumorigenesis. Biochim. Biophys. Acta 1552:27-37. [DOI] [PubMed] [Google Scholar]
- 37a.Tooze, J. 1981. DNA tumor viruses, 2nd ed., vol. 2. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 38.Wrobel, B., Y. Yosef, A. B. Oppenheim, and A. Oppenheim. 2000. Production and purification of SV40 major capsid protein (VP1) in Escherichia coli strains deficient for the GroELS chaperone machine. J. Biotechnol. 84:285-289. [DOI] [PubMed] [Google Scholar]